Abstract
A key component in the regulation of V(D)J recombination is control of the accessibility of RAG proteins to recombination signal sequences (RSS). Nucleosomes are known to inhibit this accessibility. We show here that the signal sequence itself represses accessibility by causing nucleosome positioning over the RSS. This positioning is mediated, in vitro and in vivo, by the conserved nonamer of the RSS. Consistent with this strong positioning, nucleosomes at RSSs are resistant to remodelling by nucleosome sliding. In vivo we find that consensus RSSs are preferentially protected, whereas those that lack a consensus nonamer, including some cryptic RSSs, fail to position nucleosomes. Decreased protection of these non-consensus RSSs correlates with their increased use in recombination assays. We therefore suggest that nucleosome positioning by RSSs provides a previously unanticipated level of protection and regulation of V(D)J recombination.
Keywords: chromatin remodelling/chromosomal translocation/nucleosome positioning/RAGs/V(D)J recombination
Introduction
The ability of an organism to respond to a variety of invading pathogens relies on the generation of a large antigen receptor repertoire. V(D)J recombination plays an essential role in the generation of this repertoire. In the recombination reaction, one of the many variable gene segments is joined either directly to one of the joining gene segments or is joined via one of the diversity gene segments to a joining gene segment. All of the recombining gene segments within immunoglobulin and T cell receptor loci are flanked by the same recombination signal sequences (RSS), comprising a conserved heptamer, separated by a 12- or 23-bp spacer from a conserved nonamer (reviewed in Lewis, 1994). Moreover, the same proteins, RAG1 and RAG2, initiate recombination in both B and T cells (Oettinger et al., 1990; McBlane et al., 1995). Nevertheless, V(D)J recombination is strongly regulated in a cell-, locus- and stage-specific manner. For example, immunoglobulin genes only fully rearrange in B cells and T cell receptor loci only rearrange in T cells (reviewed in Lewis, 1994; Bassing et al., 2002). A key question is how this cell and stage specificity of the recombination reaction is achieved. Moreover, tight regulation of V(D)J recombination is required to prevent inappropriate use of the estimated 10 million potential RAG targets in the mammalian genome (Lewis et al., 1997). Accessibility of the RSS in chromatin has been proposed to be the main regulatory factor (reviewed in Sleckman et al., 1996; Bassing et al., 2002).
A fundamental question, therefore, is to understand how the chromatin structure is specifically altered to achieve this regulation of V(D)J recombination. Initial studies have shown that the first level of the chromatin packaging, the nucleosome core particle, blocks RAG cutting (Kwon et al., 1998; Golding et al., 1999; McBlane and Boyes, 2000). This repression is due to the inability of RAG proteins to bind to the RSS when it is constrained by a nucleosome (McBlane and Boyes, 2000). Although nucleosomes inhibit the initiation of recombination, it is unlikely that this alone can fully explain the regulation of the recombination reaction. This is because, if nucleosomes are deposited in a sequence-independent manner, at least some of RSSs are expected to lie in linker regions between nucleosomes. Thus, it would be predicted that an additional level of regulation is required to control the use of these linker RSSs. One possibility is that higher order chromatin structure regulates accessibility to these RSSs. Indeed a number of changes of higher order chromatin structure are associated with recombination. For example, DNA methylation is decreased and histone acetylation and general DNase I sensitivity are increased in loci undergoing recombination. (reviewed in Sleckman et al., 1996; Roth and Roth, 2000). A second possibility is that the presence of RSSs in linker DNA is eliminated by the preferential positioning of nucleosomes over the RSSs. Nucleosome positioning has been found at some well-studied promoters (Fragoso et al., 1995; Svaren and Horz, 1997). However, to date, nucleosome positioning has not been reported to play a widespread role in transcription regulation.
The ability of a nucleosome to repress initiation of V(D)J recombination raises the key question of how the nucleosome-mediated repression is overcome. Although, in theory, recombination could use only those RSSs that lie in linker regions, the capacity to use all RSSs will significantly increase potential antigen receptor diversity. Currently, the mechanism of nucleosome disruption for recombination is unresolved. Evidence has been presented for a role of HMGB1 and histone acetylation in this process (Kwon et al., 1998, 2000). However, other groups saw no effect of these elements using mononucleosome templates (Golding et al., 1999; McBlane and Boyes, 2000), possibly due to the different methods of nucleosome preparation used. One likely mechanism of nucleosome disruption for recombination is via one of the energy-dependent nucleosome remodelling complexes. A number of complexes have been identified, including SWI/SNF, NURF, ACF, RSC and NuRD, and two major mechanisms of nucleosome disruption have been described: nucleosome sliding and localized DNA disruption from the nucleosome surface (reviewed in Narlikar et al., 2002).
Recently, the nucleosome-remodelling complex SWI/SNF was indeed shown to stimulate RAG cutting on nucleosome templates (Kwon et al., 2000). In these studies, SWI/SNF facilitated RAG cutting on nucleosome templates purified after remodelling by SWI/SNF (Kwon et al., 2000). Since these reconstituted templates lacked linker DNA onto which the octamer could be moved, the most probable explanation for increased RAG cutting is that SWI/SNF had caused localized nucleosome disruption. This is not necessarily the only mechanism to enhance RAG cutting on nucleosome templates. We therefore wanted to address whether a nucleosome remodelling complex that mediates nucleosome sliding also functions in this assay. We found that nucleosome sliding by NURF caused only a minimal stimulation of RAG cutting. This led us to the surprising finding that the RSS itself causes nucleosome positioning both in vitro and in vivo. Positioning of nucleosomes over the RSS will therefore significantly reduce the number of RSSs in linker regions and consequently will confer an additional level of protection from recombination. Moreover, these data illustrate a novel role for nucleosome positioning in the regulation of a nuclear process.
Results
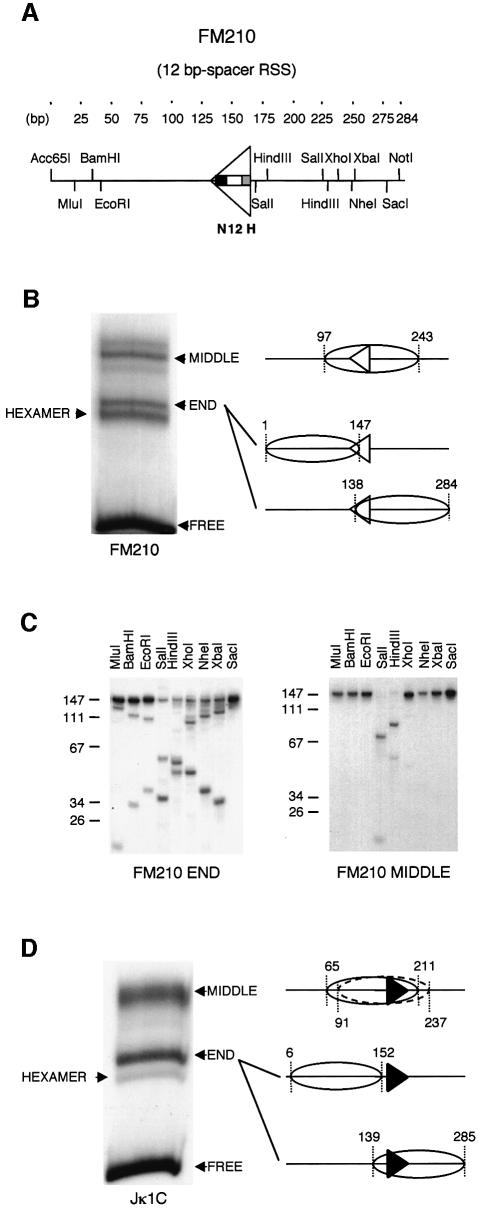
NURF efficiently remodels nucleosomes by a sliding mechanism on templates with sufficient linker DNA onto which the histone octamer can be moved (Hamiche et al., 1999). To investigate whether this class of remodelling complex stimulates RAG cutting on nucleosome templates, we reconstituted 284 and 285 bp DNA fragments carrying a 12- or 23-bp spacer RSS, respectively, into nucleosomes by salt-urea dialysis (Figure 1A). Recon stitution of the fragment carrying the 12 RSS generated three major complexes (Figure 1B). To map the nucleosome positions, the complexes were digested with micrococcal nuclease, the DNA associated with the nucleosome was isolated, end labelled and digested with restriction enzymes (Figure 1C). The sequence that was originally protected by the nucleosome was then determined from the restriction map and the fragment sizes generated (Clark and Felsenfeld, 1991; Langst et al., 1999). This showed that the predominant slower migrating complex has a nucleosome positioned over the central part of the fragment, encompassing the RSS (MIDDLE). The two faster migrating complexes (HEXAMER and END) are complexes that frequently occur as artefacts in reconstitution reactions: the HEXAMER lacks a H2A/H2B dimer, whereas the END complex is a mixture of nucleosomes positioned at the ends of the DNA fragment. Nucleosome positioning on the ends of DNA fragments is a well characterized by-product of the reconstitution reaction (Linxweiler and Horz, 1984).
Fig. 1. Mapping of nucleosomes on 12- and 23-bp spacer RSS fragments. (A) Restriction enzyme map of the 284-bp Acc65I–NotI fragment from plasmid pFM210. The triangle represents a functional 12-bp spacer RSS consisting of a nonamer (N), a 12-bp spacer (12) and a hexamer (H). (B) Native polyacrylamide gel electrophoresis of mononucleosomes reconstituted on the radiolabelled pFM210 Acc65I–NotI fragment. The nucleosomes are labelled ‘MIDDLE’ or ‘END’ according to their translational position. ‘FREE’ indicates DNA that was not reconstituted into nucleosomes. ‘HEXAMER’ indicates reconstitutes that lack one H2A/B heterodimer as determined by protection of only 105 bp following micrococcal nuclease digestion. Nucleosome positions, according to the mapping in (C), are shown in the schematic drawing to the right. (C) Restriction enzyme digestions of the 147-bp fragments obtained following micrococcal nuclease digestion of the ‘END’ or ‘MIDDLE’ reconstituted nucleosomes. (D) Native polyacrylamide gel electrophoresis of mononucleosomes reconstituted on the radiolabelled Jκ1C fragment carrying a 23-bp spacer RSS. A summary of the nucleosome position mapping is given on the right.
Reconstitution of a DNA fragment carrying a 23 RSS (from plasmid pJκ1C) generated two major complexes (Figure 1D); in the slower migrating complex, the major nucleosome position mapped to the centre of the fragment, covering the RSS. The faster migrating complex is a mixture of nucleosomes positioned equivalently over each end (data not shown).
NURF does not increase RAG cutting on nucleosome templates
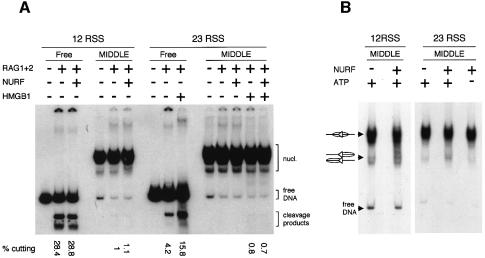
To test whether NURF stimulates RAG cutting, we used the isolated middle nucleosome complexes reconstituted onto the 12 and 23 RSSs (Figure 1B and D). In both purified reconstitutes, the RSS was completely covered by a nucleosome. As had been observed previously, RAG cutting on the nucleosome templates was barely detectable; the slight amount of cutting that was observed was most likely came from the small amount of contaminating free DNA. Addition of NURF to the 12 RSS nucleosome caused minimal, if any, increase in RAG cutting (Figure 2A). Similarly, RAG cutting on the nucleosome covering the 23 RSS was not stimulated by NURF and ATP, even in the presence of HMGB1, which has been shown to stimulate RAG cutting at 23 RSSs (Figure 2A; van Gent et al., 1997).
Fig. 2. Stimulation of V(D)J cleavage and nucleosome sliding by NURF. (A) Free DNA (Free) or the middle nucleosome complex (MIDDLE) carrying the 12-bp spacer RSS (Figure 1B) or 23-bp spacer RSS (Figure 1D) were incubated with recombinant RAG1 and RAG2 in the absence or presence of NURF. V(D)J cleavage products (161 and 123 bp) were detected using native gel electrophoresis. Addition of NURF to free DNA (lane 3) shows NURF has no effect on RAG cutting of this template. Addition of HMGB1 was required for efficient cleavage at the 23 RSS. The cleavage products for the 23 RSS DNA fragment are 162 and 123 bp. (B) Nucleosomes reconstituted into the middle position of DNA fragments carrying the 12- or 23-bp spacer RSS were incubated with NURF in the presence or absence of ATP and analysed by native gel electrophoresis. The mobilities of nucleosomes in the centre or at the ends of the fragment are shown in cartoons to the left.
One possible reason for the weak NURF effect is that optimal conditions for RAG cutting differ from those for nucleosome mobilization by NURF (McBlane et al., 1995; Hamiche et al., 1999). Therefore, we tried a variety of conditions, but none enhanced cutting above that which we had already observed. We next tested the ability of NURF simply to slide nucleosomes off the RSS. Reconstitutes were incubated with NURF under optimal conditions for nucleosome mobilization (Hamiche et al., 1999). The complexes were separated on a non-denaturing gel and the nucleosome positions compared to those without NURF. In this assay, the mobility on native gels is least when the nucleosome is centrally positioned on the DNA fragment and increases linearly as the nucleosome is positioned closer to the end. Using nucleosomes reconstituted over either the 12- or 23-bp spacer RSS, NURF caused relatively little ATP-dependent alteration in nucleosome positions (Figure 2B). This is not because the NURF preparation is inactive, since significant mobilization was observed with the same preparation on different nucleosome templates (see Figure 4C). However, the weak NURF effect does suggest that a mammalian equivalent of the NURF complex is unlikely to mediate nucleosome disruption for recombination in vivo.
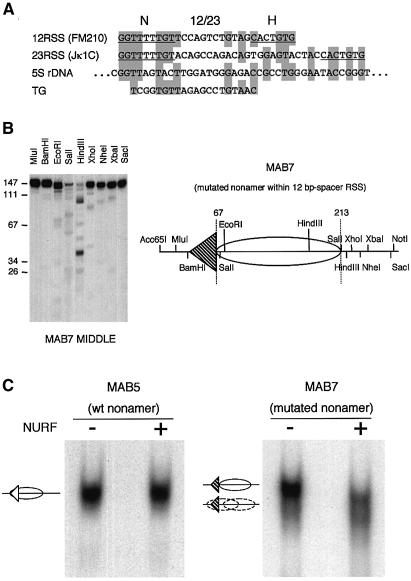
Fig. 4. Mutation of the conserved nonamer affects nucleosome positioning and mobility. (A) Alignment of the pFM210 12-bp spacer RSS and the Jκ1C 23-bp spacer RSS with the sea urchin 5S rDNA nucleosome positioning sequence and the artificial nucleosome positioning sequence TG (Shrader and Crothers, 1989). The nonamer (N) and hexamer (H) sequences are underlined. (B) Nucleosome mapping on the Acc65I–NotI fragment from pMAB7 containing a mutated nonamer sequence within the 12-bp spacer RSS was performed as described above using ‘MIDDLE’ nucleosome complex. The major position is shown schematically to the right. (C) Nucleosome sliding assay. Nucleosomes reconstituted in the MIDDLE position of the pMAB5 Acc65I–NotI or the pMAB7 Acc65I–NotI fragment were incubated with NURF and analysed by native gel electrophoresis.
Relocation of the RSS is mirrored by relocation of the nucleosome
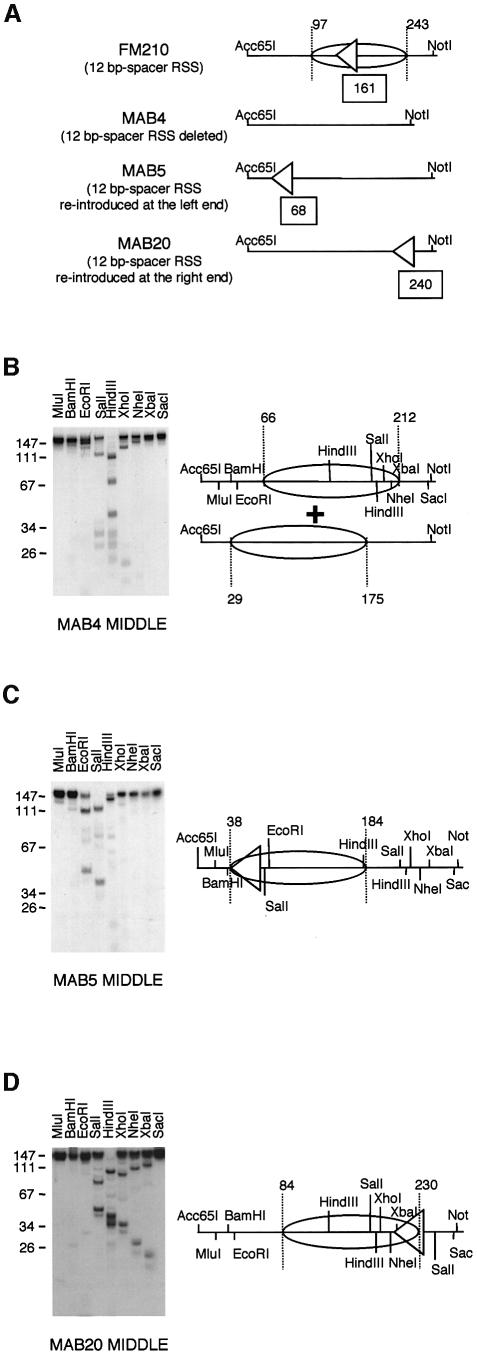
NURF mobilizes nucleosomes to the most thermally favoured position (Kang et al., 2002). We reasoned that the weak sliding effect of NURF might be due to the fact that the RSS itself confers an energetically favoured nucleosome position. Indeed, relatively little mobilization by NURF had been observed previously using nucleosomes reconstituted onto the positioning sequence from the sea urchin 5S rRNA gene (Hamiche et al., 1999). To determine whether the RSS is indeed responsible for nucleosome positioning, we deleted the 12 RSS in the context of the 284 bp fragment. Reconstitution of this fragment from pMAB4 into nucleosomes and mapping of the positions showed that two new positions exist, both of which differ from those on the fragment carrying the 12 RSS (Figure 3B).
Fig. 3. The position of nucleosomes is influenced by the position of the RSS. (A) Schematic representation of the relocation of the 12-bp spacer RSS from the centre of the Acc65I–NotI fragment in plasmid pFM210 to the left end in plasmid pMAB5, or the right end in MAB20. The RAG cutting site is indicated by a boxed number. (B–D) Mapping of nucleosome positions on the pMAB4, pMAB5, pMAB20 Acc65I–NotI fragments, respectively. Restriction enzyme analysis of the 147-bp fragments obtained following micrococcal nuclease digestion of ‘MIDDLE’ nucleosome complexes is on the left. A summary of the nucleosome positions is shown on the schematic drawing to the right.
In the original reconstitution on the wild-type 12 RSS fragment, most of the nucleosomes in the ‘end’ complex mapped to the right end of the DNA fragment (Figure 1C). To test further whether the RSS influences nucleosome positioning, we re-introduced the RSS at the potentially less favourable left end of the fragment between 40 and 68 bp (Figure 3A). Mapping of nucleosomes reconstituted onto the new 277-bp fragment from pMAB5 showed a redistribution of positions. Most of the slower migrating nucleosome complex, representing nucleosomes in the centre of the DNA fragment, mapped to a new position over the RSS (Figure 3C). Moreover, an increased proportion of the nucleosomes in the ‘end’ complex now mapped to the left side of the fragment and covered the RSS (data not shown).
To further test the ability of the RSS to position nucleosomes, we introduced the RSS into yet another position, between the original central position and the right end of the fragment. Mapping of nucleosomes reconstituted onto this fragment from pMAB20 showed that the nucleosome now adopts a further new position that covers the re-positioned RSS (Figure 3D). This suggests that the RSS indeed induces nucleosome positioning.
We next wished to examine whether other RSSs are also capable of causing nucleosome positioning. To this end, we cloned seven further RSSs (including the Jκ1 RSS) to the less favoured left end of the fragment from pMAB4. In all cases except the RSS from TCRγ5, reconstitution of nucleosomes onto these fragments resulted in the generation of a middle position very similar to that found on the fragment from pMAB5 (Supplementary figure 1, available at The EMBO Journal Online). Moreover, increased nucleosome deposition to the left end compared to the fragment from pFM210 was observed (Supplementary figure 1; data not shown). Thus, the majority of the RSSs tested are capable of causing nucleosome positioning.
The nonamer of the RSS confers nucleosome positioning
Nucleosome positioning can be influenced by a number of elements, including DNA sequences that favour nucleosome wrapping (reviewed in Richmond and Widom, 2000). Specifically, A/T bases bend more easily to compress their minor grooves and G/C sequences preferentially compress their major grooves (Drew and Travers, 1985). Thus, a preferred sequence for optimal nucleosome wrapping is (A/T)3nn(G/C)3nn (Shrader and Crothers, 1989). To gain an insight into which part of the RSS might position nucleosomes, the 12 (VκL8) and 23 (Jκ1) RSS used in the reconstitution experiments were aligned with the sea urchin 5S rRNA gene and with the artificial nucleosome positioning sequence, TG (Shrader and Crothers, 1989). The best alignment was seen with the conserved nonamer of the RSS (Figure 4A). We therefore mutated the nonamer in the context of the repositioned 12 RSS in plasmid pMAB5 to generate plasmid pMAB7. A 277-bp DNA fragment from pMAB7 was then reconstituted into a nucleosome and the positions mapped. Now, the nucleosomes were no longer positioned over the RSS but rather most mapped to one of the two positions found in the absence of an RSS (compare Figure 4B with 3B).
We predicted that if the RSS indeed confers an energetically favoured position, this might explain why NURF had a relatively small effect on sliding nucleosomes (Figure 2B). We therefore compared the sliding activity of NURF on a wild-type RSS template (from pMAB5) with a template containing a mutated nonamer (from pMAB7). Considerably greater sliding by NURF was observed on the fragment lacking a functional nonamer: all the nucleosomes are now moved from their original position (Figure 4C). This suggests that the nonamer indeed contributes to the generation of an energetically favoured nucleosome position.
To test this in a different sequence context, we mutated the nonamer in the context of the 23 RSS from Jκ1. As for the 12 RSS, this resulted in an altered nucleosome position and increased nucleosome mobility compared to that observed with a wild-type nonamer sequence (data not shown). Moreover, the RSS from TCRγ5 failed to position nucleosomes (Supplementary figure 1). This RSS lacks a consensus nonamer sequence (Table I), further supporting the idea that the consensus nonamer is instrumental in causing nucleosome positioning.
Table I. Sequence comparison of consensus and cryptic RSSs.
| Species | Locus/plasmid | Heptamer | Spacer | Nonamer |
|---|---|---|---|---|
| Consensus | CACAGTG | 12/23 | ACAAAAACC | |
| Mouse | Ig VκL8 | CACAGTG | 12 | ACAAAAACA |
| Ig Jκ1 | CACAGTG | 23 | ACAAAAACC | |
| Ig Vκ167 | CACAGTG | 12 | ACAAAAACC | |
| Ig Vκcy9 | CACAGTG | 12 | ACATAAACC | |
| Ig Vκdv36 | CACAGTG | 12 | ACAAAAACC | |
| Ig Vκ21c | CACAGTG | 12 | ACAAAAACC | |
| Ig Vκ24 | CACAGTG | 12 | ACAAAAACC | |
| TCRγ5 | CACAATG | 23 | ACTGAAGAG | |
| Human | TCRαJ34 | CACAGTG | 12 | ACAAAAACC |
| Ig Vκ2–24 | CACAGTG | 12 | ACAAAAACC | |
| TAL2 | CACTGTG | 13 | ATAAAAATA | |
| LMO2 | CACAGTA | 12 | GCAATAATT | |
| Designed | pMAB7/pMAB9 | CACAGTG | 12 (VκL8) | GGGCCCGGG |
| pMAB12 | CACAGTG | 12 (VκL8) | ATAAAAATA (TAL2) | |
| pMAB13 | CACAGTG | 12 (VκL8) | GCAATAATT (LMO2) | |
| pMAB15 | CACTGTG (TAL2) | 13 (TAL2) | ATAAAAATA (TAL2) |
The sequences of the heptamer and nonamer from various RSSs are shown. The bases that differ from the consensus are underlined. The designed sequences show the substitutions of the mutated or cryptic RSS for the consensus RSS and are named according to the plasmid in which they were constructed.
The nonamer also causes nucleosome positioning in vivo
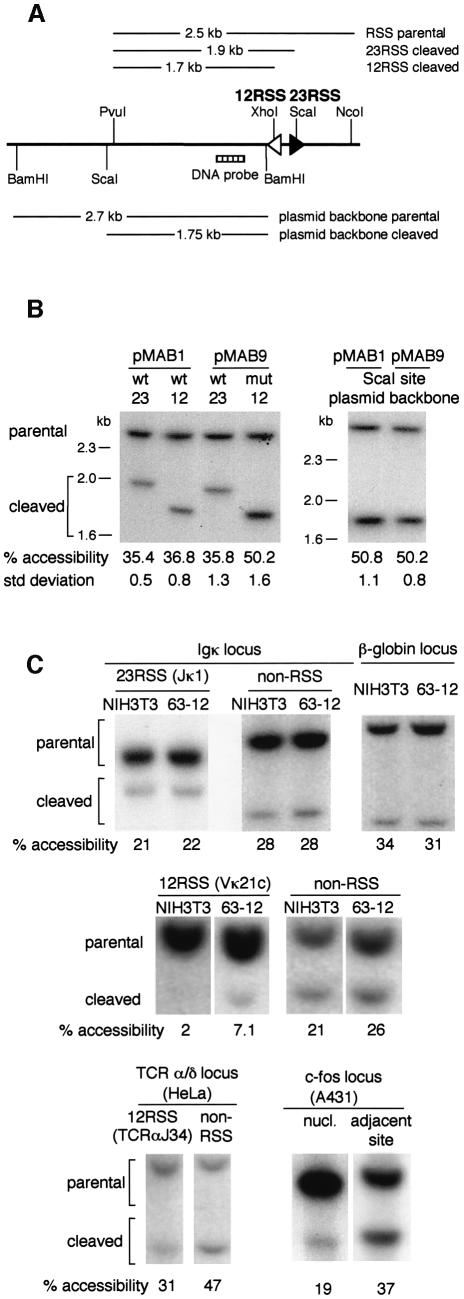
We next examined whether the RSS could also confer nucleosome positioning in vivo. Restriction enzymes cutting is considerably reduced on nucleosome templates (reviewed in Richmond and Widom, 2000) and thus they can be used to quantitatively assay nucleosome-mediated protection in vivo (Fascher et al., 1993; Svaren and Horz, 1997). We therefore used restriction enzymes to assay whether the RSS are preferentially protected. Initially, the 12 RSS within the recombination reporter plasmid, pJH200 (Lieber et al., 1987) was tagged with a restriction site for XhoI within the non-conserved spacer. A ScaI site already exists within the spacer of the 23 RSS (Figure 5A). To achieve chromatin formation on the plasmids following transient transfection, the plasmids also carry a polyoma large T antigen and origin of replication. NIH 3T3 cells were transfected with plasmids carrying either the consensus or mutated RSS nonamer; 64 h later, nuclei were prepared and the accessibility of the RSS analysed on reporter plasmids carrying the wild-type (pMAB1) or mutated 12 RSS (pMAB9) by digestion with XhoI and ScaI. The restriction enzyme had been previously titrated and an amount was used that digests the chromatin to completion. As a control, accessibility to ScaI at a site distant from the RSS was also analysed. Equivalent accessibility at the mutated 12 RSS and at this control site was observed (50% accessibility). In contrast, the wild-type 23 RSS on the same plasmid had considerably reduced accessibility (36%). Similar reduced accessibility was observed at the wild-type 12 and 23 RSS on pMAB1 (37 and 35%, respectively; Figure 5B). This suggests that the nonamer of the RSS promotes nucleosome positioning in vivo on the chromatinized plasmid templates.
Fig. 5. Effects of the nonamer sequence on the accessibility of the RSS in vivo. (A) Partial map of the plasmid recombination substrates, pMAB1 and pMAB9. The 12-bp spacer RSS (white arrowhead) and the 23-bp spacer RSS (black arrowhead) are indicated. The RSS sequences contain restriction enzyme sites for XhoI (12-bp spacer RSS) or ScaI (23-bp spacer RSS). In pMAB9 the sequence of the nonamer within the 12-bp spacer RSS is mutated. Following digestion with ScaI or XhoI in nuclei, DNA was digested with PvuI and NcoI to give the RSS parental band or with BamHI to give the parental band of the plasmid backbone. A map of the expected sizes for parental bands and digestion within the RSS is given above the plasmid diagram; that for accessibility of the plasmid backbone ScaI site is given below the plasmid diagram. The Southern probe is indicated. (B) Southern analysis of pMAB1 and pMAB9 after transfection into NIH 3T3 cells. Nuclei were prepared from transfected cells and subjected to restriction enzyme cleavage by either XhoI or ScaI as described in (A). Quantitation of the average accessibility from five experiments and the standard deviation is given below the blots. (C) Analysis of RSS accessibility at endogenous loci. Accessibility of Jκ1 23 RSS to ScaI is shown in NIH 3T3 cells and the lymphoid cell line, 63-12. This is compared to accessibility at two other ScaI sites that are not associated with an RSS: one within the kappa locus and one within the β-globin locus. Accessibility of the Vκ21c 12 RSS to StyI and the human TCRαJ34 12 RSS to BglII are compared with the accessibility of adjacent sites in NIH 3T3 and 63-12 cells and in HeLa cells, respectively. As a control, cutting at a MspI site, 25 bp within a highly positioned nucleosome at the c-fos promoter is compared with cutting at an adjacent site within the c-fos locus in A431 cells. All experiments were internally controlled: the cutting at the RSS was compared with the cutting at an adjacent site using DNA prepared from the same digested nuclei sample.
Since it is not possible to verify whether all the transfected templates are fully chromatinized, we next tested whether there is a similar change in accessibility when the RSSs are integrated into genomic DNA. The region of the plasmid carrying the RSSs was excised together with at least 1 kb of flanking DNA and was introduced stably into NIH 3T3 cells. Analysis of RSS accessibility in eight lines carrying wild type and 11 lines carrying the mutated RSS was carried out as above. On average, the wild-type RSSs (28% accessible) are 9% more protected from restriction enzyme accessibility than the mutated RSS (37% accessible; Table II). This difference was shown to be statistically significant (P = 0.016, t-test).
Table II. Analysis of accessibility of wild type and mutated 12 RSSs in stable lines.
| Cell line | Percent accessibility | |
|---|---|---|
| Wild-type 12 RSS | 1–1 | 23 |
| 1–3 | 29 | |
| 1–4 | 35 | |
| 1–5 | 28 | |
| 1–6 | 19 | |
| 1–7 | 26 | |
| 1–8 | 30 | |
| 1–9 | 34 | |
| Average | 28 (±5) | |
| Mutant 12 RSS | 9–1 | 43 |
| 9–2 | 40 | |
| 9–3 | 45 | |
| 9–4 | 22 | |
| 9–5 | 42 | |
| 9–6 | 40 | |
| 9–7 | 34 | |
| 9–8 | 31 | |
| 9–9 | 21 | |
| 9–10 | 47 | |
| 9–11 | 39 | |
| Average | 37 (±8.4) |
The percent accessibility of restriction endonucleases that cut at the RSS for individual lines is shown. The average accessibility for the wild-type and mutant RSS is given after the numbers for the individual lines.
Accessibility of endogenous RSSs is reduced
To determine whether nucleosomes are also positioned over endogenous RSSs, we made use of the ScaI restriction site that lies within the non-conserved spacer of the 23 RSS of Jκ1 (described above). Accessibility of the Jκ1 RSS was compared with an adjacent ScaI site within the kappa locus and a site within the β-globin locus (Figure 5C). Consistent with our data in stable lines, the RSS is 7–13% less accessible than the non-RSS ScaI sites. Moreover, the decreased accessibility of the RSS compared with an adjacent ScaI site in the locus indicates that it is nucleosome positioning, rather than higher order chromatin structure, that most likely mediates the protection. We find similarly that a StyI site close to the mouse Vκ21c RSS is 19% less accessible than an adjacent StyI site (Figure 5C), that a DraIII site within the consensus 12 RSS of Vκ2–24 in the human kappa locus is 7–17% less accessible than two DraIII sites within the TAL2 locus (Figure 6B) and that a BglII site at the human TCRαJ34 RSS is 16% less accessible than an adjacent site (Figure 5C). To determine the degree of protection from restriction enzyme cutting that might be expected for a positioned nucleosome, we examined the highly positioned nucleosome at the c-fos promoter in A431 cells (Herrera et al., 1997): we find 19% cutting at an MspI site 25 bp within the nucleosome and 37% accessibility at an adjacent site within the locus (Figure 5C). This compares favourably with the degree of protection we observe at RSSs. Taken together, these data suggest that RSSs are preferentially protected in vivo, most likely by the positioning of nucleosomes at the RSSs.
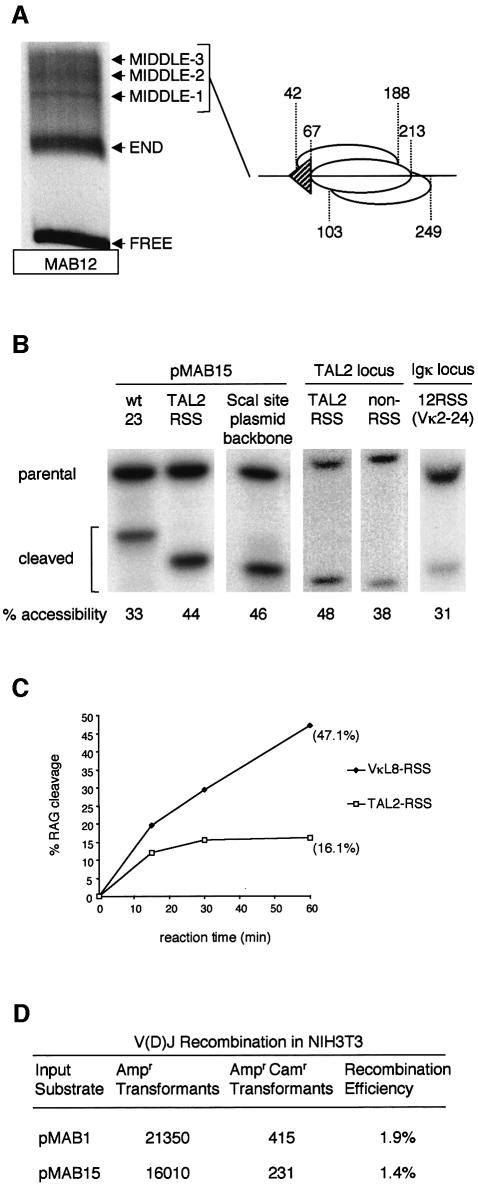
Fig. 6. Loss of nucleosome positioning at non-consensus RSSs is correlated with increased recombination. (A) Reconstitution of nucleosomes onto a 277-bp fragment where the nonamer has been substituted by the cryptic nonamer from TAL2. Free DNA and nucleosomes in the END and MIDDLE positions are indicated. The predominant complexes (middle-1, -2, -3) were excised and the nucleosome positions mapped. The various positions detected are shown diagrammatically to the left. Identical results were obtained using the LMO2 cryptic nonamer (data not shown). (B) Accessibility of the non- consensus nonamer RSSs in vivo. The TAL2 cryptic RSS was substituted for the 12 RSS in pMAB1 to generate pMAB15. Mutation of 1 bp within the linker of the cryptic RSS created an XhoI site. Accessibility of the cryptic RSS, the 23 RSS and the neutral ScaI site was analysed for the transfected, chromatinized plasmid templates as described above. Quantitation of the accessibility (an average of at least three experiments) is shown below the blot. Accessibility of the endogenous TAL2 locus was analysed in HeLa cells using a DraIII site within the heptamer of the RSS. This was compared with a neighbouring DraIII site that lies distant to the RSS and to the accessibility of a DraIII within a consensus 12 RSS at Vκ2–24. (C) RAG cutting at non-consensus and consensus RSS in vitro. Naked DNA fragments covering the TAL2 cryptic RSS and the VκL8 consensus RSS were incubated with purified RAG proteins for increasing times in vitro. The extent of RAG cutting was analysed by native gel electrophoresis and was quantitated using a PhosphorImager. (D) Recombination at non-consensus and consensus RSSs. Plasmids carrying either a consensus (pMAB1) or cryptic (pMAB15) RSS were used as substrates in a recombination assay. Reporter plasmids that have undergone recombination are both ampicillin and chloramphenicol resistant whereas those that have not are only ampicillin resistant. The number of colonies given is the sum of six experiments.
We also examined whether the accessibility of the RSSs increases in cells undergoing rearrangement. However, in neither the proB cell line, 63-12 (Figure 5C) nor in 103-BCL/2 cells that had been induced to undergo rearrangement for 12 h (Supplementary figure 2) was any significant increase in accessibility observed. We suggest that since recombination is a stochastic process, changes in accessibility at a given RSS are likely to occur in only a few cells in the population; this would not be detected by this assay.
Loss of nucleosome positioning correlates with increased recombination
We next wanted to exploit our finding that RSSs with non-consensus nonamers fail to position nucleosomes to investigate the effect of nucleosome positioning on recombination efficiency. First, we investigated whether other, functional, non-consensus nonamer RSSs indeed fail to position nucleosomes. Some of the cryptic RSSs found at the breakpoints of chromosomal translocations lack consensus nonamers (Table I). For example, the TAL2 cryptic RSS has solely A and T bases within the nonamer; such sequences are known to disfavour nucleosome positioning (Krajewski, 2002). The RSSs from the TAL2, TAL1 and LMO2 loci are, however, bona fide targets for RAG cutting in plasmid recombination assays (Raghavan et al., 2001; Marculescu et al., 2002).
To test whether these RSSs position nucleosomes in vitro, the nonamer sequences associated with TAL2 and LMO2 cryptic RSSs were substituted for the consensus nonamer in pMAB5. For both sequences, reconstitution of the equivalent 277-bp fragments into nucleosomes in vitro generated a distinct complex at the end of the fragment and a smear of complexes at a variety of positions closer to the middle of the fragment (Figure 6A). As predicted from our earlier findings with the mutated nonamer and the RSS from TCRγ5, isolation and mapping of the predominant complexes from the middle positions showed that the nucleosome positioning is lost at the non-consensus nonamer RSSs (schematic in Figure 6A; Supplementary figure 3).
To test whether the non-consensus nonamer RSSs also resulted in loss of nucleosome positioning in vivo, the entire RSS from TAL2 was substituted for the consensus 12 RSS in pMAB1. Analysis of the accessibility of the RSS following transfection into NIH 3T3 cells showed that an XhoI site within the cryptic RSS was 11% more accessible than the adjacent 23 RSS (Figure 6B). Moreover, a similar change in accessibility was observed in the genomic TAL2 locus: the accessibility of a DraIII restriction site within the cryptic RSS of TAL2 is 10% more accessible than a DraIII site distant from the RSS. Thus, in vivo, as in vitro, nucleosome positioning over the non-consensus nonamer RSS is considerably reduced.
We predicted that if non-consensus nonamer RSSs are indeed more accessible, then this should increase the probability of these RSSs being used in recombination. We therefore compared the efficiency of the TAL2-RSS and the VκL8 consensus RSS in a plasmid recombination assay. Recombination reporter vectors carrying a consensus 23 RSS and either a cryptic (pMAB15) or consensus (pMAB1) 12 RSS were transfected into NIH 3T3 cells together with a vector that constitutively expresses RAG2 (Sadofsky et al., 1995). To assay recombination on plasmids that were predominantly chromatinized, the reporter vector (which carries the polyoma origin of replication) was allowed to replicate for 36 h prior to induction of RAG1 expression (Shockett et al., 1995). Control experiments confirmed that the RSS was maximally protected at 36 h and that inducible RAG1 expression was achieved (Supplementary figure 4). Although RAG cutting is at least 3-fold less efficient at the cryptic compared to the consensus RSS in vitro (Figure 6C), recombination at the two RSSs is almost equivalent (Figure 6D). This suggests strongly that increased accessibility at the non-consensus nonamer RSSs indeed increases the probability that they are used in the recombination reaction.
Discussion
V(D)J recombination is likely to be regulated at a number of different levels. Although it is known that nucleosomes inhibit initiation of recombination, very little is known about other factors that contribute to the regulation. We show here that the RSS itself plays a key role in repressing recombination by positioning a nucleosome over the RSS. It is likely that such nucleosome positioning will have two regulatory consequences. First, it will provide an additional level of protection from recombination: the number of RSSs that lie in linker regions between nucleosomes will be significantly decreased. Thus, RAG proteins will be less likely to gain access to the RSS following disruption of higher order chromatin structure alone. Secondly, initiation of recombination will require that nucleosomes covering the RSSs are disrupted. Although the mechanism of nucleosome disruption for V(D)J recombination is currently unknown, recruitment of the proteins involved in overcoming the nucleosome-mediated repression provides another potential point at which the reaction can be regulated.
Interestingly, nucleosome positioning is lost at some non-consensus nonamer RSSs. The poorer nucleosome positioning by these RSSs implies that these sequences have a greater probability of lying within linker DNA than consensus RSSs. Indeed, in recombination assays using chromatinized templates, the use of the TAL2 non-consensus nonamer RSS was greater than predicted based on RAG cleavage alone. This strongly supports the hypothesis that the positioning of nucleosomes over consensus RSSs regulates their use in the recombination reaction.
Nucleosome positioning by RSSs is one of the few examples to date that can be attributed with a biological function. Nucleosome positioning has been described at certain promoters including MMTV and PHO5 (Fragoso et al., 1995; Svaren and Horz, 1997). In these cases, nucleosomes are positioned such that transcription factor binding can proceed in an ordered sequence (Svaren and Horz, 1997; Di Croce et al., 1999). In two other examples, positioning of a nucleosome in promoters brings together proximal and distal transcription factor binding sites to augment factor interaction (Schild et al., 1993; Lu et al., 1995). In yet another example, the strong association of nucleosomes with disease-associated triplet repeats in the myotonic dystrophy gene was suggested to inhibit transcription (Wang et al., 1994). Despite these well-studied cases, to date, nucleosome positioning has not been shown to have a widespread role in the regulation of transcription. The positioning of nucleosomes by RSSs, on the other hand, potentially provides an additional level of regulation to V(D)J recombination.
Whilst nucleosome positioning likely provides protection at many RSSs, it is unlikely that this protection, even at consensus RSSs, is absolute. This is because nucleosome positioning at RSSs in vivo also will depend on the relative strength of the nonamer positioning sequence compared with other positioning elements in the vicinity. Consistent with this, using a restriction enzyme accessibility assay, we observed some variability in the level of protection of RSSs in vivo. Nevertheless, in all cases, the consensus RSS was less accessible than adjacent sites for the same restriction enzyme.
Preferential nucleosome positioning at consensus RSSs poses the key question of how nucleosomes are mobilized for the initiation of V(D)J recombination. Since NURF mobilizes nucleosomes only poorly from energetically favoured positions (Hamiche et al., 1999), we suggest that it is unlikely that a NURF-like remodelling complex is involved. However, ACF, another ISWI-containing nucleosome remodelling complex, can slide nucleosomes to alternative positions (Kang et al., 2002). Thus, remodelling complexes such as the mammalian CHRAC/ACF (reviewed in Wu et al., 2000) are good candidates to facilitate recombination in vivo. Similarly, SWI/SNF can mobilize nucleosomes. If the preferred positions to which it mobilizes nucleosomes are different from the most thermally stable position, then mammalian SWI/SNF complexes are also potential candidates to stimulate recombination in vivo. Alternatively, nucleosomes might be remodelled by a non-sliding mechanism. Indeed, in vitro, SWI/SNF was shown to facilitate RAG cutting on templates where the nucleosome was reconstituted onto a DNA fragment of only 150 bp (Kwon et al., 2000). In this case, it is likely that RAG accessibility was gained via the ability of SWI/SNF to remodel nucleosomes by transient global disruption of histone–DNA contacts (reviewed in Narlikar et al., 2002). Exactly how recombination is stimulated in vivo, however, remains to be determined.
The positioning of a nucleosome over an RSS suggests that an important step in the reaction is the recruitment of remodelling complexes to recombining loci. Since RAGs cannot bind to the RSS when it is constrained by a nucleosome, RAG proteins bound to the RSS cannot be responsible for the recruitment of remodelling complexes as has been suggested for recruitment by transcription factors (reviewed by Cairns and Kingston, 2000). Changes in higher order chromatin structure accompany loci undergoing recombination. These include changes in nuclear localization, DNA demethylation, increased histone acetylation, increased DNase I accessibility and the presence of sterile transcripts (reviewed in Sleckman, 1996; Mostoslavsky et al., 1998; McMurry and Krangel, 2000; Kosak et al., 2002). It seems likely that the regulated opening of these chromatin domains will allow access to the remodelling complexes and thus subsequent initiation of V(D)J recombination. Exactly how the remodelling complexes are recruited, however, is unknown. Evidence exists for proteins that increase the specificity of RAGs binding to RSSs (Muegge et al., 1993; Stanhope-Baker et al., 1996). One interesting possibility is that the binding of such proteins assists in the recruitment of nucleosome remodelling complexes.
Taken together, our data show that nucleosomes are preferentially positioned at consensus RSSs. This implies that nucleosomes must be mobilized from energetically favoured positions to permit RAG cutting. The mechanism by which remodellers are recruited to their sites of action at wild-type RSSs and how they overcome the energetic barrier to nucleosome mobilization will be the subjects of future studies.
Materials and methods
DNA fragments and plasmids
Substrates for reconstitution of mononucleosomes in vitro. pFM210 was as described previously (McBlane and Boyes, 2000). The 12-bp spacer RSS is derived from VκL8, with one base of the nonamer altered to match the consensus sequence. Digestion of pFM210 with Acc65I and NotI liberates a 284-bp fragment used as a substrate for nucleosome reconstitution. The 12-bp spacer RSS within pFM210 was deleted by cutting with BstNI and SphI to generate pMAB4. An oligonucleotide containing the VκL8 RSS was inserted into BamHI–EcoRI cut pMAB4 to generate pMAB5. pMAB20 has the same oligonucleotide inserted but at the XbaI site. pMAB7 is identical to pMAB5 except that the nonamer was changed to 5′-CCCGGGCCC-3′ by insertion of a mutant oligonucleotide. The plasmids pMAB12 and pMAB13 carrying the mutated nonamer sequences found at the cryptic RSSs of the TAL2 and LMO2 genes were generated by substituting the nonamer of the wild-type RSS in pMAB5 with respective mutant nonamer sequences. pJκ1C was generated as described previously (McBlane and Boyes, 2000) except that the primers, 5′-TGCGTCTAGAGCCCAAGCGCTTCCACGCAT-3′ and 5′-ACCG TCTAGAGTATCTTTGCCTTGGAGAGTG-3′ were used. The PCR product was cut with XbaI and inserted into the XbaI site of pUC18.
Plasmids for transfection into tissue culture cells. pMAB1 was derived from the recombination substrate pJH200 (Lieber et al., 1987) by introducing a restriction enzyme cleavage site for XhoI into the VκL8 spacer. pMAB9 is identical to pMAB1 except the 12-bp spacer RSS nonamer sequence was mutated as in pMAB7. In pMAB15 the entire VκL8 RSS is substituted by the cryptic TAL2 RSS.
Cell culture and transfections
NIH 3T3 fibroblasts were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) and 10% calf serum. Transfections were performed by calcium phosphate precipitation. The cells were subjected to a glycerol shock 16 h after transfection and were harvested 64 h after transfection.
Stable lines carrying the mutated and wild-type RSSs were constructed in NIH 3T3 cells by co-transfection of a 4.2 kb EagI–XbaI fragment from pMAB1 or pMAB9 with a 4.4 kb BamHI–ClaI fragment that carries the hygromycin resistance gene from pREP7 (Invitrogen) at a ratio of 4:1. Two days after transfection, the cells were grown in selection medium containing 0.4 mg/ml hygromycin. Individual colonies, visible after ∼7 days, were expanded in hygromycin-containing medium.
63-12 cells were derived from B cells of RAG2–/– mice (Shinkai et al., 1992). They were cultured in RPMI supplemented with 10% fetal calf serum (FCS) and 50 µM β-mercaptoethanol.
HeLa and A431 cells were grown in DMEM and 10% FCS.
Proteins
RAG1 (amino acids 384–1008) and RAG2 (amino acids 1–387) were co-expressed as MBP fusion proteins, MR1 and MR2, respectively (van Gent et al., 1995). Purification was as described previously (McBlane et al., 1995).
Core histones were prepared from chicken erythrocytes (Boyes, 1999). NURF was purified from nuclear extracts of 0–12 h Drosophila embryos (Hamiche et al., 1999). Recombinant HMGB1 (truncated form) was a generous gift from Dr Kevin Hiom.
Nucleosome reconstitution and nucleosome position mapping
Twenty micrograms of 3′ end-labelled substrates containing a single RSS were reconstituted into mononucleosomes by salt-urea dialysis followed by gel purification as described previously (Boyes, 1999). The position of the histone octamers was determined by micrococcal nuclease digestion and restriction enzyme mapping as described previously (Clark and Felsenfeld, 1991) except that 100 ng of reconstitute was digested with 0.3 U of micrococcal nuclease.
RAG cleavage assay
32P 3′ end-labelled substrate (free DNA or nucleosome) (20 fmol) of was incubated with MR1 and MR2 (∼25–100 ng of each protein) for 1 h at 35°C in 1× RAG cleavage buffer (25 mM MOPS pH 7.0, 0.5 µg/µl bovine serum albumin, 3 mM MgCl2, 1 mM MnCl2, 1 mM dithiothreitol, 45 mM KCl, 1 mM ATP) in the presence or absence of 16 ng NURF and/or 10 ng HMGB1. The reaction was stopped by the addition of 6 µg of non-specific competitor DNA and cleavage products were separated on a non-denaturing 4.5% polyacrylamide gel for 4 h at 100 V.
Nucleosome mobilization assay
Nucleosome mobilization was performed essentially according to Hamiche et al. (1999) using 80 fmol of 32P-labelled mononucleosomes and 0.9 ng of NURF per reaction.
Restriction endonuclease digestion in nuclei
Nuclei from 1 × 107 cells were prepared and resuspended in 500 µl of buffer F as described previously (Boyes and Felsenfeld, 1996). Two aliquots of 250 µl were taken. For analysis of accessibility of pMAB1 or pMAB9 (or fragments from these plasmids stably integrated into NIH 3T3 cells) 100 U of XhoI or 150 U of ScaI were added and the reactions incubated at 37°C for 1 h. Analysis of the accessibility at endogenous loci was performed by digesting nuclei with ScaI for Jκ1, StyI for Vκ21c, BglII for TCRαJ34, DraIII for TAL2 and Vκ2–24 and MspI for the human c-fos promoter. The reactions were terminated by addition of EDTA to 10 mM. Preparation of DNA was performed as described previously (Boyes and Felsenfeld, 1996).
Southern blotting and hybridization
Ten micrograms of each DNA sample was digested with 60 U of each of the indicated restriction enzymes for 5 h at 37°C followed by the addition of 0.25 mg/ml proteinase K and 0.5% SDS. Following incubation at 37°C for 1 h, the DNA was extracted and run on a 0.9% agarose gel at 90 V for 4.5 h. Transfer was to NytranN membrane (Schleicher and Schuell) in 0.4 M NaOH for 1 h. The DNA probe was a 359-bp AflIII–HindIII fragment from pMAB1. Southern probes for analysis of the endogenous loci were prepared via PCR. Primer sequences are available on request. Hybridization was at 65°C overnight in 0.25 M sodium phosphate pH 7.2, 7% SDS and 1 mM EDTA. Washes were at 65°C in 20 mM sodium phosphate pH 7.2 and 1% SDS. Signal intensities were quantitated using a PhosphorImager (Molecular Dynamics).
Plasmid recombination assay
Quantitation of V(D)J recombination of extrachromosomal substrates was performed as described previously (Sadofsky et al., 1995) except that RAG1 was inducibly expressed after 36 h via the tetracycline-inducible system (Shockett et al., 1995).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank K.Hiom for the generous gift of HMGB1, D.Schatz for the pTet-tTAk vector, and M.Merkenschlager, A.Ashworth and P.Varga-Weisz for helpful comments on the manuscript. We are also grateful to L.Pearl, C.Wu and members of the Boyes laboratory for helpful suggestions. F.M. was assisted by U.Grazini. This work is supported by the MRC (UK). M.B. is a recipient of a long-term EMBO fellowship. F.M. is supported by the Italian Association for Cancer Research. J.B. gratefully acknowledges support from the Lister Institute of Preventive Medicine.
References
- Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109 Suppl., S45–S55. [DOI] [PubMed] [Google Scholar]
- Boyes J. (1999) Preparation of chromatin templates for transcription studies. In Latchman,D.S. (ed.), Transcription Factors: A Practical Approach. Oxford University Press, Oxford, UK, pp. 229–260. [Google Scholar]
- Boyes J. and Felsenfeld,G. (1996) Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J., 15, 2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R. and Kingston,R.E. (2000) The SWI/SNF family of remodelling complexes. In Elgin,S.C.R. and Workman,J.L. (eds), Chromatin Structure and Gene Expression. Oxford University Press, New York, NY, pp. 97–113. [Google Scholar]
- Clark D.J. and Felsenfeld,G. (1991) Formation of nucleosomes on positively supercoiled DNA. EMBO J., 10, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L., Koop,R., Venditti,P., Westphal,H.M., Nightingale,K.P., Corona,D.F., Becker,P.B. and Beato,M. (1999) Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell, 4, 45–54. [DOI] [PubMed] [Google Scholar]
- Drew H.R. and Travers,A.A. (1985) DNA bending and its relation to nucleosome positioning. J. Mol. Biol., 186, 773–790. [DOI] [PubMed] [Google Scholar]
- Fascher K.-D., Schmitz,J. and Horz,W. (1993) Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol., 231, 658–667. [DOI] [PubMed] [Google Scholar]
- Fragoso G., John,S., Roberts,M.S. and Hager,G.L. (1995) Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev., 9, 1933–1947. [DOI] [PubMed] [Google Scholar]
- Golding A., Chandler,S., Ballestar,E., Wolffe,A.P. and Schlissel,M.S. (1999) Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J., 18, 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Herrera R.E., Nordheim,A. and Stewart,A.F. (1997) Chromatin structure analysis of the human c-fos promoter reveals a centrally positioned nucleosome. Chromosoma, 106, 284–292. [DOI] [PubMed] [Google Scholar]
- Kang J.G., Hamiche,A. and Wu,C. (2002) GAL4 directs nucleosome sliding induced by NURF. EMBO J., 21, 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Skok,J.A., Medina,K.L., Riblet,R., Le Beau,M.M., Fisher,A.G. and Singh,H. (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science, 296, 158–162. [DOI] [PubMed] [Google Scholar]
- Krajewski W.A. (2002) Histone acetylation status and DNA sequence modulate ATP-dependent nucleosome repositioning. J. Biol. Chem., 277, 14509–14513. [DOI] [PubMed] [Google Scholar]
- Kwon J., Imbalzano,A.N., Matthews,A. and Oettinger,M.A. (1998) Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell, 2, 829–839. [DOI] [PubMed] [Google Scholar]
- Kwon J., Morshead,K.B., Guyon,J.R., Kingston,R.E. and Oettinger,M.A. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell, 6, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Langst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- Lewis S.M., Agard,E., Suh,S. and Czyzyk,L. (1997) Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol., 17, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1987) Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev., 1, 751–761. [DOI] [PubMed] [Google Scholar]
- Linxweiler W. and Horz,W. (1984) Reconstitution of mononucleosomes: characterization of distinct particles that differ in the position of the histone core. Nucleic Acids Res., 12, 9395–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath,L.L. and Elgin,S.C. (1995) The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J., 14, 4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marculescu R., Le,T., Simon,P., Jaeger,U. and Nadel,B. (2002) V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J. Exp. Med., 195, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane F. and Boyes,J. (2000) Stimulation of V(D)J recombination by histone acetylation. Curr. Biol., 10, 483–486. [DOI] [PubMed] [Google Scholar]
- McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- McMurry M.T. and Krangel,M.S. (2000) A role for histone acetylation in the developmental regulation of VDJ recombination. Science, 287, 495–498. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh,N., Kirillov,A., Pelanda,R., Cedar,H., Chess,A. and Bergman,Y. (1998) Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev., 12, 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge K., West,M. and Durum,S.K. (1993) Recombination sequence-binding protein in thymocytes undergoing T-cell receptor gene rearrangement. Proc. Natl Acad. Sci. USA, 90, 4151–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Kirsch,I.R. and Lieber,M.R. (2001) Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J. Biol. Chem., 276, 29126–29133. [DOI] [PubMed] [Google Scholar]
- Richmond T.J. and Widom,J. (2000) Nucleosome and chromatin structure. In Elgin,S.C.R. and Workman,J.L. (eds), Chromatin Structure and Gene Expression. Oxford University Press, New York, NY, pp. 1–23. [Google Scholar]
- Roth D.B. and Roth,S.Y. (2000) Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell, 103, 699–702. [DOI] [PubMed] [Google Scholar]
- Sadofsky M.J., Hesse,J.E., van Gent,D.C. and Gellert,M. (1995) RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev., 9, 2193–2199. [DOI] [PubMed] [Google Scholar]
- Schild C., Claret,F.X., Wahli,W. and Wolffe,A.P. (1993) A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J., 12, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y. et al. (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell, 68, 855–867. [DOI] [PubMed] [Google Scholar]
- Shockett P., Difilippantonio,M., Hellman,N. and Schatz,D.G. (1995) A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc. Natl Acad. Sci. USA, 92, 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T.E. and Crothers,D.M. (1989) Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA, 86, 7418–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman,J.R. and Alt,F.W. (1996) Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol., 14, 459–481. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P., Hudson,K.M., Shaffer,A.L., Constantinescu,A. and Schlissel,M.S. (1996) Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell, 85, 887–897. [DOI] [PubMed] [Google Scholar]
- Svaren J. and Horz,W. (1997) Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci., 22, 93–97. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., McBlane,J.F., Ramsden,D.A., Sadofsky,M.J., Hesse,J.E. and Gellert,M. (1995) Initiation of V(D)J recombination in a cell-free system. Cell, 81, 925–934. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Amirhaeri,S., Kang,S., Wells,R.D. and Griffith,J.D. (1994) Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science, 265, 669–671. [DOI] [PubMed] [Google Scholar]
- Wu C., Becker,P.B. and Tsukiyama,T. (2000) ATP-dependent chromatin remodelling by the ISWI complexes. In Elgin,S.C.R. and Workman,J.L. (eds), Chromatin Structure and Gene Expression. Oxford University Press, New York, NY, pp. 114–134. [Google Scholar]