Abstract
The yeast prion [URE3] is a self-propagating inactive form (the propagon) of the Ure2 protein. Ure2p is composed of two domains: residues 1–93—the prion-forming domain (PFD)—and the remaining C-terminal part of the protein, which forms the functional domain involved in nitrogen catabolite repression. Guanidine hydrochloride, and the overproduction of Ure2p 1–65 or Ure2–GFP have been shown to induce the elimination of [URE3]. We demonstrate here, two different curing mechanisms: the inhibition of [URE3] replication by guanidine hydrochloride and its destruction by Ure2p aggregation. Such aggregation is observed if PFD or Ure2–GFP are overproduced and in heterozygous URE2/URE2–GFP, [URE3] diploids. We found that the GFP foci associated with the presence of the prion were dead-end products, the propagons remaining soluble. Surprisingly, [URE3] propagated via the Ure2–GFP fusion protein alone is resistant to these two curing mechanisms and cannot promote the formation of foci. The relationship between aggregation, prion and Hsp104 gives rise to a model in which the propagon is in equilibrium with larger aggregates and functional protein.
Keywords: aggregation/amyloids/GFP/prion/Saccharomyces cerevisiae/[URE3]
Introduction
Changes in protein conformation may cause disease, as observed in a number of neurodegenerative pathologies, such as Alzheimer’s and Parkinson’s disease, cystic fibrosis and transmissible spongiform encephalopathies (Taylor et al., 2002). Some of these diseases are associated with nuclear mutations that change the biophysical properties of the protein leading to a pathogenic form. Eight inherited neurodegenerative diseases, including Huntington’s disease, are caused by polyglutamine expansions in the corresponding proteins (Perutz, 1999). One of the characteristics of these diseases is the formation of aggregates containing misfolded proteins. Although unrelated, the proteins involved in these diseases form structurally well-defined structures called amyloids. Amyloid is a general term related to particular physicochemical features like a fibrillar morphology, predominant β-sheet secondary structure and birefringence upon staining with Congo Red. The toxic species seems to be an intermediate generated during the production of amyloid fibrils (Bucciantini et al., 2002). The protein thought to be toxic in transmissible spongiform encephalopathies is remarkable in that it is thought to be the major, if not only component of the infectious agent. According to the prion theory, this agent propagates by converting the normal form of the protein into the abnormal prion form in a snow-ball mechanism (Prusiner, 1982). Several fragments of the mammalian prion protein have been demonstrated to be toxic in vitro (Ettaiche et al., 2000). Toxicity was found to be correlated with hydrophobicity rather than with tendency to form specific β-sheet structures or fibrillar aggregates. No strict correlation has been found between the toxicity of peptides and the formation of amyloid aggregates (Brown, 2000). The protease-resistant form of the mammalian prion protein has been generated in vitro, but, although similar to the infectious protein, this protein was not infectious (Kocisko et al., 1994; Hill et al., 1999). To date, no amyloids produced from the purified mammalian prion protein (Jackson et al., 1999) have been reported to be infectious and the exact nature of the transmissible agent remains unknown.
The yeast Saccharomyces cerevisiae possesses several prion proteins (for a review, see Fernandez-Bellot and Cullin, 2001). The first yeast protein to be described as a prion (Wickner, 1994) was Ure2p, a nitrogen-regulating protein that complexes with the transcription factor Gln3p in the cytoplasm, preventing the translocation of Gln3p to the nucleus (Drillien et al., 1973; Courchesne and Magasanik, 1988; Coschigano and Magasanik, 1991; Beck and Hall, 1999; Cardenas et al., 1999). The prion form of Ure2p makes it possible to take up ureidosuccinate (USA) and permits growth in selective medium. Many experiments carried out with [URE3] or [PSI] (a phenotype due to the prion form of Sup35p which is an essential translation termination factor) have concluded that there is a common mechanism of ‘prionization’ in yeast. In vitro, yeast prion proteins spontaneously form amyloid-like structures (Glover et al., 1997; King et al., 1997; Taylor et al., 1999; Thual et al., 1999), and the presence of [PSI] or [URE3] in yeast cells leads to the aggregation of Sup35p (Patino et al., 1996; Paushkin et al., 1996) and Ure2p, respectively (Schlumpberger et al., 2001). Fusions of prion proteins to GFP are also located in aggregates in the cells bearing the prion isoform (Patino et al., 1996; Edskes et al., 1999). Both [URE3] and [PSI] can therefore be seen as heritable amyloidoses (for a review, see Wickner et al., 2000). However, several recent articles have provided evidence against a model in which large prion aggregates are the infectious form (the propagons) (Fernandez-Bellot et al., 2000, 2002; Borchsenius et al., 2001). Ure2p aggregation has been observed in cells expressing both Ure2p and Ure2–GFP, a genetic situation that also leads to [URE3] elimination. However, we have demonstrated that the large aggregates observed with Ure2–GFP fusion proteins were not observed in the presence of [URE3], if this protein was stably produced in place of that encoded by the chromosomal URE2 allele. Furthermore, no specific aggregation was detected by simple centrifugation assay in [URE3] strain (Fernandez-Bellot et al., 2002).
We investigated these discrepancies further by combining genetic, biochemical and cellular approaches in conditions in which [URE3] is destabilized. The chemical compound guanidine-HCl (GdnHCl) cures [PSI] and [URE3] efficiently (Tuite et al., 1981; Wickner, 1994), probably by inhibiting Hsp104, which is thought to be essential for the propagation of these two yeast prions (Chernoff et al., 1995; Moriyama et al., 2000). We demonstrate here that GdnHCl eliminates [URE3] with the kinetics similar to those found for [PSI] elimination (Eaglestone et al., 2000), by blocking propagon proliferation. We found that [URE3] was eliminated by the prion-forming domain (PFD) of Ure2p and the Ure2–GFP hybrid, as previously described (Edskes et al., 1999). In these cases, the mechanism is different, based on the aggregation of Ure2p. These aggregates are in fact dead-end products rather than the propagons themselves. We found that the prion form of the Ure2–GFP fusion protein may arise and propagate independently of Hsp104. Overall, our results provide a more detailed model of prion formation, in which the roles of Hsp104 and the insoluble aggregates differ from those in the previous model (Moriyama et al., 2000; Speransky et al., 2001).
Results
GdnHCl and the prion domain of Ure2p cause the loss of [URE3] through different pathways
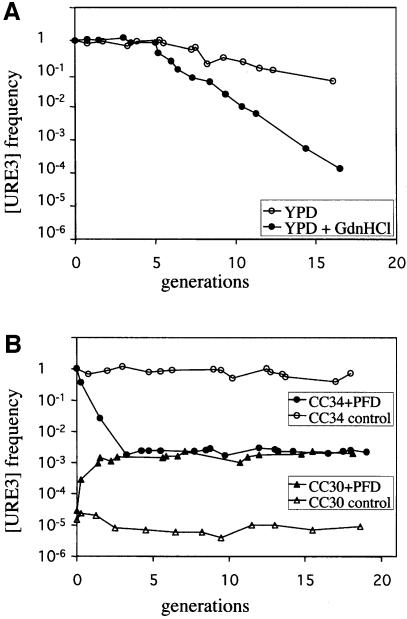
GdnHCl has been shown to lead to [PSI] loss (Tuite et al., 1981). This effect is observed only during exponential growth of the yeast cells (Eaglestone et al., 2000). The [PSI+] phenotype is assessed on the basis of the colour of the colony, and the percentage of [PSI+] cells can therefore be measured simply by determining the ratio of red to white colonies on the appropriate medium. The [URE3] phenotype permits ura2Δ strains to grow in minimal medium, without uracil, supplemented with USA. We investigated the role of GdnHCl in curing by culturing yeast cells in complete medium with or without 5 mM GdnHCl. At regular time points, cell density was determined and an aliquot of each culture was plated on minimal medium supplemented with USA or uracil. After 3–5 days at 30°C, the frequency of [URE3] was determined as the ratio ′number of colonies on USA/number of colonies on uracil′. Cell density was adjusted to ensure that it corresponds to the exponential part of the growth curve (typically between 0.1 and 2 OD600 nm). In studies of the kinetics of [URE3] loss in the presence of GdnHCl, we obtained a two-phase curve similar to that obtained for [PSI] elimination in similar conditions (Figure 1A). The second part of the curve of log ratio against number of generations is linear. This linearity is consistent with the segregation model described for [PSI], in which the percentage of yeast cells bearing the prion phenotype halves in each generation. Based on a calculation similar to that used by Eaglestone (Eaglestone et al., 2000), we determined that CC34 [URE3] strain contained ∼20 propagons. The ratio may decrease slowly if yeast cells are grown on a rich medium without GdnHCl (Figure 1A). This instability is not observed systematically (e.g. see the control, Figure 1B). When observed, the rate of [URE3] loss does not affect the rate of loss induced by curing agents beyond 17 generations, with about 10% of the cells retaining [URE3] (this value is several orders of magnitude lower in curing conditions).
Fig. 1. Kinetics of [URE3] induction and elimination following guanidine hydrochloride treatment or transient Ure2p N-terminal domain overproduction. (A) [URE3] elimination by guanidine hydrochloride is fitted by a segregational model. CC34 [URE3] cells were grown in YPD (open circle) or in YPD supplemented with 5 mM guanidine hydrochloride (closed circle). Cultures were maintained in exponential growth phase (between 0.1 and 2 OD600 nm). At regular time points over 17 generations, culture aliquots were removed, washed and plated on minimal medium supplemented with uracil or USA. [URE3] loss was estimated by calculating the frequency of Usa+ colonies (number of colonies on USA/number of colonies on uracil). (B) Kinetics of [URE3] induction and elimination by transient PFD overproduction. CC30 (URE2 [ure3°]) and CC34 (URE2 [URE3]) strains were transformed with the pYe2T control plasmid (open circle and triangle, respectively) or with pYe2T-Nter (closed circle and triangle, respectively) for overproduction of the PFD, corresponding to amino acids 1–93. Cells were cultured in galactose minimal medium supplemented with uracil and maintained in exponential growth phase. Usa+ frequency was regularly calculated from culture aliquots, which were washed and plated on glucose minimal medium supplemented with uracil or USA.
Several domains of Ure2p, when produced together with Ure2p, may promote [URE3] elimination (Edskes et al., 1999). The simplest way to test the curing properties of the various domains of Ure2p is probably to transform a [URE3] strain with a set of plasmids encoding these domains. If these domains are produced constitutively from the plasmids, then the curing efficiency is the percentage of transformants retaining [URE3]. As [URE3] may be unstable, caution is required to prevent misinterpretation of the results. Our strategy, based on the inducible GAL10-CYC1 promoter (Guarente et al., 1982), eliminates this problem. Induction of the promoter can also be used to determine the kinetic parameters of [URE3] curing by the domain considered. As expected, given that the domain of Ure2p extending from amino acid 1 to amino acid 65 cures [URE3] (Edskes et al., 1999), PFD (1–93) also efficiently cured the prion phenotype (Figure 1B). The PFD overexpression does not lead to the complete elimination of [URE3] as it also induces [URE3] (Figure 1B). The kinetics of [URE3] elimination differ remarkably from those of elimination by GdnHCl. The effect of producing the PFD is maximal after only three generations. The same curing efficiency was achieved after 12 generations in the presence of GdnHCl. The passive model of propagon inhibition cannot account for this effect and the propagons seem instead to be irreversibly destroyed by the PFD. We monitored the events leading to such elimination directly in the cell, using a fluorescent protein previously described to cure [URE3] (Edskes et al., 1999): Ure2-GFP.
Foci accumulation during the curing process
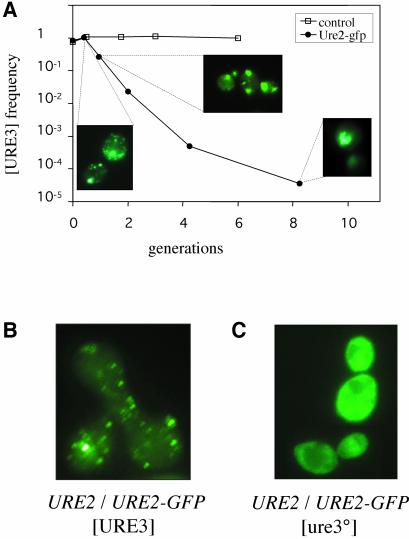
The Ure2–GFP fusion protein, when produced in large amounts (from high-copy number plasmids based on the ADH1 promoter), completely cures pre-existing [URE3] (Edskes et al., 1999). Overproduction of the chimeric Ure2–GFP protein in the CC34 strain also efficiently cured [URE3] (Figure 2A). Ure2–GFP has not been shown to induce [URE3], in contrast to the PFD. Thus, the curing effect of the fluorescent protein results in a cell population with a very low frequency of [URE3], similar to that of the background. This prion elimination is correlated with the emergence of numerous foci (Figure 2A). During the elimination process, the foci increased in size but decreased in number, from numerous foci to a single focus (for additional microscopy data, see Supplementary data, available at The EMBO Journal Online). We have demonstrated that [URE3] does not itself induce the formation of such foci (Fernandez-Bellot et al., 2002). These aggregates are therefore not due to [URE3] alone, but instead to the coexpression of Ure2–GFP and Ure2p in the presence of [URE3]. As these conditions lead to the destabilization of [URE3], these foci may correspond to an intermediate step in [URE3] elimination. We have already demonstrated that the overexpression of Ure2–GFP leads to the aggregation of this protein, and that this aggregation is not systematically associated with [URE3] formation (Fernandez-Bellot et al., 2002). To demonstrate that the aggregation observed in this study was indeed due to the destabilization of [URE3] rather than a consequence of the high level of expression, we crossed the EG12 strain (bearing a URE2-GFP allele without [URE3] and referred to as URE2-GFP [ure3°]) with the CC34 strain (URE2 [URE3]). Foci appeared in the zygotes 3 h after the cells were mixed, as soon as the mating process was completed (Figure 2B). The diploid strain resulting from this cross may have a very unstable prion phenotype. If grown on complete medium, this strain reverts to the wild-type Usa– phenotype at a very high frequency (data not shown). All of the Usa– colonies generated displayed uniform fluorescence (at least for 100 colonies tested independently; Figure 2C). This unstable [URE3] phenotype may be maintained by growth of the diploid cells in USA medium. After sporulation of this [URE3] diploid, the spores bearing the URE2-GFP allele and [URE3] (identified by growth in a medium without uracil supplemented with USA) displayed uniform fluorescence. One of these haploid strains was named EG34. Thus, no foci were observed if the URE2-GFP allele was expressed without the URE2 allele. The formation of foci required both alleles and [URE3]. This combination also resulted in [URE3] instability. We then investigated whether these foci resulted from curing activity.
Fig. 2. [URE3] curing by Ure2–GFP is related to focus formation. (A) Kinetics of [URE3] elimination following the transient overproduction of Ure2–GFP. The CC34 [URE3] strain was transformed with the pYe2T control plasmid (open square) or with pYe2T-URE2GFP (closed circle). Experimental details were as in Figure 1B. Ure2–GFP aggregation pattern is shown after 0.5 and 8 generations in galactose medium. (B) Foci formed in URE2/URE2GFP [URE3] zygotes. CC34 (URE2 [URE3]) was crossed with EG12 (URE2-GFP [ure3°]). Zygotes were observed 3 h after mixing the cells. (C) Uniform fluorescence in cured URE2/URE2-GFP diploids. CC34 X EG12 diploids from Figure 2B were grown for 15 generations on YPD. Cells were then tested on minimal medium complemented with USA. Most did not grow, indicating the loss of [URE3]. These [ure3°] cells displayed uniform fluorescence.
The frequency of cells with foci is correlated with curing frequency
As the PFD (1–93) domain of Ure2p cures [URE3] highly efficiently, we introduced this domain into the EG12 strain (URE2-GFP [ure3°]). We then allowed the PFD to be induced and tested the cell population for increases in the number of [URE3] and for the formation of foci. If these cells were plated on a glucose medium with USA but without uracil after induction, the frequency of Usa+ cells was about 10–3. Thus, PFD induces an increase in [URE3] frequency in the URE2-GFP strain similar to that observed in the URE2 strain. If a de novo [URE3] colony thus obtained was grown in galactose medium for several generations, the frequency of cells displaying foci increased from 0.1 to 1%, as previously reported (Fernandez-Bellot et al., 2002). We tested these yeast cells for [URE3] stability (by analysing growth in a glucose medium with USA and without uracil), and found that the frequency of Usa+ cells was about 90%. Two important conclusions can be drawn from this result: (i) the level of Ure2–GFP aggregation due to overproduction of the PFD was higher in presence of [URE3], but this aggregation remains anecdotal; (ii) unlike the URE2 [URE3] strain, the URE2-GFP [URE3] strain appears to be resistant to the curing effect of the PFD.
If the PFD was produced in the EG34 strain (URE2-GFP, [URE3]), its curing efficiency was also low (with about 10% of the yeast cells cured of [URE3] after 25 generations), although in the same conditions >99.9% of CC34 (URE2, [URE3]) yeast cells reverted to [ure3°]. Indeed, this resistance to curing is due to the chimeric Ure2–GFP protein, which propagates [URE3]. To understand the basis of this difference, we need to test the genetic properties of [URE3] propagated by Ure2–GFP, and to analyse the curing mechanisms of a URE2 [URE3] strain by a different method.
An aggregation process mediates the curing effect of the PFD
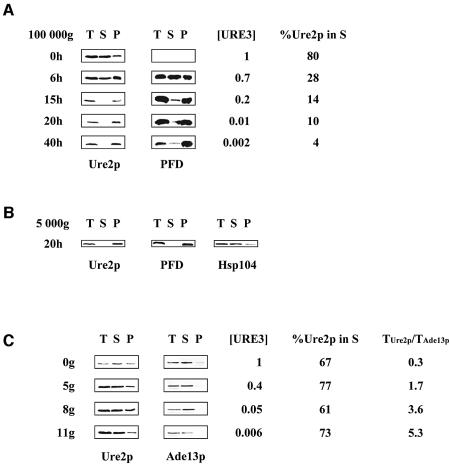
We have demonstrated that [URE3], unlike [PSI], does not cause massive aggregation that can be monitored by a simple sedimentation assay (Fernandez-Bellot et al., 2002). If there is aggregation but it leads instead to destruction of the pre-existing propagons by a combined mechanism, then Ure2p should be preferentially pelleted by centrifugation following overproduction of the PFD. This simple experiment was carried out at various time points during induction of the PFD. Nought, 6, 15, 20 and 40 h after the shift from glucose to galactose, we centrifuged an aliquot of the culture and the resulting crude extract was subjected to centrifugation at 100 000 g for 30 min. The results for the control (time 0) were consistent with those of our previous experiments, with about 80% of the protein found in the supernatant (Figure 3A). At 6 h, the cells had not yet begun to divide. The PFD was efficiently produced and was readily detected in both the pellet and the supernatant. At 15 h, the solubility of Ure2p changed dramatically, with almost all the protein found in the pellet. This huge effect also affected the solubility of the PFD. At this time point, the cells were well adapted to the new carbon source (galactose) and began to divide, the generation time being about 5 h. The same distribution of proteins (Ure2p and the PFD) between pellet and supernatant was observed at 20 and 40 h (Figure 3A). The kinetics of this specific aggregation was correlated with the kinetics of [URE3] elimination. The species formed consequently to PFD overexpression are huge aggregates since Ure2p and the PFD both pellet at 5 000 g (Figure 3B). Since Hsp104 has a major role in [URE3] propagation, we asked whether this protein was found in these aggregates. This is not the case, as Hsp104 remains soluble (Figure 3B). As the rapid elimination of [URE3] upon production of the PFD involves an aggregation mechanism, we studied what happened during the curing of [URE3] by GdnHCl. We investigated whether the differences in kinetics resulted from different mechanisms.
Fig. 3. The kinetics of Ure2p solubility following PFD overproduction and GdnHCl treatment. (A) Overproduction of the PFD leads to Ure2p aggregation. CC34 [URE3] cells transformed with pYe2T-Nter, for overproduction of the PFD, were cultured as described in Figure 1B. At various times after induction, we determined [URE3] frequency, the subcellular distribution of Ure2p and PFD, and the proportion of Ure2p in the 100 000 g supernatant fraction (%Ure2p in S), as described in Materials and methods, using an affinity-purified polyclonal anti-Ure2p antibody. T, total protein extract; S, soluble fraction; P, pellet fraction. (B) Ure2p and the PFD are found in the 5 000 g pellet. CC34 [URE3] cells transformed with pYe2T-Nter, for overproduction of the PFD, were cultured for 20 h in galactose medium (as described in Figure 1B). The total protein extract was centrifuged at 5 000 g for 30 min (see Materials and methods). The membrane was probed using an affinity-purified polyclonal anti-Ure2p antibody and a polyclonal anti-Hsp104 antibody. T, total protein extract; S, soluble fraction; P, pellet fraction. (C) GdnHCl does not affect Ure2p solubility. CC34 [URE3] cells were grown in YPD supplemented with 5 mM GdnHCl. After 0, 5, 8 and 11 generations in the presence of 5 mM GdnHCl (0g, 5g, 8g and 11g), we determined the subcellular distribution of Ure2p, [URE3] frequency and the proportion of Ure2p in the supernatant fraction as described in (A). Western blotting with a polyclonal anti-Ade13p antibody was used to determine Ade13p levels. To assess Ure2p levels in the total protein extract from the cells analysed, we calculated the ratio of Ure2p levels in total protein extract to Ade13p levels in total protein extract (TUre2p/TAde13p). %Ure2p in S; T, S, and P are as defined for (A).
[URE3] is cured without aggregation by GdnHCl
At various time points during the curing of [URE3] by GdnHCl, we tested an aliquot of the culture for both [URE3] maintenance and Ure2p solubility, as described in Figure 3. The proportion of the signal in each fraction (pellet and supernatant) remained similar (Figure 3C). Ure2p levels seemed to be higher in cured cells than in [URE3] yeast cells. The increase in Ure2p levels was assessed by quantifying the signal obtained with antibodies against Ure2p and comparing this signal with that obtained with antibodies against Ade13p, a protein not involved in prion processes. We found that the concentration of Ure2p was higher in wild-type cells than in [URE3] cells. This is not due to protein degradation during extraction process since boiling the cells directly in Laemmli buffer gave the same results (see Supplementary data).
Our results demonstrate that there are two ways to eliminate [URE3] when [URE3] is propagated by Ure2p. The first—PFD curing—linked to aggregation of the protein, irreversibly destroys [URE3]. The second—GdnHCl curing—does not destroy the propagons, but instead inhibits their replication. This mechanism was initially proposed for [PSI] (Eaglestone et al., 2000), and is probably based on Hsp104 inhibition, although it remains controversial (Ferreira et al., 2001; Wegrzyn et al., 2001; Jung et al., 2002; Ness et al., 2002). We have demonstrated that the first mechanism is inefficient if [URE3] is propagated by Ure2–GFP because EG34 (URE2-GFP [URE3]) retains [URE3] after prolonged overproduction of the PFD. So, is GdnHCl efficient for curing this strain of [URE3]?
URE2–GFP [URE3] is resistant to all curing processes and cannot promote focus formation
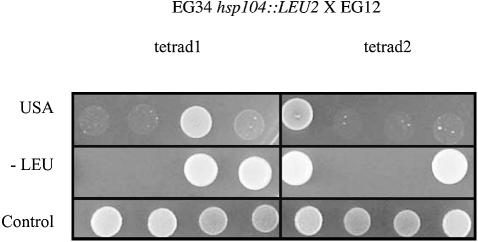
Various strains of [URE3] exist that may influence the stability of the prion phenotype (Schlumpberger et al., 2001) and possibly its sensitivity to curing agents. We tried to overcome these problems by using the EG34 strain (URE2-GFP [URE3], [URE3] introduced by mating). This strain was cultured for more than 20 generations in YPD and YPD + 5 mM GdnHCl. In both conditions, more than 50% of the yeast cells retained [URE3] (data not shown). This experiment was performed several times and was fully reproducible, indicating that the chaotropic agent has little effect on the stability of [URE3] in this strain. This chemical compound is thought to act by inhibiting Hsp104. If this is indeed the case, then the deletion of HSP104 should not lead to [URE3] elimination. This was confirmed because most of the transformants derived by deleting HSP104 in EG34 grew in USA medium. We assessed the prion properties of these hsp104Δ transformants by crossing several of them with EG12 (URE2-GFP, [ure3°]). The diploid strains retained [URE3]. Five tetrads were finally analysed and were found to display the non-Mendelian Usa+ phenotype segregation characteristic of [URE3]. Two tetrads present a (1Usa+:3Usa–) segregation, two present a (2Usa+:2Usa–) segregation, the remaining one presenting a (3Usa+:1Usa–) segregation (Figure 4). To fully demonstrate that the Usa+ phenotype of the EG34 strain is due to the presence of a prion, we deleted the URE2-GFP gene and re-introduced it later by transformation. As expected, if the phenotype is due to an autocatalytic inactivation rather than to other mechanisms, the cells were found Usa– (see Supplementary data). These results demonstrate that the fusion of GFP to the C-terminal part of Ure2p greatly changes the stability of [URE3] by increasing its resistance to known curing molecules.
Fig. 4. Non-Mendelian segregation of [URE3] in tetrads from EG34 hsp104::LEU2 X EG12 diploids. EG34 hsp104::LEU2 [URE3] was crossed with EG12 [ure3°]. After sporulation, several tetrads were analysed for [URE3] segregation. [URE3] phenotype was assessed based on growth on minimal medium without uracil, supplemented with USA. Segregation of hsp104::LEU2 was tested on minimal medium without leucine.
Discussion
The [URE3] phenotype of S.cerevisiae reflects the prion-like behaviour of Ure2p. Despite the known differences in mammalian and yeast prions, the cellular factors controlling their formation and/or propagation appear to be conserved because the same chemical compounds inhibit prion replication in both yeast and mammalian cells (Bach et al., 2003). The biochemical mechanisms involved in prion maintenance can be studied in various ways, including by cellular, biochemical and genetic approaches. The yeast [PSI] model, in which Sup35p is the yeast-encoded prion protein, is of particular interest. Very early in the yeast prion story, this model highlighted the importance of protein aggregation. Indeed, the [PSI] phenotype was found to be strictly correlated with Sup35–GFP focus formation (Patino et al., 1996) and Sup35p aggregation (monitored by sedimentation assay; Paushkin et al., 1996). More recently, it has been shown that various types of aggregate may co-exist within a given yeast cell, and that their presence is not strictly correlated with [PSI] maintenance (Zhou et al., 2001). Moreover, a decrease in Hsp104 activity, which is essential for [PSI] stability, results in an increase in the size of Sup35p aggregates and a decrease in their number (Wegrzyn et al., 2001). A Sup35p mutant protein that forms large aggregates in the presence of [PSI] also gives an unstable prion phenotype (Borchsenius et al., 2001), indicating that these foci may be dead-end products of replicating forms of prions (propagons), rather than the propagons themselves.
We recently demonstrated that the other yeast prion model, [URE3], did not cause Ure2p aggregation (Fernandez-Bellot et al., 2002). Our results conflict with previous results presenting [URE3] as an aggregated form of Ure2p and, more precisely, as a heritable amyloidosis (Wickner et al., 1995, 2000; Edskes et al., 1999; Schlumpberger et al., 2001; Speransky et al., 2001). In one case (Schlumpberger et al., 2001), Ure2p has been detected in the pellet of [URE3] crude extract. This difference may be due to differences in the methods and yeast strains used, but we were unable to obtain the strains used in Schlumpberger’s study and therefore cannot explore this issue more thoroughly. In the other studies, the Ure2p expression was modified. The Ure2p filaments have been observed only in conditions in which the protein is overproduced (Speransky et al., 2001). Ure2p aggregation has been also observed in yeast cells that express both Ure2p and Ure2–GFP fusion protein. Interestingly, in this condition, [URE3] is destabilized. We then studied the relationship between destabilization of [URE3] and Ure2p aggregation.
We first investigated the destabilization of [URE3] upon overproduction of the PFD of Ure2p (domain 1–93), or treatment with GdnHCl. The data are summarized Table I. The prion domain was defined by comparing the sequences of orthologous URE2 genes (Edskes and Wickner, 2002; Baudin-Baillieu et al., 2003), the boundary being confirmed by biochemical (Thual et al., 1999) and functional (Komar et al., 2003) experiments. [URE3] was eliminated from the cell in different ways in these two cases. The species with prion-forming capacity is referred to as the propagon. If produced in [URE3] yeast cells, the PFD may form one or several huge aggregates that contain both the PFD and Ure2p. Hsp104, which is essential for prion propagation, is not present in these aggregates. Therefore curing is not due to a sequestration of Hsp104, but rather to a destruction of the propagons through the aggregation of Ure2p. In fact, aggregated Ure2p is not involved in the prionization mechanism because the vast majority of the cells are [ure3°]. As the aggregates are dramatically large, no biochemical tools could be used for their characterization. The description of such biological material should be realized at the microscopic level.
Table I. Curing efficiencies by GdnHCl treatment and PFD over expression.
| [URE3] frequency in URE2 strain (%) | [URE3] frequency in URE2-GFP strain (%) | |||
|---|---|---|---|---|
| 3 generations | 10 generations | 3 generations | 10 generations | |
| Control | <1 | <10 | <1 | <10 |
| GdnHCl | <1 | >99.9 | <1 | <10 |
| PFD | >99.9 | >99.9 | <1 | <10 |
If GdnHCl is used to cure yeast cells of [URE3], the kinetic parameters are different and resemble those observed for [PSI] elimination. It has been suggested that GdnHCl acts by inactivating Hsp104 (Eaglestone et al., 2000). Hsp104 may be involved in the production of propagons from larger aggregates arising spontaneously (Paushkin et al., 1996; Kushnirov and Ter-Avanesyan, 1998). If this is the case, then adding GdnHCl would generate larger aggregates. We observed no such effect in our sedimentation assay. Alternatively, Hsp104 may enable the native protein to reach an intermediate state that provides an efficient substrate for prion propagation (Chernoff et al., 1995). This hypothesis is consistent with our results because a defect in Hsp104 activity would not lead to the formation of aggregates.
We then expressed another curing allele of URE2: URE2-GFP. The kinetics of [URE3] elimination upon expression of this allele are similar to those for curing with the PFD (elimination of the propagons in the first few generations). This observation suggests a common mechanism, although Ure2–GFP does not induce [URE3] as the PFD does. In both cases, the propagon species derived from Ure2p loses its prionization capacities and gives rise to aggregates that can be collected by centrifugation or observed as numerous foci. It has been suggested that Ure2p is stabilized by PFD–PFD interactions in the propagon state (Masison et al., 1997). The functional (amino acids 94–354) domain of Ure2p is also known to interact genetically (Masison and Wickner, 1995) and biochemically (Fernandez-Bellot et al., 1999) with the PFD. Introducing the PFD alone, or a PFD interacting differently with the functional domain (as for Ure2p–GFP), may then increase aggregation of the propagons. If no such increase in aggregation is observed, then, in the propagon state, the tendency towards aggregation is blocked either by trans-acting factors or by the biochemical nature of the propagon.
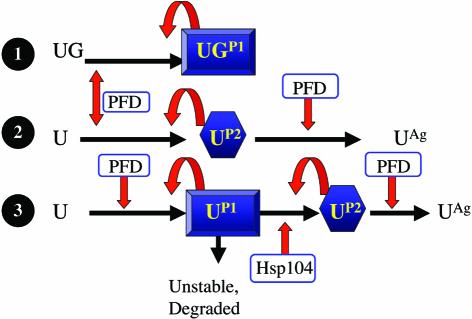
We also compared the curing of [URE3] propagated by Ure2p and by Ure2-GFP (results are summarized in Table I). We found that propagons derived from Ure2–GFP, despite having the characteristic genetic properties that distinguish between [URE3] and nucleic acid replicons, were resistant to the curing effect of the PFD and GdnHCl. The PFD induced [URE3] formation, but the resulting propagons did not aggregate and replicated stably. As these propagons were also stably maintained in yeast cells in the presence of GdnHCl, we investigated the role of Hsp104 in their propagation and clearly demonstrated that this protein was not required for [URE3] maintenance in EG34. Our work clearly demonstrates that these propagons differ from those arising from wild-type Ure2p. In both cases, the production of propagons was stimulated by the PFD. The characteristics of propagon formation are summarized in Figure 5. Lanes 1 and 2 illustrate the formation of [URE3] from Ure2–GFP (UG, the propagon form labelled as P1) and Ure2p (U, the propagon form labelled as P2), respectively. In this study, we showed that the concentration of Ure2p in crude extracts prepared from [URE3] yeast cells was lower than that in the wild type. A possible explanation would be that Ure2p, in propagon P2, may itself be unstable and more prone to degradation than cellular Ure2p. Alternatively, Ure2p may adopt an intermediate structure between cellular Ure2p and propagon P2. This intermediate structure would be unstable and quickly degraded unless Ure2p was produced in-frame with GFP, and could correspond to propagon P1. In this model (Figure 5, lane 3), Hsp104 is specifically required for propagon P2 formation.
Fig. 5. A two-step model for [URE3] stabilization. When the chimeric Ure2–GFP species (UG) sustains [URE3] replication, the propagons (UGP1) replicate without the assistance of Hsp104. Propagon production is stimulated by the PFD (1–93), which does not cure the cells of UGP1. In a wild-type yeast cell, Ure2p (U) gives rise to a propagon species (UP2) that can be induced and cured by the PFD and which also requires Hsp104 (lane 2). Its elimination by the PFD generates an aggregated form (UAg), whereas Hsp104 inhibition has no effect on the partitioning of Ure2p between soluble and insoluble fractions. We propose a simple model (lane 3), in which two kinds of propagons are generated from Ure2p. UP1 is related to UGP1. Without GFP fused to the C-terminal end of Ure2p, this propagon would be highly unstable and degraded. Hsp104 would be required to obtain the stable UP2 propagon. As UP2 is generated from UP1, the concentration of Ure2p is lower in the presence of [URE3] than in a wild-type background. The UP2 species could not be generated with Ure2–GFP.
In conclusion, our results make it possible to separate prion propagation from the aggregation observed due to the presence of the propagon. We recently showed that, although [URE3] was not conserved during evolution, prion mechanisms seem to be similar in yeast and mammals (Bach et al., 2003). The characterization of proteins involved in the appearance of both P1 and P2 is of key interest as these species may be stably produced. Their biochemical characterization will increase our understanding of the molecular mechanisms underlying prionization.
Materials and methods
Yeast strains and plasmids
The strains of Saccharomyces cerevisiae used in this study are listed Table II.
Table II. Yeast strains used in this study.
| CC30 | trp1-1, ade2-1, leu2-3,112, his3-11,15, Δura2::HIS3 |
| CC34 | trp1-1, ade2-1, leu2-3,112, his3-11,15, Δura2::HIS3, [URE3] |
| EG12 | trp1-1, ade2-1, leu2-3,112, his3-11,15, Δura2::HIS3, URE2GFP-TRP1 |
| EG34 | trp1-1, ade2-1, leu2-3,112, his3-11,15, Δura2::HIS3, URE2GFP-TRP1, [URE3] |
| EG34 Δhsp104 | trp1-1, ade2-1, leu2-3,112, his3-11,15, Δura2::HIS3, URE2GFP-TRP1, Δhsp104::LEU2, [URE3] |
[URE3] was introduced in EG34 by mating. This Usa+ spore was selected after sporulation of the CC34 X EG12 diploid on SP1 medium (1% potassium acetate, 0.1% yeast extract, 0.05% glucose, 2% agar).
The pYe plasmids used in this study contain the yeast 2µ origin of replication, the GAL10-CYC1 inducible promoter and the selective marker LEU2 (pYe2L) or TRP1 (pYe2T) (Cullin and Minvielle, 1994). The URE2 open reading frame was amplified using oligonucleotides 200 (5′-CGCGGATCCATGATGAATAACAACGGCAAC-3′), which introduces a BamHI restriction site at the 5′ end of the gene, and 66 (5′-GCGCCTTAGGTCATTCACCACGCAATGCCTTGATG-3′), which introduces a Bsu36I restriction site at the 3′ end of the gene. The resulting cassette was inserted into pYe2L, between the BamHI and Bsu36I sites, resulting in pYe2L-URE2. The URE2 prion domain, corresponding to the first 93 amino acids, was amplified using pYe2L-URE2 as the template with oligonucleotides 91 (5′-CCTTTGTAGCATAAATTAC-3′), binding to the GAL10-CYC1 promoter, and 62 (5′-CGCCTAAGGTCAATCCGAAAATGCCTG-3′), which inserts a stop codon and a Bsu36I restriction site. The PCR fragment was inserted into pYe2T, between the BamHI and Bsu36I sites, resulting in pYe2T-Nter. pYe2L-Nter was constructed by replacing the TRP1 cassette from pYe2T-Nter with the LEU2 cassette from the pTL7 marker swap plasmid, as previously described (Cross, 1997).
[URE3] curing by guanidine hydrochloride
CC34 and EG34 strains were grown in YPD (1% bacto-yeast extract, 1% bacto-peptone, 2% glucose, 20 mg/l adenine) with or without 5 mM guanidine hydrochloride. Cultures were maintained in exponential growth (between 0.1 and 2 OD600 nm). At regular time points, aliquots of culture were withdrawn, washed twice in 0.9% NaCl and plated on SD medium (0.67% yeast nitrogen base, 2% glucose, 2% agar) supplemented either with uracil (20 mg/l) or with USA (15 mg/l). Colonies were counted after incubation for 3 days at 30°C for plates containing uracil, and after 5 days for plates containing USA. [URE3] elimination was assessed by calculating the frequency of Usa+ colonies (number of colonies on USA/number of colonies on uracil).
Effect of transient Ure2p PFD overproduction on [URE3] induction or elimination
CC30 was transformed with pYe2T and pYe2T-Nter. CC34 was transformed with pYe2T, pYe2T-Nter and pYe2T-URE2GFP. EG12 and EG34 were transformed with pYe2L and pYe2L-Nter. Transformations were carried out as described previously (Gietz et al., 1995). CC34, EG12 and EG34 transformants retaining [URE3] were selected based on their ability to grow on SD + USA. Ure2p PFD overproduction was induced by growing cells in SG medium (0.67% yeast nitrogen base, 2% galactose) supplemented with uracil. Cultures were maintained in exponential growth phase (between 0.1 and 1.5 OD600 nm). Usa+ ratio was estimated regularly, as described for curing by guanidine hydrochloride.
Microscopy techniques
Cells were grown overnight on the appropriate medium and suspended in 50 µl Dabco solution [218 mM diazabicyclo 2-2-2 octane (Sigma), 25% v/v PBS, 75% v/v glycerol).
Analysis of Ure2p solubility by subcellular fractionation
Total protein extracts were prepared by lysing 50 OD units of yeast cells in exponential phase with glass beads in 200 µl of 25 mM Tris–HCl pH 7.4, 100 mM NaCl, 0.2% Triton (v/v), 1 mM PMSF, and centrifuging at 1000 g for 5 min. The resulting total protein extract was fractionated by centrifugation for 30 min at 5 and 100 kg. The resulting supernatant was recovered and replaced with an equal volume of extraction buffer to generate the pellet fraction. Identical volumes of each fraction were incubated for 10 min at 100°C in 1× loading buffer and separated by 15% SDS–PAGE. The protein bands were transferred onto nylon membranes, which were probed with specific affinity-purified polyclonal antibodies raised against Ure2p at a dilution of 1/3000, against Ade13p at a dilution of 1/2000 (kindly supplied by Bertrand Daignan-Fornier), or against Hsp104 at a dilution of 1/500 (kindly supplied by Susan Lindquist). Polyclonal, peroxidase-conjugated anti-rabbit antibodies were used as secondary antibodies. Secondary antibody binding was detected with the ECL+ reagent (Amersham-Pharmacia), and the VersaDoc Imaging System (BioRad). Ure2p, PFD and Ade13p were quantified with Quantity One software (BioRad).
HSP104 deletion
HSP104 was deleted using the plasmid pYABL5 (kindly provided by Dr Susan Lindquist). Briefly, the 4.3 kb PvuII–HindIII fragment containing the LEU2 gene was used to transform EG34. The Leu+ transformants were tested for reduced viability after prolonged incubation at 55°C.
Acknowledgments
Acknowledgements
We would like to thank S.Lindquist and B.Daignan-Fornier for providing the pYABL5 plasmid, anti-Hsp104 and anti-Ade13p antibodies, respectively. The work was supported by grants from GIS ‘Infections à Prions’ and the FRM. L.M. and L.R. were supported by FRM and BDI ‘CNRS-Région Aquitaine’ fellowships, respectively.
References
- Bach S. et al. (2003) New anti-prion drugs isolated using a yeast based assay share structural and functional similarities with drugs active against mammalian prion suggesting conservation of targets. Nat. Biotechnol., in press. [Google Scholar]
- Baudin-Baillieu A., Fernandez-Bellot,E., Reine,F., Coissac,E. and Cullin,C. (2003) Conservation of the prion properties of Ure2p through evolution. Mol. Biol. Cell., 14, 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T. and Hall,M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Borchsenius A.S., Wegrzyn,R.D., Newnam,G.P., Inge-Vechtomov,S.G. and Chernoff,Y.O. (2001) Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J., 20, 6683–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.R. (2000) Prion protein peptides: optimal toxicity and peptide blockade of toxicity. Mol. Cell. Neurosci., 15, 66–78. [DOI] [PubMed] [Google Scholar]
- Bucciantini M. et al. (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature, 416, 507–511. [DOI] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler,N.S., Lorenz,M.C., Di Como,C.J. and Heitman,J. (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev., 13, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B., Inge,V.S. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Coschigano P.W. and Magasanik,B. (1991) The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol., 11, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W.E. and Magasanik,B. (1988) Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol., 170, 708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R. (1997) ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast, 13, 647–653. [DOI] [PubMed] [Google Scholar]
- Cullin C. and Minvielle-Sebasta, L. (1994) Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast, 10, 105–112. [DOI] [PubMed] [Google Scholar]
- Drillien R., Aigle,M. and Lacroute,F. (1973) Yeast mutants pleiotropically impaired in the regulation of the two glutamate dehydrogenases. Biochem. Biophys. Res. Commun., 53, 367–372. [DOI] [PubMed] [Google Scholar]
- Eaglestone S.S., Ruddock,L.W., Cox,B.S. and Tuite,M.F. (2000) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K. and Wickner,R.B. (2002) Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full- length protein. Proc. Natl Acad. Sci. USA, 99 (Suppl. 4), 16384–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K., Gray,V.T. and Wickner,R.B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA, 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettaiche M., Pichot,R., Vincent,J.P. and Chabry,J. (2000) In vivo cytotoxicity of the prion protein fragment 106–126. J. Biol. Chem., 275, 36487–36490. [DOI] [PubMed] [Google Scholar]
- Fernandez-Bellot E. and Cullin,C. (2001) The protein-only theory and the yeast Saccharomyces cerevisiae: the prions and the propagons. Cell Mol. Life Sci., 58, 1857–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet,E., Baudin-Baillieu,A., Gaumer,S., Komar,A.A. and Cullin,C. (1999) Characterization of the interaction domains of Ure2p, a prion-like protein of yeast. Biochem. J., 338, 403–407. [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet,E. and Cullin,C. (2000) The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J., 19, 3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet,E., Ness,F., Baudin-Baillieu,A., Ripaud,L., Tuite,M. and Cullin,C. (2002) The [URE3] phenotype: evidence for a soluble prion in yeast. EMBO Rep., 3, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.C., Ness,F., Edwards,S.R., Cox,B.S. and Tuite,M.F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydro chloride is the result of Hsp104 inactivation. Mol. Microbiol., 40, 1357–1369. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl,R.H., Willems,A.R. and Woods,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS–DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Kowal,A.S., Schirmer,E.C., Patino,M.M., Liu,J.J. and Lindquist,S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum,R.R. and Gifford,P. (1982) A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl Acad. Sci. USA, 79, 7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.F., Antoniou,M. and Collinge,J. (1999) Protease-resistant prion protein produced in vitro lacks detectable infectivity. J. Gen. Virol., 80, 11–14. [DOI] [PubMed] [Google Scholar]
- Jackson G.S. et al. (1999) Reversible conversion of monomeric human prion protein between native and fibrilogenic conformations. Science, 283, 1935–1937. [DOI] [PubMed] [Google Scholar]
- Jung G., Jones,G. and Masison,D.C. (2002) Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl Acad. Sci. USA, 99, 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.Y., Tittmann,P., Gross,H., Gebert,R., Aebi,M. and Wuthrich,K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA, 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D.A., Come,J.H., Priola,S.A., Chesebro,B., Raymond,G.J., Lansbury,P.T. and Caughey,B. (1994) Cell-free formation of protease-resistant prion protein. Nature, 370, 471–474. [DOI] [PubMed] [Google Scholar]
- Komar A.A., Lesnik,T., Cullin,C., Merrick,C., Trachsel,H. and Altmann,M. (2003) Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J., 22, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V. and Ter-Avanesyan,M.D. (1998) Structure and replication of yeast prions. Cell, 94, 13–16. [DOI] [PubMed] [Google Scholar]
- Masison D. and Wickner,R.B. (1995) Prion-inducing domain of yeast ure2p and protease resistance of ure2p in prion-containing cells. Science, 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Masison D.C., Maddelein,M.L. and Wickner,R.B. (1997) The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl Acad. Sci. USA, 94, 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes,H.K. and Wickner,R.B. (2000) [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol., 20, 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F., Ferreira,P., Cox,B.S. and Tuite,M.F. (2002) Guanidine hydrochloride inhibits the generation of prion ′seeds′ but not prion protein aggregation in yeast. Mol. Cell. Biol., 22, 5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M.M., Liu,J.J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Perutz M.F. (1999) Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci., 24, 58–63. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1982) Novel proteinaceous infectious particles cause scrapie. Science, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M., Prusiner,S.B. and Herskowitz,I. (2001) Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol., 21, 7035– 7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speransky V.V., Taylor,K.L., Edskes,H.K., Wickner,R.B. and Steven,A.C. (2001) Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J. Cell Biol., 153, 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.L., Cheng,N., Williams,R.W., Steven,A.C. and Wickner,R.B. (1999) Prion domain initiation of amyloid formation in vitro from native Ure2p. Science, 283, 1339–1343. [DOI] [PubMed] [Google Scholar]
- Taylor J.P., Hardy,J. and Fischbeck,K.H. (2002) Toxic proteins in neurodegenerative disease. Science, 296, 1991–1995. [DOI] [PubMed] [Google Scholar]
- Thual C., Komar,A.A., Bousset,L., Fernandez-Bellot,E., Cullin,C. and Melki,R. (1999) Structural characterization of Saccharomyces cerevisiae prion-like protein Ure2. J. Biol. Chem., 274, 13666– 13674. [DOI] [PubMed] [Google Scholar]
- Tuite M.F., Mundy,C.R. and Cox,B.S. (1981) Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics, 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn R.D., Bapat,K., Newnam,G.P., Zink,A.D. and Chernoff,Y.O. (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol., 21, 4656–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Masison,D.C. and Edskes,H.K. (1995) [PSI] and [URE3] as yeast prions. Yeast, 11, 1671–1685. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Taylor,K.L., Edskes,H.K., Maddelein,M.L., Moriyama,H. and Roberts,B.T. (2000) Prions of yeast as heritable amyloidoses. J. Struct. Biol., 130, 310–322. [DOI] [PubMed] [Google Scholar]
- Zhou P., Derkatch,I.L. and Liebman,S.W. (2001) The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol. Microbiol., 39, 37–46. [DOI] [PubMed] [Google Scholar]