Abstract
To understand the mechanism of nucleotide excision repair (NER), one of the major human DNA repair pathways, we have set up a DNA repair system in which a linear damaged DNA substrate is immobilized by its terminus. By isolating functionally active intermediate complexes, our data dissect the ordered arrival and displacement of NER factors in the progress of the dual incision step. We describe (i) the role of ATP in remodelling the NER-initiating complex of XPC/TFIIH/damaged DNA as a prerequisite for the recruitment of the next NER factors; (ii) the coordination between damage removal and DNA resynthesis and the release of XPC-HR23B, TFIIH and XPA upon arrival of XPG and XPF-ERCC1, respectively; (iii) how RPA remains associated with the excised DNA initiating the assembly of resynthesis factors such as PCNA; (iv) the recycling of XPC-HR23B, TFIIH and XPA in the NER; and the shuttling of TFIIH between NER and transcription. Thus, our findings define multiple functions of NER factors to explain the molecular basis of human NER disorders.
Keywords: DNA resynthesis/dual incision/NER/recycling
Introduction
Maintaining the integrity of the genome to allow proper cellular functioning is the objective of DNA repair pathways. The highly conserved nucleotide excision repair (NER) eliminates a broad variety of DNA lesions caused by UV light and chemical mutagens by two related subpathways (Lindahl and Wood, 1999). While the general global genome repair (GGR) removes DNA damage from the entire genome, the cell has set up a more specialized transcription-coupled repair (TCR) subpathway that corrects DNA lesions located on the actively transcribed strand (Mullenders and Berneburg, 2001). The biological consequences of NER defects are apparent from the inherited multisystem disorders xeroderma pigmentosum (XP), trichothiodystrophy (TTD) and Cockayne syndrome (CS) that are characterized by hyperphotosensitivity and a wide spectrum of neurodevelopmental abnormalities. XP patients show in addition a high incidence of UV-related skin cancers (reviewed in Berneburg and Lehmann, 2001).
Following genotoxic attacks that block transcription and might result in cell cycle arrest (Zhou and Elledge, 2000), NER takes place after chromatin remodelling (Ura and Hayes, 2002) by eliminating a damage-containing oligomer (dual incision) and resynthesizing the excised DNA fragment (DNA resynthesis). In TCR, the elimination of the DNA damage is initiated by stalled RNA polymerase II (Mellon and Hanawalt, 1989), whereas in GGR the damage-induced DNA distortion is recognized by factors such as XPC-HR23B. In GGR, the six core NER factors XPC-HR23B, TFIIH, XPA, replication protein A (RPA), XPG and XPF-ERCC1 act in concert (Aboussekhra et al., 1995) to remove the damaged oligonucleotide, which spans 24 to 32 residues depending on the lesion and its surrounding sequences (Huang et al., 1992; Moggs et al., 1996).
Although controversially discussed, it is now widely accepted that these NER factors assemble sequentially at sites of DNA damage (Guzder et al., 1996) rather than being recruited as a preformed repairosome complex (Svejstrup et al., 1995) or as part of the RNA pol II holoenzyme (Maldonado et al., 1996). While numerous in vitro studies have demonstrated potential roles in damage recognition not only for XPC-HR23B (Sugasawa et al., 1998), but also for XPA (Asahina et al., 1994), RPA (Reardon and Sancar, 2002) and a XPA–RPA complex (Wakasugi and Sancar, 1999), recent in vivo data support a model in which pre-incision complex formation is triggered by the binding of XPC-HR23B to the DNA damage (Volker et al., 2001). In a later step of NER, the XPB and XPD helicases of the transcription/repair factor TFIIH unwind the DNA around the lesion and provide an adequate substrate for the XPG and XPF-ERCC1 endonucleases (Evans et al., 1997), which cut the damaged DNA strand 3′ and 5′ to the lesion, respectively (O’Donovan et al., 1994; Sijbers et al., 1996). Subsequently, the gapped DNA is refilled by DNA Pol δ/ε, replication factor C (RFC), PCNA, RPA and DNA ligase I (Shivji et al., 1995). Attempts to investigate the sequential assembly of the dual incision complex in vitro and in vivo did neither document the functional relevance of observed complex intermediates nor reveal how the NER factors move upon damage removal to hand over the gapped DNA to the resynthesis machinery.
In order to understand how gene expression can cope with the detrimental effects of DNA damage, we set up an elaborate system based on a mono-cisplatin-damaged DNA substrate immobilized on magnetic beads, which upon addition of selected NER factors allows investigation of the various phases of the dual incision reaction. We were able to determine the ATP-modulated sequential assembly of the NER factors resulting in active complex intermediates as well as the movement of the factors along the progress of the NER reaction including the targeting of the damaged DNA, the DNA opening and the recruitment of XPG and XPF-ERCC1 nucleases, which excise the damaged DNA fragment and finally allow the coordinated handing over to the subsequent DNA resynthesis step.
Results
ATP-independent initiation of NER by XPC-HR23B/TFIIH
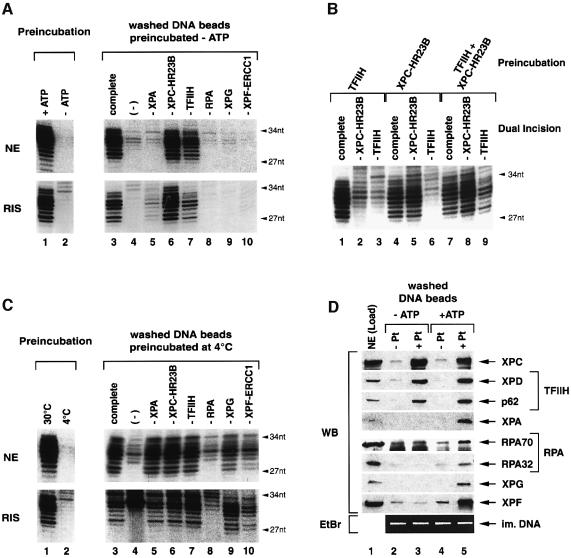
To characterize the various steps that direct the elimination of DNA damage, we designed an assay in which the DNA substrate containing a single cisplatin lesion is immobilized on magnetic beads and pre-incubated with either repair-competent HeLa nuclear extract (NE) or a reconstituted dual incision system (RIS) consisting of highly purified recombinant XPC-HR23B, TFIIH, XPA, RPA, XPG and XPF-ERCC1. In the absence of ATP no dual incision is observed (Figure 1A, lane 2). On the contrary, the addition of ATP mediates the binding of all the NER factors that participate to the incision/excision reaction as shown by the presence of radiolabelled 27–34 nt dual incision products (Figure 1A, lane 1).
Fig. 1. Binding of NER factors to damaged DNA. (A) Immobilized damaged DNA was pre-incubated with either NE or highly purified NER factors (RIS) in the presence (lane 1) or absence of ATP (lane 2) and aliquots were analysed for dual incision, which is indicated by radiolabelled 27 to 34-nt products. Proteins bound to the damaged DNA beads in the absence of ATP were analyzed by single NER factor omission in a reconstituted dual incision assay as indicated at the top (lanes 3–10). (B) Damaged DNA beads were pre-incubated with TFIIH and XPC-HR23B either alone or in combination. Proteins bound to the washed DNA beads were analyzed in a reconstituted dual incision assay lacking either XPC-HR23B (lanes 2, 5 and 8) or TFIIH (lanes 3, 6 and 9). Control reactions included the complete set of NER factors (lanes 1, 4 and 7). (C) Damaged DNA beads were pre-incubated with either NE or RIS in the presence of ATP at either 30 or 4°C and aliquots were analyzed for dual incision (lanes 1 and 2). DNA beads pre-incubated at 4°C were washed and subjected to dual incision assays lacking single NER factors (lanes 3–10). (D) Immobilized undamaged (–Pt) and damaged (+Pt) DNA was pre-incubated with NE (Load) either in the absence (lanes 2 and 3) or in the presence of ATP (lanes 4 and 5). Upon washing, DNA-bound proteins were analyzed by western blot (WB) using antibodies against indicated NER factors. Immobilized DNA was visualized by ethidium bromide staining (EtBr) after separation on agarose gel.
To investigate which step of the reaction requires ATP, the cisplatinated DNA beads were first pre-incubated with either NE or RIS in the absence of ATP (Figure 1A, lanes 3–10). The immobilized DNA was subsequently rinsed with buffer containing 50 mM KCl and 0.02% NP40 to remove non-specifically bound proteins. The factors associated with the washed DNA beads were analyzed either functionally in a dual incision assay (Figure 1A, lanes 3–10) or by western blot (Figure 1D, lanes 1–3). When pre-incubated with NE in the absence of ATP, the cisplatinated DNA specifically retained XPC and TFIIH according to the detection of its XPD and p62 subunits, but no other NER factors (Figure 1D, lanes 2 and 3). When added to a complementation assay reconstituted with the purified NER factors (RIS), we observed that XPC(-HR23B) and TFIIH bound to the DNA beads participate in the incision reaction (Figure 1A, lanes 6 and 7). Using the RIS for the pre-incubation step, we additionally detected traces of XPA activity associated with the damaged DNA beads (Figure 1A, lower panel, lane 5), suggesting a possible interaction between XPA and the damage recognition complex XPC-HR23B–TFIIH (Nocentini et al., 1997).
To discriminate which NER factor binds the DNA damage first, XPC-HR23B and TFIIH either alone or in combination were pre-incubated with the cisplatinated DNA beads in the absence of ATP. After washing, the adsorbed proteins were analyzed in a reconstituted dual incision assay that lacks either XPC-HR23B or TFIIH. Under these conditions, XPC-HR23B alone can bind the damaged DNA resulting in the NER complex 1 (NER-C1) (Figure 1B, lane 5). TFIIH does not interact with the damaged DNA unless XPC-HR23B is added (Figure 1B, lanes 3 and 9). Addition of ATP in the pre-incubation mixture did not enhance the XPC-HR23B–TFIIH binding (Figure 1D, lanes 3 and 5).
The binding of XPC-HR23B and TFIIH resulting in the NER complex 2 (NER-C2) represents the initial step of NER that occurs independently of ATP.
ATP-dependent recruitment of NER factors to damaged DNA
The dual incision reaction is ATP-dependent (Figure 1A, lanes 1 and 2) and the XPB and XPD helicases of TFIIH require ATP to open the DNA around the lesion (Schaeffer et al., 1993; Evans et al., 1997). Thus, we wondered whether ATP mediates the arrival of additional NER factors to NER-C2. We pre-incubated the immobilized damaged DNA together with either HeLa NE or RIS in the presence of ATP at 4°C so as to prevent DNA incision by XPG and XPF-ERCC1 nucleases (Figure 1C, lanes 1 and 2) (Wakasugi and Sancar, 1998). Following a washing step, the NER factors associated with NER-C2 were analyzed (Figure 1C and D). Under these conditions (+ ATP), all the NER factors including XPC-HR23B, TFIIH, XPA, RPA, XPG and XPF-ERCC1 are present on the cisplatinated DNA beads (Figure 1D, lane 5) and all of them, except RPA, are active in dual incision (Figure 1C, lanes 5–10). Remarkably, although two of the three RPA subunits are found associated with the damaged DNA when ATP is present (Figure 1D, lane 5), its activity was not detected (Figure 1C, lane 8). However, we cannot exclude that this is due to the lack of the third RPA14 subunit that could not be analyzed by western blot, because we failed to generate a corresponding antibody. Addition of recombinant RPA to the characterized damaged DNA–protein complex restored the incision reaction (data not shown). Importantly, the omission of XPC-HR23B from pre-incubations in the presence of ATP completely abolished the binding of any NER factor (data not shown).
Sequential arrival of NER factors to the pre-incision complex
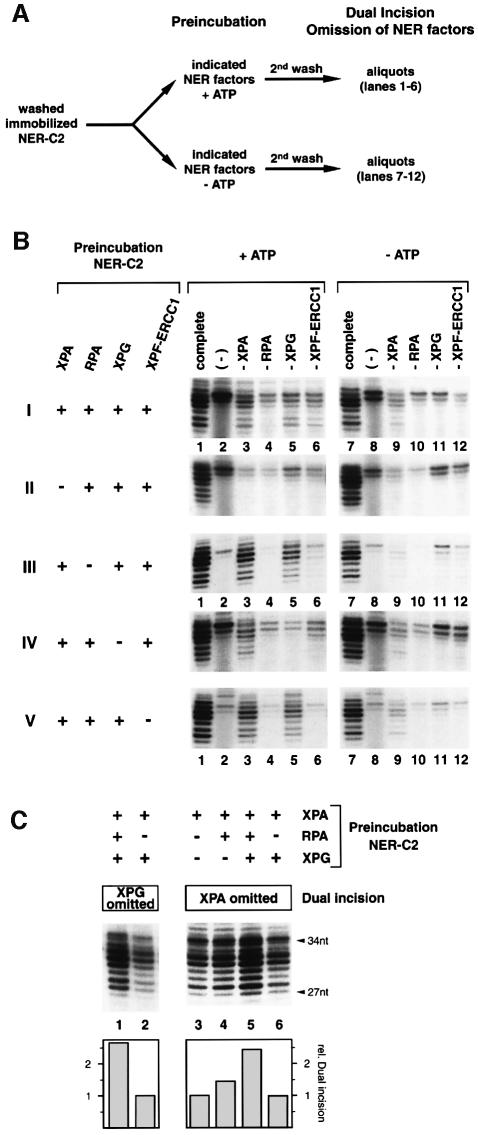
We then investigated the chronological order of ATP-dependent XPA, RPA, XPG and XPF-ERCC1 recruitment to the NER-C2 containing both XPC-HR23B and TFIIH. The pre-assembled and washed NER-C2 was pre-incubated in the presence or absence of ATP with different combinations of XPA, RPA, XPG and XPF-ERCC1 (Figure 2A) as indicated at the left of Figure 2B. Following a second wash (2nd wash), the resulting complexes were analyzed by factor omission in a dual incision reaction (Figure 2B).
Fig. 2. Sequential arrival of NER factors to the pre-incision complex. (A) NER-C2 was assembled on immobilized damaged DNA by incubation with XPC-HR23B and TFIIH in the absence of ATP, washed, and pre-incubated with different combinations of XPA, RPA, XPG and XPF-ERCC1 in the presence (+ATP) or absence of ATP (–ATP). After a second wash (2nd wash) aliquots were analyzed in dual incision assays. (B) Immobilized NER-C2 was pre-incubated with NER factors indicated in the left of the figure in the presence (lanes 1–6) or absence of ATP (lanes 7–12). The resulting complexes were tested in a dual incision reaction lacking one of the NER factors as indicated at the top. (C) NER-C2 was pre-incubated in the presence of ATP with combinations of XPA, RPA and XPG as indicated. Upon washing, bound XPG (lanes 1 and 2) and XPA (lanes 3–6) activity was analyzed in a dual incision assay. The quantification by phosphorimager (lower panel) was normalized to lanes 2 and 3, respectively.
First, none of the four additional NER factors significantly bound the preformed NER-C2 in the absence of ATP (Figure 2B, panels I–V, lanes 9–12). When pre-incubated together in the presence of ATP, all of them, except RPA, associated with NER-C2 to participate in the dual incision reaction (Figure 2B, panel I, lanes 3–6). As demonstrated above, although we did not detect RPA activity (Figure 2B, lane 4), it might be present in the complexes.
Secondly, the removal of XPA from the pre-incubation completely abolished the binding of any additional factor to NER-C2 (Figure 2B, panel II), suggesting that association of XPA with NER-C2 results in the subsequent NER-C3 complex.
Thirdly, in the absence of RPA, XPF-ERCC1 could not join the preformed NER-C2, whereas XPG (as well as XPA) still bound to the damaged DNA (Figure 2B, panel III, compare lane 3 with 9 and lane 5 with 11, respectively). To define the role of RPA in the incision reaction, we analyzed quantitatively the effect of RPA on XPG (and XPA) binding. In these experiments, RPA was pre-incubated together with XPG in the presence of NER-C2 and XPA (Figure 2C). After the second wash, the resulting nucleoprotein complexes were tested in a dual incision assay in which either XPG or XPA were omitted (Figure 2C, left and right panels). Although RPA is not required for the binding of XPG to NER-C3 (Figure 2B, panel III), its presence strongly conditions the association of XPG to the dual incision complex (Figure 2C, lanes 1 and 2); therefore RPA forms the NER-C4 complex. We however noticed that when added together, XPA, RPA and XPG mutually stabilize on the NER-C2 (Figure 2C, lanes 3–5).
Fourthly, in the absence of XPG, XPF-ERCC1 could not bind to the damaged DNA (Figure 2B, panel IV, lane 6). On the contrary, the removal of XPF-ERCC1 from pre-incubation still allows XPG binding to form the NER-C5 (Figure 2B, panel V, lane 5). Thus, XPF-ERCC1 is the last factor arriving to the DNA damage resulting in the mature pre-incision complex NER-C6.
The movement of NER factors upon dual incision
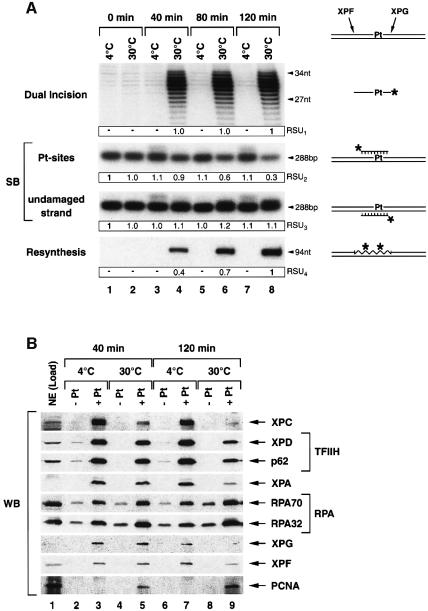
To investigate which NER factors might remain associated with the DNA once the damaged oligonucleotide is excised, we incubated the immobilized cisplatinated DNA with HeLa NE and analyzed over time the progress of the NER reaction (Figure 3A) as well as the NER factors bound to the DNA beads (Figure 3B). Some reactions were performed at 4°C to prevent DNA incision by XPG/XPF-ERCC1.
Fig. 3. Progress of the dual incision reaction. (A) Immobilized damaged DNA was incubated with NE in the presence of ATP either at 4 or 30°C and aliquots were analyzed over time for NER as illustrated on both sides of the figure. Dual incision was monitored as in preceding experiments. Southern blot (SB) membrane was probed with either a 24-nt radiolabelled oligonucleotide complementary to the cisplatin damage (Pt-sites) or a 24-nt oligonucleotide corresponding to the excised damaged DNA patch (undamaged strand). Resynthesis is indicated by a radiolabelled 94-nt product. For resynthesis, the DNA polymerases δ/ε inhibitor aphidicolin was deprived from reactions. Quantification was performed on a phosphorimager (Relative Scanning Units, RSU1 – 4). Data were normalized either to lane 8 (RSU1+4) or to lane 1 (RSU2+3), respectively. (B) Undamaged (–Pt) and damaged (+Pt) DNA beads were incubated with NE (Load) in the presence of ATP at either 4 or 30°C for 40 and 120 min, respectively. DNA bound proteins were analyzed after washing by western blot. The experiment has been repeated four times.
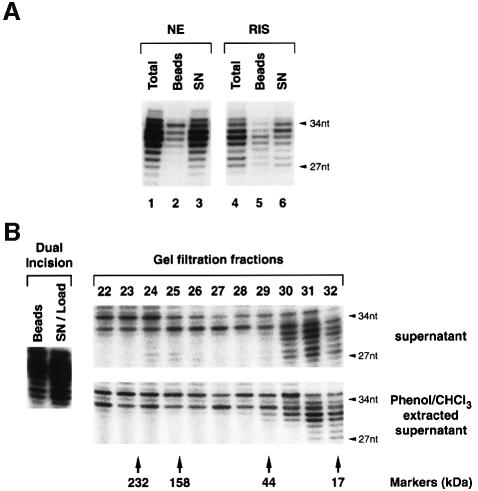
Monitoring over time the dual incision, we found a maximum of the excision product after 40 min of incubation at 30°C (Figure 3A, upper panel, lane 4). We further investigated the removal of platin sites (Pt-sites) from the damaged DNA by Southern blot (Figure 3A, middle panels) probing the DNA substrate either with an oligonucleotide complementary to the excised damaged DNA fragment to reveal the remaining Pt-sites on the DNA (Figure 3A, middle panel, Pt-sites) or with an oligonucleotide corresponding to the excised damaged DNA to quantify the undamaged strand (Figure 3A, middle panel, undamaged strand). After 40 min of incubation at 30°C, only a small portion of Pt-sites have been removed from the DNA (Figure 3A, Pt-sites, lane 4: 10%), whereas after 120 min the majority (up to 70%) of damage sites have been eliminated (Figure 3A, Pt-sites, lane 8). In correlation with this, we also found an increase of resynthesis of the excised damaged DNA fragment up to 120 min of incubation when DNA polymerase δ/ε inhibitor aphidicolin was deprived from the reaction (Figure 3A, lower panel). Remarkably, both kinetics illustrating the damage removal and the DNA resynthesis present the same slope, suggesting coordination between these two steps of the NER reaction. The observed difference in the progress of damage removal when quantifying either the excised damaged DNA fragment (Figure 3A, upper panel, dual incision) or the amount of remaining Pt-sites on the substrate (Figure 3A, middle panel, Pt-sites) most likely reflects a concomitant degradation of the excised DNA fragment being released to the supernatant (Figure 4A, lanes 1–3) by nucleases present in the NE.
Fig. 4. Characterization of excised damaged DNA fragments. (A) Immobilized damaged DNA was incubated with NE or RIS and aliquots were analyzed for dual incision products either directly (lanes 1 and 4) or after separating DNA beads (lanes 2 and 5) from the corresponding supernatants (lanes 3 and 6). (B) Immobilized NER-C2 was incubated with RIS and the supernatant (SN/Load) was separated from the damaged DNA beads and loaded either directly (supernatant) or after phenol–chloroform extraction (Phenol/CHCl3 extracted) on a gel filtration column. The collected fractions were analyzed for dual incision products. The elution of catalase (232 kDa), aldolase (158 kDa), ovalbumin (43 kDa) and myoglobin (17 kDa) as markers is indicated.
In parallel to the excision of the damaged oligonucleotide (Figure 3A, middle panel, Pt-sites), most of the factors that were present in the readily assembled NER complex when pre-incubated at 4°C (Figure 3B, lanes 3 and 7) progressively leave the immobilized DNA after 40 and 120 min, once dual incision takes place at 30°C (Figure 3B, lanes 5 and 9). RPA known to be involved in the resynthesis step (Shivji et al., 1995) was the only NER factor that remained stably associated with the incised DNA even after 120 min of incubation at 30°C (Figure 3B, lanes 3, 5 and 9). Correlated with the removal of the damage, we also observed an accumulation of PCNA on the formerly cisplatinated DNA (Figure 3B, lanes 5 and 9).
The excised damaged DNA fragment is free of NER factors
Having observed the release of NER factors upon dual incision, we asked whether any of these factors might be associated with the excised damaged DNA fragment. Following the dual incision by either HeLa NE or RIS, the immobilized DNA was separated from the soluble supernatant and the excised damaged oligonucleotides from both fractions were end-labelled. In each case, most (90 and 70%, respectively) of the excised damaged DNA fragment was found in the supernatant (SN) (Figure 4A, lanes 3 and 6) demonstrating that the minimal set of NER factors (RIS) is sufficient to promote not only dual incision, but also excision (the release) of the damaged DNA fragment.
When the soluble supernatant of a RIS reaction was loaded on a gel filtration column, the excised damaged DNA detected by post-column end-labelling eluted with a retention volume corresponding to 20 kDa (Figure 4B, upper panel). This excised DNA fragment was protein-free, since phenol chloroform extraction of the supernatant before loading did not shift the retention volume (Figure 4B, lower panel).
Release of XPC-HR23B and TFIIH from the NER complex
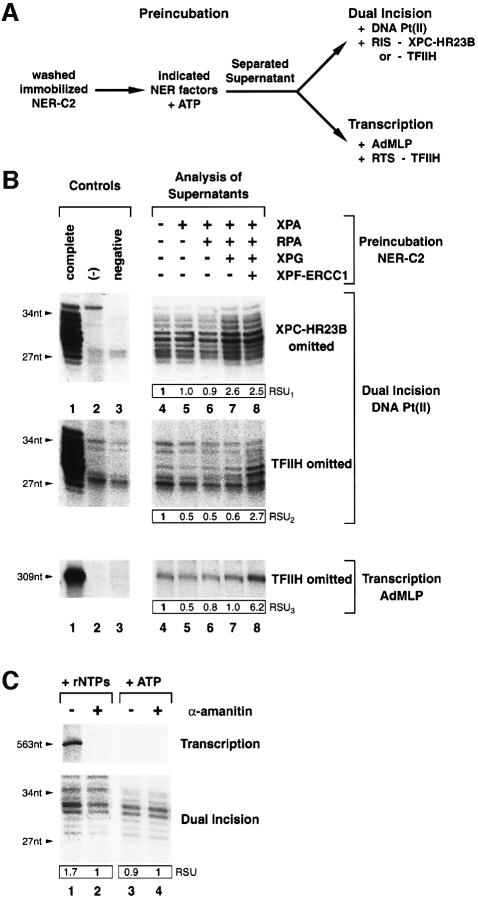
To analyze the movement of XPC-HR23B and TFIIH along the dual incision reaction, immobilized NER-C2 was pre-incubated in the presence of ATP with combinations of XPA, RPA, XPG and XPF-ERCC1 as indicated at the top of Figure 5B. Supernatants were then separated from the damaged DNA beads and analyzed for the presence of XPC-HR23B and/or TFIIH either in a dual incision reaction using a second damaged DNA substrate Pt(II) or in a reconstituted transcription assay (Figure 5A).
Fig. 5. Release of XPC-HR23B and TFIIH; shuttling of TFIIH. (A) NER-C2 immobilized on damaged DNA Pt(I) was pre-incubated in the presence of ATP with indicated NER factors. Resulting supernatants were separated from DNA beads and tested for XPC-HR23B and TFIIH activity in a dual incision assay using damaged DNA substrate Pt(II) and saturating amounts of NER factors (RIS). The use of damaged DNA Pt(II) prevents interference of dual incision products. TFIIH release was additionally monitored by its omission in a basal transcription assay with a DNA template containing the AdMLP (Gerard et al., 1991). (B) Supernatants obtained by pre-incubating NER-C2 with indicated NER proteins were analyzed either by dual incision assays in which XPC-HR23B (top panel) or TFIIH (middle panel) were omitted or in a basal transcription assay with a 309-nt run-off product (lower panel). Control reactions contained the complete set of NER or transcription factors (lane 1), no NER or transcription factors (lane 2) or were lacking only either XPC-HR23B or TFIIH (negative, lane 3). Quantifications by phosphorimager (RSU) were normalized to lane 4. (C) Washed pre-initiation complexes were assembled on immobilized AdMLP DNA template with purified transcription factors. Transcription was then initiated by the addition of rNTPs in the presence of damaged DNA and NER factors lacking TFIIH (lanes 1 and 2). When indicated, rNTPs were substituted by ATP (lanes 3 and 4) and/or RNA pol II inhibitor α-amanitin was added (lanes 2 and 4). The transcription run-off product (563 nt) and dual incision of damaged DNA (27–34 nt) were monitored. The experiment has been reproduced five times using various transcription templates.
Although pre-incubation of NER-C2 with XPA either alone or in combination with RPA did not promote the release of XPC-HR23B and TFIIH to the supernatant (Figure 5B, upper panel, compare lanes 5 and 6 with lane 4), the pre-incubation of NER-C2 together with XPA, RPA and XPG resulting in NER-C5 significantly increased (2.6×) the release of XPC-HR23B (Figure 5B, lane 7). Moreover, further addition of XPF-ERCC1, which generates the NER-C6 and allows dual incision, directed the release (2.7×) of TFIIH (Figure 5B, middle panel, lane 8).
These data suggest that (i) XPC-HR23B leaves the premature NER-C5 upon XPG arrival; (ii) although XPC-HR23B mediates TFIIH association with the damaged DNA, it is not required to maintain TFIIH in a higher-order complex until XPF-ERCC1 arrival; and (iii) the released XPC-HR23B and TFIIH are still active in NER.
TFIIH shuttles between transcription and DNA repair
In addition to its role in NER, TFIIH is an essential factor in RNA pol II transcription (Gerard et al., 1991). Therefore we investigated whether TFIIH is able to shuttle between NER and transcription. We observed that TFIIH released from the dual incision complex by pre-incubating NER-C2 together with XPA, RPA, XPG and XPF-ERCC1 substantially (6.2×) supports RNA synthesis when added in a reconstituted transcription assay containing in addition to an adenovirus major late promoter (AdMLP) template, RNA pol II, TFIIA, TFIIB, TBP, TFIIE and TFIIF transcription factors (Figure 5B, lower panel, lane 8); demonstrating that TFIIH having been involved in the NER reaction can subsequently participate in RNA pol II transcription.
Knowing that TFIIH incorporated in RNA pol II pre-initiation complexes is released from the DNA template while RNA pol II enters the elongation phase (Zawel et al., 1995; Spangler et al., 2001), we investigated whether such a released TFIIH might subsequently participate in the removal of DNA damage. Purified pre-initiation complexes were generated by pre-incubation of immobilized AdMLP transcription template with RNA pol II and the basal transcription factors and subsequent washing with a buffer containing 50 mM KCl and 0.05% sarkosyl to remove non-specifically bound proteins. The transcription pre-initiation complexes were then incubated in the presence of all four rNTPs together with a damaged DNA substrate and the purified NER factors (RIS) except TFIIH. When indicated (Figure 5C), the transcription inhibitor α-amanitin was added and/or rNTPs were substituted by ATP. Occurrence of RNA synthesis promotes the release of TFIIH that further participates in dual incision (Figure 5C, lane 1). On the contrary, addition of the α-amanitin transcription inhibitor prevents transcription, the release of TFIIH, and consequently its participation in dual incision (Figure 5C, lane 2). α-Amanitin per se did not inhibit the dual incision reaction (Figure 5C, lanes 3 and 4). Together our data demonstrate that in purified systems TFIIH is able to switch from transcription to DNA repair and vice versa.
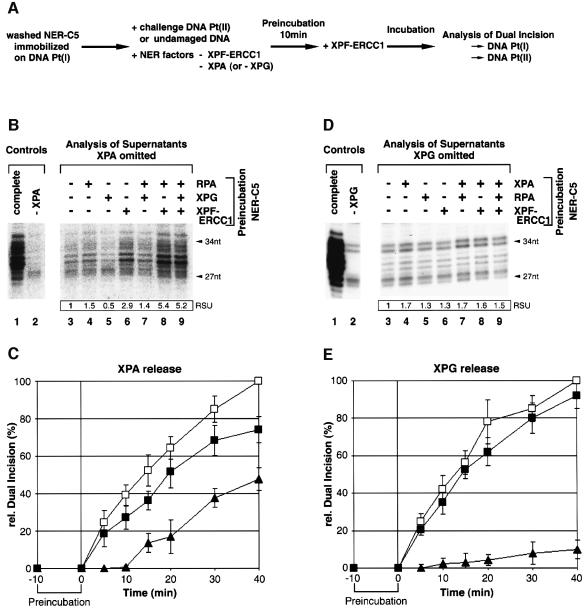
The release of XPA and XPG from dual incision complexes was analyzed by pre-incubating the immobilized and purified NER-C5 complex was pre-incubated in the presence of ATP with XPF/ERCC1 either alone or in combination with XPG and RPA in excess as indicated at the top of Figure 6B. Similar to the scheme in Figure 5A, supernatants were then separated and analyzed in dual incision reactions lacking XPA. Pre-incubation of NER-C5 with RPA or XPG either alone or in combination did not promote XPA release to the supernatant (Figure 6B, lanes 4, 5 and 7). On the contrary, addition of XPF-ERCC1 was sufficient to promote the release of XPA from NER-C5 and its participation in a subsequent dual incision reaction (Figure 6B, lane 6). The release of XPA was even increased when RPA was added to pre-incubation with XPF/ERCC1 (Figure 6B, lanes 8 and 9), possibly highlighting the role of RPA as the relay for further NER steps.
Fig. 6. Release of XPA and XPG. (A) Experimental design of NER substrate challenge assays. NER-C5 was pre-assembled by incubating immobilized DNA Pt(I) with purified XPC-HR23B, TFIIH, XPA, RPA and XPG in the presence of ATP. Washed NER-C5 complexes were pre-incubated for 10 min in the presence of ATP with XPC-HR23B, TFIIH, RPA and either XPG (for C) or XPA (for E). Pre-incubations included either challenge DNA Pt(II) or undamaged DNA. After pre-incubation, XPF-ERCC1 was added to the reaction and dual incision of both substrates Pt(I) and Pt(II) was monitored over time. (B) Washed NER-C5 was pre-incubated in the presence of ATP with the indicated combinations of NER factors (top of the figures). Resulting supernatants were separated from DNA beads and analyzed by omitting XPA in a dual incision assay with DNA Pt(II). (C) Substrate challenge assay: XPA release. Dual incision products were resolved on denaturing polyacrylamide gels and quantified by phosphorimager. Data were normalized to the dual incision of Pt(I) substrate measured after 40 min incubation in the presence of undamaged competitor DNA. Open squares: dual incision of Pt(I) in the presence of undamaged control DNA; closed squares: dual incision of Pt(I) in the presence of Pt(II) challenge DNA; closed triangles: dual incision of challenge DNA Pt(II). (D) XPG release corresponding to (B). (E) Substrate challenge assay: XPG release. Experiments in panels (B–E) have been performed at least three times.
We extended our study on the release of XPA from the dual incision complex by a DNA substrate challenge assay outlined in Figure 6A. NER-C5 immobilized on damaged DNA Pt(I) was pre-incubated in the presence of ATP with either undamaged DNA or damaged DNA Pt(II), XPC/HR23B, TFIIH, RPA and XPG in the absence of XPA and XPF-ERCC1. After pre-incubation, XPF-ERCC1 was added and dual incision of both, DNA Pt(I) and DNA Pt(II), was monitored over time. Dual incision of DNA Pt(II) indicates XPA release from DNA Pt(I). We observed that dual incision of DNA Pt(I) starts readily with the addition of XPF-ERCC1 approaching a maximum at t = 40 min (Figure 6C, open and closed squares). The presence of the challenge DNA Pt(II) instead of undamaged DNA reduced the dual incision of DNA Pt(I) (Figure 6C, closed squares). The reduction in dual incision of DNA Pt(I) becomes more pronounced when the released XPA joins the DNA Pt(II) substrate to initiate the excision of its damaged DNA fragment (Figure 6C, closed triangles).
When similar experiments were performed to analyze the release of XPG (Figure 6D and E), we did not observe any displacement of functional active XPG from dual incision complexes in the highly purified NER system (RIS). It thus appears likely that XPG release requires proteins such as resynthesis factors present in the NE (Figure 3B).
Discussion
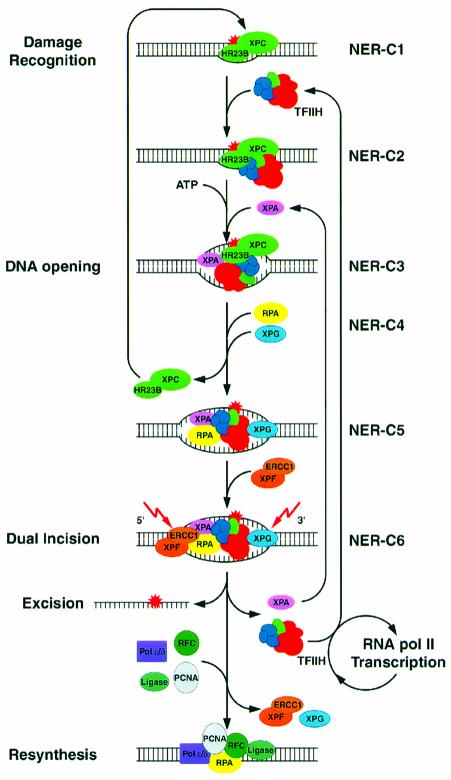
To dissect the various steps of the dual incision reaction and to characterize the dynamics of the NER complex we analyzed subsequent NER intermediate (sub-) complexes starting from damage recognition, passing the elimination of the DNA lesion by dual incision and preparing the onset of the resynthesis process. In the light of the present study in which we associate the presence and the function of each of the NER factors, we are in a position to present the following scheme (Figure 7) highlighting the importance of the ordered coming and going of each factor that results in the elimination of the DNA damage.
Fig. 7. Sequential complexes in NER. XPC-HR23B interacts with the damaged DNA structure resulting in the NER-C1 complex. TFIIH joins NER-C1 to form NER-C2. In the presence of ATP, XPB and XPD helicases in TFIIH unwind the damaged DNA allowing the sequential association of XPA, RPA and XPG to NER-C5. The arrival of XPG mediates the release and the recycling of XPC-HR23B. The recruitment of XPF-ERCC1 resulting in NER-C6 triggers dual incision, excision of the protein-free damaged oligonucleotide, and the release of XPA and TFIIH that re-join new incision complexes. Released TFIIH may switch to RNA pol II transcription in the absence of further DNA damage. RPA and eventually XPG and XPF-ERCC1 remain bound to the incised DNA. With the arrival of resynthesis factors XPG and XPF-ERCC1, if still present, are released from the DNA whereas RPA remains associated as part of resynthesis apparatus.
The arrival of NER factors
The DNA structure induced by the cisplatin adduct is first recognized by XPC-HR23B resulting in the NER-C1 complex (Figure 7). Although several in vitro and in vivo studies identified XPC-HR23B as the first factor that targets DNA damaged by UV irradiation or cisplatin and NA-AAF treatment (Sugasawa et al., 1998; Batty et al., 2000; Volker et al., 2001), others have proposed that NER complex formation is initiated by either XPA, RPA or the pre-assembled XPA–RPA (Asahina et al., 1994; Wakasugi and Sancar, 1999; Reardon and Sancar, 2002). In the latter case, besides having identified the interaction, no functional relevance has been demonstrated. It however remains that some of these discrepancies might be explained by differences in the experimental conditions.
TFIIH is the second factor to join the damaged DNA giving rise to the NER-C2(a) complex. The interaction of TFIIH with damage-bound XPC-HR23B does not require ATP (Figure 1). In a second step, the presence of ATP allows XPB and XPD helicases to unwind the DNA (Roy et al., 1994) generating the NER-C2(b) complex. This finding determines exactly at which stage of the NER reaction ATP is required to activate both TFIIH helicases, which remodel the NER-C2 complex preparing the arrival of the forthcoming factors (Figure 1C), the first one being XPA.
XPA recruitment resulting in NER-C3 is directed by both the modified DNA structure within NER-C2(b) and protein–protein interactions with TFIIH (Park et al., 1995; Nocentini et al., 1997) that may occur, although weakly, even in the absence of ATP (Figure 1A). In NER-C3, XPA might act as a wedge to keep the DNA structure ready for the arrival of RPA, XPG and finally XPF-ERCC1. The single-stranded DNA binding protein RPA interacts with XPA and enhances its interaction with DNA (Stigger et al., 1998; Wang et al., 2000) stabilizing not only XPA within NER-C4 but also XPG in NER-C5 (de Laat et al., 1998). Although RPA clearly supported dual incision complex formation (Figure 2), we had difficulties with detecting RPA within NER intermediate complexes by its function. Possibly, this lack of function can be explained either by substoichiometric incorporation of RPA or partial dissociation of RPA subunits in washed dual incision complexes. Knowing that XPG associates with sites of DNA damage in XPA-deficient cells (Volker et al., 2001), we do not exclude that XPA, RPA and XPG might bind the damaged DNA rather synergistically than sequentially in vivo, since they also mutually stabilize their association within NER-C5 in vitro (Figure 2C).
Finally we demonstrated that the functional binding of XPF-ERCC1 to NER-C5 depends on a multitude of sequential prerequisites including the ATP-dependent opening of the DNA by TFIIH and the binding of XPA, RPA and XPG. Indeed, XPF-ERCC1 can interact specifically with XPA as well as RPA (Bessho et al., 1997; de Laat et al., 1998). Although no direct interaction of XPF-ERCC1 with XPG has been reported, XPG might contribute to XPF-ERCC1 recruitment by inducing a structural change in NER-C5 (Evans et al., 1997).
The release of dual incision factors
The present study points out how in our in vitro system the various events of the NER are coordinated, thus mimicking the in vivo situation. Indeed, the removal of the cisplatinated oligonucleotide perfectly parallels the refilling of the DNA gap (Figure 3A). Both the removal and the resynthesis kinetics have exactly the same slope, strongly suggesting that as soon as the damage is eliminated, the DNA strand is resynthesized. In parallel, we observed that some NER factors progressively leave the DNA substrate possibly preparing the resynthesis step.
We have demonstrated that XPC-HR23B is excluded from the premature dual incision complex with the arrival of XPG, explaining the difficulties in detection of XPC-HR23B association with damaged DNA in the presence of TFIIH, XPA, RPA and XPG in gel shift experiments (Wakasugi and Sancar, 1998). Such a displacement of XPC-HR23B from the cisplatinated DNA could be coordinated with XPA and RPA, both present in NER-C4/5 (You et al., 2003). Importantly, the disengaged XPC-HR23B remained functionally active to initiate a new round of dual incision.
Upon the final arrival of XPF-ERCC1, TFIIH is released from the NER complex and remains functionally active to participate not only in a new round of productive NER, but also in RNA pol II transcription. In this respect, we noticed that the specific activity of TFIIH released from the dual incision complexes was not significantly altered when compared with the TFIIH used for NER-C2 formation (data not shown). Importantly, we also demonstrated that TFIIH being involved in transcription initiation is able to switch to DNA repair after having been excluded from the elongating complexes (Figure 5C). Thus, our experiments indicate firstly that in well defined reconstituted systems at least part of TFIIH is able to shuttle between DNA repair and mRNA synthesis, and secondly that TFIIH is not switched on/off by any modification during NER or transcription to support only one of these cellular processes. Recently, a shuttling of TFIIH was also suggested by some in vivo data (Hoogstraten et al., 2002). Such a mechanism grants the immediate removal of transcription blocking lesions and the rapid resumption of transcription upon NER, thus being possibly significant for TCR. Interestingly, challenge studies suggested that TFIIH might exhibit a higher affinity to NER than to transcription (Vichi et al., 1997; You et al., 1998).
At this stage of our investigations we wondered whether the arrival of XPF-ERCC1 (or the departure of XPC), could initiate a break up of the dual incision complex. Indeed, upon dual incision we also observed a specific release of XPA, which could be recycled (Figure 6B and C). On the contrary, we did not observe any release of functionally active XPG in such conditions (RIS) (Figure 6D and E). The release of XPG when experiments were performed with NE suggests the requirement of additional factors for its release upon dual incision. We did not investigate whether XPF-ERCC1 was released either by its own or by DNA resynthesis factors (Figure 3B). However, in vivo studies suggested that XPF-ERCC1, TFIIH and XPA might be simultaneously released from DNA damage (Hoogstraten et al., 2002).
The passage from dual incision to DNA resynthesis
Our data strongly suggest that dual incision goes hand in hand with DNA resynthesis. It is obvious that NER factors not being released upon dual incision could contribute to the gap filling DNA synthesis as a transient nucleoprotein complex comprising RPA and eventually XPG and/or XPF-ERCC1 to prime DNA resynthesis.
We found that RPA remains associated with the incised DNA, whereas all the other NER factors are released either along the dual incision reaction (e.g. XPC-HR23B, TFIIH, and XPA) or upon the arrival of resynthesis factors (Figure 3B). As an integral component of both processes, RPA might act upon dual incision firstly, by protecting the undamaged strand from inadvertent nuclease attacks (de Laat et al., 1998), and secondly by promoting the arrival of PCNA [our data and Gomes and Burgers (2001)] and RFC (Yuzhakov et al., 1999) to initiate DNA resynthesis (Shivji et al., 1995). Although it has been shown that XPG can interact with and possibly recruits PCNA to the gapped DNA (Miura et al., 1996; Gary et al., 1997), we did not observe any interaction of PCNA with XPG within the pre-incision complex (Figure 3B).
Our work underlines the complexity of the mechanism that drives the process from damage removal to DNA resynthesis. Knowing that mutations in six out of the seven genes involved in the dual incision step of the NER originate XP disorders, our data provide important insights to understand how distinct mutations might affect the NER reaction and to establish biochemically the genotype/phenotype relationships.
Materials and methods
Damaged DNA substrates
Covalently closed circular DNA Pt(I) containing a single 1,3-intrastrand d(GpTpG) cisplatin–DNA crosslink was prepared as described (Shivji et al., 1999) based on the 105.TS plasmid (Frit et al., 2002). A second DNA repair substrate Pt(II) with modified sequence of 36 bp around the Pt-GTG site (5′-GAGCTCTTCTTAATTAACTCGTGCACACTAC ATCAC-3′) was prepared in the same way.
Immobilized damaged DNA was generated by digesting the DNA Pt(I) plasmid with HincII and BanI resulting in a 288 bp fragment with the cisplatin-adduct located 87 bp from the HincII site. The non-damaged strand was biotinylated at the BanI site by Klenow fill-in reaction with Bio-dUTP and purified after separation on agarose gel with QIAEX gel extraction kit (Qiagen). Undamaged DNA was prepared in the same way. Purified biotinylated DNA (40 fmol) was bound to 10 µg magnetic streptavidin beads (Dynabeads M-280 Streptavidin, Dynal) and equilibrated with dual incision assay buffer prior to use. For further analysis, the biotinylated DNA was eluted from the streptavidin beads by phenol chloroform extraction followed by ethanol precipitation.
Dual incision assay
Reconstituted dual incision reactions (25 µl) were carried out in a buffer containing 50 mM HEPES–KOH pH 7.6, 20 mM Tris–HCl pH 7.6, 50 mM KCl, 2.5 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, 10% glycerol, 0.02% NP40 and 2 mM ATP. NER factors were purified as described (Araujo et al., 2000) and used in saturating amounts: XPC-HR23B (10 ng), TFIIH HAP fraction 5 (50 ng) (Gerard et al., 1991), XPA (30 ng), RPA (200 ng), XPG (50 ng) and XPF-ERCC1 (10 ng) per reaction. The NER factors were incubated with 40 fmols of either free or immobilized damaged DNA substrate, for 40 min at 30°C in the presence of ATP. Reactions were stopped by boiling for 5 min after addition of 9 ng 32 nt oligonucleotide complementary to the excised DNA fragment with a 5′-extension of four extra G residues. After annealing the oligonucleotide to the excised DNA fragment, excision products were radiolabelled by extension with 0.15 U of Sequenase version 2.0 polymerase (U.S.B.) and 2 µCi of [α-32P]dCTP (3000 Ci/mmol). Labelled excision products were separated on a denaturing 14% polyacrylamide gel and visualized by autoradiography.
Dual incision reactions with HeLa NE (30 µg) (Dignam et al., 1983) were performed in the presence of 15 µM wortmannin, 60 µM aphidicolin and 2% DMSO; reactions were pre-incubated for 10 min at 30°C before DNA substrate addition and further incubated at 30°C for 40 min or indicated times. DNA–PK inhibitor wortmannin prevents the binding of Ku antigen to linearized DNA and thus improves NER efficiency (Frit et al., 2000).
DNA resynthesis assay
DNA Pt(I) (40 fmol) was incubated with 30 µg of HeLa NE in dual incision buffer in the absence of aphidicolin (Shivji et al., 1995). Reactions were substituted with 5 µM of each dATP, dGTP, dTTP, 1 µM dCTP and 2.5 µCi [α-32P]dCTP (3000 Ci/mmol). After incubation for the indicated times at 30°C, DNA was purified and digested with EcoRI and NdeI resulting in a 94 nt fragment containing the resynthesized DNA patch. Restriction reactions were loaded on a denaturating 8% polyacrylamide gel and visualized by autoradiography.
Protein binding studies on immobilized DNA
Several reaction mixtures each containing 40 fmol immobilized DNA, purified NER factors or HeLa nuclear extract were incubated together under dual incision assay conditions at 30°C or 4°C for 40 min if not stated otherwise. After incubation, magnetic beads were collected on a magnetic particle concentrator and washed 5× by resuspending in four volumes dual incision buffer. Washed and aliquoted beads were further analysed for bound proteins either functionally or by western blot. Functional protein binding studies were carried out with the equivalent of one, western blot analysis with the equivalent of six dual incision reactions.
Immunoblotting
Proteins were separated on 10% SDS–PAGE gels and transferred to nitrocellulose membranes. Mouse monoclonal (MAb) and rabbit polyclonal (PAb) antibodies used as primary antibodies were: XPC: PAb (Sugasawa et al., 1996); XPD: MAb 2F6; p62: MAb 3C9; cdk7: MAb 2F8; XPA: MAb 1E11 raised against peptide aa 242–261; RPA32/70: PAb N2.2 (Henricksen et al., 1994); RPA70: PAb J3 raised against a mixture of peptides aa 112–126 and aa 127–141 (a gift from J.Imbert); XPG: MAb 1B5 raised against peptide aa 1167–1186; XPF: MAb Ab-5 (NeoMarkers); PCNA: MAb (PC10):sc-56 (Santa Cruz Biotechnology).
Gel filtration chromatography
The supernatant of two reconstituted dual incision reactions with an immobilized DNA substrate was injected onto the Superose6 column (PC3.2/30, SMART system, Amersham Pharmacia Biotech; flow rate 20 µl/min, fraction size 50 µl) at 4°C, equilibrated with dual incision reaction buffer. Fractions were analyzed for excision product as described for the dual incision assay.
Acknowledgments
Acknowledgements
We acknowledge R.Wood, J.H.J.Hoeijmakers and S.Clarkson for providing some recombinant baculoviruses, antibodies and their help in setting up the in vitro NER assays. We thank F.Coin for fruitful discussions. We are grateful to I.Kolb-Cheynel and J.L.Weickert for production of recombinant baculoviruses and J.M.Chipoulet and A.Larnicol for their high technical expertise. These studies were supported by grants from the Association pour la Recherche sur le Cancer, The Commissariat à l’Energie atomique and the EEC (QLG1-1999 and QLRT-1999–02002). T.R. is a recipient of a Fondation pour la Recherche Médicale grant (FdT).
References
- Aboussekhra A. et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- Araujo S.J., Tirode,F., Coin,F., Pospiech,H., Syvaoja,J.E., Stucki,M., Hubscher,U., Egly,J.M. and Wood,R.D. (2000) Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH and modulation by CAK. Genes Dev., 14, 349–359. [PMC free article] [PubMed] [Google Scholar]
- Asahina H., Kuraoka,I., Shirakawa,M., Morita,E.H., Miura,N., Miyamoto,I., Ohtsuka,E., Okada,Y. and Tanaka,K. (1994) The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat. Res., 315, 229–237. [DOI] [PubMed] [Google Scholar]
- Batty D., Rapic’-Otrin,V., Levine,A.S. and Wood,R.D. (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol., 300, 275–290. [DOI] [PubMed] [Google Scholar]
- Berneburg M. and Lehmann,A.R. (2001) Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet., 43, 71–102. [DOI] [PubMed] [Google Scholar]
- Bessho T., Sancar,A., Thompson,L.H. and Thelen,M.P. (1997) Reconstitution of human excision nuclease with recombinant XPF–ERCC1 complex. J. Biol. Chem., 272, 3833–3837. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Sugasawa,K., Weterings,E., Jaspers,N.G. and Hoeijmakers,J.H. (1998) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev., 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Martin,P.L., Shastry,B.S. and Roeder,R.G. (1983) Eukaryotic gene transcription with purified components. Methods Enzymol., 101, 582–598. [DOI] [PubMed] [Google Scholar]
- Evans E., Moggs,J.G., Hwang,J.R., Egly,J.-M. and Wood,R.D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frit P., Li,R.Y., Arzel,D., Salles,B. and Calsou,P. (2000) Ku entry into DNA inhibits inward DNA transactions in vitro. J. Biol. Chem., 275, 35684–35691. [DOI] [PubMed] [Google Scholar]
- Frit P., Kwon,K., Coin,F., Auriol,J., Dubaele,S., Salles,B. and Egly,J.M. (2002) Transcriptional activators stimulate DNA repair. Mol. Cell, 10, 1391–1401. [DOI] [PubMed] [Google Scholar]
- Gary R., Ludwig,D.L., Cornelius,H.L., MacInnes,M.A. and Park,M.S. (1997) The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem., 272, 24522–24529. [DOI] [PubMed] [Google Scholar]
- Gerard M., Fischer,L., Moncollin,V., Chipoulet,J.M., Chambon,P. and Egly,J.M. (1991) Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem., 266, 20940–20945. [PubMed] [Google Scholar]
- Gomes X.V. and Burgers,P.M. (2001) ATP utilization by yeast replication factor C.I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem., 276, 34768–34775. [DOI] [PubMed] [Google Scholar]
- Guzder S.M., Sung,P., Prakash,L. and Prakash,S. (1996) Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem., 271, 8903–8910. [DOI] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- Hoogstraten D., Nigg,A.L., Heath,H., Mullenders,L.H., van Driel,R., Hoeijmakers,J.H., Vermeulen,W. and Houtsmuller,A.B. (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell, 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Huang J.C., Svoboda,D.L., Reardon,J.T. and Sancar,A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Maldonado E. et al. (1996) A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature, 381, 86–89. [DOI] [PubMed] [Google Scholar]
- Mellon I. and Hanawalt,P.C. (1989) Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature, 342, 95–98. [DOI] [PubMed] [Google Scholar]
- Miura M., Nakamura,S., Sasaki,T., Takasaki,Y., Shiomi,T. and Yamaizumi,M. (1996) Roles of XPG and XPF/ERCC1 endonucleases in UV-induced immunostaining of PCNA in fibroblasts. Exp. Cell Res., 226, 126–132. [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Yarema,K.J., Essigmann,J.M. and Wood,R.D. (1996) Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem., 271, 7177–7186. [DOI] [PubMed] [Google Scholar]
- Mullenders L.H. and Berneburg,M. (2001) Photoimmunology and nucleotide excision repair: impact of transcription coupled and global genome excision repair. J. Photochem. Photobiol. B, 65, 97–100. [DOI] [PubMed] [Google Scholar]
- Nocentini S., Coin,F., Saijo,M., Tanaka,K. and Egly,J.-M. (1997) DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J. Biol. Sci., 272, 22991–22994. [DOI] [PubMed] [Google Scholar]
- O’Donovan A., Davies,A.A., Moggs,J.G., West,S.C. and Wood,R.D. (1994) XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature, 371, 432–435. [DOI] [PubMed] [Google Scholar]
- Park C.H., Mu,D., Reardon,J.T. and Sancar,A. (1995) The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem., 270, 4896–4902. [DOI] [PubMed] [Google Scholar]
- Reardon J.T. and Sancar,A. (2002) Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol. Cell. Biol., 22, 5938–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Schaeffer,L., Humbert,S., Vermeulen,W., Weeda,G. and Egly,J.M. (1994) The DNA-dependent ATPase activity associated with the class II transcription factor BTF2/TFIIH. J. Biol. Chem., 269, 9826–9832. [PubMed] [Google Scholar]
- Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H.J., Chambon,P. and Egly,J.M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science, 260, 58–63. [DOI] [PubMed] [Google Scholar]
- Shivji M.K.K., Podust,V.N., Hubsher,U. and Wood,R.D. (1995) Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC and RPA. Biochemistry, 34, 5011–5017. [DOI] [PubMed] [Google Scholar]
- Shivji M.K., Moggs,J.G., Kuraoka,I. and Wood,R.D. (1999) Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol., 113, 373–392. [DOI] [PubMed] [Google Scholar]
- Sijbers A.M. et al. (1996) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Spangler L., Wang,X., Conaway,J.W., Conaway,R.C. and Dvir,A. (2001) TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc. Natl Acad. Sci. USA, 98, 5544–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigger E., Drissi,R. and Lee,S.H. (1998) Functional analysis of human replication protein A in nucleotide excision repair. J. Biol. Chem., 273, 9337–9343. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Masutani,C., Uchida,A., Maekawa,T., van der Spek,P.J., Bootsma,D., Hoeijmakers,J.H. and Hanaoka,F. (1996) HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol., 16, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng,J.M., Masutani,C., Iwai,S., van der Spek,P.J., Eker,A.P., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q., Wang,Z., Feaver,W.J., Wu,X., Bushnell,D.A., Donahue,T.F., Friedberg,E.C. and Kornberg,R.D. (1995) Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell, 80, 21–28. [DOI] [PubMed] [Google Scholar]
- Ura K. and Hayes,J.J. (2002) Nucleotide excision repair and chromatin remodeling. Eur. J. Biochem., 269, 2288–2293. [DOI] [PubMed] [Google Scholar]
- Vichi P., Coin,F., Renaud,J.P., Moras,D. and Egly,J.M. (1997) UV and cisplatin lesions lure basal transcription factor TFIID/TBP. EMBO J., 16, 7444–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wakasugi M. and Sancar,A. (1998) Assembly, subunit composition and footprint of human DNA repair excision nuclease. Proc. Natl Acad. Sci. USA, 95, 6669–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M. and Sancar,A. (1999) Order of assembly of human DNA repair excision nuclease. J. Biol. Chem., 274, 18759–18768. [DOI] [PubMed] [Google Scholar]
- Wang M., Mahrenholz,A. and Lee,S.H. (2000) RPA stabilizes the XPA-damaged DNA complex through protein–protein interaction. Biochemistry, 39, 6433–6439. [DOI] [PubMed] [Google Scholar]
- You Z., Feaver,W.J. and Friedberg,E.C. (1998) Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26. Mol. Cell. Biol., 18, 2668–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J.S., Wang,M. and Lee,S.H. (2003) Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem., 278, 7476–7485. [DOI] [PubMed] [Google Scholar]
- Yuzhakov A., Kelman,Z., Hurwitz,J. and O‘Donnell,M. (1999) Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J., 18, 6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Kumar,K.P. and Reinberg,D. (1995) Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev., 9, 1479–1490. [DOI] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]