Abstract
Methylation of histone tails and CpG methylation are involved in determining heterochromatin structure, but their cause and effect relationship has not been resolved as yet in mammals. Here we report that Lsh, a member of the SNF2 chromatin remodeling family, controls both types of epigenetic modifications. Lsh has been shown to be associated with pericentromeric heterochromatin and to be required for normal CpG methylation at pericentromeric sequences. Loss of Lsh, in Lsh-deficient mice, results in accumulation of di- and tri-methylated histone 3 at lysine 4 (H3-K4me) at pericentromeric DNA and other repetitive sequences. In contrast, di- or tri-methylation of H3-K9 and distribution of HP1 appear unchanged after Lsh deletion, suggesting independent regulatory mechanisms for H3-K4 or K9 methylation. Experimental DNA demethylation with 5′-azacytidine results in a similar increase of H3-K4me. These results support the model that loss of CpG methylation caused by Lsh deficiency antecedes elevation of H3-K4me. Thus, Lsh is crucial for the formation of normal heterochromatin, implying a functional role for Lsh in the regulation of transcription and mitosis.
Keywords: chromatin/DNA methylation/histone/Lsh/SNF2
Introduction
The genome is packaged into ‘active’ euchromatin and ‘silent’ heterochromatin domains. Covalent histone tail modifications including acetylation or methylation, regulate these different states of chromatin configuration (Bird, 2002; Richards and Elgin, 2002). Euchromatin usually contains single copy genes and packages regions that are transcribed. Heterochromatin is frequently composed of repetitive sequences and regulates many diverse functions, including gene silencing, normal centromere function and nuclear organization. Euchromatin is characterized by hypomethylated DNA sequences and methylation of histone 3 at lysine 4 residues (H3-K4) (Bernstein et al., 2002; Lachner and Jenuwein, 2002). Conversely, heterochromatin is composed of hypermethylated DNA and shows low methylation at H3-K4 and high methylation at lysine residue 9 (H3-K9). The specific methylation mark at lysine 9 in histone 3 serves as recognition signal for binding of the heterochromatic protein 1 (HP1) (Bannister et al., 2001; Lachner et al., 2001; Peters et al., 2001).
Lsh (lymphoid specific helicase) is a member of the SNF2/helicase family, that is involved in chromatin remodeling (Jarvis et al., 1996; Geiman et al., 1998). ATP-dependent chromatin remodeling complexes disrupt histone–DNA interactions and allow for sliding of nucleosomes along the DNA (Becker and Horz, 2002; Peterson, 2002). Thus, they alter the accessibility of chromatin to transcription factors or other DNA binding factors. Members of the SNF2 chromatin remodeling family have been implicated in the control of CpG methylation (Jeddeloh et al., 1999; Gibbons et al., 2000; Dennis et al., 2001; Meehan et al., 2001; Bourc’his and Bestor, 2002). Lsh regulates DNA methylation levels in mice (Dennis et al., 2001). Targeted deletion of Lsh results in perinatal lethality, with abnormal lymphoid development and renal defects (Geiman and Muegge 2000; Geiman et al., 2001). Thus, Lsh is a non-redundant member of the SNF2 family and is essential for normal murine development and survival. DNA derived from Lsh–/– tissue shows substantial loss of CpG methylaton throughout the genome. This suggests that the presumed chromatin remodeling ability of Lsh is crucial for setting methylation patterns.
Though CpG methylation as well as histone methylation appear to play an important role in chromatin formation, their cause and effect relationship is less well understood in the mammalian species. Here we examine how Lsh and genomic hypomethylation caused by 5′-azacytidine treatment influence heterochromatin structure and specifically modulate the methylation status of histone 3 at pericentromeric heterochromatin.
Results
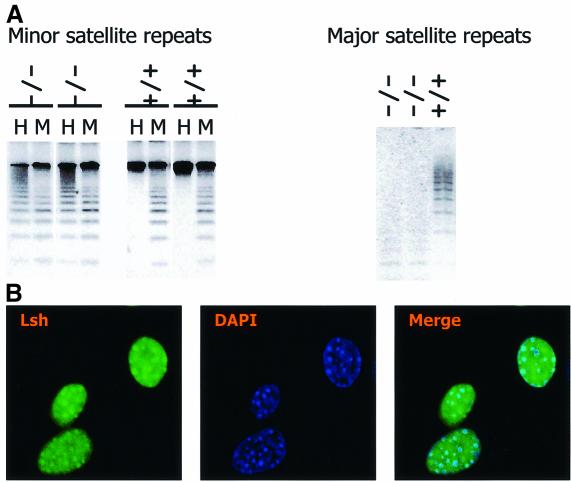
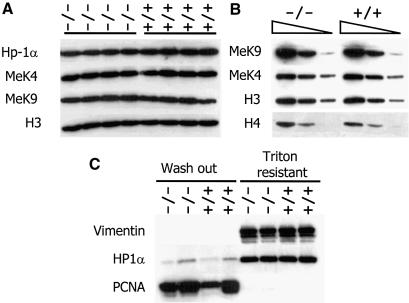
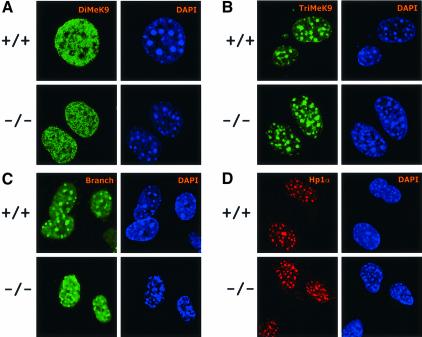
Targeted deletion of Lsh in murine tissues results in methylation defects at repetitive sequences (Dennis et al., 2001). Figure 1A demonstrates that the hypomethylation of minor and major satellite sequences is stably propagated in murine embryonal fibroblasts (MEF) derived from Lsh-deficient embryos. These satellite sequences are major components of pericentromeric DNA. When Lsh was transiently expressed as GFP-tagged protein (Yan et al., 2003) or when Lsh was under control of an inducible promoter (Figure 1B), a direct association of Lsh with DAPI-rich regions was observed. DAPI-rich regions are indicative for murine centromeric DNA. Furthermore, Lsh co-localizes with heterochromatin protein 1 (HP1) α and can be co-precipitated with HP1α during chromatin immunprecipitation (Q.Yan and K.Muegge, unpublished) suggesting that Lsh is directly involved in the formation of pericentromeric heterochromatin. Thus, we sought to investigate whether Lsh determines histone tail modifications at heterochromatin. Silent heterochromatin consists of low H3-K4me and high H3-K9me, the latter serving as ‘anchor’ for HP1 recruitment (Bannister et al., 2001; Lachner et al., 2001). DDM1 mutants, a Lsh homolog in Arabidopsis thaliana, showed a disturbed pattern of histone methylation with H3-K9me being largely replaced by H3-K4me (Gendrel et al., 2002; Johnson et al., 2002; Soppe et al., 2002). In order to determine a role for Lsh in histone methylation, we first examined global histone methylation levels by western analysis. No differences in global levels of H3-K4me, H3-K9me and HP1α in embryos or MEFs were detected (Figure 2A and B). Next immunofluorescence analysis was performed to detect the distribution pattern of histone tail modifications. MEFs derived from Lsh+/+ and Lsh–/– mice were stained with antiserum directed against distinct forms of methylated H3-K9. Staining with anti-dimethyl H3-K9 revealed a coarse pattern all over the nucleus (Figure 3A). Antiserum directed against trimethylated H3-K9 detected a focal stain that co-localizes with DAPI (Figure 3B), suggesting an enrichment for trimethylated H3-K9 at pericentromeric DNA. As shown in Figure 3C, a similar co-localization with DAPI-rich heterochromatin was observed using the branched peptide antiserum that recognizes dimethylated H3-K9 in a more compact higher-order chromatin organization (Peters et al., 2001). All three stains for H3-K9me were indistinguishable between Lsh wild-type and Lsh-deficient cells indicating no substantial loss of H3-K9 di- or tri-methylation. The resistance of HP1 to Triton X-100 washes (Taddei et al., 2001) was not altered in Lsh-deficient MEFs (Figure 2C), suggesting that the asssociation of Hp1 with chromatin was not disturbed in the absence of Lsh. Furthermore, the HP1α immunofluorescence staining pattern in Lsh- deficient MEFs was indistinguishable from wild-type cells showing a normal co-localization of HP1α with pericentromeric heterochromatin (Figure 3D). Thus, despite the hypomethylation of pericentric DNA sequences, H3-K9me levels and HP1 recruitment were not disturbed in the absence of Lsh.
Fig. 1. Lsh associates with pericentromeric heterochromatin. (A) Lsh controls CpG methylation at pericentromeric regions in mice. Southern analysis of minor and major satellite sequences utilizing the CpG-sensitive restriction enzyme HpaII (H) and the methylation-insensitive restriction enzymes MspI (M) (for minor) or MaeII (for major) comparing Lsh+/+ and Lsh–/– MEFs. (B) Lsh localizes to pericentromeric heterochromatin. Immunofluorescence staining of Flag-tagged Lsh after induction in 3T3 fibroblasts utilizing an anti-Flag antibody. Nuclei are stained with DAPI.
Fig. 2. Lsh does not regulate global histone methylation levels. (A) Loss of Lsh does not alter H3-K9me, H3-K4me or Hp1 protein levels. Western analysis of nuclear extracts derived from four Lsh-deficient embryos (gestation day 14) and their wild-type littermates. (B) Titration of nuclear extracts of one wild-type and one Lsh-deficient embryo. (C) Loss of Lsh does not alter Hp1 binding to chromatin. Lsh-deficient and Lsh wild-type MEFs are compared for resistance to Triton X-100 washes followed by western analysis to detect HP1 or PCNA and Vimentin as control.
Fig. 3. Loss of Lsh does not alter H3-K9 methylation or HP1 association at pericentromeric chromatin. (A) Lsh-deficient and wild-type MEFs are immunostained for detection of dimethylated H3-K9 and counterstained with DAPI. (B) The same as in (A) but antiserum raised against trimethylated H3-K9 peptide was used. (C) The same as in (A) but antiserum raised against the branched dimethylated H3-K9 peptide was used. (D) Lsh-deficient and wild-type MEFs are immunostained for detection of HP1 and counterstained with DAPI.
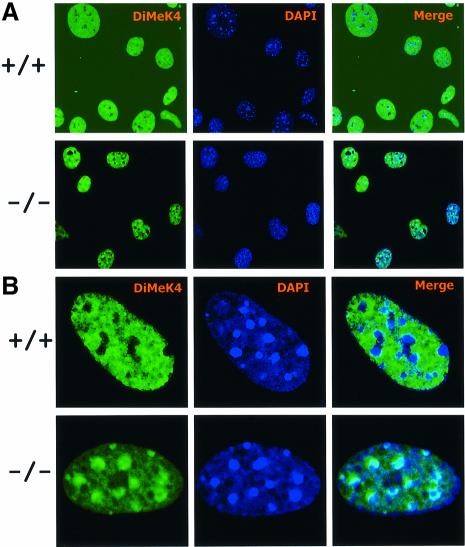
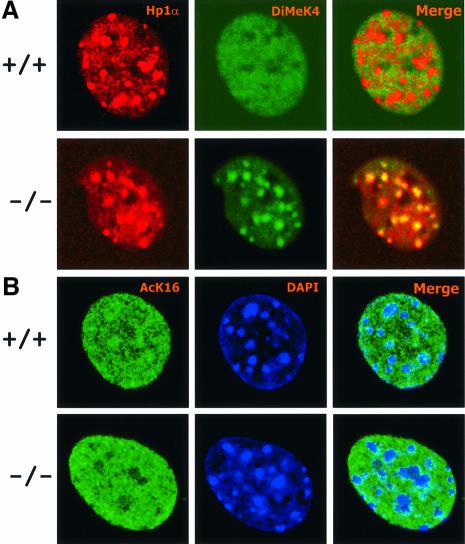
This result was in striking contrast with the staining pattern provided by antibodies raised against H3-K4me. Wild-type MEFs (six cell lines were examined) probed with anti-dimethylated H3-K4 showed diffuse nuclear speckles that spared the DAPI intense regions, a property characteristic of euchromatic staining patterns (Figure 4). Conversely, a large proportion of Lsh-deficient cells revealed a reverse staining pattern in which the dimethylated H3-K4 localized to pericentromeric heterochromatin as evidenced by co-localization with DAPI (Figure 4) and HP1α (Figure 5A). Among six Lsh-deficient cell lines examined, the proportion of cells that showed the ‘speckled’ H3-K4 methylation pattern varied from 20 to 33% in fresh fibroblasts cultures, down to 5% in long-term cultures. Possibly the raise in H3-K4 methylation may give a growth disadvantage and cells may be negatively selected against in longer cultures. This result of a raise in H3-K4 methylation is distinct from Arabidopsis in which mutation of DDM1 caused decreased levels of H3-K9me and no change in H3-K4me at pericentromeric heterochromatin (Johnson et al., 2002; Soppe et al., 2002). As a further distinction between mammals and plants, no alteration in staining for H4-K16 acetylation was detected in Lsh-deficient cells (Figure 5B), which had been reported to accumulate at heterochromatic regions in DDM1 mutants (Soppe et al., 2002). Recently, the trimethylated form of H3-K4 was found to be closely associated with the transcription state of a gene. Whereas the dimethylated form of H3-K4 was present at transciptionally active or inactive euchromatic genes, the trimethylated form was only found at active sites (Santos-Rosa et al., 2002). Figure 6A shows that the stain for the trimethylated form of H3-K4 was also enhanced at pericentromeric regions in the absence of Lsh. In addition, staining of condensed metaphase chromosomes for dimethylated H3-K4, as shown in Figure 6B, supports the observation that the increase in lysine 4 methylation occurred predominantly at the centromeric chromosomal regions. Since a global change in methylation of H3-K4 was not observed, the marked elevation at pericentromeric regions suggests a redistribution of methylated histones to heterochromatin. These results point to a crucial role of Lsh in maintenance of normal H3-K4me pattern in mammalian cells.
Fig. 4. Absence of Lsh leads to accumulation of dimethylated H3-K4 at pericentromeric heterochromatin. (A) Lsh-deficient and wild-type MEFs are immunostained for detection of dimethylated H3-K4 and counterstained with DAPI. (B) The same as in (A), but at higher magnification.
Fig. 5. Co-localization of methylated H3-K4 with HP1. (A) Lsh-deficient and wild-type MEFs are double stained for detection of dimethylated H3-K4 and HP1. (B) Lsh-deficient and wild-type MEFs are immunostained for detection of acetylated H4-K16 and counterstained with DAPI.
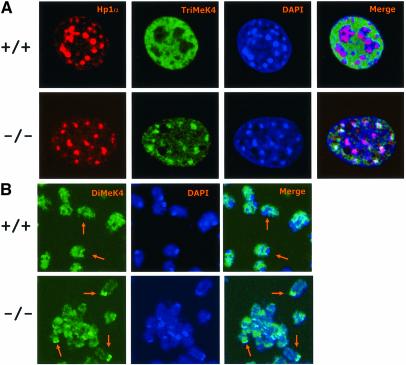
Fig. 6. Lsh deficiency raises trimethylated H3-K4 at pericentromeric regions. (A) Lsh-deficient and wild-type MEFs are double stained for detection of trimethylated H3-K4, HP1 and counterstained with Dapi. (B) Lsh-deficient and wild-type MEFs are treated with colcemid and the metaphase spread immunostained for detection of dimethylated H3-K4 and counterstained with DAPI.
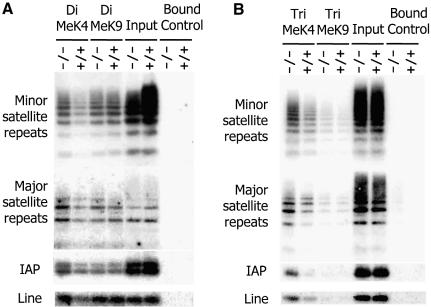
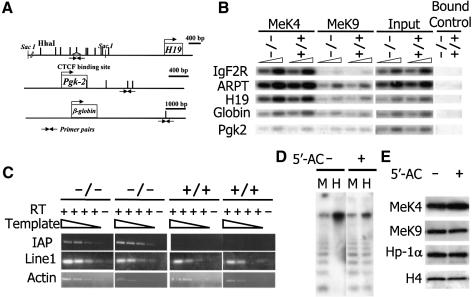
In order to confirm the increase of H3-K4me at pericentromeric regions and to examine changes at other genomic loci, ChIPs analysis was performed. The presence of minor and major satellite sequences was enhanced in the anti-dimethylated H3-K4 as well as anti-trimethylated H3-K4 precipitates derived from Lsh–/– MEFs, confirming an increase in both types of H3-K4 methylation in the absence of Lsh at pericentromeric DNA (Figure 7). Furthermore, there was a detectable raise of dimethylated H3-K4 as well as trimethylated H3-K4 at intracisternal A particle (IAP) as well as the Line element indicating that other heterochromatic sites also showed perturbed histone methylation. In contrast, the amount of satellite DNA sequences and IAP or Line elements in either dimethylated or trimethylated H3-K9me precipitates were not significantly altered comparing wild-type and Lsh-deficient cells, supporting the results generated by H3-K9me immunofluorescence analysis (Figure 7). Next we sought to determine if H3-K4 methylation at single copy sequences was affected by Lsh. As shown in Figure 8A and B, none of the single copy genes examined were altered at either dimethylated H3-K4me or dimethylated H3-K9me, despite the previously reported DNA methylation differences at globin, Pgk2 or H19 loci. Thus, Lsh specifically regulates H3-K4 methylation at repetitive heterochromatin.
Fig. 7. Loss of Lsh increases H3-K4 methylation at repetitive sequences. (A) Chromatin immunoprecipitation with specific antibodies raised against dimethylated H3-K4 and dimethylated H3-K9 were performed using nuclear extracts derived from Lsh-deficient and wild-type MEFs, followed by PCR analysis for detection of minor and major satellite sequences, the IAP and Line elements. (B) ChIPs was performed as in (A) with specific antibodies raised against trimethylated H3-K4 and trimethylated H3-K9.
Fig. 8. Derepression of IAP transcripts in the absence of Lsh. (A) Diagram of the regions for single copy genes that have been previously examined by Southern analysis and found to be hypomethylated (H19, globin, pgk-2) or unaltered (Igf2R, ARPT). Triangles indicate the position of the primers used in ChIps experiments located at methylation-sensitive HhaI sites. (B) Chromatin immunoprecipitation with specific antibodies raised against dimethylated H3-K4 and dimethylated H3-K9 were performed using nuclear extracts derived from Lsh-deficient and wild-type MEFs followed by PCR analysis for detection of the indicated single copy genes. (C) RT–PCR analysis of cDNA derived from Lsh-deficient and wild-type MEFs for detection of IAP and Line1 transcripts with actin as a control. For better comparison cDNA was titrated in dilution steps of 1:3. Amplification in the absence of reverse transcriptase served as control. (D) 5′-azacytidine treatment reduces CpG methylation at pericentromeric DNA. Southern analysis of minor satellite sequences utilizing the CpG sensitive restriction enzyme HpaII (H) and the methylation-insensitive restriction enzyme MspI (M) comparing untreated 3T3 fibroblasts and cells treated for 5 days with 5′-azacytidine. (E) 5′-azacytidine does not alter global H3 methylation levels. Western analysis of nuclear extracts derived from untreated and 5′-azacytidine-treated 3T3 fibroblasts for detection of methylated H3 and Hp1.
It has been reported that H3-K4me can facilitate subsequent H3 acetylation and that H3-K4me precludes the association of the repressor complex NURD with the H3 tail, thus activating transcription (Wang et al., 2001; Nishioka et al., 2002). In order to determine the effect of perturbed histone methylation pattern at repetitive sequences we measured reactivation of silenced elements in the absence of Lsh. Whereas IAP transcripts were derepressed in Lsh–/– MEFs, there was no evidence for a further increase of Line1 transcripts (Figure 8C). Thus, the loss of CpG methylation and the elevation in H3-K4me correlated only in selective repetitive elements such as IAP with a derepression of transcription.
To test the hypothesis whether CpG hypomethylation in Lsh-deficient cells was responsible for H3-K4me elevation or alternatively whether Lsh would directly influence H3-K4me, we treated wild-type MEFs (Q.Yan, J.Huang and K.Muegge, unpublished) or 3T3 fibroblasts with the demethylating agent 5′-azacytidine (Figure 8D). Although 5′-azacytidine treatment did not change global H3-K4me levels (Figure 8E) it led to an elevation of H3-K4me at pericentromeric heterochromatin (Figure 9A and B). In order to discriminate between the possibilities that Lsh deficiency acts via CpG hypomethylation on H3-K4me, or alternatively, that Lsh dissociates from heterochromatin after CpG hypomethylation and thus mediates an effect more directly on H3 methylation, we analysed Lsh localization in 5′-azacytidine-treated cells. As shown in Figure 9E, Lsh still co-localized with DAPI in demethylated cells. The observation that Lsh still localizes to pericentromeric heterochromatin in demethylated cells supports a model in which Lsh exerts its effects on H3-K4 methylation via CpG methylation. Furthermore, Lsh co-localized with increased H3-K4me staining patterns. The co-stain of Lsh and elevated H3-K4me also supports the idea that Lsh deficiency does not simply unmask a binding site and facilitate H3-K4me detection, but leads to an actual increase in the H3-K4me signal. Cells treated with 5′-azacytidine retained normal HP1α staining patterns (Taddei et al., 2001; Q.Yan and K.Muegge, unpublished) and did not affect staining with antibody raised against methylated branched H3-K9 peptides (Figure 9C). Since methylation of histone tails is thought to be a rather stable protein modification, we extended the treatment time with 5′-azacytidine to 5 days but still could not observe any change in the distribution of trimethylated H3-K9 (Figure 9D). Thus, azacytidine faithfully mimicked the results of Lsh–/– MEFs, resulting in unaltered H3-K9me and Hp1 association and a redistribution of H3-K4me pattern. These observations suggest that Lsh acts upstream of CpG methylation and that CpG methylation may control H3-K4 methylation levels in mammals.
Fig. 9. Global demethylation causes redistribution of methylated H3-K4 at pericentromeric heterochromatin. (A and B) Demethylation by 5′-azacytidine increases H3-K4 methylation. 3T3 fibroblasts were treated for 3 days with 5′-azacytidine and immunostained for detection of dimethylated H3-K4 and counterstained with DAPI. (C) Same as in (A), but antiserum raised against the branched dimethylated H3-K9 peptide was used. (D) Prolonged treatment of 3T3 fibroblasts (for 5 days) with 5′-azacytidine followed by immunostaining for detection of HP1, trimethylated H3-K9 and counterstain with DAPI. (E) Same as in (D) but Flag-tagged Lsh was induced over the last 24 h and cells were then immunostained using antiserum for detection of the Flag-tag Lsh protein or detection of trimethylated H3-K4.
Discussion
Histone 3 modifications are frequently inversely regulated with high K4 methylation accompanied by low K9 methylation levels of euchromatin and low K4 and high K9 methylation of repressed heterochromatin in plants as well as in mammalian cells (Lachner and Jenuwein, 2002; Richards and Elgin, 2002). Since both histone modifications inhibit each other in vitro, the regulation is thought to be interdependent. For example, methylation of H3-K4 impairs Suv39h1-mediated methylation at K9 suggesting an interdependence in the regulation of H3 tail modifications (Nishioka et al., 2002). Here we report that loss of Lsh in mice results in redistribution of di- and tri-methylated H3-K4 to heterochromatin without apparent effect on K9 di- or tri-methylation at pericentromeric regions. Our results indicate that independent regulatory mechanisms at pericentromeic repeats occur in mammals that modify lysine residues on different sets of H3 molecules. Similarly, it has been reported that trichostatin A (TSA) treatment can modify K9 acetylation at histone 3 without modifying K9 methylation. The independent regulation and dissociation of acetylation and methylation may also involve distinct histone molecules in vivo (Taddei et al., 2001; Maison et al., 2002).
Histone 3 K4 methylation is generally associated with transcriptionally active regions (Bernstein et al., 2002; Santos-Rosa et al., 2002) and we observed in at least one case (IAP) a correlation of elevated K4 methylation and derepression of transcription. Although the precise mechanism of how elevated K4 augments transcription is not known, it has been reported that H3-K4 methylation affects subsequent histone acetylation by the histone acetylase p300 (Wang et al., 2001). As an alternative mechanism, unmodified H3 tails and K9 methylated H3 associate with NuRD a transcriptional repressive complex (Nishioka et al., 2002). Methylation of histone H3-K4 precludes the association of NuRD with the H3 tail. In this model it is not the level of H3-K9 but rather the amount of K4 methylation that determines the transcriptional repression by NURD. Consequently, a reduction in CpG methylation and increase in H3-K4 methylation in the case of IAP is sufficient for transcriptional activation.
Another effect of histone methylation may be the control of centromere function and mitosis. Centromeric heterochromatin structure affects the deposition of Rad21 and the cohesion of sister chromatids (Bernard et al., 2001). Subsequent defects in chromosome segregation may arise during mitosis. We have found a mitotic defect in Lsh-deficient MEF cells, with micronuclei, signs of abnormal spindle formation and centrosome amplification (Fan et al., 2003). Thus, CpG methylation may contribute to intact centromere function by proper H3-K4 methylation. Also, hypomethylation at centromeric regions in ICF patients that carry a dnmt3b mutation, may lead to disturbed H3-K4 methylation patterns and contribute to chromosomal instability—a hypothesis waiting to be tested.
The link between CpG methylation and histone methylation has not been resolved as yet (reviewed in Richards, 2002). In lower organisms there are several lines of evidence pointing to a role of H3-K9 methylation in regulating DNA methylation or vice versa. For example, mutants of H3-K9 methyltransferase (such as dim-5 or KYP) lower cytosine methylation in Neurospora crassa or A.thaliana (Tamaru and Selker, 2001; Jackson et al., 2002). A recent report suggests that trimethylation and not dimethylation of H3-K9 is important for DNA methylation (Tamaru et al., 2003). There are many differences in DNA methylation in comparing lower organisms with mammalian species. For example, Arabidopsis shows methylation predominantly at CNG motifs or CNN, as opposed to CG motifs. Also hyperacteylation in Neurospora affects CpG methylation, a phenomenon unreported in mammals (Selker, 1998). The possibility that the reverse relationship exists, that is whether CpG methylation influences H3-K9 methylation, has been disputed in Arabidopsis (Johnson et al., 2002; Soppe et al., 2002). We suggest a model in mammalian species in which Lsh, via a reduction of CpG methylation, controls H3-K4 methylation levels, but not H3-K9me. Loss of CpG and methyl-DNA binding proteins may alter the access of Set7/9 to chromatin sites (Wang et al., 2001; Nishioka et al., 2002). Set7/9 is to date the only histone methyltransferase (HMT) known to specifically methylate H3-K4 in vitro; however, only marginal activity has been reported on mononucleosomal or oligosomal targets. Alternatively CpG methylation may regulate the deposition of methylated histones. It has been suggested recently that the deposition rate of H3.3 in Drosophila melanogaster is independent of replication, supporting the idea that deposition of histones may occur during altered transcription rates (Ahmad and Henikoff, 2002). Thus, CpG hypomethylation could reactivate transcription, which in turn influences replacement of methylated K4 histones. Since the reverse effect has also been reported, that H3-K4 methylation facilitates transcription, we can speculate that the relationships of CpG hypomethylation, transcription and methylated H3-K4 may reinforce each other.
The organization of the genome into active and silent regions is crucial for normal differentiation, development and maintenance of genomic stability. Lsh is a modulator of two pivotal epigenetic modifications, including histone and DNA methylation. The results of this study suggest a model in which Lsh, acting upstream by demethylating CpG, augments the methylation of H3-K4 at heterochromatin, which in turn can control gene transcription.
Materials and methods
Tissue culture and transfections
Embryos from crosses between Lsh+/– mice were removed at day 13.5 of gestation and genotyped by PCR as described before (Geiman et al., 2001). Primary MEF cells were cultured as described (Fan et al., 2003) and were grown in DMEM (GIBCO) supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine and antibiotics (GIBCO). NIH3T3 cells were treated with 5-aza-2-deoxycytidine (Sigma) at 10 µM for 3 days or with 2.5 µM for 5 days. The metaphase spreads were prepared after treatment of MEFs for 3 h with 100 ng/ml colcemid as described elsewhere (McDowell et al., 1999). For HpaII digestion, genomic DNA was extracted using a standard proteinase-K protocol under the manufacturer’s instruction of Dneasy Tissue Kit (QIAGEN), and digested for 16 h with HpaII at 37°C. Southern analysis was performed as described elsewhere (Dennis et al., 2001). Cloning of full length Lsh into an inducible expression vector and use of the GenSwitch System (Invitrogen), a mifepristone-regulated expression system for mammalian cells, has been described in detail elsewhere (Q.Yan, J.Huang, T.Fan, S.Lockett and K.Muegge, submitted). NIH3T3 cells were stably transfected using 2–4 µg of Flag-tagged Lsh vector using SuperFect Transfection Reagent (Qiagen). RT–PCR analysis was performed as previously described (Dennis et al., 2001) using the following oligonucleotides as primers. For detection of IAP: 5′-GAAGCAGG TGAAGAATCCTCC-3′ and 5′-TCCTTCAAAGACCGGAAATGC-3′; for detection of Line1, 5′-GCAATGGCTTGTGCTGTAAG-3′ and 5′-TGCATTTCCCTGATGATTAAG-3′.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described in detail elsewhere (Huang et al., 2001). In brief, cells were crosslinked with 1% formaldehyde, lysed and sonicated on ice to generate DNA fragments with an average length of ∼200–500 bp. After pre-cleaning, 10% of each sample was saved as input fraction. Immunoprecipitation was performed using specific antibodies against the indicated peptides or IgG used as control. The complex was eluted and the crosslinking reversed. After purification, the DNA concentration was determined and PCR performed as described (Huang et al., 2001). Amplification conditions were as follows: 94°C for 3 min; 94°C for 1 min, 62°C for 2 min, 72°C for 3 min (10 cycles for major satellite sequences, 18 cycles for minor satellite sequences and IAP, 25 cycles for single copy genes); 72°C for 7 min. The following primer pairs were used for sequence detection: IAP (accession no. NM_010490) 5′-(463–484 bp) 3′-(741–762 bp) probe (689–709 bp); major satellite (accession no. X06899) 5′-(9–30 bp) 3′-(113–132 bp) probe (53–72 bp); minor satellite (accession no. Z22164) 5′-(113–132 bp) 3′-(220–242 bp) probe (166–189 bp); IgF2R (accession no. L06446) 5′-(2021–2040 bp) 3′-(2144–2163 bp) probe (2049–2068 bp); ARPT (accession no. M11310) 5′-(2486–2506 bp) 3′-(2568–2589 bp) probe (2520–2539 bp); H19 (accession no. U19619) 5′-(2609–2630 bp) 3′-(2745–2766 bp) probe (2658–2681 bp); β-globin (accession no. J00413) 5′-(4885–4904 bp) 3′-(4966–4995 bp) probe (2658–2681 bp); Pgk-2 (accession no. M17299) 5′-(1564–1585 bp) 3′-(1694–1715 bp) probe (1586–1609 bp).
Western blotting and antibodies
Protein extracts were separated on NuPAGE Bis–Tris Pre-Cast Gels (for small to mid-size molecular weight proteins), or NuPAGE Tris–Acetate Gels (for larger proteins) by electrophoresis and transferred onto nitrocellulose. Following the blocking, the membrane was incubated with primary antibodies against distinct proteins. After incubation with secondary HRP-coupled antibodies (Amersham Pharmacia), detection of HRP was performed using the ECL western blotting detection reagents according to the manufacturer’s instructions and visualized on Hyperfilm ECL chemiluminescent film. The following antibodies were used for detection of proteins as the primary antibodies: histone 3 (05–499), histone 4 (07–108), PCNA (PC10) (05–347), dimethyl-histone 3 lysine 9 (07–212), dimethyl-histone 3 lysine 4 (07–030), and acetyl-histone 4 lysine 16 (06–762) were from Upstate Biotechnology; trimethyl-histone 3 lysine 9 (ab8898) was from Abcam Limited; trimethyl-histone 3 lysine 4 (ab8580) was from Abcam Limited; HP1α (2G9) (MAB3446) was from Chemicon, Vimentin (V2258, Sigma). Branched antibody of histone 3 lysine 9 methylation (Peters et al., 2001) was a kind gift from Dr Jenuwein. Cell extraction with Triton X-100 was performed as described (Reyes et al., 1997).
Indirect immunofluorescence studies
MEFs were seeded on glass coverslips to 50–60% confluency. Cells were fixed in 2% paraformaldehyde solution and permeabilized with 0.5% Triton X-100 solution. Coverslips were blocked with 5% goat serum for 30 min at room temperature and then incubated with primary antibodies in a humidifying chamber at room temperature for 1 h or at 4°C overnight. After washing coverslips five times, coverslips were incubated with Cy5 and FITC-conjugated secondary antibodies (Jackson Immunoresearch) in PBS with 1.0 mM MgCl2 at room temperature for 1 h. After extensive washing, coverslips were mounted onto glass microscope slides using mounting reagent (Molecular Probes). The cells were observed by 3D confocal microscopy (LSM 310 confocal microscope, Zeiss).
Acknowledgments
Acknowledgements
We thank Dr Joost Oppenheim, Dr Hiltrud Brauch, Dr David Munroe and Dr Howard Young for critical reading of the manuscript. We are grateful to Edward Cho and Dr Stephen Locket for their advice on the confocal studies. The branched antibody of histone 3 lysine 9 methylation (Peters et al., 2001) was a kind gift from Dr Thomas Jenuwein. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This project has been funded in whole or part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-C0-12400.
References
- Ahmad K. and Henikoff,S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell, 9, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Becker P.B. and Horz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Humphrey,E.L., Erlich,R.L., Schneider,R., Bouman,P., Liu,J.S., Kouzarides,T. and Schreiber,S.L. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA, 99, 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- Bourc’his D. and Bestor,T.H. (2002) Helicase homologues maintain cytosine methylation in plants and mammals. BioEssays, 24, 297–299. [DOI] [PubMed] [Google Scholar]
- Dennis K., Fan,T., Geiman,T.M., Yan,Q. and Muegge,K. (2001) Lsh, a member of the SNF2 family, is required for genome wide methylation. Genes Dev., 15, 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Yan,Q., Huang,J., Austin,S., Cho,E., Ferris,D. and Muegge,K. (2003) Lsh deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res., 63, 4677–4683. [PubMed] [Google Scholar]
- Geiman T.M. and Muegge,K. (2000) Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc. Natl Acad. Sci. USA, 97, 4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman T.M., Durum,S.K. and Muegge,K. (1998) Characterization of gene expression, genomic structure, and chromosomal localization of Hells (Lsh). Genomics, 54, 477–483. [DOI] [PubMed] [Google Scholar]
- Geiman T.M., Tessarollo,L., Anver,M.R., Kopp,J.B., Ward,J.M. and Muegge,K. (2001) Lsh, a SNF2 family member, is required for normal murine development. Biochim. Biophys. Acta, 1526, 211–220. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman,Z., Yordan,C., Colot,V. and Martienssen,R.A. (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science, 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gibbons R.J., McDowell,T.L., Raman,S., O’Rourke,D.M., Garrick,D., Ayyub,H. and Higgs,D.R. (2000) Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet., 24, 368–371. [DOI] [PubMed] [Google Scholar]
- Huang J., Durum,S.K. and Muegge,K. (2001) Cutting edge: histone acetylation and recombination at the TCR gamma locus follows IL-7 induction. J. Immunol., 167, 6073–6077. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth,A.M., Cao,X. and Jacobsen,S.E. (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 416, 556–560. [DOI] [PubMed] [Google Scholar]
- Jarvis C.D., Geiman,T., Vila-Storm,M.P., Osipovich,O., Akella,U., Candeias,S., Nathan,I., Durum,S.K. and Muegge,K. (1996) A novel putative helicase produced in early murine lymphocytes. Gene, 169, 203–207. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao,X. and Jacobsen,S. (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol., 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Lachner M. and Jenuwein,T. (2002) The many faces of histone lysine methylation. Curr. Opin. Cell Biol., 14, 286–298. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly,D., Peters,A.H., Quivy,J.P., Roche,D., Taddei,A., Lachner,M., Jenuwein,T. and Almouzni,G. (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet., 30, 329–334. [DOI] [PubMed] [Google Scholar]
- McDowell T.L. et al. (1999) Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA, 96, 13983–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R.R., Pennings,S. and Stancheva,I. (2001) Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes Dev., 15, 3231–3236. [DOI] [PubMed] [Google Scholar]
- Nishioka K., Chuikov,S., Sarma,K., Erdjument-Bromage,H., Allis,C.D., Tempst,P. and Reinberg,D. (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev., 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H. et al. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. (2002) Chromatin remodeling enzymes: taming the machines. EMBO Rep., 3, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.C., Muchardt,C. and Yaniv,M. (1997) Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell. Biol., 137, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. (2002) Chromatin methylation: who’s on first? Curr. Biol., 12, R694–R695. [DOI] [PubMed] [Google Scholar]
- Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider,R., Bannister,A.J., Sherriff,J., Bernstein,B.E., Emre,N.C., Schreiber,S.L., Mellor,J. and Kouzarides,T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature, 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Selker E.U. (1998) Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl Acad. Sci. USA, 95, 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova,Z., Houben,A., Kakutani,T., Meister,A., Huang,M.S., Jacobsen,S.E., Schubert,I. and Fransz,P.F. (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J., 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Maison,C., Roche,D. and Almouzni,G. (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol., 3, 114–120. [DOI] [PubMed] [Google Scholar]
- Tamaru H. and Selker,E.U. (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature, 414, 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru H., Zhang,X., McMillen,D., Singh,P.B., Nakayama,J.I., Grewal,S.I., Allis,C.D., Cheng,X. and Selker,E.U. (2003) Tri methylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet., 34, 75–79. [DOI] [PubMed] [Google Scholar]
- Wang H., Cao,R., Xia,L., Erdjument-Bromage,H., Borchers,C., Tempst,P. and Zhang,Y. (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell, 8, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Yan Q., Cho,E., Lockett,S. and Muegge,K. (2003) Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]