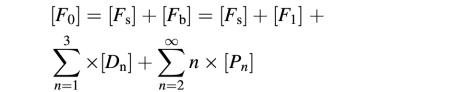Abstract
Single-molecule fluorescence imaging was used to investigate assembly of Staphylococcus aureus LukF and HS monomers into pore-forming oligomers (γ-hemolysin) on erythrocyte membranes. We distinguished the hetero-oligomers from the monomers, as indicated by fluorescence resonance energy transfer between different dyes attached to monomeric subunits. The stoichiometry of LukF (donor) and HS (acceptor) subunits in oligomers was deduced from the acceptor emission intensities during energy transfer and by direct acceptor excitation, respectively. Based on populations of monomeric and oligomeric intermediates, we estimated 11 sequential equilibrium constants for the assembly pathway, beginning with membrane binding of monomers, proceeding through single pore oligomerization, and culminating in the formation of clusters of pores. Several stages are highly cooperative, critically enhancing the efficiency of assembly.
Keywords: association constants/cell membranes/oligomeric intermediates/pore assembly/single-FRET
Introduction
Assembly of large macromolecular complexes such as membrane channels and cytoskeletal elements is essential for cell function. A crucial problem of protein complex assembly is to understand mechanisms of assembly processes by elucidating information on the beginning, intermediate and final stages involved (Alberts et al., 2002). Heterogeneous populations of intermediate states, however, are not readily analyzed using ensemble-averaged data. In contrast, single-molecule imaging methods provide direct information about individual intermediate states (Funatsu et al., 1995; Ishijima et al., 1998; Schütz et al., 2000). Recently, individual protein–protein interactions at very low concentrations have been observed in vitro under total internal reflection fluorescence microscopy (TIRF microscopy) (Mendelsohn et al., 1999; Taguchi et al., 2001). At high concentrations of proteins, however, dimers cannot be distinguished from crowded monomers using these techniques. Fluorescence resonance energy transfer (FRET) between single pairs of acceptor and donor fluorophores (single-FRET) has allowed observation of the dimerization even at high concentrations because the acceptor only emits fluorescence if located within several nanometers of the donor (Ha et al., 1996; Sako et al., 2000; Ha, 2001). Oligomers consisting of more than two molecules are also of great interest in protein assembly, although they have not been analyzed yet using single-molecule imaging.
Pore-forming toxins of bacteria are excellent models for studying the nature of assembly for oligomeric molecules on membranes because of the high stability of recombinant monomeric subunits in solution (Olson et al., 1999; van der Goot, 2001). Staphylococcus aureus leukocidin fast fraction (LukF) and γ-hemolysin second component (HS) are water-soluble proteins that assemble into hetero-oligomeric pores of γ-hemolysin on membranes of human red blood cells (Tomita and Kamio, 1997). Using the powerful single-FRET method, hetero-oligomeric toxins consisting of two distinct components are more advantageous to study than homo-oligomeric toxins because the individual components can be treated as specific fluorescence donors and acceptors, respectively. The crystal structure of LukF monomer in solution and the pore structures observed by electron microscopy suggest beginning and ending stages of pore assembly. Nevertheless, an HS structure is not yet available, and little information about intermediates has been reported so far (Olson et al., 1999; Nguyen et al., 2002).
Here, we directly observed the assembly of single LukF and HS monomers into pore-forming oligomers on erythrocyte membranes under the TIRF microscope. As LukF and HS lack cysteine residues, we created single-cysteine LukF and HS mutants which are specifically labeled with donor and acceptor dyes, respectively. We developed a method to calculate the number of subunits in individual oligomers based on the intensity of FRET and direct acceptor signals. We distinguished multiple species of intermediate oligomers, and measured equilibrium association constants of sequential intermediate stages. Equilibrium constants associated with several of the stages were much higher than others, indicating cooperative assembly which resulted in great numbers of pores formed on the membranes.
Results
Fluorescence labeled proteins and model of pore assembly
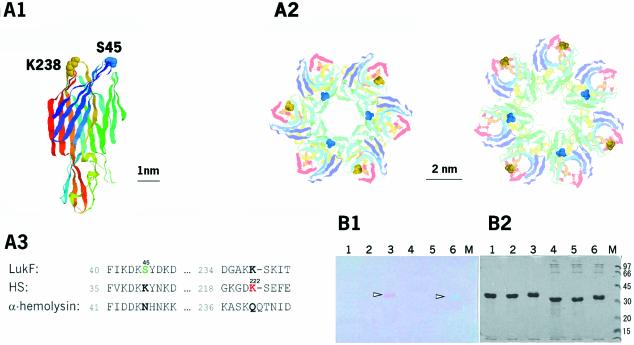
We designed single-cysteine mutants of LukF (S45C) and HS (K222C) in which introduced cysteines were located on the top of the respective cap domains (Figure 1A). Fluorophore-labeled LukF-S45C–TMR (LukF–TMR) and HS-K222C–IC5 (HS–IC5) were purified with labeling degrees of >95% and were almost free of unbound dyes (Figure 1B). Hemolytic activity of the labeled mutants was the same as that of wild-type proteins [LukF (3 nM) and HS (30 nM) caused 50% hemolysis against 6 × 1010 HRBC/l], indicating that fluorophore labeling of both mutants did not inhibit membrane binding and pore oligomerization. We detected sequential stages beginning with the binding of monomers to the membranes, then the assembly of dimers, small oligomers and single pores, and finally the formation of clusters of pores, and measured their equilibrium constants as described in the following results (Figure 6B).
Fig. 1. Structures and labeling of LukF and HS. (A) S45 (blue) of LukF and K222 (yellow) of HS (corresponding to K238 of LukF) were mutated to Cys, shown using the LukF structure (A1), the modelled γ-hemolysin complex in hexamers or heptamers (A2), and the amino acid sequence alignment between LukF, HS, and α-hemolysin (A3). (B) SDS–PAGE gels of 10 µg of fluorescently labeled proteins, unstained (B1) and stained (B2) with Coomassie brilliant blue. Samples included: lane 1, LukF treated with TMR-maleimide; lane 2, LukF-S45C; lane 3, LukF–TMR (arrowhead); lane 4, HS treated with IC5-maleimide; lane 5, HS-K222C; and lane 6, HS–IC5 (arrowhead).
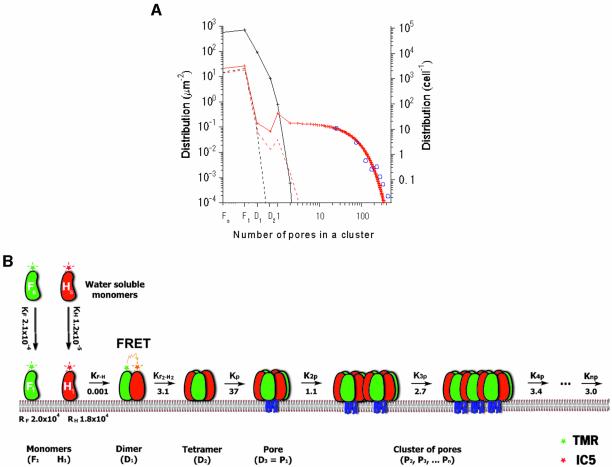
Fig. 6. Simulations and models for pore assembly. (A) Experimental and theoretical distribution of intermediates at given protein concentrations (Knp is assumed to be ∼K3p or K4p). The blue circles were measured from 237 spots of data (as for Figure 5C), and were fitted by the theoretical red line. The dotted and solid lines represent theoretical distributions at [Fo] and [Ho] of 25 and 1000 µm–2, respectively. The red lines represent a fully cooperative process, similar to natural conditions. The black lines represent non-cooperative processes (K of all stages are ∼KF·H). (B) Cartoon model for pore assembly of LukF and HS. Water-soluble LukF (green) and HS (red) monomers bind to putative binding sites on the membranes. Sequentially, the membrane-bound monomers assemble into small oligomers (e.g. dimers and tetramers), then into single pores and clusters of pores. The pore is represented as a hexamer, although formation of hexameric and/or heptameric pores is possible. The FRET signals indicate oligomers. The blue lines indicate the transmembrane domains inserting through lipid bilayers upon pore formation. The numbers indicate equilibrium binding and association constants (µm2).
Cooperative binding of HS in the presence of LukF
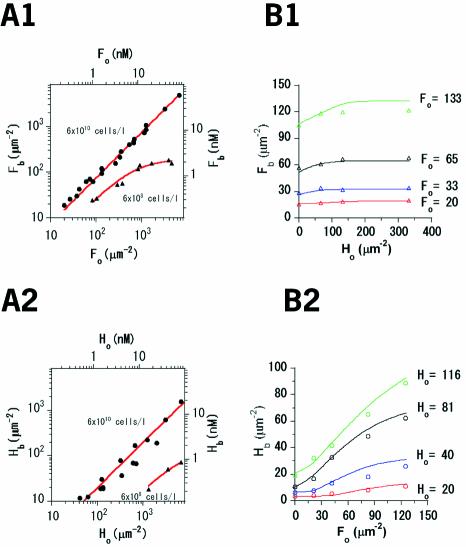
We verified binding constants (KF, KH) and the number of binding sites per µm2 of erythrocyte membranes (RF, RH) for individual components. Concentrations of LukF–TMR or HS–IC5 bound to the membranes ([Fb], [Hb]) were estimated by subtracting the free, unbound protein in the supernatant from the initial protein concentration ([Fo], [Ho]) applied to a significant number of cells (e.g. 6 × 108 or 6 × 1010 cells/l). The concentrations of initial and unbound proteins were measured using a spectrofluorometer (Okada and Hirokawa, 1999). Both components quickly bound to the membranes, giving the same levels at 1 and 30 min, confirming that the standard incubation time of 10 min is long enough for equilibrium binding of LukF or HS. KF, RF, KH and RH were calculated from the fitted curves shown in Figure 2A. LukF bound strongly to membranes with a KF of 2.1 × 10–4 µm2 and a large number of binding sites (RF) of 2.0 × 104 µm–2. In contrast, a lower binding constant (KH, 1.2 × 10–5 µm2) combined with a similar number of binding sites (RH, 1.8 × 104 µm–2) indicated a 15-fold decrease in the extent of binding of HS compared with that of LukF.
Fig. 2. Membrane binding of LukF and HS. (A) Binding of LukF–TMR (A1) or HS–IC5 (A2). [Fb] or [Hb] were plotted against [Fo] or [Ho] at 6 × 1010 (filled circles) and at 6 × 108 (filled triangles) HRBC/l. The red lines represent the fitting of data (Appendix 1). (B) Binding of both LukF–TMR and HS–IC5 to 6 × 1010 HRBC/l. At four values of [Fo] (B1) and [Ho] (B2), the relative [Fb] and [Hb] were measured in the presence of the other at different concentrations. The lines represent theoretical values of Fb (Hb), calculated based on the binding and association constants.
The binding ability of one component in the presence of the other was also determined. At a given [Fo], [Fb] slightly increased even in the presence of HS at 5-fold higher concentrations than LukF (Figure 2B1). Meanwhile the binding of HS was obviously enhanced by LukF: [Hb] at a given [Ho] was increased with [Fo]. With the same [Fo] and [Ho], HS bound ∼4 times more than it did without LukF (Figure 2B2). Combining all these data, we demonstrate that both LukF and HS can spontaneously bind to HRBC membranes, but with different binding constants, and that LukF obviously enhances the membrane binding of HS.
Individual monomers and dimers on the membranes
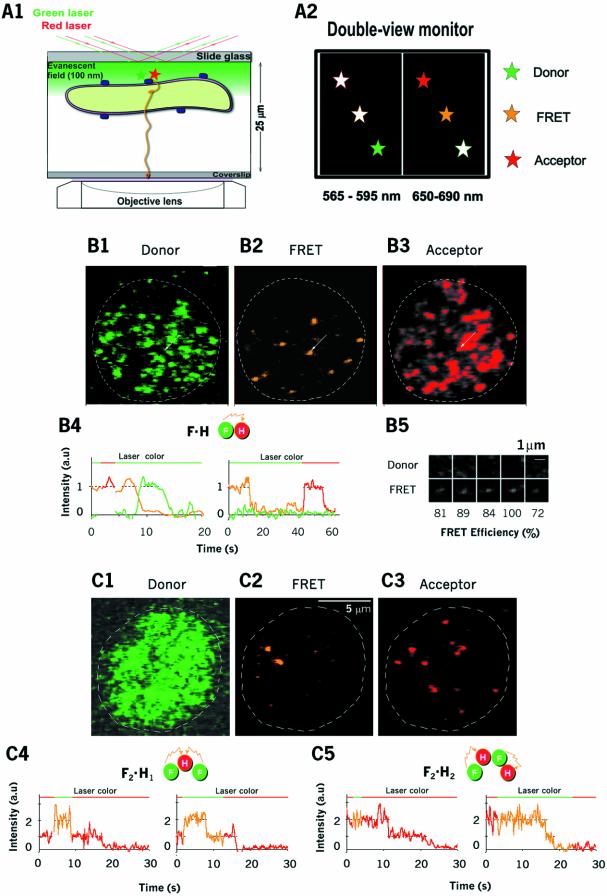
We visualized assembly of LukF–TMR and HS–IC5 on the membranes, at equilibrium, under a TIRF microscope equipped with a double-view unit to observe images simultaneously at two wavelengths, 565–595 nm (the TMR signal) and 650–690 nm (the IC5 signal) (Figure 3A). Using very low concentrations of the two proteins (75 pM LukF and 750 pM HS), at which hemolysis did not occur, Figure 3B1 and B2 show TMR and FRET-IC5 signals on the same cell, excited by the green laser. Figure 3B3 shows IC5 signals on that same cell, excited by the red laser. The spots on Figure 3B1 and B3 represent membrane-bound LukF–TMR and HS–IC5, and the spots on Figure 3B2 represent LukF–TMR assembling into oligomers with HS–IC5. Only a few punctate FRET-IC5 spots, ranging from 0 to 10, were observed on each cell.
Fig. 3. Visualization of small oligomers of LukF and HS. (A) Arrangement of the TIRF microscope for observation of oligomerization on the membranes (A1). Fluorescence signals near the basal membrane appeared on the double-view monitor: the left is for the donor, the right is for FRET and the acceptor (A2). (B and C) Images of dimers formed by LukF–TMR and HS–IC5 on HRBC membranes at low concentrations of proteins (75 and 750 pM, respectively) (B1–B3), and at higher concentrations of LukF–TMR (300 pM) and lower HS–IC5 (200 pM) (C1–C3). TMR, FRET and IC5 signals are shown after excitation by the green laser (B1 and B2; C1 and C2; time 0), and by the red laser (B3 and C3; time 0), respectively. B4 shows time traces of TMR (green), FRET (orange) and IC5 (red) emission corresponding to dimers (F·H = LukF–TMR HS–IC5). The left trace (eight frame-averaged) indicates that IC5 photobleaches first, and the right trace (30 ms interval) indicates that TMR photobleaches first. Five images of dual signals of TMR and FRET acquired from the same spots (B5). Thirty millisecond interval time traces of acceptor emission on anticorrelated excitation by the green and red lasers for short (∼3 s) or long times (>10 s), showing trimers (F2·H1; C4) and tetramers [(F·H)2; C5).
To calculate the numbers of LukF–TMR (m) and HS–IC5 (n) in individual FRET-IC5 oligomers (Fm·Hn), the occurrence of stepwise decays in the fluorescence intensity was measured. An acceptor (orange line at 8 s in Figure 3B4, left panel) was suddenly photobleached, and at the same time the intensity of donor was increased (green line). In another case, the donor was photobleached first (at 12 s in the right panel of Figure 3B4) as the FRET signal suddenly dropped without an increase in donor emission. This was confirmed by the fact that the acceptor signal detected by direct excitation with the red laser remained until its own photobleaching at 57 s (Figure 3B4, right panel). All of the stepwise photobleachings were classified into these two groups (acceptor photobleached first, number of spots = 31; donor photobleached first, number of spots = 14). The absence of anticorrelated fluctuations between the donor and acceptor indicated the flexible rotations of the labeled dyes during video recording (33 ms), allowing us to exclude the effect of polarization of dyes in deducing the distance between donor and acceptor from FRET efficiency. This behavior proves that single FRET spots are dimers containing one LukF and one HS (F·H).
We measured the FRET-IC5 intensity (IFRET-IC5) and the FRET efficiency (number of spots = 45) on the dimers. IFRET-IC5 values were well fitted in a Gaussian distribution with a mean of 0.87 ± 0.25 relative to IC5 intensities excited by the red laser. Five representative images (Figure 3B5) showed large FRET efficiencies, ranging from 72 to 100%. FRET efficiency was 87 ± 10% relative to the TMR intensity in dimers, indicating a distance between LukF–TMR and HS–IC5 in a dimer of ∼4.5 nm, estimated from a Förster distance (R0) of 5.4 nm between TMR and IC5 (R0 between TMR and Cy5 is 5.3 nm; Ha et al., 1996). This distance is consistent with predicted distances between IC5 and the two neighbouring TMR moieties, as shown in Figure 1A2.
The equilibrium association constants for dimerization KF·H = ([F·H] × [F1]–1 × [H1]–1) were also estimated, based on the concentrations of monomers ([F1] = 3.8 and [H1] = 6.9 µm–2) and dimers ([F·H] = 0.026 µm–2) on the cell membranes (number of cells = 37), to be 0.0010 ± 0.0003 µm2 (Table I). In this experiment, the ghost cells were incubated with low concentrations of proteins as above, but without washing off free proteins in solution. [F·H] in both the washed preparations (0.026 ± 0.004 µm–2) and the non-washed ones (0.027 ± 0.008 µm–2) was similar, confirming that dissociation of F·H into monomers is negligibly slow in our conditions. Therefore, in further experiments, we washed off the unbound proteins to reduce the background from monomers in FRET-IC5 detection and to analyze the oligomers more precisely.
Table I. Binding and association constants of intermediate stages in the pore assembly pathway.
| Data in Figures |
Constants |
Concentrations of monomeric and oligomeric intermediates (µm–2) |
|||
| 2A | KF | 2.1 × 10–4 | |||
| KH | 1.2 × 10–5 | ||||
| RF | 2.0 × 104 | ||||
| RH | 1.8 × 104 | ||||
| 3B | KFH | 0.0010 ± 0.0003 | [F1] = 3.8 ± 0.6 | [H1] = 6.9 ± 1.1 | [F·H] = 0.026 ± 0.004 |
| 3C | KF2·H | 0.081 ± 0.032 | [F1] = 3.75 ± 0.05 | [F·H] = 0.056 ± 0.006 | [F2·H1] = 0.017 ± 0.003 |
| KF·H2 | 0.12 ± 0.084 | [H1] = 0.27 ± 0.02 | [F·H] = 0.056 ± 0.006 | [F1·H2] = 0.0017 ± 0.0012 | |
| KF2·H2 | 3.82 ± 1.23 | [F·H] = 0.056 ± 0.006 | [F·H] = 0.056 ± 0.006 | [F2·H2] = 0.012 ± 0.003 | |
| 4 | KF2·H2 | 3.1 ± 1.1 | [F·H] = 0.065 ± 0.0092 | [F·H] = 0.065 ± 0.0092 | [F2·H2] = 0.013 ± 0.0041 |
| Kp | 37 ± 14 | [F·H] = 0.065 ± 0.0092 | [F2·H2] = 0.013 ± 0.0041 | [F3·H3] = 0.032 ± 0.0063 | |
| 5A and B | K2p | 1.1 ± 0.22 | [F3·H3] = 0.24 ± 0.019 | [F3·H3] = 0.24 ± 0.019 | [F6·H6] = 0.066 ± 0.010 |
| K3p | 2.7 ± 0.68 | [F3·H3] = 0.24 ± 0.019 | [F6·H6] = 0.066 ± 0.010 | [F9·H9] = 0.043 ± 0.0080 | |
| K4p | 3.4 ± 1.1 | [F3·H3] = 0.24 ± 0.019 | [F9·H9] = 0.043 ± 0.0080 | [F12·H12] = 0.035 ± 0.0075 | |
| 6A | Knp | 3.0 | |||
Binding and association constants are in units of µm2; numbers of binding sites (RF, RH) are in µm–2. Values are given as mean ± error, where error = mean × n–0.5, and n is the number of measured spots. The values were calculated from the equations described in the Appendix.
To confirm that LukF and HS do not dimerize in solution before incubation with cells, we incubated the two components in aqueous solution at 6-fold higher concentrations than those used in the presence of cells, then observed these in the same system. However, there is no observable FRET signal, confirming that LukF and HS are monomers in solution and oligomerize only on the membranes.
To test for the possibility of homodimer oligomerization (F·F or H·H) on the membranes, we observed each individual component labeled with both dyes, that is, mixtures of LukF–TMR and LukF–IC5, or of HS–TMR and HS–IC5, on the membranes. And to test whether FRET actually is an indicator of direct interaction between the two components, a couple of mutants of LukF (LukF33–TMR) and HS (HS29–IC5) that failed to form oligomers were used as a negative control for FRET. In both cases, no FRET-IC5 was detected even at concentrations 6-fold higher than those used for the heterodimer of LukF–TMR and HS–IC5, confirming that only the LukF and HS couple have a high affinity for direct interaction to form oligomers on the membranes.
Tetramerization: the dimer–dimer interaction
To achieve assembly of LukF and HS into dimers and other small oligomers, the concentration of LukF (Figure 3C) was increased to four times that of Figure 3B, and the concentration of HS–IC5 monomers was decreased. Most of the individual FRET intensities were equal to or twice the value of the single-FRET intensities measured in Figure 3B. The FRET-IC5 intensity of oligomers should be proportional to the number of donors if the acceptors are close to the donors and excitations of two or more donors do not overlap due to the low excitation power of the green laser (Lakowicz, 1999). The requirement for close distances between the acceptors and neighbouring donors is likely met, as supported by the evidence that single HS–IC5 absorbed nearly 90% of the energy emitted by single LukF–TMR within dimers. The FRET intensity of heterodimers was proportional to the intensity of the green laser up to 10 times higher than our standard illumination, indicating that the excitations of the donors did not overlap under our condition. m and n in Fm·Hn were deduced from the number of steps observed during photobleaching of FRET-IC5 (orange lines) and IC5 signals (red lines), respectively (Figure 3C4 and C5). The left panel of Figure 3C4 shows representative photobleaching steps of trimers: single IC5 intensity on excitation by the red laser up to 2 s indicates a single HS (H1). Then, on switching to the green laser, the intensity of the FRET-IC5 signal doubled, indicating the presence of two molecules of LukF (F2). Switching back to the red laser at ∼15 s caused the single photobleaching of HS–IC5, confirming H1. Similarly, dimers, trimers and teteramers were characterized and shown in representative images and profiles of FRET and acceptor intensities, such as F·H (Figure 3C2), F2·H1 (Figure 3C4), F1·H2 (data not shown), and (F·H)2 (Figure 3C5).
A tetramer could be formed by two pathways: (i) tetramerization of two dimers [F·H + F·H → (F·H)2] or (ii) ‘step-by-step’ oligomerization of monomers [F·H + F → F2·H1 + H → (F·H)2 or F·H + H → F1·H2 + F → (F·H)2]. To test which is the main pathway, we measured the association constants for each stage from the concentrations of oligomeric intermediates (KF2·H, KF·H2 and K(F·H)2 in Table I; Appendix 2). In observations from 20 cells (number of spots = 104), even though [F1] was ∼10–35 times higher than [H1], the number of F2·H1 complexes was almost equal to that of (F·H)2 complexes, and much fewer than the number of F·H heterodimers, indicating that monomers prefer to oligomerize into dimers and tetramers rather than into trimers. Association constants for ‘dimer–dimer’ tetramerization, K(F·H)2 = 3.8 µm2, were >30 times those for the step-by-step processes, KF2·H = 0.081 µm2 and KF·H2 = 0.12 µm2. This result could be interpreted as the step-by-step pathway being of far less significance than the dimer–dimer pathway. Moreover, we did not find any spot corresponding to F3·H1 or F4·H1, confirming that the complementary side-by-side interaction between LukF and HS is specific for dimerization and that oligomers containing multiple F·H pairs are stable.
Assembly into a single pore: a cooperative step
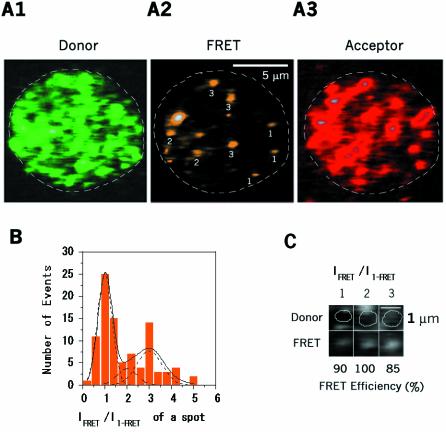
To analyze the next oligomerization steps, we raised the concentrations of both proteins so that various intermediates from dimers to larger oligomers could be obtained (Figure 4). Figure 4A2 shows various intense spots scattered on the membrane. Individual FRET intensities were ∼1, ∼2 and ∼3 times the single-FRET intensity of F·H (Figure 4A2 and B). The multi-FRET efficiencies of representative spots was also ∼90% (Figure 4C), indicating a close distance between LukF and HS components in larger oligomers.
Fig. 4. Visualization of intermediate oligomers and single pores. LukF–TMR and HS–IC5 were incubated with HRBC at intermediate concentrations of 400 pM and 4 nM, respectively. (A) TMR (A1; time 0), FRET (A2; time 0) and IC5 signals (A3; time 44 s) are on the same cell. White numbers on A2 indicate m in Fm·Hn. (B) Population histogram of intermediate oligomers: the dashed and solid lines indicate Gaussian distribution peaks for F·H, (F·H)2, F3·H3–4 and for the total population, respectively. (C) Three images of dual signals of TMR and FRET-IC5, showing single and multi-molecule FRET efficiencies between LukF–TMR and HS–IC5 in dimers and larger oligomers.
Because the major pathway for tetramerization was via dimer–dimer interaction, the spots at relative intensities of ∼2 should be tetramers consisting of pairs of dimers. Considering heterodimers to be of high stability, the spots at an intensity of ∼3 could be resolved as hexamers of three heterodimers. However, it is also possible to resolve these spots as heptamers consisting of 3F and 4H because the FRET-IC5 intensity of oligomers is proportional to the number of donors if the acceptors are close to the donors. For statistical analysis, the histogram of FRET intensities was fitted as the sum of three Gaussian distribution curves (Figure 4B). The spots at intensity ∼3 were more abundant than those at ∼2, and ∼4. Since the ratio of initial concentrations of F:H applied to the cells is 1:10, the population of spots at intensity ∼4 presenting heptamers of 4F:3H were low. The possible octameric formation of 4F:4H also appeared minor. Assuming the area under each Gaussian curve is proportional to the number of oligomers, F·H, (F·H)2 and F3·H3–4, we estimated sequential association constants K(F·H)2, and KF3·H3–4 to be 3.1 and 37 µm2, respectively (Table I). As single γ-hemolysin pores have been reported as hexamers and/or heptamers (Sugawara et al., 1997; Comai et al., 2002; Sugawara-Tomita et al., 2002), KF3·H3–4 was assigned as the association constant for single a pore, Kp. Obviously, Kp was more than 10 times higher than K(F·H)2, suggesting that LukF prefers assembling with HS into hexameric and/or heptameric pores F3·H3–4 rather than into tetramers (F·H)2.
Assembly into clusters of pores
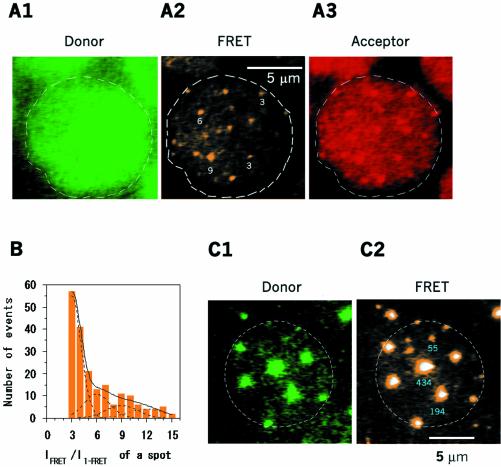
The final experiment was conducted to analyze distributions of large oligomers by increasing concentrations of LukF–TMR and HS–IC5 to achieve 15–100% hemolysis. At higher concentrations (∼15% hemolysis), we lowered the excitation powers of the green and red lasers by 25% to stay within the linear intensity range of the camera. This resulted in an ability to resolve FRET signals of about twice the value of single-FRET. Therefore, we could reliably observe FRET signals which were equal to or higher than three times the single-FRET signal, corresponding to a pore. The number of pores incorporated into each FRET spot was deduced from the FRET intensity of the spot divided by three times the single-FRET intensity. The pores did not scatter randomly on the membranes but aggregated into small clusters consisting of 2–3 pores, as estimated from the intensity of the spots (Figure 5A2). The occurrence of single pores and clusters on the membranes was plotted as a histogram in Figure 5B, indicating that single pores were abundant compared to clusters of pores. Association constants of single pores into two (K2p), three (K3p) and four pores (K4p) were 1.1, 2.7 and 3.4 µm2, respectively, as shown in Table I. Those increasing values indicate that single pores tend to assemble into 3-pore or 4-pore clusters rather than into 2-pore clusters.
Fig. 5. Visualization of clusters of pores. LukF–TMR and HS–IC5 were incubated with HRBC at high concentrations of 1.5 and 15 nM (A), or of 15 and 150 nM (C), respectively. LukF–TMR and HS–IC5 were excited by two lasers at 25% power (A), and at 2.5% power (C), compared to that used for single FRET observation. TMR (A1 and C1) and FRET (A2 and C2) signals show clusters of pores scattered on the membranes. IC5 signals (A3) are on the same cell with A1, A2. White numbers on A2 indicate m in Fm·Hn. Blue numbers on C2 indicate the number of pores in each large spot. (B) Histogram of populations of Fm·Hn corresponding to single pores and groups of pores at 15% hemolysis. The dotted and solid lines indicate Gaussian distribution peaks at single, two, three and four pores, and the total population, respectively.
When LukF–TMR and HS–IC5 were increased over the concentrations that start causing 100% hemolysis (Figure 5C), the power of the green excitation was reduced to 2.5%. Very highly intense domains of multi-molecular FRET-IC5, probably clusters of pores, could be observed. The domains were far from being randomly distributed on the cell, but appeared with an occurrence of 7–15 domains/cell. We plotted a distribution of clusters with various numbers of pores on the membranes in Figure 6A (blue circles). The distribution dropped gradually with the sizes of clusters. The FRET efficiency values of those domains were about 73 ± 13%. These values were slightly lower than that of single-FRET or multi-FRET of a pore, presumably due to co-localization of LukF–TMR monomers in areas of the clusters.
Calculating the population of intermediate states
Taking the four binding (KF, KH, RF, RH) and five association (KF·H, K(F·H)2, Kp, K2p, Knp) constants (Table I; Appendix sections 2 and 3), we estimated the theoretical distribution of intermediates and the total number of pores at given concentrations of proteins. Assuming that Knp (n > 2) values are similar to K3p and K4p (3 µm2), at certain [Fo] and [Ho], for example 25 and 1000 µm–2, the distribution of monomers and oligomers could be calculated. At low concentrations of LukF and HS (25 mm–2) close to those used in Figure 4, the calculated distribution of dimers, tetramers and single pores (Figure 6A, dotted red line) is similar to the distribution data shown in Figure 4B. The total number of pores is calculated to be Σn × [Pn] ∼ 0.036 µm–2. Using 1000 µm–2 each of LukF and HS (Figure 6A, red solid line), almost the same concentrations used in Figure 5C, the total concentration of pores (Σn × [Pn]) was ∼300 µm–2. Populations of clusters of pores (Figure 6A, blue circles) measured from 237 spots on cell membranes at the concentration used in Figure 5C fits well with the theoretical red line, confirming the assumption that Knp (n > 2) = 3 µm2.
To explain the enhancement in membrane binding of HS induced by LukF, the total concentrations of HS on the membranes ([Hb]) for applied [Ho] in Figure 2B2 were calculated from the association constants in Table I. The calculated values (lines) gave a good fit to the experimental results (circles) for HS binding in the presence of LukF (Figure 2B2). At very high concentrations of LukF, most of the HS bound to the membranes. The effect of LukF on membrane binding of HS was probably due to the sequential oligomerizations that shifted the membrane association–dissociation balance of HS in the direction of association.
Discussion
Advantages of single-FRET in the detection of intermediate oligomers
Ensemble methods such as equilibrium analytical ultracentrifugation have been used to measure concentration averages of assumed monomers, tetramers and octamers during cooperative association of an oligomeric DNA-binding protein (Daugherty et al., 1999). However, the oligomers are not well separated and the concentrations of monomers required are high (µM). Transmission electron microscopy at the level of single molecules cannot allow for the observation of the oligomerization process from monomers to tetramers due to limited spatial resolution. Our single molecule observation technique offers the ability to distinguish intermediates under physiologically-relevant conditions. The FRET measurement for single and multiple molecules is effective in distinguishing oligomers from crowded monomers. Moreover, we developed a novel method to deduce the number of LukF (donor) and HS (acceptor) molecules in small oligomers, such as trimers and tetramers, by measuring the stepwise photobleaching of FRET and acceptor signals, respectively.
Cooperative assembly of monomers into oligomers
The membrane binding affinity of each component of γ-hemolysin has been unclear and controversial because of low resolution in detecting protein concentrations (1 nM) (Kaneko et al., 1997; Ferreras et al., 1998). Here, we demonstrate that both LukF and HS individually bind to membranes as monomers, even at 20 pM. LukF binds to membranes with a 20-fold higher affinity than does HS (KF = 10 × KH), consistent with the data reported by Kaneko and colleagues in 1997. The higher KF for LukF supports a previous suggestion regarding stable interactions between aromatic amino acids of the rim domain of LukF and the phospholipid head groups on the outer membranes (Olson et al., 1999). The lower KH for HS suggests different properties of putative binding sites for HS or different membrane-binding surfaces compared with those of LukF.
At the dimerization stage, stepwise photobleaching of single-FRET in the presence of LukF–TMR and HS–IC5 on the membranes proved the existence of heterodimer F·H (Figure 3B4). However, the lack of observable FRET of the couples LukF–TMR and LukF–IC5, and HS–TMR and HS–IC5 proves that there is no possibility of F·F and H·H homodimer formation. The heterodimer F·H is formed not in solution but on the membranes because no FRET was observed in solution. The specific formation of heterodimer on the membranes may be promoted by two dimensional side-by-side collisions between LukF and HS, as confirmed by negative FRET evidence using LukF33–TMR and HS29–IC5, the point mutants at interaction surfaces lacking oligomerization ability. We suggest that structural changes of LukF or HS induced at membrane binding strengthen the side-by-side interactions between the two subunits.
Membrane-bound trimers and tetramers were observed for the first time. The abundance of dimers compared to trimers, together with the observation of similar concentrations of trimers and tetramers, even when the membrane-bound LukF monomer concentrations are ∼10–35 times those of HS monomers, prove that monomers have a tendency to oligomerize into dimers and tetramers, rather than into trimers. Thus, dimer–dimer interaction is the major pathway for formation of tetramers (Figure 6B). It is possible that dimerization triggers structural changes at the interaction sides of their subunits for further cooperative tetramerization.
In the assembly of pores and clusters of pores, the required high HS concentration did not allow us to discern an HS oligomer from crowded HS monomers under direct acceptor excitation. Therefore, we were obliged to assume that the ratio of F to H in an oligomer is close to 1:1, Fm·Hm or Fm·Hm+1, as suggested previously from stability of the multi-dimer oligomers. Concerning the subunit stoichiometry of single pores, an exact number has not been determined yet as the fine crystal structure of γ-hemolysin purified from membranes is not available. Recent reports by Comai et al., and Sugawara et al., respectively, suggest that six and seven subunits of LukF and HS make up a single pore (Sugawara et al., 1997; Comai et al., 2002; Sugawara-Tomita et al., 2002). However, the latter group also observed ∼25% hexameric pores in addition to the heptameric pores, suggesting the coexistence of hexamers and heptamers (N.Sugawara, T.Tomita and Y.Kamio, unpublished data). In another report, Miles et al. proposed an octameric structure for the staphylococcal leukocidin pore of LukF and LukS formed on lipid bilayers, based on the results obtained from gel shift electrophoresis and site-specific chemical modification during single-channel recording, but not by direct imaging (Miles et al., 2002). However, their results are also consistent with the heptameric stoichiometry with 3:4 or 4:3 compositions of LukF and LukS (Sugawara-Tomita et al., 2002). In view of this, we assume that the 3× single FRET spots, which could be resolved as either hexamers (F·H)3 or heptamers F3·H4, represent single pores, and that the spots exhibiting intensity with a common multiple (n times) of three LukF molecules (F3·H3–4)n present clusters of pores. Cell lysis started occurring at the protein concentrations (Figure 4) applied, confirming that functional pores were formed, although we have not been able to correlate the 3× single-FRET spots with the direct imaging of pore opening. Actually, in modelling the pathway of pore assembly (Figure 6B), whether the pore is assumed to be composed of six or seven subunits, the further calculation of association constants and final fitting of intermediate populations (Figure 6A) are not affected because the populations of groups and clusters of pores were simply deduced based on the value of 3× single FRET. Here, the F·H dimers, (F·H)2 tetramers and F3·H3–4 single pores presented major macroscopic intermediates in single pore oligomerization (Figure 6B). The high value of KF3·H3–4 or Kp (37 µm2) is an indicator of the cooperative assembly of subunits to generate single pores. The closed, ring-shaped structure of a pore would be very stable for minimizing the free energy at surface regions. At the pore formation step in Figure 6B, we depict the transmembrane domains (blue lines) inserting into lipid bilayers because previous data report that the transmembrane domain of LukF inserts into membranes to form a barrel-shaped channel on conversion from a pre-pore to a functional hemolytic pore (Nguyen et al., 2002).
γ-hemolysin pores have a greater tendency to condense into clusters as the concentration of toxin is increased, and may assemble into ≥3 pores because K3p and K4p were ∼3 times higher than K2p. This result is supported by the previous data showing that large clusters of pores could be observed at higher concentrations of proteins (1 µM) under a transmission electron microscope (TEM) (Sugawara et al., 1997). Since clusters of pores are also observed in many other toxins, even in lipid vesicles (Wallace et al., 2000), we suppose that the aggregation of pores is not only driven by the distribution of their receptors on the membranes, but also by non-covalent linkages between amino acids located at the outer surfaces of the pores. One advantage of pore aggregation could be the switching of the equilibrium balance of single pore assembly toward association, thereby increasing the total number of pores per cell. The other advantage would be that large clusters of pores on the membranes weaken the cell membranes and enhance the cell bursting. Cell membranes were recently found to be prominently disrupted in areas surrounding the clusters, as observed under TEM (N.Sugawara, T.Tomita and Y.Kamio, unpublished data).
Self-assembly of macrostructures and association constants
We estimated the distributions of intermediate oligomers over a wide concentration range of toxin applied to the cells. The cooperative assemblies for tetramers, single pores and clusters of pores greatly enhance the total concentration of pores (Σn × [Pn] ≈ 300 µm–2) up to ∼2.5 × 105 times of that in a non-cooperative assembly (Σn × [Pn] ≈ 0.0122 µm–2) for which the assembly for all stages is assumed to be ∼KF·H of 0.0010 µm2 (Figure 6, black solid line). Such overall cooperativity is, indeed, critical for γ-hemolysin to burst the erythrocytes in a human body at low concentrations of the toxin. With this new understanding of the cooperative assembly, we may interfere with the toxicity of γ-hemolysin by suppressing cooperative stages. For example, a chemical reagent that binds specifically to an interaction surface of either LukF or HS could effectively inhibit tetramerization and pore formation.
Other toxins from pathogenic bacteria, such as aerolysin, streptolysin and perfringolysin O which form homo-oligomeric pores on human target cell membranes (van der Goot, 2001), may also cooperatively assemble in a mechanism similar to that of γ-hemolysin. Outside of the toxin field, there are many kinds of ring-shaped membrane channels and pumps made up of multiple subunits. Selective, cooperative interactions between distinct subunits may establish structures, distributions and functions of these membrane proteins (Alberts et al., 2002). Notably, cooperative assembly is also fundamental to the linear polymerization of actin filaments and microtubules, wherein the nucleation mechanism is similar to cooperative single pore oligomerization (Oosawa and Kasai, 1962; Alberts et al., 2002).
In conclusion, the three cooperative stages (dimer–dimer interaction, single pore assembly and aggregation of pores) substantially enhance the efficiency of assembly of oligomeric pores. Here, the results present the first application of single molecule fluorescence microscopy to the observation of cooperative oligomeric pore assembly for a bacterial toxin on target cell membranes. The detection of heterogenous oligomeric populations, based on single and multi-FRET analysis, is applicable for the study of individual protein interactions in live cells using conjugates fused with fluorescent proteins including CFP, GFP and YFP. In general, single-molecule observations and statistical analysis of populations of intermediates will be useful in understanding how single molecules are brought together into macromolecular complexes in cells.
Materials and methods
Design and preparation of proteins
The residues for cysteine substitutions in LukF (S45) and HS (K222) were selected based on: (i) the LukF structure (Olson et al., 1999); (ii) the related staphylococcal α-heptameric pore (Song et al., 1996); and (iii) the amino acid sequence homology between LukF, HS and α-hemolysin (Gouaux et al., 1997). We modelled the structure of γ-hemolysin pore in hexamers or heptamers by fitting six or seven units of LukF into the structure of α-heptameric pores. The distances between S45 and K222 were estimated to be ∼2.2 and 3.0 nm in the hexamer (Figure 1A2, left) and ∼3.0 nm in the heptamer (Figure 1A2, right). The glutathione S-transferase (GST) fusion pGEX-4T-1 plasmids GST–LukF-S45C and GST–HS-K222C, were transformed into Escherichia coli DH5α for expression of GST–LukF-S45C and GST–HS-K222C, respectively. GST–LukF-S45C and GST–HS-K222C bound to glutathione agarose were labeled with a 10-fold excess of TMR-6-maleimide (Molecular Probes) and IC5-PE-maleimide (Dojindo) for 30 min at room temperture, and then washed free of unreacted dyes. LukF–TMR and HS–IC5 were finally eluted after incubation with 40 U of thrombin.
The degrees of labeling were determined by percent of fluorophore-labeled proteins (10 µg) on SDS–PAGE (12%), using NIH Image Scion 4.02. These agreed with measurements of the absorbance of TMR and IC5 at 546 and 650 nm, respectively. The labeled mutants electrophoresed (Figure 1B, lanes 3 and 6) more slowly than did the non-labeled (lanes 2 and 5). No bands of non-labeled proteins were observed in lanes 3 and 6, confirming that the labelling of both mutants was >95%. The specificity of the cysteine labeling was confirmed by negative controls of wild-type LukF and HS (lanes 1 and 4). The spectral FRET overlap between TMR emission and IC5 absorption is 4.6 × 1015 nm4M–1cm–1. The extinction coefficient of IC5 (ε = 220 000 M–1cm–1) is slightly smaller than that of Cy5 (ε = 250 000 M–1cm–1) while the spectral profile of IC5 is similar, but shifted to a wavelength 8 nm shorter than that of Cy5. The Förster distance between TMR and IC5 was 5.4 nm.
To make mutants as negative controls for interactions observed by FRET signals, Ser33 of LukF and Thr29 of HS located at the side-by-side interaction surface (Olson et al., 1999) were mutated to His and Asp, respectively. Such point mutations dramatically inhibit interaction of LukFS33H with HST29D, resulting in no oligomer or pore formation, and an absence of hemolytic activity, although the mutants could bind well to the membranes. These mutants were combined with single-cysteine mutations (LukFS33H/S45C and HST29D/K222C) to label the thiol groups with TMR (LukF33–TMR) and IC5 (HS29–IC5), respectively, and were tested in the same system with LukF–TMR and HS–IC5.
Hemolytic titration assay
Hemolytic assays of LukF–TMR and HS–IC5 in phosphate-containing saline buffer (PBS) at pH 7.4 were performed as described previously with the wild-type proteins (Kaneko et al., 1997), except that 0.1% bovine serum albumin (BSA) was added to prevent the binding of proteins to the tubes, the incubation temperature was 25°C, and the concentration of HRBC was 6 × 1010 cells/l (1% cells/vol).
Ensemble membrane binding assay of each component
Sample preparation. LukF–TMR or HS–IC5 at concentrations ranging from 250 pM to 100 nM were incubated with either 6 × 108 or 6 × 1010 HBRC/l, in PBS containing 0.1% BSA at 25°C for: (i) 10 min to measure equilibrium binding constants; or (ii) for different periods of time (1–30 min) to measure binding rates. Unbound proteins in the supernatant (Fs, Hs) were obtained by centrifuging the cells at 2000 g, and measured using a spectrofluorometer. When both components were incubated with the cells for 15 min, Fs and Hs were measured in a similar way. The initial concentrations of LukF–TMR (Fo) and HS–IC5 (Ho) applied to cells were determined by performing parallel incubations in the absence of HBRC.
Spectrofluorometer set-up. LukF–TMR and HS–IC5 in PBS containing 0.1% BSA were measured in a spectrofluorometer (FP-750, Jasco) under excitation of 532 nm (green) and 635 nm (red), respectively. Cut-off filters at 560 nm and 650 nm (Omega) were inserted in front of the fluorescence detector to reduce the light scattering of green and red, respectively. Oxygen scavengers (Funatsu et al., 1995) were used to reduce photobleaching. Experiments were performed in triplicate and the detectable resolution was ∼10 pM fluorophores.
Single-molecule imaging of oligomerization
Sample preparation. To visualize LukF and HS monomers, ∼20 pM of LukF–TMR or 400 pM of HS–IC5 were individually incubated with HRBC (6 × 1010cells/l) for 5 min at 25°C. To visualize the dimerization of LukF and HS, we incubated the ghost cells with low protein concentrations (75 pM of LukF–TMR and 750 pM HS–IC5), and then detected monomers and dimers in the presence of soluble proteins at equilibrium. However, as the crowded monomers prevented the precise analysis of oligomers, this method was applied only in measuring the association constant of dimerization. In the other experiments, both components were incubated with HRBC at 25°C for 15 min to reach equilibrium (oligomerization efficiencies at 15 and 30 min were similar). As LukF bound to HRBC more strongly than did HS, the ratio between LukF–TMR and HS–IC5 was also set at 1:10 to lower the background of TMR signals interfering with the detection of FRET-IC5 spots. The cells were then washed with PBS once to remove unbound toxins, homogenized in 5 mM Tris buffer, resealed to generate ghost cells using PBS, and finally adhered on l-polylysine-coated glass chamber in PBS containing oxygen scavengers.
TIRF microscopy set-up and imaging. Samples were observed under a TIRF microscope (Figure 3A; Funatsu et al., 1995; Ishii et al., 1999). LukF–TMR was excited by a Nd:YAG laser (TIM-6222, Transverse) at 532 nm, and HS–IC5 was excited by a diode laser (F44-30M, Coherent) at 635 nm. In Figure 3C, to distinguish spots corresponding to dimers, trimers and tetramers, the two lasers were switched on and off in an anticorrelated fashion. Emission signals were collected using an objective (plan Apo ×60, N.A 1.4; Olympus), split by a dichroic mirror (610 nm DM; Ashahi), and passed through barrier filters (580DF30 for TMR; 670DF40 for IC5, Omega). The fluorescence images were captured by a SIT camera (C2400-08, Hamamatsu Photonics) with an image intensifier (VS4-1845, Video Scope). Images were, in some cases, integrated for eight frames in tracing the intensity of fluorescence (Figure 3B4, left), and in other cases for 32 frames in measuring the intensity of fluorescence to improve the ratio of signal-to-noise. Thus, the spatial and temporal resolution of this system is 80 nm (pixel size) and 266 ms or 1.1 s, respectively. Adobe Photoshop was used for multi-color featured imaging: white spots showing signals for TMR, FRET-IC5 and IC5 were filled with green, orange and red, respectively. Offset was made with errors up to 10% in every image for easy detection of FRET spots from background. NIH Image with a custom macro program was used for analysis of photobleaching steps.
Data analysis. For the visualization of each component, mean intensity values of LukF–TMR (n = 97) and HS–IC5 (n = 106) molecules were 1.0 ± 0.39 and 1.0 ± 0.42 arbitrary units (a.u.), respectively. Those single spots emitted almost constant intensities with average life times of 10–30 s, then photobleached in one step, proving that membrane-bound LukF–TMR and HS–IC5 were monomers. The monomers exhibited two styles of motion on the membranes, restricted movements (D ≈ 1–5 × 10–2 µm2/s; 16 per total 29 spots for LukF–TMR, and 16 per total 27 spots for HS–IC5) and free bilateral diffusion (D ≈ 2–8 × 10–1 µm2/s; 13 spots for LukF–TMR and 11 spots for HS–IC5).
For FRET efficiency measurements of dimers (single-FRET of F·H), the 1 s-averaged images were used to analyze the TMR (ITMR) and IC5 (IFRET-IC5) intensities on TMR excitation. Measuring the average intensities was appropriate in our case because random anticorrelated fluctuation between the donor and acceptor was not observed. The single-FRET efficiency was calculated as follows: FRET efficiency = IFRET-IC5/(IFRET-IC5 + γITMR) where γ is the correction factor which accounts for detection efficiency by the established microscope with the two dyes, determined to be 1.0 based on the method proposed by Deniz et al. (1999).
FRET-IC5 and IC5 intensities of spots representing trimers and tetramers were also measured from the 1 s-integrated images. Photobleaching steps of IC5 were analyzed from 30 ms interval images to critically confirm the reliability of the intensity deduction. In the assembly of larger oligomers, we estimated Fm·Hn based only on the FRET from multiple IC5 fluorophores in the oligomers (multi-FRET). The above equation for single-FRET efficiency was also applied to multi-FRET efficiency. Tracking the oligomers on the membranes, oligomers appeared to exhibit restricted motion, remaining essentially stationary, with D ≈ 1–3.5 × 10–2 µm2/s (dimers, trimers, tetramers), and there were a few freely-diffusing oligomers. The larger the oligomers/clusters were, the slower and more restricted their diffusion was observed to be.
Acknowledgments
Acknowledgements
We thank Y.Okada and L.Poole for critical reading of the manuscript. This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.H. and Y.K.). V.T.N. is the recipient of a scholarship from the Ministry.
APPENDIX
1. Membrane binding constants
Unit conversions for concentrations of proteins from molar to µm–2 membranes:
[Fo] = [F′o]/(120 × [C′])
Where [Fo] and [F′o] are initial LukF concentrations applied to cells in µm–2 and M, respectively. 120 µm2 is the membrane area of one erythrocyte. [C′] is the cell concentration in M (6 × 1010 cells/l = 0.1 pM). The same definition was also applied to [Fb] which is the concentration of membrane-bound LukF.
Membrane binding constants for each component (KF and KH in µm2) were calculated as follows:
KF = [Fb] × ([Fo] – [Fb])–1 × (RF – [Fb])–1
where RF is concentration of binding sites for LukF in µm–2. The above calculation was also applied to HS.
2. Association constants
KF·H, KF2·H1, KF1·H2, K(F·H)2, KF3·H3–4(Kp), K2p, K3p, K4p (µm2) were estimated based on concentrations of intermediates at equilibrium. For example,
KF·H = [F·H] × [F1]–1 × [H1]–1 orKp = [F3·H3–4] × [(F·H)2]–1 × [F·H]–1
where [F1] and [H1] (µm–2) were measured by dividing the total fluorescence intensity by the intensity of single monomers. [F·H], [(F·H)2], [F3·H3–4], [F6·H6–8], [F9·H9–12] and [F12·H12–16] are numbers of dimers, tetramers, single pores, two pores, three pores and four pores per µm2, estimated based on the FRET-IC5 intensity spots corresponding to 1, 2, 3, 6, 9 and 12 times IFRET-IC5. [F1] and [H1] were corrected by subtracting the number of LukF or HS assembled into oligomers from the previously measured values.
3. Theoretical distribution of intermediates at certain protein concentrations
Basic equations for each intermediate are presented given approximations of [Fb] << RF and [Hb] << RH as follows (Oosawa and Kasai, 1962):
[Fs] = [F1]/(KF × RF);[Hs] = [H1]/(KH × RH)
[D1] = K1 × [F1] × [H1];
[D2] = K2 × [D1]2
[D3] or [P1] = Kp × K2 × [D1]3
[Pn] = K2p × Knpn–2 × [D3]n; n =2, 3, 4…
where [Fs] is the concentration of unbound protein in solution ([F0] – [F1]), D1 and D2 are shorthand for F·H and (F·H)2, respectively. The theoretical values shown in Figures 2B and 6A were calculated using KaleidaGraph. The definition for each intermediate is as described in Table I and Figure 6B.
References
- Alberts B., Alexander,J., Lewis,J., Raff,M., Roberts,K. and Walter,P. (2002) Protein. In Albert,B. (ed.), Molecular Biology of the Cell, 4th edn. Garland Science, New York, NY, pp. 147–162. [Google Scholar]
- Alberts B., Alexander,J., Lewis,J., Raff,M., Roberts,K. and Walter,P. (2002) The cytoskelton. In Albert,B. (ed.), Molecular Biology of the Cell, 4th edn. Garland Science, New York, NY, pp. 907–915. [Google Scholar]
- Comai M. et al. (2002) Protein engineering modulates the transport properties and ion selectivity of the pores formed by staphylococcal γ-hemolysins in lipid membranes. Mol. Microbiol., 44, 1251–1267. [DOI] [PubMed] [Google Scholar]
- Daugherty M.A., Brenowitz,M. and Fried,M.G. (1999) The TATA-binding protein from Saccharomyces cerevisiae oligomerizes in solution at micromolar concentrations to form tetramers and octamers. J. Mol. Biol., 285, 1389–1399. [DOI] [PubMed] [Google Scholar]
- Deniz A.A. et al. (1999) Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc. Natl Acad. Sci. USA, 96, 3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras M., Frank,H., Serra,M.D., Colin,D.A., Prevost,G. and Menestrina,G. (1998) The interaction of Staphylococcus aureus bi-component γ-hemolysins and leucocidins with cells and lipid membranes. Biochim. Biophys. Acta, 1414, 108–126. [DOI] [PubMed] [Google Scholar]
- Funatsu T., Harada,Y., Tokunaga,M., Saito,K. and Yanagida,T. (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature, 374, 555–559. [DOI] [PubMed] [Google Scholar]
- Gouaux E., Hobaugh,M.R. and Song,L. (1997) α-hemolysin, γ-hemolysin and leukocidin from Staphylococcus aureus: distant in sequence but similar in structure. Protein Sci., 6, 2631–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. (2001) Single-molecule fluorescence methods for the study of nucleic acids. Curr. Opin. Struct. Biol., 11, 287–292. [DOI] [PubMed] [Google Scholar]
- Ha T., Enderle,T., Ogletree,D.F., Chemla,D.S., Selvin,P.R. and Weiss,S. (1996) Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA, 93, 6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Yoshida,T., Funastu,T., Wazawa,T. and Yanagida,T. (1999) Fluorescence resonance energy transfer between single fluorophores attached to a coiled-coil protein in aqueous solution. Chem. Phys., 247, 136–173. [Google Scholar]
- Ishijima A. et al. (1998) Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell, 23, 161–171. [DOI] [PubMed] [Google Scholar]
- Kaneko J., Ozawa,T., Tomita,T. and Kamio,Y. (1997) Sequential binding of Staphylococcal γ-hemolysin to human erythrocytes and complex formation of the hemolysin on the cell surface. Biosci. Biotechnol. Biochem., 61, 846–851. [DOI] [PubMed] [Google Scholar]
- Lakowicz J.R. (1999) Time-resolved energy transfer and conformational distributions of biopolymers. In Lakowicz,J.R. (ed.), Principles of Fluorescence Spectroscopy. 2nd edn. Kluwer Academic/Plenum Publishers, NewYork, NY, pp. 395–424. [Google Scholar]
- Mendelsohn A.R. and Brent,R. (1999) Protein interaction methods: toward an endgame. Science, 284, 1948–1950. [DOI] [PubMed] [Google Scholar]
- Miles G., Movileanu,L. and Bayley,H. (2002) Subunit composition of a bicomponent toxin: Staphylococcal leukocidin forms an octameric transmembrane pore. Protein Sci., 11, 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.V., Higuchi,H. and Kamio,Y. (2002) Controlling pore assembly of staphylococcal γ-hemolysin by low temperature and by disulfide bond formation in double-cysteine LukF mutants. Mol. Microbiol., 45, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Okada Y. and Hirokawa,N. (1999) A processive single-headed motor: kinesin superfamily protein KIF1A. Science, 283, 1152–1157. [DOI] [PubMed] [Google Scholar]
- Olson R., Nariya,H., Yokota,K., Kamio,Y. and Gouaux,E. (1999) Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat. Struct. Biol., 6, 134–140. [DOI] [PubMed] [Google Scholar]
- Oosawa F. and Kasai,M. (1962) A theory of linear and helical aggregations of macromolecules. J. Mol. Biol., 4, 10–21. [DOI] [PubMed] [Google Scholar]
- Sako Y., Minoghchi,S. and Yanagida,T. (2000) Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol., 2, 168–172. [DOI] [PubMed] [Google Scholar]
- Schütz G.T., Kada,G., Pastushenko,V.P. and Schindler,H. (2000) Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J., 19, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Hobaugh,M.R., Shustak,C., Cheyley,A., Bayley,H. and Gaoux,E. (1996) Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science, 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Tomita,T. and Kamio,Y. (1997) Assembly of Staphylococcus aureus γ-hemolysin into a pore-forming ring-shaped complex on the surface of human erythrocytes. FEBS Lett., 410, 333–337. [DOI] [PubMed] [Google Scholar]
- Sugawara-Tomita N., Tomita,T. and Kamio,Y. (2002) Stochastic assembly of two-component staphylococcal γ-hemolysin into heteroheptameric transmembrane pores with alternate subunit arrangements in ratios of 3:4 and 4:3. J. Bacteriol., 184, 4747–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi H., Ueno,T., Tadakuma,H., Yoshida,M. and Funatsu,T. (2001) Single-molecule observation of protein-protein interactions in the chaperonin system. Nat. Biotechnol., 19, 861–865. [DOI] [PubMed] [Google Scholar]
- Tomita T. and Kamio,Y. (1997) Molecular biology of the pore-forming cytolysins from Staphylococcus aureus α-hemolysin and γ-hemolysins and leukocidin. Biosci. Biotechnol. Biochem., 61, 565–572. [DOI] [PubMed] [Google Scholar]
- van der Goot G. (ed.) (2001) Pore-forming toxins. Springer-Verlag, Berlin, Heidelberg, Germany. [Google Scholar]
- Wallace A.J. et al. (2000) E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell, 100, 265–276. [DOI] [PubMed] [Google Scholar]