Abstract
RecQ family helicases catalyze critical genome maintenance reactions in bacterial and eukaryotic cells, playing key roles in several DNA metabolic processes. Mutations in recQ genes are linked to genome instability and human disease. To define the physical basis of RecQ enzyme function, we have determined a 1.8 Å resolution crystal structure of the catalytic core of Escherichia coli RecQ in its unbound form and a 2.5 Å resolution structure of the core bound to the ATP analog ATPγS. The RecQ core comprises four conserved subdomains; two of these combine to form its helicase region, while the others form unexpected Zn2+-binding and winged-helix motifs. The structures reveal the molecular basis of missense mutations that cause Bloom’s syndrome, a human RecQ-associated disease. Finally, based on findings from the structures, we propose a mechanism for RecQ activity that could explain its functional coordination with topoisomerase III.
Keywords: helicase/RecQ/structure/winged helix/Zn2+ binding
Introduction
RecQ DNA helicases are ubiquitous enzymes in bacteria and eukaryotes that carry out important functions in genome biology (Cobb et al., 2002; Wu and Hickson, 2002). Like other DNA helicases, RecQ proteins are ATP-dependent molecular motors that unwind double-stranded (ds) DNA. Members of the RecQ family differ from other helicases, however, in their unusual breadth of activities, including roles in DNA replication, recombination, and repair. With their central activities in genome maintenance, it is not surprising that mutation of many known recQ genes leads to genome instability and, in the case of three human recQ genes, to disease. Despite the importance of RecQ proteins, the structural basis of their function has remained poorly defined.
The founding RecQ family member was discovered in Escherichia coli where it functions in the RecF recombination pathway (Nakayama et al., 1984; Umezu et al., 1990). This pathway participates in homologous recombination and repair of ultraviolet light-induced DNA damage (Kowalczykowski et al., 1994). RecQ and other proteins in the RecF pathway appear to catalyze reactions that help repair replication forks that have stalled at sites of DNA damage (Courcelle et al., 1997; Courcelle and Hanawalt, 1999; Courcelle et al., 1999; 2003). In addition, E.coli RecQ helps suppress illegitimate recombination in cells (Hanada et al., 1997). Although the precise cellular substrates on which RecQ proteins act remain a mystery, several reactions catalyzed by E.coli RecQ have been reconstituted in vitro, including ATP-dependent DNA unwinding (Umezu et al., 1990), recombination initiation with RecA and single-stranded (ss) DNA binding protein (SSB) (Harmon and Kowalczykowski, 1998), and plasmid DNA catenation with topoisomerase III (Topo III) and SSB (Harmon et al., 1999). Coordinated activities with recombinases and topoisomerases, as well as DNA unwinding functions, are conserved in a number of eukaryotic RecQ proteins (reviewed in Wu and Hickson, 2001), implying a functional homology among RecQ family members.
Three diseases (Bloom’s, Werner’s, and Rothmund–Thompson syndromes) can be attributed to mutations of human genes that encode homologs of E.coli RecQ (BLM, Ellis et al., 1995; WRN, Yu et al., 1996; RECQ4, Kitao et al., 1999). While each of these syndromes has distinct clinical manifestations, they all share a predisposition to cancer. A common feature of these disorders is pronounced genomic instability, which is characterized by heightened levels of homologous recombination and/or chromosomal deletions (reviewed in Karow et al., 2000). Several disease-linked nonsense and frame-shift mutations in WRN, BLM, and RECQ4, as well as all known BLM missense mutations associated with Bloom’s syndrome, map to regions that are conserved across the RecQ family of enzymes.
Three regions conserved among bacterial and eukaryotic RecQ proteins have been identified by sequence analysis: helicase, RecQ-conserved (RecQ-Ct), and Helicase-and-RNaseD-C-terminal (HRDC) domains (Figure 1A; Morozov et al., 1997). The helicase and RecQ-Ct regions are the best conserved regions among RecQ proteins, with some RecQs lacking an identifiable HRDC domain altogether. The RecQ family helicase region contains sequence motifs that are broadly conserved among DNA unwinding enzymes (Gorbalenya and Koonin, 1993). These motifs link the energy of nucleoside triphosphate binding and hydrolysis to DNA unwinding. The structure and function of the RecQ-Ct region is more poorly understood since it appears to be unique to RecQ proteins. However, evidence indicates that this region is essential for RecQ function. Two disease-causing BLM missense mutations map to the enzyme’s RecQ-Ct region (Ellis et al., 1995; Foucault et al., 1997) and abolish BLM ATPase and helicase activities in vitro (Bahr et al., 1998). Mutation and deletion analyses in the Saccharomyces cerevisiae RecQ homolog (Sgs1) have also shown that this region is essential for in vivo function (Mullen et al., 2000; Onoda et al., 2000; Ui et al., 2001). Together, the helicase and RecQ-Ct regions form the core, conserved sequence that defines the RecQ family of enzymes.
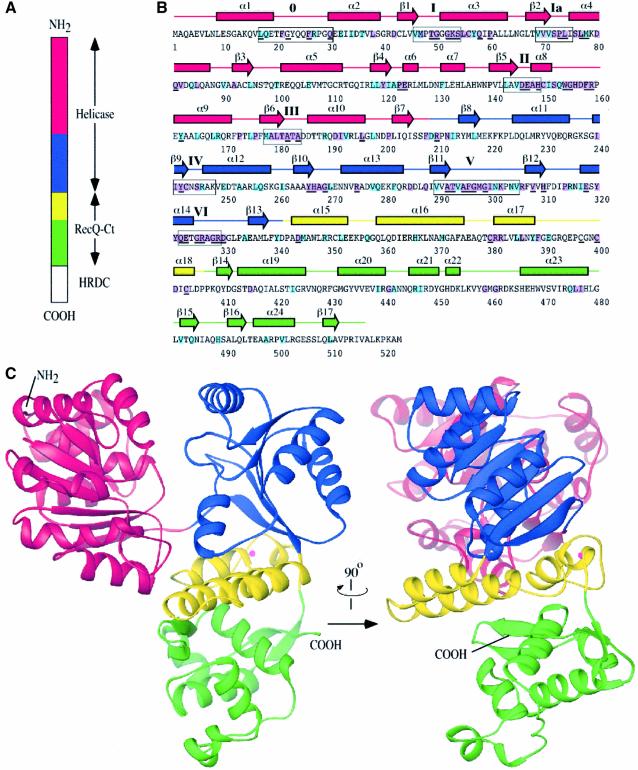
Fig. 1. Structure of the E.coli RecQ catalytic core. (A) Schematic diagram of E.coli RecQ. Three conserved regions, helicase, RecQ-conserved (RecQ-Ct), and Helicase-and-RNaseD-C-terminal (HRDC) (Morozov et al., 1997), are labeled. The catalytic core of E.coli RecQ (RecQΔC) includes only the helicase and RecQ-Ct regions, and comprises four apparent subdomains in the structure: residues 1–208 in red, 209–340 in blue, 341–406 in yellow, and 407–516 in green. (B) Sequence and secondary structure of RecQΔC. Helices (boxes) and β-strands (arrows) are shown above the sequence and labeled sequentially. Color coding is the same as in (A). Conserved helicase motifs (motif 0, Bernstein and Keck, 2003; and motifs I-VI, Gorbalenya and Koonin, 1993) are labeled and enclosed in boxes. Residues that are invariant among 65 bacterial RecQ proteins are underlined, and residues that are invariant or highly conserved with a subset of eukaryotic RecQ proteins (human WRN, BLM, and S.cerevisiae Sgs1) are highlighted in purple or light-blue boxes, respectively. (C) Orthogonal views of a ribbon diagram of the crystal structure of RecQΔC, color-coded as in (A). A bound Zn2+ ion is shown as a magenta sphere.
Domain mapping experiments with E.coli RecQ have demonstrated that the conserved helicase and RecQ-Ct regions fold together to form a single 59 kDa structural domain (RecQΔC) (Bernstein and Keck, 2003). The isolated RecQΔC domain is active as a DNA-dependent ATPase and can unwind DNA with essentially the same specific activity as full-length E.coli RecQ, indicating that it is the catalytic core of the enzyme (Bernstein and Keck, 2003). The 9 kDa HRDC domain appears to modulate DNA binding activity of the catalytic core but is not necessary for enzyme function in vitro (Bernstein and Keck, 2003). Similarly, an S.cerevisiae Sgs1 variant that lacks its C-terminal HRDC domain can complement the increased DNA damage sensitivity phenotype of sgs1Δ cells (Mullen et al., 2000; Mullen et al., 2001), implying that the HRDC domain is dispensable for at least some activities in vivo. In addition, a recent study has shown that a catalytic core fragment of the human BLM protein that includes just its helicase and RecQ-Ct regions is active in vitro (Janscak et al., 2003). These findings demonstrate that the evolutionarily conserved domain shared among RecQ proteins also forms the structural and catalytic core of the RecQ enzyme family.
Beyond low-resolution domain mapping, a number of other studies have revealed important structural features of RecQ helicases. First, sequence analysis places RecQ family members in the Superfamily 2 (SF2) group of enzymes (Gorbalenya and Koonin, 1993). While structurally similar to Superfamily 1 (SF1) enzymes, SF2 helicases are more similar to each other in sequence within their helicase domains than they are to SF1 enzymes. Secondly, the NMR structure of the S.cerevisiae Sgs1 HRDC domain has been determined (Liu et al., 1999), revealing a domain that shares structural similarity with auxiliary DNA-binding domains in E.coli Rep and Bacillus stearothermophilus PcrA helicases (Subramanya et al., 1996; Korolev et al., 1997). Although this domain is not required for E.coli RecQ function and is not preserved in all RecQ family members, solution of this structure makes an important link to other well-studied helicases. Finally, a number of contradictory studies have investigated the active oligomeric state of RecQ helicases. Three findings indicate that RecQ proteins form oligomeric structures that may be important for their function: (i) E.coli RecQ hydrolyzes ATP with a Hill coefficient of 3 when unwinding DNA, implying that multiple ATPase sites interact during DNA strand separation (Harmon and Kowalczykowski, 2001); (ii) the human BLM protein forms homooligomeric ring structures (Karow et al., 1999); and (iii) Drosophila melanogaster RecQ5 forms homooligomers (Kawasaki et al, 2002). However, other recent findings may indicate that RecQ proteins can operate as monomers. These include a biochemical analysis of E.coli RecQ in centrifugation and kinetic experiments where it was concluded that the enzyme functions as a monomer (Xu et al, 2003), and biochemical experiments with catalytically active recombinant fragments of the human BLM protein that no longer forms hexameric rings but instead exist as monomers (Janscak et al., 2003). Thus the question of the active oligomeric state of RecQ proteins remains unresolved.
To examine the physical basis of RecQ function, we have determined two high-resolution X-ray crystal structures of the conserved catalytic core of E.coli RecQ. The core domain comprises four apparent subdomains; two of the subdomains combine to form the helicase region and the remaining two join to form the RecQ-Ct region of RecQ. The RecQ-Ct subdomains include one that folds as a platform of helices that binds a Zn2+ ion using four conserved cysteine residues and another that forms an unexpected winged-helix (WH) subdomain. The WH motif shares significant structural similarity with other DNA binding proteins, thus identifying a likely DNA binding site in E.coli RecQ. Based on the structures, we propose a mechanism of action that could explain the coordination of RecQ proteins with Topo III. Furthermore, our structures reveal the molecular basis of missense mutations that cause Bloom’s syndrome. Together, these structures reveal the first high-resolution images of the enzymatic core conserved among RecQ family members.
Results and discussion
Structure determination of the RecQΔC domain
We first attempted to crystallize full-length E.coli RecQ protein, but extensive crystallization trials yielded no crystals. This result could have been due to flexibility that has been observed between the catalytic core and HRDC domains of E.coli RecQ (Bernstein and Keck, 2003) leading to structural heterogeneity that would hinder crystallization. Since we had characterized the catalytic core domain (RecQΔC) and found that it has essentially the same specific activities as full-length E.coli RecQ in ATPase and helicase assays (Bernstein and Keck, 2003), we subjected RecQΔC to crystallization trials as well and were able to crystallize it in a form that diffracted to 1.8 Å resolution (Table I). The crystal form contained one molecule per asymmetric unit. Crystals of selenomethionine-substituted RecQΔC were used to determine the structure of the domain using single-wavelength anomalous dispersion (SAD) phasing methods (Table I). The final refined model includes both the helicase and RecQ-Ct regions of the protein (residues 1 to 516 of RecQΔC, Figure 1B and C).
Table I. Data collection, phasing, and refinement.
| Data collection | RecQΔC | ATPγS-bound RecQΔC | Phasing statistics | Refinement | RecQΔC | ATPγS-bound RecQΔC | |
|---|---|---|---|---|---|---|---|
| Wavelength (Å) | 0.9791 | 0.9000 | Figure of merit | 0.293 | Resolution (Å) | 20.0–1.8 | 20.0–2.5 |
| Resolution (last shell) (Å) | 74–1.8 (1.89–1.8) | 21–2.5 (2.63–2.5) | (after density modification) | 0.809 | Rwork/Rfreeb | 19.3/24.4 | 21.0/29.8 |
| Measured reflections/unique | 353 669/51 478 | 47 206/17 529 | R.m.s.d. bond lengths (Å) | 0.006 | 0.012 | ||
| Rsyma (last shell) (%) | 8.1 (44.2) | 4.8 (13.3) | R.m.s.d. bond angles (°) | 1.7 | 3.2 | ||
| I/σ (last shell) | 16.3 (4.6) | 12.8 (7.1) | PDB code | 1OYW | 1OYY | ||
| Completeness (last shell) (%) | 99.7 (99.7) | 92.1 (92.1) |
aRsym = ΣΣj|Ij – <I>|/ΣIj, where Ij is the intensity measurement for reflection j and <I> is the mean intensity for multiply recorded reflections.
bRwork, free = Σ||Fobs| – |Fcalc||/|Fobs|, where the working and free R-factors are calculated using the working and free reflection sets, respectively. The free reflections (5% of the total) were held aside throughout refinement.
The RecQΔC structure reveals a modular, tri-lobed molecule that is comprised of four apparent structural subdomains (Figure 1C). The molecule is roughly 40 × 60 × 70 Å in size and forms a Y-shaped structure with major clefts apparent on its surface. The two N-terminal subdomains form distinct lobes of the molecule that combine to make the helicase region. The remaining two subdomains form the RecQ-Ct region, which is the third lobe of the molecule.
Helicase region of RecQ
The two N-terminal subdomains of RecQΔC abut to form a deep cleft in the protein (Figure 1C). Conserved helicase sequence motifs (Gorbalenya and Koonin, 1993) line the cleft walls in an arrangement similar to that observed in other helicase structures (Caruthers and McKay, 2002) (Figure 2A). The RecQΔC helicase region is most similar in structure to helicase domains in other SF2 helicases, including three RNA helicases [Methanococcus jannaschii putative RNA helicase (Story et al., 2001), S.cerevisiae eIF4a (Caruthers et al., 2000), hepatitis C virus NS3 helicase (Yao et al., 1997)], and Thermotoga maritima RecG DNA helicase (Singleton et al., 2001), as well as the helicase domain in Archaeoglobus fulgidus reverse gyrase (Rodriguez and Stock, 2002). The RecQΔC structure also shares significant but lesser structural homology with SF1 DNA helicases and other ATP-binding proteins (see discussion below).
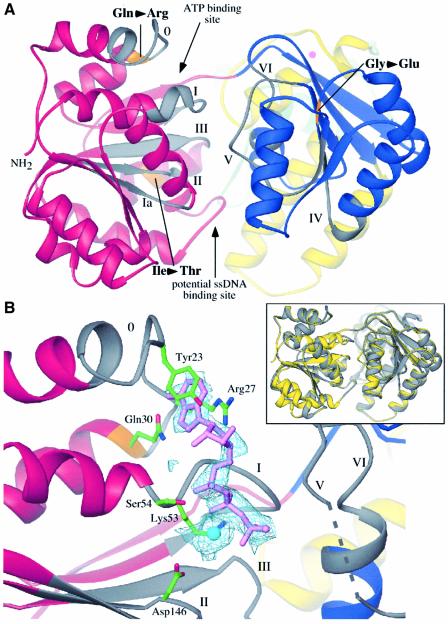
Fig. 2. Features of the helicase region of RecQΔC. (A) View into the cleft formed by the two helicase subdomains. Color coding is the same as Figure 1A, except that helicase motifs are labeled and colored in grey, and residues corresponding to known BLM missense mutations are labeled and colored in orange. Sites where nucleotide and ssDNA have been observed to bind in other helicase structures are indicated. (B) Structure of ATPγS/Mn2+-bound RecQΔC. (Inset) overlay of the apo- and ATPγS-bound RecQΔC structures in yellow and grey, respectively. A slight relative rotation of the second helicase lobe is seen in the structure with bound nucleotide. (Main panel) Fo–Fc difference electron density (light blue) is contoured at 1.5 σ. Motif I is in an open conformation relative to its position in Figure 2A. The ATPγS (lavender) adenine moiety is sandwiched between Tyr23 and Arg27, and hydrogen bonds are formed between the N6 and N7 atoms of the adenine and the side chain of Gln30. The triphosphate is bound by interactions with Lys53 and backbone amides from motif I. A Mn2+ ion (cyan) is bound by Ser54 from motif I and Asp146 from motif II. Helicase motif labels and color-coding are the same as in Figure 2A. Electron density was not observed for residues 296 to 299 (dashed line).
Based upon structural features of the RecQΔC helicase region that are conserved in other helicases, we hypothesized that ATP and ssDNA would bind to distinct sites within the helicase region, as indicated in Figure 2A. Consistent with this prediction, a second crystal structure of RecQΔC soaked in ATPγS, an ATP analog, and Mn2+ showed clear Fo–Fc difference electron density for ATPγS and a metal ion in the predicted ATP binding site (Figure 2B). The adenine moiety is packed between Tyr23 and Arg27 side chains and hydrogen bonds are formed between the N6 and N7 atoms of the adenine and Gln30 of RecQ motif 0. This adenine binding mode likely explains the preference of ATP as a cofactor in E.coli RecQ reactions (Umezu et al., 1990). Structural aspects of adenine binding by PcrA helicase (Subramanya et al., 1996) are strikingly similar to those observed in RecQΔC. The ATPγS triphosphate is bound to RecQΔC by Lys53 and several backbone amides in motif I, and through a Mn2+ ion that makes water-mediated contacts with Ser54 of motif I and Asp146 of motif II.
The presence of a ssDNA binding site between the RecQΔC helicase subdomains is predicted by its structural similarity to PcrA (Velankar et al., 1999), Rep (Korolev et al., 1997), and NS3 (Kim et al., 1998) helicases solved with bound nucleic acid. In these structures, ssDNA binds to the two helicase lobes at a cleft that is on the opposite face from the ATP binding site. The interactions with ss nucleic acid are predominantly through either hydrophobic association with the DNA bases (Velankar et al., 1999), ionic interactions with the substrate backbone (Kim et al., 1998), or a combination of hydrophobic and ionic interactions with the nucleic acid substrate (Korolev et al., 1997). However, an alternative DNA binding mechanism has recently been suggested based on the T.maritima RecG helicase structure. In the RecG model, the helicase domain is proposed to bind to dsDNA and to act as a dsDNA translocase (Singleton et al., 2001). Evidence supporting this model are that the path of dsDNA in the DNA-bound RecG structure extrapolates to the helicase domain (Singleton et al., 2001) and that RecG’s ATPase activity is significantly more sensitive to stimulation by dsDNA than ssDNA (Whitby and Lloyd, 1998). While the RecQΔC helicase region is structurally similar to that in RecG, E.coli RecQ’s ATPase activity is ssDNA-dependent with only low levels of ATPase activity stimulated by dsDNA (Umezu et al., 1990), and ssDNA-binding features similar to those found in PcrA are present in RecQΔC (see discussion below). Although we cannot rule out a RecG-type dsDNA translocase mechanism based on the structure of RecQΔC, the available evidence does not support this type of reaction. We currently favor a model for RecQΔC DNA binding where ssDNA binds between the two helicase subdomains as has been observed in PcrA, Rep, and NS3 helicases. Elucidation of the structural basis of RecQ DNA binding will require further experimental characterization.
RecQ-Ct region of RecQ
The function of the RecQ-Ct region in RecQ-catalyzed reactions has not been well defined. RecQ-Ct sequences are not found in proteins outside of the RecQ family and previous functional characterization of this region in S.cerevisiae Sgs1 (Mullen et al., 2000; Onoda et al., 2000; Ui et al., 2001), as well as in murine (Bahr et al., 1998) and human (Janscak et al., 2003) BLM proteins has been limited to mutational analyses. These studies indicate the importance of the RecQ-Ct region, but have not resolved its structure or function(s).
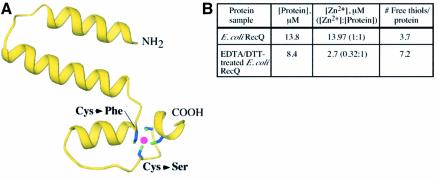
The RecQΔC structure shows that the RecQ-Ct region comprises two conserved subdomains. The first forms a platform of helices that is sandwiched between the helicase subdomains of RecQΔC and the rest of the RecQ-Ct region (Figures 1C and 3A). Inspection of the electron density map revealed that the side chains from four highly conserved cysteine residues at the ends of two of these helices were arrayed around a single intense electron density peak. This peak was modeled as a Zn2+ ion since X-ray fluorescence experiments demonstrated that Zn2+ was present in the crystals in spite of its exclusion from any solutions used to purify or crystallize RecQΔC (data not shown). A second indication that this electron density peak identified a bound Zn2+ ion came from biochemical analysis of E.coli RecQ (Figure 3B). In these experiments, the Zn2+ content and the number of solvent-accessible thiol groups in E.coli RecQ were analyzed. The results indicated that E.coli RecQ copurifies with stoichiometric amounts of Zn2+. In addition, dialysis of E.coli RecQ against DTT and EDTA removed ∼2/3 of the bound Zn2+ and uncovered an additional 3–4 solvent-accessible thiol groups, consistent with the Zn2+ binding site being comprised of 3–4 cysteines near the surface of RecQ. These biochemical data correlate well with the observed binding site in the structure.
Fig. 3. Features of the Zn2+-binding subdomain in RecQΔC. (A) View of the Zn2+-binding subdomain (residues 341–406) extracted from the rest of RecQΔC. Four highly conserved cysteine residues (Cys380, Cys397, Cys400, Cys403) are arrayed around a Zn2+ ion (magenta). Two BLM missense mutations alter individual cysteines in this array as labeled. The N- and C-terminal residues that connect to the helicase and WH subdomains, respectively, are labeled. (B) Biochemical evidence that full-length E.coli RecQ binds Zn2+ in a site that includes 3–4 cysteine residues. [Zn2+] and [thiolfree] were determined for E.coli RecQ and E.coli RecQ that was dialyzed against 10 mM EDTA, 1 mM DTT to extract bound Zn2+.
Given the strong conservation of the Zn2+-binding site among RecQ proteins (Figure 1B), it is perhaps not surprising that functional analyses have shown this region to be important for RecQ function. BLM missense mutation sites in the RecQ-Ct region mutate codons for cysteine residues in BLM protein that are analogous to residues 380 or 397 in E.coli RecQ (Ellis et al., 1995; Foucault et al., 1997; see discussion below). In addition, mutated S.cerevisiae SGS1 genes fail to complement the DNA-damage sensitivity and hyperrecombination phenotypes in an sgs1Δ strain when any of the analogous Zn2+-binding site cysteine residues have been altered (Onoda et al., 2000; Ui et al., 2001). Finally, a number of point mutations in residues analogous to RecQΔC Zn2+-liganding cysteines in either the human BLM protein catalytic core (Janscak et al., 2003) or in E.coli RecQ (M.C.Zittel and J.L.Keck, unpublished observation) result in variants that are difficult to purify due perhaps to rapid proteolysis, indicating that Zn2+ binding to this region may stabilize the protein. The RecQ Zn2+-binding motif is unlike previously observed zinc finger folds (Krishna et al., 2003), and does not have substantial structural similarity with any other known proteins. Future experiments will therefore be needed to determine its specific function in RecQ. Potential functions could include thermodynamic stabilization of the protein and/or acting as a DNA or protein binding site, all of which have been observed for Zn2+-binding domains in other proteins (Berg and Shi, 1996).
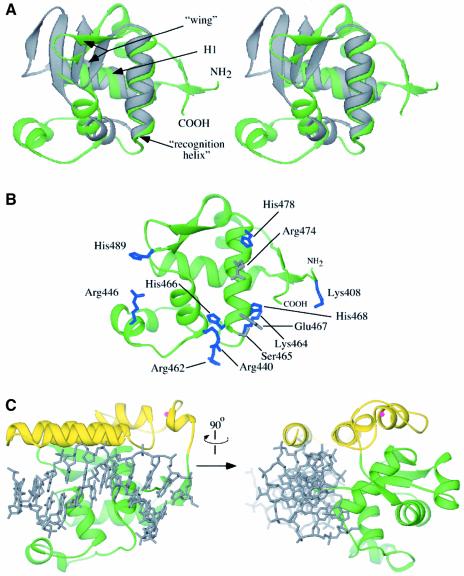
The second RecQ-Ct subdomain forms a specialized helix–turn–helix fold called a WH subdomain, which has been found to act as a DNA-binding motif in many proteins (Gajiwala and Burley, 2000; Figure 4A). The RecQ WH fold is structurally homologous to WH domains found in reverse gyrase (Rodriguez and Stock, 2002) and other type-IA topoisomerases (Lima et al., 1994; Mondragon and DiGate, 1999) as well as to several other DNA binding proteins, including the E.coli catabolite gene activator protein (CAP) (Schultz et al., 1991). The intersection of the RecQ WH and Zn2+-binding helical subdomains creates a large cleft on the surface of the protein. Several features of this cleft suggest that it can serve as a binding site for dsDNA: (i) the expected dsDNA binding face of the WH fold, including the putative DNA binding ‘recognition helix’, forms an exposed wall of the cleft; (ii) several residues that mediate WH/DNA phosphate interactions in other proteins are conserved in the RecQ WH domain (Figure 4B); and (iii) superposition of the dsDNA from the CAP/DNA co-crystal structure onto the WH RecQ fold demonstrates that the cleft could accommodate dsDNA without needing to significantly alter the structure of the RecQ-Ct region (Figure 4C). In support of this dsDNA binding model, we have found that a recombinant HRDC domain fragment from E.coli RecQ, which would be localized near the putative dsDNA binding site on the RecQ-Ct region in the full-length protein, has a strong binding preference for dsDNA over ssDNA (D.A.Bernstein and J.L.Keck, unpublished observation). dsDNA binding by the HRDC domain could serve as a clamp to stabilize dsDNA binding to the catalytic core. This model is consistent with previous observations of different DNA binding stabilities between E.coli RecQ and RecQΔC (Bernstein and Keck, 2003).
Fig. 4. Features of the WH subdomain in RecQΔC. (A) Stereo diagram of the superposition of the WH subdomain extracted from the RecQΔC structure (green, residues 407–516) with the DNA binding WH domain of CAP (Schultz et al., 1991) (grey). The two major helices of the fold (H1 and recognition helix) and the wing elements are labeled. The N-terminal residue of the RecQΔC WH fold, which connects to the Zn2+ binding region, and C-terminal residue are labeled. (B) Potential DNA binding residues in the RecQΔC WH subdomain. Residues that are structurally conserved with DNA binding residues in CAP (Schultz et al., 1991) are shown in grey and other residues with positive charge potential on the same putative DNA binding face are shown in blue. (C) Orthogonal views of a model of dsDNA binding to the RecQ-Ct region of RecQ. dsDNA from the CAP/DNA complex structure (grey) (Schultz et al., 1991) is overlayed onto the RecQ-Ct region. A minor opening of the helical Zn2+-binding region relative to the WH region would create a cleft sufficiently wide for dsDNA binding.
Comparison of RecQΔC to SF1 and SF2 helicases
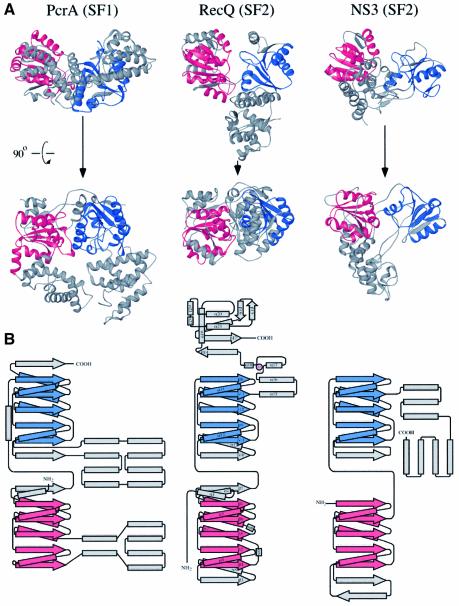
Sequence analysis of RecQ proteins have placed them among the SF2 group of helicases (Gorbalenya and Koonin, 1993), which includes several members with known structures. Consistent with its sequence alignment with other SF2 enzymes, the RecQΔC structure is more closely related to other SF2 family members in structure than to SF1 enzymes. For example, the two RecQΔC helicase lobes align better with hepatitis C virus NS3 helicase (Yao et al., 1997), an SF2 enzyme (DALI Z-score = 14.9 for 224 Cα atoms) than with PcrA helicase (Subramanya et al., 1996), the SF1 enzyme that is most structurally similar to RecQΔC (Z-score = 9.8 for 244 Cα atoms) (Holm and Sander, 1993) (Figure 5). Nevertheless, RecQΔC’s similarity with PcrA is still informative. As mentioned above, RecQΔC’s mode of ATP binding is quite similar to that seen in PcrA. In addition, a patch of conserved aromatic and charged residues in RecQΔC (residues 156–159, C-terminal to helicase motif II in RecQΔC) map to the same relative surface location as similar residues in PcrA that are used in DNA binding (residues 257–260, C-terminal to motif III in PcrA) (Velankar et al, 1999). This structural conservation between RecQΔC and PcrA could indicate that the enzymes use conserved mechanisms of DNA binding and unwinding. RecQΔC primarily differs from other helicases by its novel accessory elements in the RecQ-Ct domain (Figure 5). These features will most likely prove critical to the specialized functions of RecQ proteins.
Fig. 5. Structural comparison of RecQΔC to representative helicases belonging to the SF1 and SF2 helicase superfamilies. (A) Structures of PcrA (Subramanya et al., 1996) and NS3 (Yao et al., 1997), SF1 and SF2 helicases, respectively, were superimposed on RecQΔC using the structurally related Cα atoms in their N-terminal most helicase subdomains (in red) and then translated to allow visual comparison. Red and blue colors mark the conserved RecA-like folds found in the helicase subdomains of each protein, while grey coloration indicates sequences outside of the RecA fold. An orthogonal view is shown of each protein below the top row. (B) Topology diagrams of PcrA, RecQΔC and NS3. Colors are shown as in (A), with RecQΔC’s Zn2+ in magenta. Helices and β-strands in RecQΔC are labeled as in Figure 1B. Diagrams of PcrA and NS3 were adapted from Caruthers and McKay (2002).
Structural basis of Bloom’s syndrome missense mutations
Several recessive mutations in the human BLM, WRN, and RECQ4 genes are known to cause Bloom’s, Werner’s, and Rothmund–Thompson syndromes, respectively. Many of the mutations in BLM and all known disease-associated alleles in WRN and RECQ4 are nonsense or frame-shift mutations that would express truncated versions of the proteins (Kitao et al., 1999). In WRN protein, these mutations lead to mislocalization in the cell due to failure to express the C-terminal nuclear localization sequence (Matsumoto et al., 1997). A similar rationale might also explain the phenotypes of BLM nonsense alleles (Matsumoto et al., 1998). In addition to nonsense and frame-shift mutations, seven missense mutations in BLM leading to single amino acid changes in the protein are known to cause Bloom’s syndrome (Ellis et al., 1995; Foucault et al., 1997; Barakat et al., 2000; Rong et al., 2000). Three of these mutations have been shown to inactivate the murine BLM protein in vitro (Bahr et al., 1998), implying that the missense mutations cause disease as a result of enzymatic failure. All seven known disease-linked BLM missense mutations map to the RecQ family catalytic core domain, with five of them occurring in residues identically conserved within the E.coli RecQ sequence. Given the ability of a catalytically active fragment of human BLM protein to complement mutation of the recQ gene in E.coli (Janscak et al., 2003), the RecQΔC structure provides an excellent model for rationalizing the biophysical basis underlying known BLM missense mutations.
Five of the seven BLM missense mutations map to the helicase region of the protein: Gln672Arg, Ile841Thr, Cys878Arg, Gly891Glu, and Cys901Tyr (Ellis et al., 1995; Barakat et al., 2000; Rong et al., 2000). Three of these positions are conserved in RecQΔC (Gln30, Ile192 and Gly239), whereas the two cysteines are poorly conserved among RecQ helicases. The RecQΔC structures reveal plausible effects of these missense mutations that could impair enzyme function. The first mutation, Gln→Arg, would likely diminish ATP binding to RecQ, since the equivalent Gln in RecQΔC (Gln30) contacts the adenine moiety of the nucleotide cofactor directly through hydrogen bonds (Figure 2B). Mutation of this conserved Gln results in loss of ATPase and helicase activity in the murine BLM protein (Bahr et al., 1998), consistent with its observed role in ATP binding. The other BLM helicase mutations are likely to affect protein folding or stability (Figure 2A). Both the Ile→Thr and Gly→Glu mutations introduce polar side chains into the hydrophobic core of the helicase subdomains, which would likely destabilize folded BLM protein and could inactivate the enzyme.
The two remaining disease-linked BLM missense mutations, Cys1036Phe and Cys1055Ser (Ellis et al., 1995; Foucault et al., 1997), alter residues analogous to two that directly bind Zn2+ in RecQΔC (Figure 3A). These mutations introduce either a large hydrophobic group (Phe) or a poor Zn2+ ligand (Ser) into the structure. Either mutation could reduce Zn2+ binding, thus altering the structure of this region of the enzyme. Previous studies have shown that these mutations inactivate the murine BLM protein in vitro (Bahr et al., 1998), thus highlighting the importance of the Zn2+-binding region to proper RecQ family function.
Model for RecQ catalytic function
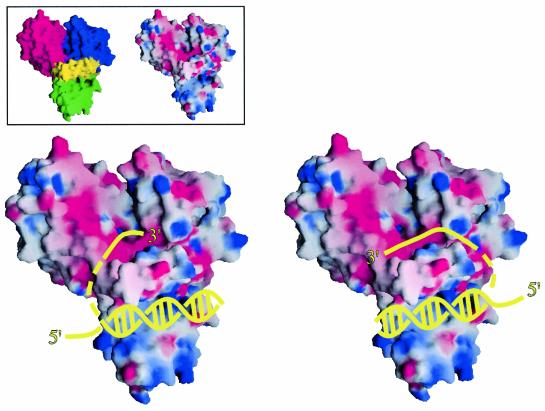
Using structural information from the RecQΔC structures, we have constructed a general model for RecQ activity. Two likely DNA binding sites are present in RecQΔC: a ssDNA binding site that is similar to those observed in other helicases (Figure 2A), and a putative dsDNA binding site in the RecQ-Ct region (Figure 4C). An arrangement in which the RecQ-Ct region binds dsDNA and ssDNA is bound between the helicase subdomains therefore seems plausible in RecQ (Figure 6). We predict that the 3′ ssDNA end of unwound DNA would be bound to the helicase region, since RecQ proteins unwind dsDNA in a 3′→5′ direction (Umezu et al., 1990). RecQ helicase activity would produce 3′ and 5′ ss segments of unwound DNA as potential substrates for enzymatic partners of RecQ that require ssDNA, such as RecA (Harmon and Kowalczykowski, 1998) or Topo III (Harmon et al., 1999). Moreover, conserved surface features of RecQ, such as the Zn2+-binding subdomain, could provide protein–protein interaction sites or additional DNA binding surfaces. Since our structure is of a monomer of RecQΔC, but full-length E.coli RecQ might function as an oligomer in DNA unwinding (Harmon and Kowalczykowski, 2001), it is not clear whether the ds and ss portions of DNA being unwound by RecQ would bind to the same or different protomers in a RecQ oligomer. In addition, DNA binding could alter the domain arrangement in RecQΔC as has been observed for PcrA helicase (Velankar et al, 1999). Future studies will be required to investigate the structural dynamics associated with DNA binding in E.coli RecQ.
Fig. 6. Models for DNA binding to E.coli RecQΔC. Surface representations of E.coli RecQΔC are shown (color-coded as in Figure 1, inset left) bound to cartoon diagrams of a partially unwound DNA molecule in two possible orientations (yellow). The RecQΔC surface is colored by its electrostatic surface potential (inset right and main panel) at + or –6 kBT/e for positive (blue) or negative (red) charge potential using the program GRASP (Nicholls et al., 1991). Since E.coli RecQ may unwind DNA as an oligomer (Harmon and Kowalczykowski, 2001), it is not clear whether ssDNA bound by the helicase region would come from dsDNA bound by the same protomer or another protomer in the oligomer during unwinding. As such, the connection linking the dsDNA to the 3′ ssDNA is shown as a dashed line. In addition, relative subdomain orientations could change in the DNA-bound form.
Insights into the nature of RecQ functions with topoisomerase III
Important functional interactions between RecQ and Topo III homologs have been reported for many organisms (reviewed in Wu and Hickson, 2001; Oakley and Hickson, 2002; Wang, 2002), but a model explaining their joint activity has been hampered by the lack of structural information on RecQ. DNA unwinding by a helicase such as RecQ creates domains of (+) and (–) supercoiled DNA (Liu and Wang, 1987; Peter et al., 1998). The ssDNA product from DNA unwinding is a substrate for Topo III, which selectively relaxes (–) supercoiled DNA (DiGate and Marians, 1988). Thus, the net product of RecQ/Topo III activities would likely be (+) supercoiled DNA. As previously proposed, this reaction would constitute a ‘reverse gyrase’ activity (Gangloff et al., 1994). Reverse gyrases have thus far been found only in thermophilic eubacteria and archaea, and they contain both helicase and type-IA topoisomerase domains (reviewed in Champoux, 2001). The RecQΔC structure reveals an important clue as to how RecQ might have evolved to support a reverse gyrase-like activity, since its major subdomains are structurally homologous to the helicase and WH regions of A.fulgidis reverse gyrase (Rodriguez and Stock, 2002). In addition, RecQΔC has a Zn2+-binding domain, an element that is lacking in bacterial and yeast Topo III homologs but present in other type-IA topoisomerases, such as reverse gyrase (Champoux, 2001). We propose that RecQ and Topo III could function together in a manner similar to reverse gyrase using functional domains supplied by both proteins. Interaction between RecQ and Topo III could position Topo III in close proximity to ssDNA produced by the RecQ helicase domain, thereby providing Topo III with substrate. The joint action of RecQ and Topo III may not rely on processive DNA unwinding but could involve a local melting of DNA by RecQ and relaxation by Topo III. This reverse gyrase-like activity could be an important common step in many RecQ-mediated DNA remodeling functions in cells.
The structures of the catalytic core of E.coli RecQ presented here yield insights into the molecular origins of human diseases that stem from mutations of recQ genes and offer new clues that will help elucidate the biochemical mechanisms used by the RecQ family of DNA helicases. These findings help pave the way for uncovering the precise functions of RecQ proteins in cellular genome maintenance reactions.
Materials and methods
Protein expression and purification
T7-overexpression plasmids encoding either full-length E.coli RecQ or RecQΔC (residues 1–523) preceded by a His6 affinity-purification tag were constructed, and the proteins were overexpressed and purified as previously described (Bernstein and Keck, 2003). Selenomethionine-incorporated RecQΔC was expressed as described in Van Duyne et al. (1993), and was purified as for the unsubstituted protein, except that 0.8 mM Tri(2-carboxyethyl) phosphine hydrochloride (TCEP) was added to all buffers, the NaCl concentration of all buffers was adjusted to 500 mM, and the ion exchange purification step was eliminated from the purification procedure.
Crystallization of RecQΔC
Native or selenomethionine-incorporated RecQΔC was dialyzed against buffer containing 10 mM Tris pH 8.0 and 500 mM ammonium acetate. Crystals of native RecQΔC were grown by hanging drop vapor diffusion by mixing 0.9 µl of 20 mg/mL protein, 0.2 µl of 100 mM MnCl2, and 0.9 µl of well solution [0.5% polyethylene glycol (PEG) 1000, 2–4% PEG400, 50 mM MES pH 6.5, 50 mM ammonium sulfate] and equilibrating at room temperature for several days. Crystals measuring ∼200 µm × ∼100 µm × ∼50 µm of symmetry P21 with unit cell length a = 65, b = 54 and c = 78 Å, β = 110.7° were formed. This crystal form has one molecule of RecQΔC per asymmetric unit. Isomorphous selenomethionine-incorporated RecQΔC crystals were grown by microseeding with native crystals. Crystals were stabilized by transferring into a cryoprotectant solution (3% PEG1000, 50 mM HEPES pH 7.5, 10 mM MnCl2, 25% ethylene glycol) and frozen in liquid nitrogen. Crystals with ATPγS bound were obtained by soaking native RecQΔC crystals in 2.5% PEG1000, 50 mM HEPES pH 7.5, 10 mM ATPγS, 5 mM MgCl2, for 3 days at room temperature, and were then transferred to cryoprotectant buffer and frozen in liquid nitrogen.
SAD phasing and model refinement
Selenomethionine-incorporated crystals were solved by SAD phasing to 1.8 Å (Table I). Data were indexed and scaled with MOSFLM (Leslie, 1992) and SCALA (Kabsch, 1988). Selenium sites were determined with SHELX (Schneider and Sheldrick, 2002) and refined with MLPHARE (Otwinowski, 1991). Solvent flattening with DM (Cowtan, 1994) resulted in interpretable experimental electron density maps for model building. Over 90% of the model was built automatically using ARP/wARP (Lamzin and Wilson, 1993), with the remainder built manually using the program O (Jones et al., 1991) and improved by rounds of refinement with REFMAC/ARP (Lamzin and Wilson, 1993; Murshudov et al., 1997) and manual rebuilding. The final model contains residues 1–516. No bond angles for this model fall into disallowed regions of Ramachandrian space.
Molecular replacement solution of nucleotide-bound RecQΔC
The ATPγS/Mn2+-bound RecQΔC structure was solved by molecular replacement using AMORE (Navaza, 2001) with the unbound RecQΔC structure as a search model. The molecular replacement solution was refined with REFMAC/ARP (Lamzin and Wilson, 1993; Murshudov et al., 1997) and by manually rebuilding the model using O (Jones et al., 1991). Since the nucleotide soaking solution contained 5 mM MgCl2 but the cryoprotectant used to stabilize the crystals prior to freezing included 10 mM MnCl2, the identity of the bound metal was ambiguous. We therefore attempted to refine the structure with either a Mg2+ or a Mn2+ ion in the binding site. Placing a Mn2+ ion in the site satisfied the observed scattering, whereas using a Mg2+ ion left substantial positive difference electron density and refined the atomic B-factor of the metal to an unrealistically low value. Therefore, the metal was modeled as a Mn2+ ion. E.coli RecQ has essentially the same level of ATPase activity using either Mg2+ or Mn2+ as a cofactor (Umezu et al., 1990). The final model contains residues 1–516 excluding residues 296–299 for which electron density was not observed. No bond angles for this model fall into disallowed regions of Ramachandrian space.
Quantitation of Zn2+ bound to E.coli RecQ
The Zn2+ content of purified full-length E.coli RecQ was measured using 4-(2-pyridylazo)resorcinol (PAR), a reporter dye that absorbs light at 490 nm when bound to Zn2+ (Hunt et al., 1984). Zn2+ was extracted from a sample of purified RecQ by dialysis against 20 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 10 mM EDTA, 1 mM DTT, at 4°C overnight. One hundred microliters of untreated RecQ or Zn2+-extracted RecQ (in 10% glycerol, 300 mM NaCl, 20 mM Tris–HCl pH 8.0) was incubated with 60 µg Proteinase K at 37°C for 1 h. Two microliters of 5 mM PAR was then added to each sample and their absorbances at 490 nm were measured and compared to a standard curve of ZnCl2 samples at a range of concentrations using an identical sample preparation procedure as described above, but omitting RecQ. RecQ concentrations were determined as in Bernstein and Keck, (2003).
Quantitation of free thiol groups in E.coli RecQ
[thiolsfree] in E.coli RecQ and Zn2+-extracted RecQ were determined by measuring the absorbance at 412 nm following reaction of 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB) with thiols in the protein as described in Riddles et al. (1983). Ten microliters of 10 mM DTNB (in 0.1 M NaPhosphate pH 8.0) was added to 10 µl of untreated or Zn2+-extracted E.coli RecQ (in 10% glycerol, 300 mM NaCl, 20 mM Tris–HCl pH 8.0) and diluted into 300 µl of 0.1 M NaPhosphate pH 8.0. Samples were incubated at room temperature for 1 h and their absorbances at 412 nm were measured. These values were converted to [thiolsfree] as described in Riddles et al. (1983).
Accession numbers
Model coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 1OYW and 1OYY.
Acknowledgments
Acknowledgements
We thank James Holton (Advanced Light Source beamline 8.3.1) and the Advanced Photon Source beamline staff (BioCARS beamline 14-BMC) for assistance in data collection, and Bruker AXS for generous use of their instrumentation. R.J.Bennett, J.M.Berger, D.A.Brow, E.A.Craig, J.E.Dahlberg and E.Shor contributed helpful discussions. This work was supported by a grant from the NIH to J.L.K. (GM068061), start-up funds from the University of Wisconsin, and a grant from the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools. D.A.B. was supported in part by an NIH training grant in Molecular Biophysics (GM08293).
References
- Bahr A., De Graeve,F., Kedinger,C. and Chatton,B. (1998) Point mutations causing Bloom’s syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene, 17, 2565–2571. [DOI] [PubMed] [Google Scholar]
- Barakat A., Ababou,M., Onclercq,R., Dutertre,S., Chadli,E., Hda,N., Benslimane,A. and Amor-Gueret,M. (2000) Identification of a novel BLM missense mutation (2706T>C) in a Moroccan patient with Bloom’s syndrome. Hum. Mutat., 15, 584–585. [DOI] [PubMed] [Google Scholar]
- Berg J.M. and Shi,Y. (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science, 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Bernstein D.A. and Keck,J.L. (2003) Domain mapping of Escherichia coli RecQ defines the roles of conserved amino- and carboxyl-terminal regions in the RecQ family. Nucleic Acids Res., 31, 2778–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers J.M., Johnson,E.R. and McKay,D.B. (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA, 97, 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers J.M. and McKay,D.B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol., 12, 123–133. [DOI] [PubMed] [Google Scholar]
- Champoux J.J. (2001) DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem., 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek,L. and Gasser,S.M. (2002) RecQ helicases: at the heart of genetic stability. FEBS Lett., 529, 43–48. [DOI] [PubMed] [Google Scholar]
- Courcelle J., Carswell-Crumpton,C. and Hanawalt,P.C. (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Crowley,D.J. and Hanawalt,P.C. (1999) Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol., 181, 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Donaldson,J.R., Chow,K.H. and Courcelle,C.T. (2003) DNA damage-induced replication fork regression and processing in Escherichia coli. Science, 299, 1064–1067. [DOI] [PubMed] [Google Scholar]
- Courcelle J. and Hanawalt,P.C. (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet., 262, 543–551. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) Joint CCP4 ESF-EACBM Newsletter. Protein Crystallogr., 31, 34. [Google Scholar]
- DiGate R.J. and Marians,K.J. (1988) Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem., 263, 13366–13373. [PubMed] [Google Scholar]
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- Foucault F., Vaury,C., Barakat,A., Thibout,D., Planchon,P., Jaulin,C., Praz,F. and Amor-Gueret,M. (1997) Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum. Mol. Genet., 6, 1427–1434. [DOI] [PubMed] [Google Scholar]
- Gajiwala K.S. and Burley,S.K. (2000) Winged helix proteins. Curr. Opin. Struct. Biol., 10, 110–116. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- Hanada K., Ukita,T., Kohno,Y., Saito,K., Kato,J. and Ikeda,H. (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G., DiGate,R.J. and Kowalczykowski,S.C. (1999) RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell, 3, 611–620. [DOI] [PubMed] [Google Scholar]
- Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G. and Kowalczykowski,S.C. (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem., 276, 232–243. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hunt J.B., Neece,S.H., Schachman,H.K. and Ginsburg,A. (1984) Mercurial-promoted Zn2+ release from Escherichia coli aspartate transcarbamoylase. J. Biol. Chem., 259, 14793–14803. [PubMed] [Google Scholar]
- Janscak P., Garcia,P.L., Hamburger,F., Makuta,Y., Shiraishi,K., Imai,Y., Ikeda,H. and Bickle,T.A. (2003) Characterization and mutational analysis of the RecQ core of the Bloom syndrome protein. J. Mol. Biol., 330, 29–42. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. D, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1988) Evaluation of single-crystal X-ray diffraction from a position sensitive detector. J. Appl. Crystallogr., 21, 916–924. [Google Scholar]
- Karow J.K., Newman,R.H., Freemont,P.S. and Hickson,I.D. (1999) Oligomeric ring structure of the Bloom’s syndrome helicase. Curr. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- Kawasaki K., Maruyama,S., Nakayama,M., Matsumoto,K. and Shibata,T. (2002) Drosophila melanogaster RECQ5/QE DNA helicase: stimulation by GTP binding. Nucleic Acids Res., 30, 3682–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat. Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- Korolev S., Hsieh,J., Gauss,G.H., Lohman,T.M. and Waksman,G. (1997) Major domain swivelling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S.S., Majumdar,I. and Grishin,N.V. (2003) Structural classification of zinc fingers. Nucleic Acids Res., 31, 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin V.S. and Wilson,K.S. (1993) Automated refinement of protein models. Acta Cryst. D, 49, 129–147. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1992) Newsletter on Protein Crystallography No. 26, Daresbury Laboratory.
- Lima C.D., Wang,J.C. and Mondragon,A. (1994) Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature, 367, 138–146. [DOI] [PubMed] [Google Scholar]
- Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Macias,M.J., Bottomley,M.J., Stier,G., Linge,J.P., Nilges,M., Bork,P. and Sattler,M. (1999) The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins Structure, 7, 1557–1566. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Imamura,O., Goto,M. and Furuichi,Y. (1998) Charaterization of the nuclear localization sequence in the DNA helicase involved in Werner’s syndrome. Int. J. Mol. Med., 1, 71–76. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Shimamoto,A., Goto,M. and Furuichi,Y. (1997) Impaired nuclear localization of defective DNA helicases in Werner’s syndrome. Nat. Genet., 16, 335–336. [DOI] [PubMed] [Google Scholar]
- Mondragon A. and DiGate,R. (1999) The structure of Escherichia coli DNA topoisomerase III. Struc. Fold Des., 7, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Morozov V., Mushegian,A.R., Koonin,E.V. and Bork,P. (1997) A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem. Sci., 22, 417–418. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V. and Brill,S.J. (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics, 154, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nakayama H., Nakayama,K., Nakayama,R., Irino,N., Nakayama,Y. and Hanawalt,P.C. (1984) Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet., 195, 474–480. [DOI] [PubMed] [Google Scholar]
- Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D, 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Oakley T.J. and Hickson,I.D. (2002) Defending genome integrity during S-phase: putative roles for RecQ helicases and topoisomerase III. DNA Repair (Amst.), 1, 175–207. [DOI] [PubMed] [Google Scholar]
- Onoda F., Seki,M., Miyajima,A. and Enomoto,T. (2000) Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom’s syndrome gene. Mutat. Res., 459, 203–209. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1991) Isomorphous Refinement and Anomalous Scattering. Proceedings of the CCP4 Study Weekend, 80–86.
- Peter B.J., Ullsperger,C., Hiasa,H., Marians,K.J. and Cozzarelli,N.R. (1998) The structure of supercoiled intermediates in DNA replication. Cell, 94, 819–827. [DOI] [PubMed] [Google Scholar]
- Riddles P.W., Blakeley,R.L. and Zerner,B. (1983) Reassessment of Ellman’s reagent. Methods Enzymol., 91, 49–60. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.C. and Stock,D. (2002) Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J., 21, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S.B., Valiaho,J. and Vihinen,M. (2000) Structural basis of Bloom syndrome (BS) causing mutations in the BLM helicase domain. Mol. Med., 6, 155–164. [PMC free article] [PubMed] [Google Scholar]
- Schneider T.R. and Sheldrick,G.M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D, 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Scaife,S. and Wigley,D.B. (2001) Structural analysis of DNA replication fork reversal by RecG. Cell, 107, 79–89. [DOI] [PubMed] [Google Scholar]
- Story R.M., Li,H. and Abelson,J.N. (2001) Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl Acad. Sci. USA, 98, 1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- Ui A., Satoh,Y., Onoda,F., Miyajima,A., Seki,M. and Enomoto,T. (2001) The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics, 265, 837–850. [DOI] [PubMed] [Google Scholar]
- Umezu K., Nakayama,K. and Nakayama,H. (1990) Escherichia coli RecQ protein is a DNA helicase. Proc. Natl Acad. Sci. USA, 87, 5363–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Wang J.C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol., 3, 430–440. [DOI] [PubMed] [Google Scholar]
- Whitby M.C. and Lloyd,R.G. (1998) Targeting Holliday junctions by the RecG branch migration protein of Escherichia coli. J. Biol. Chem., 273, 19729–19739. [DOI] [PubMed] [Google Scholar]
- Wu L. and Hickson,I.D. (2001) RecQ helicases and topoisomerases: components of a conserved complex for the regulation of genetic recombination. Cell. Mol. Life Sci., 58, 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. and Hickson,I.D. (2002) RecQ helicases and cellular responses to DNA damage. Mutat. Res., 509, 35–47. [DOI] [PubMed] [Google Scholar]
- Xu H.Q., Deprez,E., Zhang,A.H., Tauc,P., Ladjimi,M.M., Brochon,J.-C., Auclair,C. and Xi,X.G. (2003) The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem., (published on line, June 12, 2003). [DOI] [PubMed] [Google Scholar]
- Yao N., Hesson,T., Cable,M., Hong,Z., Kwong,A.D., Le,H.V. and Weber,P.C. (1997) Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol., 4, 463–467. [DOI] [PubMed] [Google Scholar]
- Yu C.E. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]