Abstract
p300 and CBP are large nuclear acetyltransferases exhibiting a complex multi-domain structure. Mouse embryos nullizygous for either p300 or Cbp die at midgestation, while heterozygotes are viable but in part display defects in neurulation or bone morphogenesis. To directly examine the contribution of the acetyltransferase (AT) activity to mouse development, we have abrogated this function by a knock-in approach. Remarkably, a single AT-deficient allele of p300 or Cbp leads to embryonic or neonatal lethality, indicating that the mutant alleles are dominant. Formation of the cardiovascular system, the lung and the small intestine are strongly impaired in p300 AT and to a much lesser extent in Cbp AT mutant embryos, a difference that is also reflected by the defects in gene expression. Embryonic stem cells homozygous for either the p300 AT or a p300 null mutation respond differently to BMP2 stimulation, indicating that the two alleles are not equivalent. Unexpectedly, the p300 AT-mutant cells upregulate BMP-inducible genes to levels similar or even higher than observed in wild-type cells.
Keywords: BMP/CBP/coronary vasculature/ES cells/villus formation
Introduction
The covalent modification status of histones and other chromosomal proteins is an important and dynamically regulated level of control for gene expression. Depending on the nature of the modification, transcription is either enhanced or repressed (Turner, 2000; Jenuwein and Allis, 2001). Originally proposed by Allfrey and colleagues (Allfrey et al., 1964), it is now well documented that hyperacetylation of core histones at specific lysine residues within their N-terminal tails facilitates transcription in general. Detailed studies of the β-globin locus in differentiating erythrocytes have shown that moderately hyperacetylated histones are present throughout the entire locus and that acetylation levels are maximal around actively transcribed genes and their regulatory elements (Hebbes et al., 1994; Litt et al., 2001). However, increased histone acetylation itself is often insufficient for transcriptional activation (Hebbes et al., 1994; Gregory et al., 1999). Efficient gene expression requires cooperation between acetyltransferases and other chromatin modifying activities such as ATP-dependent remodelling complexes (Hassan et al., 2001; Narlikar et al., 2002).
The cloning of transcription-related histone acetyltransferases (ATs) (reviewed in Roth et al., 2001) made it possible to study the precise role of individual enzymes by genetic and biochemical means. In yeast, mutation of gcn5 causes relatively small proliferation defects under normal growth conditions and reduces histone acetylation around the transcription start site of specific promoters (Kuo et al., 1998; Wang et al., 1998). However, deletion of gcn5 or of the MYST family member esa1 also leads to a global reduction in histone acetylation, indicating that these two enzymes fulfill gene-specific as well as genome-wide functions (Reid et al., 2000; Vogelauer et al., 2000).
In higher eukaryotes, several additional AT families exist, one of which is composed of the multifunctional transcriptional co-regulators p300 and CBP (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Goodman and Smolik, 2000). Under certain circumstances, p300 and CBP are able to bind other acetyltransferases such as GCN5 and PCAF, thus assembling protein complexes harboring two or more acetyltransferases (Yang et al., 1996). p300 and CBP are in part recruited to chromatin by their ability to interact with many sequence-specific transcription factors participating in signal transduction, cellular proliferation or differentiation programs.
Gene disruption experiments in mice have begun to provide evidence that the various ATs are likely to play distinct roles in vivo. Embryos lacking p300 arrest development and die between E8.5 and E11 (Yao et al., 1998), while those lacking Cbp die between E10 and E12 (Oike et al., 1999; Kung et al., 2000; Tanaka et al., 2000). Gcn5 is essential for mouse development and formation of several mesodermal tissues while Pcaf is dispensable (Xu et al., 2000; Yamauchi et al., 2000). Around midgestation, p300 is involved in the formation of the heart (Yao et al., 1998), a tissue in which Gcn5 is not expressed at that time of development.
Several aspects of development are very sensitive to the dosage of p300/CBP. Inactivation of a single allele of Cbp results in bone malformations (Tanaka et al., 1997) and monoallelic deletion of p300 causes a partially penetrant neural tube closure defect (Yao et al., 1998). In humans, haploinsufficiency of Cbp leads to Rubinstein–Taybi Syndrome (RTS) (Petrij et al., 1995), a congenital disease characterized by malformation of facial bones and digits as well as an increased incidence of heart defects (Stevens and Bhakta, 1995).
Since p300 and CBP encompass multiple conserved domains, the specific role of the AT activity in gene activation and development is unknown. To address this question, we have introduced an inactivating substitution mutation into the AT domain of murine p300 or CBP proteins using a knock-in approach. A single AT-mutant allele of p300 or Cbp resulted in embryonic or neonatal lethality of mice. In particular the p300 AT mutation caused multiple defects in heart, lung and intestine formation, indicating a major function for p300 AT activity in organogenesis. The transcriptional response to BMP2 is differentially altered in p300 AT-deficient relative to p300 null cells, thus pointing to multiple mechanisms by which p300 converts an extracellular signal into an appropriate transcriptional read-out. Remarkably, mutant mice and ES cells show evidence of ectopic or enhanced gene transcription, implying that p300 AT activity is required not only for activation but also repression.
Results
Lethality of mice harboring a single mutant allele of p300 or Cbp lacking AT activity
To investigate the role of the AT activity of p300 or CBP in mice, we have introduced into the mouse germline an amino acid substitution mutation abrogating the AT activity of these two proteins. The targeting vector harbored a floxed neomycin cassette in the intron preceding the exon with the WY to AS amino acid substitution (Roth et al., 2003). Mutant mice could only be maintained as long as the floxed neomycin cassette, transcribed in the opposite direction relative to the p300 or Cbp gene, suppressed expression of the mutant allele/protein (Roth et al., 2003). Thus, the mutant alleles containing the neo cassette (named p300+/AS–neo or Cbp+/AS–neo) resemble null alleles. To remove the neo cassette and allow expression of the AT-deficient p300 or CBP protein (Roth et al., 2003), p300+/AS–neo or Cbp+/AS–neo mice were mated with two general deleter strains harboring either a CMV-Cre (Schwenk et al., 1995) or EIIA-Cre (Lakso et al., 1996) transgene.
With either deleter strain, no live p300+/AS progeny were encountered at weaning, following genotyping of more than 300 offspring. Timed matings indicated that more than a third of the p300+/AS embryos died between E12.5 and E15.5 (42 dead/70 alive). The p300+/AS embryos developing to term were either born dead or became cyanotic within minutes after birth due to respiratory failure (see also Figures 3 and 4). Therefore, abrogation of the AT activity of a single p300 allele results in complete embryonic or perinatal lethality. In contrast to p300+/AS embryos, only ∼10% of Cbp+/AS embryos died between E12.5 and E16.5 and several heterozygous neonates were born alive. However, with the exception of three animals (3/107), these neonates did not survive beyond 1 or 2 days following birth. Accordingly, since mice heterozygous for p300 or Cbp null alleles are viable, the AT-mutant alleles are dominant-negative at the level of the organism.
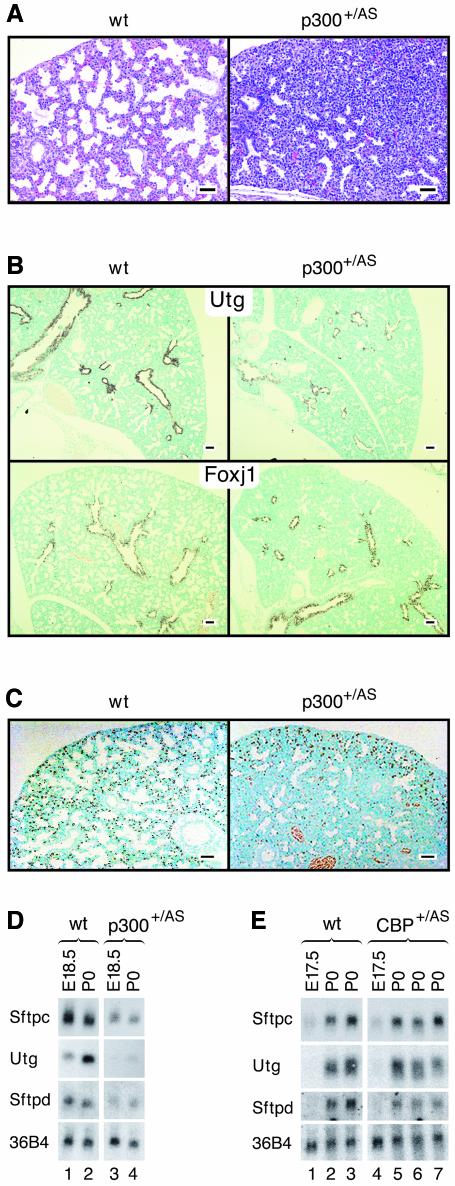
Fig. 3. Defective saccule formation and impaired proximal as well as distal epithelial cell differentiation in p300+/AS lungs. (A) HE staining of lung sections from wild-type and p300+/AS embryos at E18.5, showing condensed tissue with little evidence for saccule formation in the mutant. (B) In situ hybridization for proximal markers Utg (top panels) and Foxj1 (bottom panels). (C) Immunohistochemistry for the distal endoderm marker protein surfactant protein C showing that positive cells are confined to the periphery of p300+/AS mutant lungs. Two blood vessels are visible near the bottom part of the right panel. (D and E) Northern blot analysis of the indicated epithelial marker transcripts in p300+/AS and Cbp+/AS lungs. 36B4 served as internal standard. Bars = 100 µm.
Fig. 4. Intact BMP4 and Shh pathways and ectopic α-smooth muscle actin expression in the periphery of p300+/AS lungs. (A) Northern blot analysis of Bmp4, Shh, Ptc1 and Nkx2.1 transcript levels in wild-type and p300+/AS lungs at E14.4, E16.5 and E18.5. 36B4 served as an internal standard. (B) In situ hybridization for Bmp4 (top) and Shh (bottom) transcripts at E16.5. The arrowheads mark high levels of Bmp4 RNA in distal epithelial buds. Bars = 100 µm. (C) Immunohistochemistry for α-smooth muscle actin (SMA) showing staining around proximal airways (filled arrowheads in top panel) and blood vessels (open arrowheads). Ectopic SMA expression around peripheral (distal) respiratory saccules and in the mesothelium is shown in bottom panel. All bars = 50 µm.
Defects in heart formation and coronary vascularization in p300+/AS embryos
Most p300+/AS but not Cbp+/AS embryos were ∼20% smaller in size than their wild-type littermates (Figure 1Aa and b). At E14.5 and E15.5, all p300+/AS embryos suffered from an edema (Figure 1Ac) which was often accompanied by peripheral hemorrhage (Figure 1Ad and e). Interestingly, most embryos aged E16.5 to E18.5 no longer exhibited edema and hemorrhage, indicating that they were capable of overcoming the developmental defects underlying these malformations. Cbp+/AS embryos rarely presented edema and hemorrhage.
Fig. 1. Appearance and heart defects of p300+/AS and Cbp+/AS embryos. (A)(a–c) Wild-type (left) and mutant (right) littermates of indicated age. The edema of the p300+/AS embryo in panel (c) is marked by an arrowhead. (d and e) Dorsal region of a normal+/+ (d) and a hemorrhagic p300+/AS embryo (e) at E15.5. Bars = 1 mm. (B)(a–d) Transverse sections of E14.5 mouse hearts of wild-type (a) and p300+/AS littermates (b) showing a thin ventricular compact layer (open arrowhead) of right (RV) and left (LV) ventricular chambers, a ventricular septum closure defect (VSD) and an underdeveloped atrioventricular septum and leaflet (filled arrowhead). (c) Ruptured ventricular wall (arrow) with blood leakage (left side of arrow). (d) p300+/AS heart with a VSD and an underdeveloped leaflet of the mitral valve (marked by asterisk), but normal ventricular compact layer. (e and f) Transverse section of E14.0 hearts of wild-type (e) and Cbp+/AS (f) littermates. The mutant heart exhibits a reduced thickness of the ventricular compact layer, but a closed ventricular septum and fully formed AV valvuloseptal structures. (g and h) Transverse section of E15.5 wild-type (g) and p300+/AS (h) hearts. The mutant heart exhibits a thin myocardium, VSD and atrial septa closure defects (ASD) and a hypoplastic AV septum including the leaflets of the mitral valve (filled arrowhead). Bars = 250 µm, except in (Bc) where bar = 60 µm.
A comparison of heart sections from E11 to E16.5 wild-type and mutant littermates revealed multiple cardiac malformations in p300+/AS and to a much lesser extent in Cbp+/AS embryos. With variable severity, ∼80% of the p300+/AS embryos exhibited a clear reduction in the thickness of the myocardial compact layer of both ventricular chambers (Figure 1B, compare Ba and g with Bb and h). In some of the mutant hearts, blood leakage through ruptures in the ventricular wall was apparent (Figure 1Bc). Despite a reduced muscle mass, mutant hearts were beating normally and stained positive for myosin heavy chain (MHC) or α-actinin as early as E9 (data not shown), indicating that cardiomyocytes were able to differentiate. At E14.5, a ventricular septum closure defect (VSD) accompanied by underdeveloped or malformed valve leaflets was apparent in 7/8 p300+/AS embryos and at E15.5 in 5/6 (Figure 1Bb, d and h). Hearts displaying little myocardial thinning also exhibited underdeveloped leaflets and a VSD which, however, was restricted to the membranous (i.e. non-muscular) part located at the tip of the septum (Figure 1Bd). In addition, three embryos showed an atrial septum closure defect (ASD), associated with an underdeveloped atrioventricular (AV) septum (Figure 1Bb and h). Taken together, these malformations suggest that the full dose of p300 AT activity is required for the development of AV cushion-derived structures (valve leaflets, membranous part of ventricular septum and AV septum) and ventricular myocardium.
The majority of the p300+/AS embryos surviving to E16.5 displayed a closed ventricular septum, suggesting that the VSD is partly due to a delay of 2 to 3 days in completing this morphogenetic process which is normally finished by E13.5 to E14. In contrast, in all except one (1/14) Cbp+/AS embryos, the ventricular septum was closed as early as E14. The compact layer of the ventricles was only moderately thinner than in wild-type hearts (Figure 1Be and f), and malformed valve leaflets or AV septa were never observed. The mild phenotype of Cbp+/AS hearts correlates with rare formation of edema and little lethality of these embryos.
The reduction in thickness of the p300+/AS myocardium became first apparent around E11 and primarily affected the ventricular but not the atrial wall (Figure 2Aa and b). Wild-type hearts of this age were surrounded by a contiguous sheet of epicardial cells which can be distinguished from myocardial cells by their expression of the Wilms tumor protein WT-1 (Moore et al., 1999) (Figure 2Ac and e). p300+/AS mutant hearts had not completed epicardium formation and were surrounded by few scattered cells staining for WT-1 (Figure 2Ad and f). By E12.5, the epicardium was formed around most mutant hearts, but few subepicardial mesenchymal cells (SEMCs) were present (Figure 2Ag and h). Normally, SEMCs delaminate from the epicardium, enter the myocardium and juxtapose to assemble coronary capillaries. SEMCs expressing WT-1 protein entered the myocardium in wild-type but not mutant hearts at E12.5 (Figure 2Ai and j). By E13.5, a clear delay or reduction in the development of the coronary vessel network became apparent, as revealed by PECAM-1 (platelet/endothelial cell adhesion molecule 1) staining (Figure 2B). In addition, in approximately one third of E13.5 p300+/AS embryos, blood leakage around ventricles was detected (Figure 2C), suggesting that newly formed vessels and/or the myocardium are not tight.
Fig. 2. Delayed formation of epicardium and coronary vasculature. (A)(a and b) Saggital sections at the level of the dextrodorsal outflow tract showing the ventricular (V) and atrial (A) chambers of a wild-type (a) and a p300+/AS (b) heart at E11.0. The arrowhead in (b) points to the thin wall of the mutant ventricular chamber. (c–f) Saggital sections of E11.0 ventricular wall stained with hematoxylin/eosin (HE) (c and d) or with Wilms tumor 1 protein (WT-1) antibodies (e and f). Arrowheads point to epicardial cells. (g–j) Transverse sections of E12.5 ventricular wall stained with HE (g and h) or WT-1 antibodies (i and j). The arrowhead in (g and h) highlights delaminating subepicardial cells. The arrow in panel (i) shows WT-1 positive epicardial cells having migrated into the myocardium. Bars = 60 µm, except (Ba and b) where bars=125 µm. (B)(a–d) PECAM1 immunostaining of E13.5 hearts showing strong (b) or moderate (d) reduction in coronary capillary formation on the dorsal side of p300+/AS mutant hearts. (C)(a and b) Ventral view of E13.5 p300+/AS heart showing bleeding (arrow) around the left ventricle.
Most p300+/AS embryos surviving past E16.5 exhibited a dense coronary capillary plexus and a ventricular wall of increased thickness (data not shown). Thus, we propose that delayed coronary vasculature formation is a major cause for the retarded development of the myocardium. In around half of the p300+/AS embryos, the cardiovascular defects are transient and moderate enough so that they can be overcome at later stages of embryogenesis, thereby explaining the observation that all embryos were edematous at E14.5 while at E16.5 some were not. As some p300+/AS hearts exhibited a thin ventricular wall prior to the onset of vascularization (Figure 2Ab), an additional defect, unrelated to capillary formation, is likely to contribute to the reduced thickness of the myocardium. In summary, our data suggest that p300 AT activity is required in the heart for the development of both valvuloseptal structures and coronary vasculature.
Respiratory failure and defective proximal and distal epithelial differentiation of p300+/AS lungs
p300+/AS neonates became cyanotic within minutes following birth, suggesting a respiratory failure. Visual inspection of neonatal lungs indicated that they failed to inflate. Analysis of 45 mutant lungs dissected between E14.5 and birth showed correct lobe, esophagus and trachea formation (data not shown), irrespective of the extent of cardiac malformation. For the following analysis, embryos presenting little or no edema were chosen.
Histological examination suggested that most p300+/AS but not Cbp+/AS lungs were developmentally arrested around the early saccular stage since air saccules failed to form efficiently (Figure 3A and data not shown). This observation indicates that lung remodeling at the saccular stage specifically requires the full dose of p300 AT activity.
Examination of the differentiation status of proximal air-conducting bronchiolar and distal respiratory epithelium, both of which are endoderm-derived, revealed several abnormalities. Expression of uteroglobin (Utg, also known as SP-CC or CC10), a proximal marker expressed by Clara secretory cells, was significantly reduced in p300+/AS lungs, particularly in peripheral bronchioles (Figure 3B, top) and failed to be upregulated at birth, as measured by northern blotting (Figure 3D). By contrast, expression of the forkhead box transcription factor Foxj1, marking ciliated cells, another proximal cell type, was slightly overexpressed in p300 AT-mutant lungs (Figure 3B, bottom). Distal respiratory epithelial cells, characterized by surfactant protein C (Sftpc) expression, were largely confined to the periphery of mutant lobes, while they were uniformly distributed in normal lobes (Figure 3C). Overall, Sftpc mRNA levels were reduced by approximately a factor of four (Figure 3D). Surfactant protein D (Sftpd) transcripts, synthesized by proximal as well as distal epithelia, were also diminished in p300+/AS lungs (Figure 3D). Cbp+/AS lungs showed a moderate reduction in Sftpd and normal Utg or Sftpc transcript levels (Figure 3E). These results suggest that either formation or differentiation of proximal as well as distal epithelial cell types is impaired in the absence of the full dosage of p300 AT activity, while that of CBP is less critical. Since two of the transcription factors driving proximal epithelial differentiation, Foxj1 and Nkx2.1 (Figure 4A), are expressed, the block in proximal differentiation is likely to occur downstream of these two transcription factors.
Formation of distal respiratory epithelium and branching of proximal airways is directed by BMP4 which is expressed at the tip of elongating epithelial buds (Weaver et al., 1999). In situ hybridization and northern blotting revealed correct localization and comparable expression levels of Bmp4 RNA in wild-type and p300+/AS pulmonary lobes (Figures 4B, top and 4A). Moreover, mutant and wild-type lungs contained a similar number of proximal epithelial tubules consisting of columnar or cuboidal cells, indicating that airway branching is not impaired. Distal epithelial cells, squamous in morphology, were present both near the center and along the entire periphery of p300+/AS E18.5 lobes (data not shown). We conclude that distal cells are specified and hence, the defect in Sftpc expression reflects impaired differentiation. The identity of the signal(s) triggering distal cell differentiation is unknown. Our analysis suggests that several aspects of BMP4 signaling are largely intact in p300+/AS lungs.
Sonic hedgehog (Shh) is another key morphogen for lung organogenesis, regulating epithelial branching morphogenesis as well as proliferation and differentiation of certain mesenchymal cell types (Litingtung et al., 1998; Pepicelli et al., 1998; reviewed in Warburton et al., 2000). The fly ortholog of p300/CBP, dCBP, has been reported to function as a coactivator in the hedgehog pathway (Akimaru et al., 1997). Based on these data, we assessed the functionality of this pathway in p300 AT mutant lungs. In situ and northern hybridizations showed that Shh is correctly expressed throughout the epithelium at E16.5 (Figure 4B, bottom), and that Shh RNA levels were comparable in wild-type and p300+/AS lungs at three different developmental stages (Figure 4A). Importantly, the Shh mesenchymal target gene patched1 (Ptc1) was present at normal levels in p300 AT mutant lungs (Figure 4A). Expression of α-smooth muscle actin (α-Sma) around proximal airways also depends on Shh signaling (Pepicelli et al., 1998). α-SMA was readily detectable in p300+/AS lungs (Figure 4C, top). These data indicate that Shh signaling in the lung is not ostensibly compromised by a reduction in p300 AT activity.
Strikingly, at E18.5, we frequently observed areas of ectopic expression of α-SMA near the periphery, which also included the mesothelium of mutant lobes (Figure 4C, bottom). Since there was no evidence for ectopic Shh expression in the mesothelium (Figure 4B and data not shown), we do not attribute this ectopic α-SMA expression to altered Shh signaling (see Discussion).
In summary, our analysis of lung development reveals specific and restricted alterations in gene expression in p300 AT mutant lungs, affecting both endoderm and mesoderm. Several aspects of Shh and BMP4 signaling appear to be intact. Interestingly, while the expression of several transcripts is impaired in p300 AT mutant lungs, we have also seen enhanced or ectopic gene activity for Foxj1 and α-Sma, respectively. The latter observation suggests that p300 AT activity might also be required, directly or indirectly, to maintain repression.
Delayed villi formation and increased mesenchymal cell proliferation in p300+/AS small intestine
Formation of intestinal villi begins around E14 by a combination of invagination of mesenchymal cells into the overlying epithelial cell layer and evagination of the epithelium (Roberts, 2000) (Figure 5Aa). By E16.5, mesenchymal invaginations underneath a layer of stratified epithelium, forming nascent villi, are well visible (Figures 5Ac and e). In p300+/AS mutant midgut, no mesenchymal invagination was observed at E14.5 and E16.5 (Figure 5Ab, d and f), while that of Cbp+/AS displayed nascent villi at E15.5 (Figure 5C). Interestingly, the epithelial cell layer of p300+/AS midguts showed clear signs of evagination and was greatly expanded in thickness (Figures 5Ad and f). By E18.5, nascent villi with prominent mesenchymal invagination were also present in p300+/AS embryos (Figure 6A). However, even at E18.5, areas with little or no mesenchymal invagination persisted in the duodenum and jejunum of some p300+/AS embryos (see e.g. Figure 6B, top). These data suggest that villi formation is delayed by 2 to 4 days in p300+/AS embryos.
Fig. 5. Delayed villi formation and expression of Bmp4 in p300+/AS small intestine. (A) HE staining of small intestine sections from wild-type (a, c and e) and p300+/AS (b, d and f) embryos at E14.5 (a and b) and E16.5 (c–f); (e and f) show higher magnification of the boxed regions in (c and d). Mesenchymal invagination in wild-type intestine is marked by arrowheads. The intestines analyzed above were from embryos exhibiting a normal myocardium with little (E14.5, b) or no (E16.5, d and f) edema. (B) In situ hybridization detecting Bmp4 expression. The arrowheads point to the Bmp4 signal in the submucosa and the arrows indicate high Bmp4 expression levels in the invaginating mesenchymal tip. (C) HE staining of small intestine sections from wild-type (left) and Cbp+/AS (right) embryos at E15.5 showing normal villi formation. All bars = 80 µm.

Fig. 6. Proliferation and differentiation in p300+/AS intestine. (A) Immunohistochemistry for the proliferation marker Ki67 on E18.5 intestinal sections. The closed arrowheads mark proliferating epithelial cells in the prospective crypt stem cell compartment. Proliferating mesenchyme is highlighted near the base of villi (open arrowheads) and the tip (arrows). Bars = 80 µm. (B) Alcian Blue staining for goblet cells (top panel, arrowheads) and alkaline phosphatase staining for enterocytes (bottom). The villus shown in the p300+/AS panel shows very little mesenchymal invagination. The arrowhead (bottom panel) indicates a p300+/AS mutant villus manifesting slightly reduced AP secretion. Bars = 30 µm (top) and 60 µm (bottom). (C) Immuno histochemistry for α-smooth muscle actin expression in circumferential smooth muscle layers of small intestine. lm: longitudinal muscle, cm: circular muscle, me: mesenchyme. Bars = 80 µm.
In situ hybridization with a Bmp4 probe revealed a close correlation between BMP4 transcript levels in the mesenchyme underlying the epithelium and villi formation (Figure 5B). In particular, high levels of Bmp4 expression in the tip of the invading mesenchyme were associated with villi formation in the wild-type intestine (Figure 5Ba and c). In some of the E16.5 p300+/AS mutants, elevated Bmp4 expression marked the leading tip of mesenchymal areas beginning to invaginate (Figure 5Bd). We conclude that in the intestine p300 AT activity is required for the correct timing of Bmp4 induction which, in turn, appears to contribute to villi formation by facilitating mesenchymal invagination.
Proliferation and differentiation is well coordinated in the intestine (van de Wetering et al., 2002). Staining with the proliferation marker Ki67 revealed enhanced mesenchymal cell proliferation in p300+/AS intestine at E18.5, while epithelial proliferation was restricted to the stem cell compartment at the base of villi, as expected (Figure 6A). The increased proliferation of mutant mesenchyme was particularly prominent at the mesenchymal base and tip of villi, overlapping with high Bmp4 expression, implying that Bmp4 might exert, directly or indirectly, a mitogenic function on this tissue. In contrast, differentiation of the epithelium was not overtly impaired in p300 AT mutant intestine. Similar numbers of mucous secreting goblet cells (Figure 6B, top) and phosphatase secreting enterocytes (Figure 6B, bottom) were present in wild-type and p300+/AS intestine at E18.5. Thus, unlike in the lung, epithelial differentiation in the midgut is not significantly impaired by the p300 AT mutation. In addition, the two circumferential smooth muscle layers, a longitudinal outer and a circular inner layer, were fully formed in p300+/AS intestine (Figure 6C), showing that this aspect of mesenchymal differentiation can take place.
p300 AT-deficient and p300 nullizygous ES cells respond differently to BMP: increased gene activity in cells lacking p300 AT activity
To assess the importance of the AT activity for the overall function of p300 at the cellular level, we have compared the ability of p300 AT-deficient and p300 null ES cells to induce BMP-dependent gene expression. p300/CBP have been reported to be essential coactivators in this pathway (Waltzer and Bienz, 1999; see also review by Itoh et al., 2000). We focused on the Inhibitors of differentiation Id1 and 2 genes, that are both inducible by BMP2 or BMP4 in serum-starved ES cells, even in the presence of the translation inhibitor cycloheximide, suggesting that the inductive effect of BMPs is direct and does not require additional protein synthesis (Hollnagel et al., 1999).
The cells were propagated in two types of medium, one with fetal calf serum (referred to as ‘serum’) and one with defined composition (referred to as ‘SR’, serum replacement) in order to determine if the serum plays a role in Id1 gene induction. 36B4 mRNA expression was monitored as internal standard. Unexpectedly, p300 AT-deficient cells propagated in either serum- or SR-containing medium exhibited higher BMP2-induced Id1 transcript levels compared with wild-type or p300–/– cells (Figure 7A and B). In addition, basal Id1 levels were elevated in AT-deficient cells cultured in serum-containing medium (Figure 7A, lane 4). These results were reproducible as shown in Supplementary figures S1A and S1B, available at The EMBO Journal Online. Thus, rather than impairing gene expression as one would expect when an AT activity is abrogated (Kuo et al., 1998; Wang et al., 1998), loss of p300 AT activity leads to elevated basal and/or induced Id1 transcript levels. Furthermore, p300–/– cells grown in SR- but not serum-containing medium were compromised in upregulating Id1, suggesting that different types of serum differentially influence Id1 inducibility in p300 null versus wild-type cells, presumably by activating distinct sets of intracellular signaling cascades.

Fig. 7. BMP induction of Id1 and Id2. (A) Northern blot of Id1. Wild-type (wt), p300 AT-deficient (AS/AS) and p300 null (–/–) ES cells grown in ‘serum’ medium were starved for 18 h and left untreated (–) or stimulated for 1 or 2 h with 30 ng/ml of BMP2. The bar diagram shows Id1 transcript levels after normalization against the internal control 36B4 mRNA (bottom). Comparable results have been obtained in three independent experiments. Fold induction relative to the basal level of each genotype are: wt: 1 h 1.6, 2 h 2.8; AS/AS: 1 h 1.5, 2 h 2.3; –/–: 1 h 1.7, 2 h 2.4. (B) BMP2 induction of Id1 (top) and Id2 (bottom) transcripts in ES cells cultured in ‘serum replacement’ medium. Fold induction relative to the basal level of each genotype are for Id1: wt: 1 h 1.3, 2 h 3.4; AS/AS: 1 h 2.7, 2 h 5.9; –/–: 1 h 1, 2 h 1.8 and for Id2: wt: 1 h 1.2, 2 h 3.4; AS/AS: 1 h 1.9, 2 h 6.7; –/–: 1 h 1, 2 h 1.2. (C) BMP4 induction of Id1 transcripts in ES cells propagated in ‘serum’ medium. Fold induction relative to the basal level of each genotype are: wt: 1 h 11.6, 2 h 15.5; AS/AS: 1 h 16.1, 2 h 11.4; –/–: 1 h 6.1, 2 h 7.1.
To extend this analysis to another gene and an additional BMP family member, we investigated Id2 induction by BMP2 and Id1 stimulation by BMP4. Unlike observed for Id1 transcripts, Id2 mRNA was induced by BMP2 to comparable levels in p300 AT-deficient and wild-type cells (Figure 7B, compare lanes 3 and 6 of Id2 panel). Accordingly, the precise role exerted by p300 AT activity varies from gene to gene. Id1 induction by BMP4 was assayed in cells cultured in medium supplemented with serum. As expected from Figure 7A, basal Id1 levels were elevated in p300 AT-deficient compared with wild-type cells (Figure 7C, lanes 4 and 1). Induced Id1 transcripts reached similar levels in AT-deficient and wild-type cells. However, they were lower in p300–/– cells (Figure 7C), suggesting that one or more p300 activities other than the AT function normally contribute to the BMP4 response.
In conclusion, these results demonstrate that the AT and the null mutation of p300 are not equivalent since they differentially alter the response to BMP, thereby strongly suggesting that AT activity is only one of several functions of p300 (see also Discussion). Moreover, loss of AT activity enhances basal and/or induced transcript levels in certain cases, implying that p300 AT activity is also involved in repression, particularly at the Id1 promoter. Finally, the precise roles of p300 and its AT activity depend on the overall signaling conditions.
Discussion
Essential function of p300 or CBP AT activity in mouse development
This study shows that inactivation of just one allele of p300 or CBP AT activity is sufficient to cause embryonic or neonatal lethality, indicating a striking dependence of mouse development on the full dose of p300 or CBP AT activity. In fact, monoallelic abrogation of AT activity perturbs embryogenesis and formation of several organs more than a monoallelic null mutation in either gene (Tanaka et al., 1997; Yao et al., 1998). Thus, the AT-deficient p300 and CBP proteins are dominant-negative at the level of the whole organism and in several aspects of organogenesis. Due to their dominant phenotype, the AT-deficient alleles serve as sensitive indicators for the participation of p300 and CBP in specific processes in vivo. Furthermore, since the p300 AT mutation affects organogenesis and gene expression more severely than that of CBP, the two proteins are likely to act at distinct functional levels. Phenotypic differences were recently reported for a KIX domain mutation which only caused hematopoietic defects in p300 but not CBP mutant mice (Kasper et al., 2002).
Our results also suggest that other acetyltransferases known to bind p300/CBP such as PCAF and GCN5 (Yang et al., 1996) are unable to substitute for the loss of p300 or CBP AT activity. Similarly, transgenes encoding an AT-deficient CBP cannot rescue the early embyonic lethality caused by a CBP knock-down or null allele in either worms (Victor et al., 2002) or flies (Ludlam et al., 2002). These observations favor the view that the p300/CBP branch of AT enzymes carries out specific and non-redundant functions.
Dominant phenotype of AT-mutant alleles: implications for p300 and CBP function
Dominant mutations usually occur in genes whose protein products assemble in one or several homo- or heteromeric multi-protein complexes. p300 and CBP are part of transcription factor complexes formed on enhancer and promoter elements (reviewed in Goodman and Smolik, 2000). AT-deficient proteins could exert dominant effects by ‘freezing’ target promoters in a partially activated state if for example p300/CBP auto-acetylation is required to trigger their dissociation, and to allow access of subsequently acting proteins. Another possibility, not mutually exclusive with the first one, is that the perturbation of the intramolecular balance of p300/CBP results in a dominant phenotype. If p300/CBP harbor additional enzymatic activities which are in a balance with AT activity, the loss of the latter will lead to an unbalanced exposure of chromatin to the other activities. Most relevant in this context is the recent description of an intrinsic ubiquitin E3-ligase activity localized at the N-terminus of p300 (Grossman et al., 2003). Acetylation and ubiquitylation sometimes target the same lysine residues and therefore can be antagonistic, as demonstrated for the Smad7 repressor (Grönroos et al., 2002). Hence, it is conceivable that the AT mutation leads to aberrant turnover and/or modification of chromatin components. Depending on whether the chromatin component has an activating or repressing function, this could lead to impaired or enhanced gene activity.
Evidence that histones are not always the most relevant targets of p300 AT activity
Loss of AT activity is in general associated with decreased gene activity (see e.g. Kuo et al., 1998; Wang et al., 1998). Consistent with this notion, we have indeed observed several genes, such as intestinal BMP4 and pulmonary surfactant protein transcripts, whose expression was reduced or delayed in p300 AT mutant compared with wild-type embryos. However, we have also observed ectopic expression of smooth muscle actin and increased Foxj1 mRNA levels in AT-mutant embryos. Similarly, in p300 AT-deficient ES cells, basal as well as BMP2-induced levels of Id1 were significantly higher than in wild-type cells. The enhanced gene activity in both p300 AT mutant mice and cells does not support the view that p300 AT activity primarily and directly acts only as a HAT, i.e. that histones are functionally the most significant substrate of p300 AT activity. Rather, our data are more consistent with the hypothesis that p300 AT activity is required to maintain and/or regulate repression on certain promoters, such as Id1, by modifying non-histone substrates. This hypothesis might explain why abrogation of p300 AT activity leads to the unexpected finding of elevated gene expression. It remains to be determined whether p300 AT activity regulates repression directly or indirectly.
Essential role of p300 AT activity in organogenesis and specific epithelial–mesenchymal signaling interactions
The reduction in p300 AT dosage strongly affects morphogenetic and differentiation events associated with the late phase of heart, lung and intestine organogenesis as well as formation of several facial and oral structures such as palate closure (N.Shikama and R.Eckner, unpublished data). A common denominator of the impaired processes is that they are driven by complex reciprocal epithelial–mesenchymal signaling interactions. During heart organogenesis, epicardium and myocardium are known to depend on each other and to communicate in multiple ways for coronary vasculature formation and compact layer expansion (Moore et al., 1999; Tevosian et al., 2000; reviewed in Reese et al., 2002). In the intestine and the lung, the epithelia are of endodermal origin while the mesenchyme is mesodermal. Signaling between these two tissue layers is essential for morphogenesis and cell differentiation in the intestine (reviewed in Roberts, 2000) and the lung (reviewed in Warburton et al., 2000). The delay in villi formation in the intestine on the one hand and the impaired saccule development and epithelial differentiation in the lung on the other are consistent with perturbed interactions between the endodermal and mesodermal cell layers of these organs caused by the p300 AT mutation.
Which signaling pathways are perturbed by the reduction in p300 AT activity? In the lung, several developmental processes requiring SHH or BMP4 signaling are intact. The results of the ES cell induction experiments also support the notion that at least BMP4-dependent gene induction (Figure 7c) is not severely compromised in p300 AT-deficient cells. In the intestine, Bmp4 RNA is upregulated with a delay, but following its induction, villi formation takes place, implying that BMP4 signaling itself is not greatly inhibited. Thus, p300 AT activity is required upstream of Bmp4 and further study of Bmp4 gene regulation is necessary to pinpoint the defect.
In contrast, there is some evidence that aspects of TGFβ signaling are altered in p300+/AS lungs. The ectopic expression phenotype of α-SMA in the mesothelium and around distal epithelium bears a striking resemblance to that of lungs treated with TGFβ1 in organ explant culture (Bragg et al., 2001) or overexpressing TGFβ1 in distal epithelia (Zhou et al., 1996). Moreover, α-SMA is known to be inducible by TGFβ (Hautmann et al., 1997). Accordingly, the ectopic α-SMA expression in p300+/AS lungs might reflect either the presence of ectopic TGFβ activity leading to the conversion of mesothelial cells to myofibroblasts, or derepression of α-Sma. In either case, the lung phenotype caused by the p300 AT mutation is consistent with an impaired repression of genes in the TGFβ pathway, thereby mimicking ectopic activation of this pathway.
In summary, the analysis of p300 or CBP AT-mutant embryos has revealed a major role of p300 AT activity during organogenesis, while that of CBP is less critical. Our results demonstrate that p300 is intimately involved in heart formation through its AT activity which is required for both coronary vascularization and valvuloseptal morphogenesis. Our data also provide the first in vivo evidence that p300 is involved in lung and intestine development. In the latter organ, a close correlation exists between p300 AT-dependent induction of Bmp4 transcripts and villi formation.
Materials and methods
Generation of p300+/AS and cbp+/AS mice
p300+/AS and Cbp+/AS mice were generated as described (Roth et al., 2003). For histological and in situ hybridization analysis, p300+/AS; cre and p300+/+; cre embryos were compared. Details including PCR primers for genotyping are available upon request.
ES cell culture and BMP stimulation
Generation of p300 AT deficient (p300–/–, these cells correspond to p300AS–neo/AS–neo cells) described in (Roth et al., 2003). For BMP stimulation studies, ES cells were cultured without feeder cells on gelatin-coated dishes in DMEM containing 4.5 g/l glucose, 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycine, 20 mM Hepes buffer pH 7.0, 0.085 mM β-mercaptoethanol, 500 U/ml LIF, supplemented with 15% FBS (fetal bovine serum)-defined (Lot no. AKG12143B, Hyclone) or 15% Knockout–SR serum replacement for embryonic stem cells (GIBCO). The cells were starved for 18 h in the medium supplemented with 0.5% FBS or Knockout–SR in the absence of LIF prior to stimulation by BMP2 (30 ng/ml) or BMP4 (10 ng/ml).
Histological analysis and immunohistochemistry
Embryos were fixed in 4% paraformaldehyde/PBS, embedded in paraffin and sectioned at 3–6 µm. Sections were stained with haematoxylin and eosin for histological analysis or used for immunohistochemistry with the following antibodies: SP-C (sc-7705, Santa Cruz), WT1 (sc-192, Santa Cruz), α-smooth muscle actin (A2547, Sigma) and Ki67 (NCL-L-Ki67 MM1, Novocastra).
Northern blot analysis and in situ hybridization
Northern blot analysis was performed according to standard protocols. Total RNA was isolated with Trizol reagent (Invitrogen) from mouse tissues or ES cells. In situ hybridization was carried out using digoxigenin-labelled antisense RNA probes as described (Wilkinson, 1992). The Sftpd cDNA was obtained from the genome resource center RZPD, Germany (EST BG518956).
Whole mount PECAM staining
Heart whole mount PECAM staining was performed according to Ehler et al. (Ehler et al., 1999) with the following modifications. Hearts were fixed in 4% paraformaldehyde/PBS for 1.5 h, treated with hyaluronidase (1 mg/ml, Sigma) in PBS for 45 min, permeabilized with PBS containing 0.2% Triton-X100 for 45 min, and bleached with 5% H2O2 for 2–3 h at room temperature. Following bleaching, the hearts were washed with PBT (PBS containing 0.002% Triton X-100), blocked in 5% goat serum/1% BSA in PBS and incubated with anti-PECAM antibody (MEC 13.3, Pharmingen) overnight at 4°C. On the next day, they were washed several times in PBT and incubated with rat IgG antibody coupled to horseradish peroxidase (Jackson Immuno Research) overnight at 4°C. After several washes in PBT, the peroxidase developing reaction was carried out using DAB as a substrate.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank Dr Birgit Ledermann for blastocyst injection of CBP mutant ES cells, and Anita Nussbaumer and Marianne Antony for maintenance of the mouse colony. We are grateful to Drs R.Benezra, A.Erkner, E.Hara, B.Hogan, C.-c.Hui, A.McMahon, M.Scott and G.Suske for providing hybridization probes, to Owen Sansom for advice on processing of intestinal samples, to Drs Jean-Claude Perriard and Elisabeth Ehler for discussion of cardiac malformations, and to Fritz Ochsenbein for the artwork. This work was supported by a postdoctoral fellowship of the Roche Research Foundation to N.S., a grant from the Forschungskommission of the University of Zurich to J.-F.R., a grant of the Julius Klaus Foundation to R.K., and by a START fellowship and grant from the Swiss National Science Foundation to R.E.
References
- Akimaru H., Chen,Y., Dai,P., Hou,D.X., Nonaka,M., Smolik,S.M., Armstrong,S., Goodman,R.H. and Ishii,S. (1997) Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature, 386, 735–738. [DOI] [PubMed] [Google Scholar]
- Allfrey V.G., Faulkner,R. and Mirsky,A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bragg A.D., Moses,H.L. and Serra,R. (2001) Signaling to the epithelium is not sufficient to mediate all of the effects of transforming growth factor β and bone morphogenetic protein-4 on murine embryonic lung development. Mech. Dev., 109, 13–26. [DOI] [PubMed] [Google Scholar]
- Ehler E., Rothen,B.M., Hämmerle,S.P., Komiyama,M. and Perriard,J.-C. (1999) Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J. Cell Sci., 112, 1529–1539. [DOI] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Munsterkotter,M. and Horz,W. (1999) Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J., 18, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönroos E., Hellman,U., Heldin,C.-H. and Ericsson,J. (2002) Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell, 10, 483–493. [DOI] [PubMed] [Google Scholar]
- Grossman S.R., Deato,M.E., Brignone,C., Chan,H.M., Kung,A.L., Tagami,H., Nakatani,Y. and Livingston,D.M. (2003) Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science, 300, 342–344. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Neely,K.E. and Workman,J.L. (2001) Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell, 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Hautmann M.B., Madsen,C.S. and Owens,G.K. (1997) A transforming growth factor beta (TGFβ) control element drives TGFβ-induced stimulation of smooth muscle alpha -actin gene expression in concert with two CArG elements. J. Biol. Chem., 272, 10948–10956. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollnagel A., Oehlmann,V., Heymer,J., Ruther,U. and Nordheim,A. (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem., 274, 19838–19845. [DOI] [PubMed] [Google Scholar]
- Itoh S., Itoh,F., Goumans,M.J. and Ten Dijke,P. (2000) Signaling of transforming growth factor-beta family members through smad proteins. Eur. J. Biochem., 267, 6954–6967. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Boussouar,F., Ney,P.A., Jackson,C.W., Rehg,J., van Deursen,J.M. and Brindle,P.K. (2002) A transcription-factor-binding surface of coactivator p300 is required for hematopoiesis. Nature, 419, 738–743. [DOI] [PubMed] [Google Scholar]
- Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.E., Sieff,C.A., Livingston,D.M. and Yao,T.P. (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- Kuo M.-H., Zhou,J., Jambeck,P., Churchill,M.E.A. and Allis,D.C. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M., Pichel,J.G., Gorman,J.R., Sauer,B., Okamoto,Y., Lee,E., Alt,F.W. and Westphal,H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. USA, 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y., Lei,L., Westphal,H. and Chiang,C. (1998) Sonic hedgehog is essential to foregut development. Nat. Genet., 20, 58–61. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Recillas-Targa,F., Prioleau,M.-N. and Felsenfeld,G. (2001) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J., 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlam H.W., Taylor,M.H., Tanner,K.G., Denu,J.M., Goodman,R.H. and Smolik,S.M. (2002) The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosphila melanogaster. Mol. Cell. Biol., 22, 3832–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.W., McInnes,L., Kreidberg,J., Hastie,N.D. and Schedl,A. (1999) YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development, 126, 1845–1857. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.-Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Oike Y. et al. (1999) Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood, 93, 2771–2779. [PubMed] [Google Scholar]
- Pepicelli C.V., Lewis,P.M. and McMahon,A.P. (1998) Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr. Biol., 8, 1083–1086. [DOI] [PubMed] [Google Scholar]
- Petrij F. et al. (1995) Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature, 376, 348–351. [DOI] [PubMed] [Google Scholar]
- Reese D.E., Mikawa,T. and Bader,D.M. (2002) Development of the coronary vessel system. Circ. Res., 91, 761–768. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Iyer,V.R., Brown,P.O. and Struhl,K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of ESA1 histone acetylase. Mol. Cell, 6, 1297–1307. [DOI] [PubMed] [Google Scholar]
- Roberts D.J. (2000) Molecular mechanisms of development of the gastrointestinal tract. Dev. Dyn., 219, 109–120. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Roth J.-F. et al. (2003) Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J., 22, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron,U. and Rajewski,K. (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res., 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C.A. and Bhakta,M.G. (1995) Cardiac abnormalities in the Rubinstein–Taybi syndrome. Am. J. Med. Genet., 59, 346–348. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse,I., Hongo,T., Xu,M.-J., Nakahata,T., Maekawa,T. and Ishii,S. (2000) Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev., 95, 133–145. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse,I., Maekawa,T., Masuya,H., Shiroishi,T. and Ishii,S. (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein–Taybi syndrome. Proc. Natl Acad. Sci. USA, 94, 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian S.G., Deconinck,A.E., Tanaka,M., Schinke,M., Litovsky,S.H., Izumo,S., Fujiwara,Y. and Orkin,S.H. (2000) FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell, 101, 729–739. [DOI] [PubMed] [Google Scholar]
- Turner B. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- van de Wetering M. et al. (2002) The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell, 111, 241–250. [DOI] [PubMed] [Google Scholar]
- Victor M., Bei,Y., Gay,F., Calvo,D.,C.,M. and Shi,Y. (2002) HAT activity is essential for CBP-1-dependent transcription and differentiation in Caenorhabditis elegans. EMBO Rep., 3, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1999) A function of CBP as a transcriptional co-activator during Dpp signalling. EMBO J., 18, 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu,L. and Berger,S.L. (1998) Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev., 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., Schwarz,M., Tefft,D., Flores-Delgado,G. Anderson,K.D. and Cardoso,W.V. (2000) The molecular basis of lung development. Mech. Dev., 92, 55–81. [DOI] [PubMed] [Google Scholar]
- Weaver M., Yingling,J.M., Dunn,N.R., Bellusci,S. and Hogan,B.L.M. (1999) Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development, 126, 4005–4015. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G. (1992) In Situ Hybridization: A Practical Approach. Oxford University Press, Oxford, UK. [Google Scholar]
- Xu W., Edmondson,D.G., Evrard,Y.A., Wakamiya,M., Behringer,R.R. and Roth,S.Y. (2000) Loss of Gcn512 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet., 26, 229–232. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Yamauchi,J., Kuwata,T., Tamura,T., Yamashita,T., Bae,N., Westphal,H., Ozato,K. and Nakatani,Y. (2000) Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl Acad. Sci. USA, 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Yao T.-P., Oh,S.P., Fuchs,M., Zhou,N.-D., Ch’ng,L.-E., Newsome,D., Bronson,R.T., Livingston,D.M. and Eckner,R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- Zhou L., Dey,C.R., Wert,S.E. and Whitsett,J.A. (1996) Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-β 1 chimeric gene. Dev. Biol., 175, 227–238. [DOI] [PubMed] [Google Scholar]