Abstract
The mechanism of prokaryotic chromosome segregation is not known. MreB, an actin homolog, is a shape-determining factor in rod-shaped prokaryotic cells. Using immunofluorescence microscopy we found that MreB of Escherichia coli formed helical filaments located beneath the cell surface. Flow cytometric and cytological analyses indicated that MreB-depleted cells segregated their chromosomes in pairs, consistent with chromosome cohesion. Overexpression of wild-type MreB inhibited cell division but did not perturb chromosome segregation. Overexpression of mutant forms of MreB inhibited cell division, caused abnormal MreB filament morphology and induced severe localization defects of the nucleoid and of the oriC and terC chromosomal regions. The chromosomal terminus regions appeared cohered in both MreB-depleted cells and in cells overexpressing mutant forms of MreB. Our observations indicate that MreB filaments participate in directional chromosome movement and segregation.
Keywords: actin/chromosome segregation/MreB/oriC/terC
Introduction
In eukaryotic cells, the mitotic spindle apparatus segregates sister chromatids to daughter cells; however, it is largely unknown how prokaryotic cells segregate their chromosomes. Early on, the replicon model suggested that newly replicated sister chromosomes are attached to centrally located sites on the cell membrane that move towards opposite cell poles in parallel with cell elongation (Jacob et al., 1963). In this model, the process of chromosome segregation is essentially passive. In support of the replicon model, it was observed that nucleoids segregated slowly concomitantly with cell elongation (Van Helvoort and Woldringh, 1994; Woldringh et al., 1994). Based on these reports it was suggested that the coupling between transcription and translation of genes specifying co-translationally secreted proteins transiently anchors DNA to the membrane, which, in turn, slowly pulls the chromosomes apart as the cell elongates (Norris, 1995; Woldringh, 2002). In contrast, it has also been suggested that spooling of DNA through a replication factory located at mid-cell might be an important driving force in chromosome segregation (Gordon and Wright, 1998, 2000; Lemon and Grossman, 2000, 2001). RNA polymerase has also been proposed as a driving force for chromosome segregation (Dworkin and Losick, 2002). Recent experiments employing fluorescence microscopic techniques demonstrated that replicated copies of the oriC region moved rapidly apart from mid-cell to predetermined sites closer to the cell poles, and that this movement was independent of cell growth (Glaser et al., 1997; Gordon et al., 1997; Lewis and Errington, 1997; Webb et al., 1997, 1998; Niki and Hiraga, 1998; Teleman et al., 1998; Jensen and Shapiro, 1999; Niki et al., 2000). These observations are consistent with the existence of a bacterial mitotic-like apparatus, which provides the force and directionality required for the accurate segregation of chromosomal origin DNA (Sharpe and Errington, 1999; Draper and Gober, 2002). Thus, three different contemporary models have attempted to explain chromosome segregation in prokaryotic cells.
Several protein candidates have been suggested to accomplish chromosome segregation. For example, Escherichia coli MukB mutants exhibited a severe chromosome segregation defect (Niki et al., 1991), but later reports showed the MukB protein to be involved in supercoiling-dependent chromosome condensation (Weitao et al., 1999; Sawitzke and Austin, 2000). Most bacterial chromosomes encode homologs of plasmid-borne partitioning loci (Gerdes et al., 2000; Hayes, 2000; Yamaichi and Niki, 2000). For example, soj and spo0J of Bacillus subtilis are homologs of the P1 parAB genes. Soj, an oscillating protein, and Spo0J, a DNA binding protein, have many of the characteristics that might be expected for components of a chromosome segregation machinery. However, Spo0J was not needed for rapid oriC movement and may instead be involved in condensation and/or organization of the oriC proximal region (Lin and Grossman, 1998; Webb et al., 1998; Wu and Errington, 2002). Some γ-proteobacteria, such as E.coli and Haemophilus influenzae, do not carry parAB homologous loci on their chromosomes (Gerdes et al., 2000), yet these bacteria exhibit proper chromosome segregation. Thus, despite considerable effort, a single cellular machinery responsible for rapid movement of bacterial chromosomes has not yet been identified.
Plasmid-encoded partitioning loci (par) are of two types, those that encode an ATPase homologous to the oscillating Soj protein of B.subtilis and those that encode an actin-like ATPase (Gerdes et al., 2000). Recently, we obtained evidence that the DNA segregation machinery encoded by E.coli plasmid R1 specifies a simple prokaryotic analog of the eukaryotic spindle apparatus. The plasmid-encoded ParM protein, an actin homolog, formed F-actin-like filaments that were responsible for the active movement of plasmid copies to opposite cell poles (Jensen and Gerdes, 1999; Moller-Jensen et al., 2002; van den Ent et al., 2002). Interestingly, ParM belongs to the actin superfamily of ATPases encompassing also the prokaryotic, filament-forming MreB protein (Bork et al., 1992; Jones et al., 2001; van den Ent et al., 2001). Database analyses using MreB as a query sequence revealed plasmid-encoded partitioning proteins, in both Gram-negative and Gram-positive bacteria, that are more related to the MreB family of proteins than to each other (Moller-Jensen et al., 2002). The plasmid-borne mreB homologs are encoded by par loci, indicating that they, as in the case of parM of R1, may encode filament-forming proteins that segregate plasmid DNA (Moller-Jensen et al., 2002).
MreB is present in all rod-shaped bacteria and is required to maintain the non-spherical shape of the cells. Thus, the disruption of mreB in E.coli and B.subtilis was associated with a change in cell morphology from the normal rod shape to a rounded, inflated form (Wachi et al., 1987; Doi et al., 1988; Jones et al., 2001). mreB is the first gene in the mreBCD operon in both E.coli and B.subtilis (Figure 1A). MreB of B. subtilis formed helical filamentous structures lying beneath the cell surface (Jones et al., 2001). Furthermore, MreB from Thermotoga maritima adopted the canonical actin fold and formed filaments in vitro (van den Ent et al., 2001).
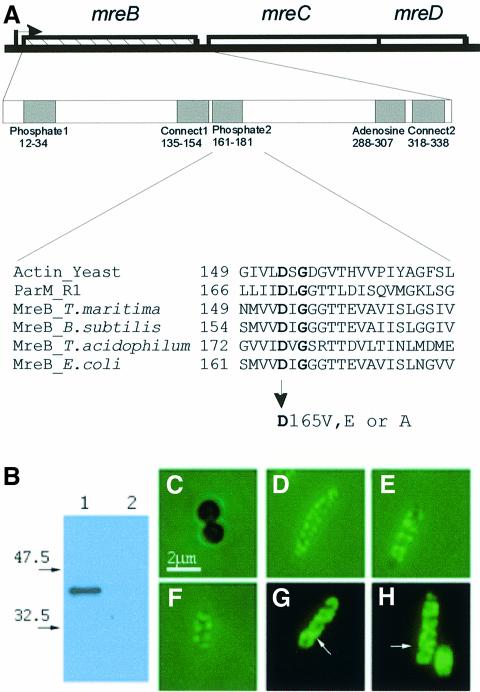
Fig. 1. Subcellular localization of the MreB protein in E.coli. (A) Schematic drawing of the mreBCD locus of E.coli. The mreB gene (347 codons) is shown as a hatched box, mreC (367 codons) and mreD (162 codons) as open boxes. The broken arrow pointing right-wards indicates the mreBCD promoter. The first enlargement shows the five regions in MreB that exhibit similarity to analogous regions present in all proteins with the actin fold (Bork et al., 1992). Numbers are amino acids in MreB. The second enlargement shows an alignment of the phosphate-2 regions of actin (from yeast), ParM from plasmid R1, MreBs from T.maritima (a deep branching eubacterium), B.subtilis, Thermoplasma acidophilum (an archaeon) and E.coli. Conserved amino acids are in bold. Amino acid changes at position +165 in the phosphate-2 region of MreB of E.coli are indicated. (B) Immunoblot showing the specificity of the affinity purified anti-MreB antibodies. Lane 1, protein extract from wild-type cells (MC1000); lane 2, MC1000ΔmreB. Molecular weight markers are shown to the left. (C–H) Subcellular localization of MreB visualized by combined phase-contrast and immunofluorescence microscopy using affinity purified anti-MreB antibodies. (C) Cells of MC1000ΔmreB; (D–F) cells of MC1000; (G and H) cells of MC1000 visualized by fluorescence microscopy in the absence of phase-contrast. Arrows point to helical structures. Cells were grown at 30°C in AB glycerol medium supplemented with 0.025% casamino acids.
Here, we investigated the role of MreB in cell-shape determination and chromosome segregation in E.coli. We found that MreB formed helical filaments that extended along the long axis of the cells. Flow-cytometric analyses of cells lacking MreB indicated that, in spherical cells, chromosomes segregated in pairs. Direct cytological localization of oriC and terC in round cells was consistent with paired segregation of chromosomes. Overexpression of wild-type MreB inhibited cell division but did not perturb nucleoid morphology or DNA segregation. In contrast, overexpression of mutant forms of MreB in wild-type cells inhibited cell division and concomitantly caused aberrant MreB filament morphology, impaired nucleoid segregation and severe mislocalization of the chromosomal origin and terminus regions. Thus, our results raise the possibility that MreB is directly or indirectly involved in chromosome movement and segregation.
Results
MreB forms helical structures
Polyclonal antibodies were raised against purified, affinity-tagged MreB of E.coli and their specificity verified by immunoblotting. In extracts of wild-type cells, the anti-MreB antibodies specifically recognized a single protein of the expected size (Figure 1B, lane 1). In extracts prepared from cells carrying an in-frame deletion in mreB (ΔmreB), no protein was detected (Figure 1B, lane 2). Control immunofluorescence microscopy (IFM) performed on ΔmreB cells also confirmed the specificity of the antiserum (Figure 1C). In accordance with previous results (Wachi et al., 1987; Doi et al., 1988), E.coli cells lacking MreB were spherical (Figure 1C) and 4′,6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI) staining of DNA showed that 1–2% of the spheres were anucleate (data not shown).
IFM of wild-type cells using the MreB-specific antibodies revealed a range of distinct structures that included transverse bands and pairs of dots that were distributed in a criss-cross pattern extending along the long axis of the cells (Figure 1D–H). Reduction in light intensity enabled the visualization of structures that were clearly helical (Figure 1G and H). These results suggested that the MreB structures located close to the cell surface. As MreB is a cytosolic protein, the helices probably located to the inner surface of the cytoplasmic membrane. The helical pitch based on measurements of 95 clearly defined structures was estimated to be 0.46 ± 0.08 µm.
To assess whether a sufficient number of MreB molecules were present to account for the formation of these helical structures, the cellular amount of MreB was determined by quantitative immunoblotting. The amount of MreB was estimated as ∼17 000 molecules/cell in slowly growing cells and ∼40 000 molecules/cell in rapidly growing cells (data not shown). Assuming a longitudinal monomer spacing of 51 Å within MreB filaments (van den Ent et al., 2001), there seemingly is sufficient MreB to form a filament that could encircle the cell several times. We conclude that MreB monomers assemble into, or adhere to, helical filamentous structures that extend along the longitudinal axis of E.coli cells just beneath the surface of the cytoplasmic membrane.
In B.subtilis, depletion of MreB led to a rounded, inflated cell morphology and eventually to cell lysis (Jones et al., 2001). Thus, in B.subtilis, mreB is essential. Using a conditional mreB expression system, we found that depletion of MreB conferred a spherical cell morphology and a significant reduction (5-fold) in cell plating efficiency (to be published elsewhere).
MreB deficient cells contain even numbers of replication origins
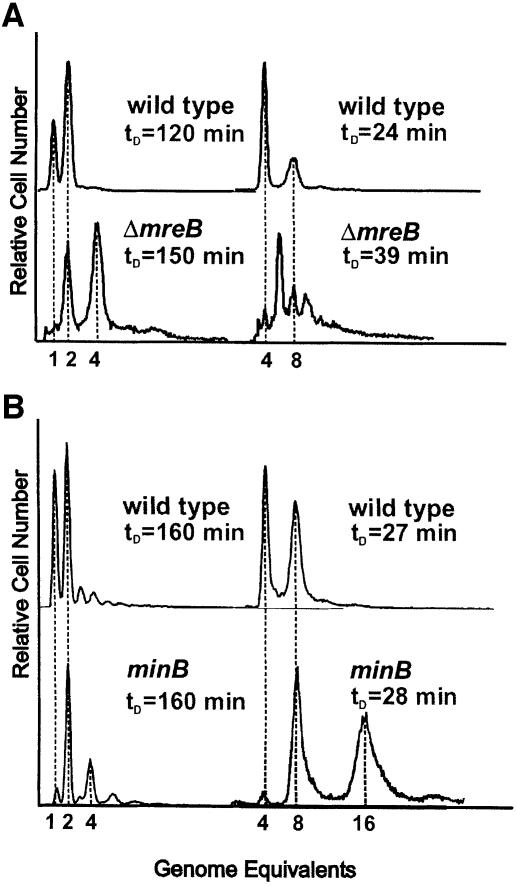
Numbers of replication origins in wild-type and ΔmreB cells were determined by flow cytometry. The method used takes advantage of the fact that rifampicin stops new rounds of replication initiations at oriC but allows ongoing replication forks to finish. Wild-type cells grown in minimal medium had a doubling time of 120 min and contained predominantly one or two origins (Figure 2A). When grown in rich medium, wild-type cells had a doubling time of 24 min and contained four or eight origins. This is in agreement with previously published results (Skarstad et al., 1986). In contrast, ΔmreB cells grown in minimal medium had a doubling time of 150 min and contained two or four origins (Figure 2A). When grown in rich medium, ΔmreB cells had a doubling time of 39 min and contained 2, 4, 6, 8, 10, 12 or even 14 origins. The predominant classes of cells had 4, 6, 8 or 10 origins. These observations are consistent with the proposal that, in ΔmreB cells, the origins segregated in pairs. Even more importantly, the distribution indicates that the paired chromosomes segregated randomly.
Fig. 2. Number of replication origins in single cells counted by flow cytometry. (A) Shown are flow cytometric histograms of E.coli strains MC1000 (wild type) and MC1000ΔmreB obtained after 4 h of treatment with rifampicin. Rifampicin was added to inhibit new replication initiations at oriC and to allow for run-out synthesis of ongoing rounds of DNA replication. DNA was stained with ethidium bromide. Left, cells were grown in AB glycerol minimal medium at 37°C. Right, the cells were grown in LB medium. The cell mass doubling times at the times of addition of rifampicin are indicated. (B) Flow-cytometric histograms of PB103 (wild type) and PB114 (minB) cells grown as described above.
Cell size distributions obtained by flow cytometry showed that the average size of ΔmreB cells was approximately two times that of wild-type cells in rich and poor medium. Thus, both cell size and DNA content were elevated in ΔmreB cells. Mini-cell producing strains (minB) also had an ∼2-fold increased cell size (de Boer et al., 1989; this work). We investigated the oriC distribution pattern in PB103 (wild type) and PB114 (minB) by flow cytometry (Figure 2B). Slowly growing minB cells had two or four origins, whereas rapidly growing minB cells had 4, 8 or 16 origins, but never 6, 10 or 12. Thus, minB cells exhibited the 2n distribution of replication origins similar to that of wild-type cells, indicating that the chromosome segregation machinery was operational even with an increased number of chromosomes. We conclude that the even number of replication origins of MreB-depleted cells is not a simple consequence of an increased number of chromosomes and cell volume.
Subcellular localization of oriC and terC in ΔmreB cells
To determine the subcellular localization of specific chromosomal regions, the bacteriophage P1 partitioning site, parS, was inserted near oriC (in the bglF gene at 84 min) or terC (in the relBE operon at 34 min) in both wild-type and ΔmreB cells (see Materials and methods). The ParB protein of plasmid P1 binds to the parS site and spreads outward to cover adjacent sequences (Rodionov et al., 1999). When a Gfp–ParB fusion protein was expressed from a co-resident plasmid, the DNA region harboring parS formed a bright fluorescent signal, which thus allowed visualization of parS containing DNA segments (Li et al., 2002). Gfp–ParB synthesis was induced for 3 h of growth before inspection by fluorescence microscopy.
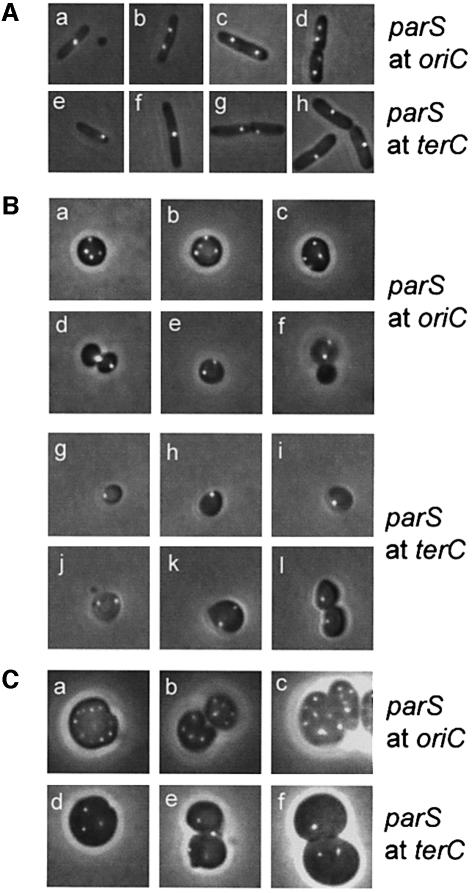
In minimal medium, wild-type cells had either one oriC focus at mid-cell or two foci near the one-quarter and three-quarter positions (Figure 3Aa and b). In rich medium, newborn wild-type cells had two oriC foci located at the one-quarter and three-quarter positions, whereas predivisional cells had four foci evenly distributed (Figure 3Ac and d). Newborn cells invariably had one terC focus, in both minimal and rich media, indicating that terminus segregation and septum formation occur in parallel. In minimal medium, the focus was located near a cell pole (Figure 3Ae), whereas in rich medium, the terC focus was always located at mid-cell (Figure 3Ah). Predivisional cells grown in minimal medium either had one centrally located terC focus (Figure 3Af) or two foci located close to each other at mid-cell (Figure 3Ag). These patterns of oriC and terC localization in wild-type cells are consistent with previous observations (Niki et al., 2000; Li et al., 2002), thus validating the Gfp–ParB/parS-based method of DNA detection used here.
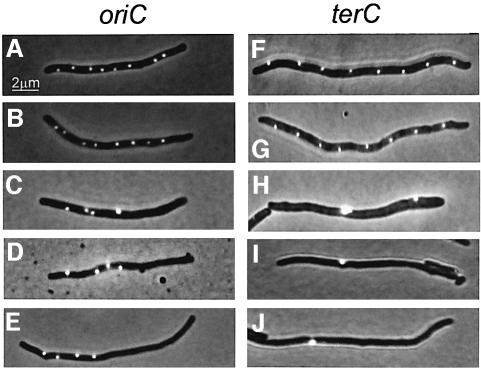
Fig. 3. Subcellular localization of oriC and terC. oriC and terC regions were visualized by Gfp–ParB nucleation on parS sites inserted in bglF (near oriC) or in relBE (near terC). All strains shown contained plasmid pTK536 that expressed a Gfp–ParB fusion protein. Cells were grown at 37°C in AB glycerol or LB medium with a doubling time of 145 or 27 min (wild type) and 174 or 44 min (ΔmreB), respectively. (A) Expression of Gfp–ParB was induced with 0.2% arabinose 180 min before inspection. (a and b) MC1000 with parS inserted near oriC grown in AB glycerol. (c and d) Same strain grown in LB medium. (e–g) MC1000 with parS inserted near terC grown in AB glycerol. (h) Same strain grown in LB medium. (B) MC1000ΔmreB cells with parS near oriC (a–f) or terC (g–l) grown in AB glycerol. (C) MC1000ΔmreB cells with parS inserted at oriC (a–c) or terC (d–f) grown in LB medium.
Grown in minimal medium, ΔmreB cells invariably had two (68%) or four (31%) oriC foci (Figure 3Ba–f), consistent with the number of replication origins counted by flow cytometry (Figure 2A) and with the inference that the chromosomes segregated in pairs. Predivisional cells usually exhibited a non-symmetrical, random oriC localization pattern (Figure 3Ba–c) or even clustering at the site of division (Figure 3Bd). New-born cells with two oriC foci were also seen (Figure 3Be and f). The presence of spheres without an oriC focus indicated loss of the entire chromosome during cell division (Figure 3Bf). Spherical cells grown in rich medium had many, highly irregularly spaced oriC foci in a pattern too complex to be quantified (Figure 3Ca–c). Many cells had more than 14 origins. These cells were not detected by flow cytometry (Figure 2A), probably due to their disintegration during processing of the cell samples.
Interestingly, slowly growing ΔmreB cells only had one (71%) or two terC foci (28%) (Figure 3Bg–l). Despite thorough examination, such cells never had more than two terC foci. In these cells, growing with a 174 min doubling time, we determined the replication time for the chromosome (C-period) to be 70 ± 5 min. Thus, cells had ample time to finish chromosome replication within one doubling time and therefore contained four copies of oriC as well as terC at the time of cell division. It is therefore conceivable that each terC focus in predivisional cells represents two terC regions in close proximity. Fast growing ΔmreB cells had an irregular terC focus pattern, too complex to be quantified (Figure 3Cd–f). However, these cells clearly had fewer terC than oriC foci, also consistent with cohesion of the terminus regions.
Mutant MreB proteins perturb wild-type MreB filament morphology
Next, we investigated MreB filament morphology in wild-type cells expressing wild-type or mutant MreBs ectopically. The conserved aspartic acid residue at position 165 in MreB is located in the phosphate-2 domain that forms part of the ATP binding domain in all members of the actin ATPase family (Bork et al., 1992; see Figure 1A). Mutations at the similar position in ParM of plasmid R1 inactivates the par locus and the mutant ParM protein exhibits trans-dominance (Jensen and Gerdes, 1997). Using site-directed mutagenesis, the aspartic acid residue was changed to valine (a change to an uncharged amino acid of similar size), to glutamate (a conservative change) and to alanine (a change to a small uncharged amino acid). The mutated and native mreB genes were cloned downstream of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pA1/O3/O4-promoter in the low-copy-number expression vector pNDM220 (Gotfredsen and Gerdes, 1998). To verify that the mutant forms of MreB were actually defective, we tested whether the proteins were able to complement the cell-shape defect conferred by the ΔmreB deletion. Although a wide range of IPTG concentrations were used for induction, even the wild-type mreB allele did not fully complement the ΔmreB mutation (data not shown). The reason for this lack of complementation is not known, but a similar lack of complementation was seen in the case of B.subtilis mreB (Jones et al., 2001). To circumvent this problem, we cloned the entire mreBCD operon into our expression vector. In this case, transcription of the wild-type mreBCD operon fully complemented the defects of the ΔmreB mutation (data not shown). Transcription of the mreBCD operon encoding the mutant forms of MreB described above, however, did not complement the ΔmreB deletion (data not shown). Thus, the single amino acid changes at +165 in all cases inactivated MreB.
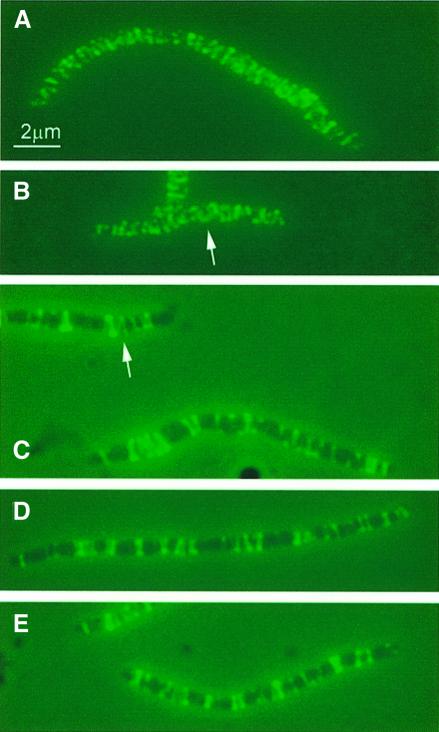
Wild-type cells carrying wild-type or mutant forms of mreB under the control of the IPTG-inducible promoter were grown at an intermediate level of induction (100 µM IPTG) for five mass doublings. For convenience, the strains were denoted: wild type/mreB165D, wild type/mreB165V, wild type/mreB165E and wild type/mreB165A, respectively. Combined phase-contrast and IFM were performed on the merodiploid cells using MreB-specific antibodies. Overexpression of the wild-type form of MreB resulted in the formation of elongated cells, indicating that cell division was inhibited (Figure 4B). This effect was in agreement with previous findings (Wachi and Matsuhashi, 1989). MreB formed helical structures that were present over the entire length of the elongated cells, very similar to the MreB filaments seen in wild-type cells treated with the cell division inhibitor cephalexin (Figure 4A). Wild-type cells expressing mutant MreB proteins ectopically also resulted in the formation of elongated cells, again indicating a defect in cell division (Figure 4C–E). MreB filament formation was, however, perturbed in these cells. It was still possible to detect clearly defined fluorescent structures as dots, transverse bands and even helices, but the helices were unevenly distributed, leaving parts of the cells with little or no detectable MreB signal (helical structures are indicated with arrows in Figure 4B and C). Thus, ectopic expression of mutant MreB proteins in a wild-type background interfered with MreB filament formation and cell division while the rod shape of the cells was maintained.
Fig. 4. MreB localization in elongated cells visualized by immuno-fluorescence microscopy. Subcellular localization of MreB in wild-type cells treated with cephalexin or in wild-type cells expressing native or mutated forms of mreB present in the low-copy-number expression vector pNDM220. Cells were grown at 30°C in AB glycerol medium supplemented with 0.025% casamino acids. Cells were induced with 100 µM of IPTG for five mass doublings. Localization of MreB was performed using affinity purified anti-MreB antibodies and combined phase-contrast and immunofluorescence microscopy. Arrows point to unmistakable helical structures. (A) Cells of MC1000 (mreBCD+) were treated with 10 µg/ml cephalexin for three mass doublings prior to microscopy. (B) MC1000/pTK505 (wild-type mreB); (C) MC1000/pTK509 (mreB165V); (D) MC1000/pTK510 (mreB165E); (E) MC1000/pTK511 (mreB165A).
Interestingly, the diameter of the elongated cells expressing the mutant forms of MreB varied significantly (Figures 4–6). Thus, wild-type cells treated with cephalexin and wild-type cells expressing native MreB all had an average diameter of 0.83 ± 0.04 µm. In contrast, wild-type cells expressing MreB165E and MreB165A had significantly smaller diameters (0.69 ± 0.03 and 0.60 ± 0.06 µm, respectively), whereas cells expressing MreB165V had a significantly larger diameter (1.09 ± 0.07 µm). Thus, MreB controls not only cell length but also cell width.
Fig. 6. Defect localization of oriC and terC in cells expressing mutant forms of MreB. Localization of oriC and terC by Gfp–ParB labeling in wild-type cells treated with cephalexin or in wild-type cells expressing native or mutated forms of mreB present in expression vector pNDM220. Cells were grown at 30°C in AB glycerol medium supplemented with 0.025% casamino acids. MreB synthesis was induced with IPTG (100 µM) for five mass doublings and Gfp–ParB synthesis (from pTK536) with 0.2% arabinose for 180 min. (A–E) Cells with parS inserted into bglF (near oriC); (F–J) cells with parS inserted into relBE (near terC). (A) MC1000bglF::parS/pTK536 cells treated with 10 µg/ml cephalexin for three mass doublings; (B) MC1000bglF::parS/pTK536/pTK505 (wild-type mreB); (C) MC1000bglF::parS/pTK536/pTK509 (mreB165V); (D) MC1000bglF::parS/pTK536/pTK510 (mreB165E); (E) MC1000bglF::parS/pTK536/pTK511 (mreB165A); (F) MC1000relBE:: parS/pTK536 treated with 10 µg/ml cephalexin; (G) MC1000relBE:: parS/pTK536/pTK505 (wild-type mreB); (H) MC1000relBE::parS/pTK536/pTK509 (mreB165V); (I) MC1000relBE::parS/pTK536/pTK510 (mreB165E); (J) MC1000relBE::parS/pTK536/pTK511 (mreB165A).
Interference with MreB filament formation results in aberrant nucleoid morphology and positioning
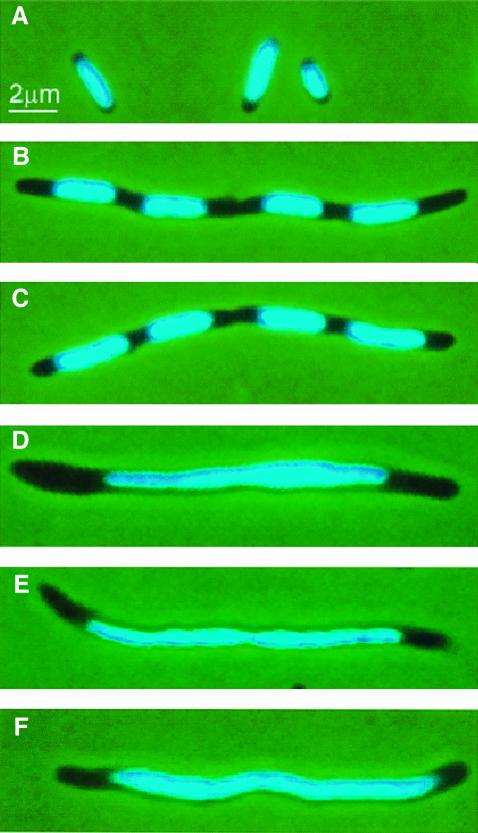
Nucleoid morphology was visualized by DAPI staining. Slowly growing wild-type cells contained one centrally located nucleoid (Figure 5A). Treatment of these cells with cephalexin for two to three mass doublings resulted in filamentous cells with nucleoids that were well defined and evenly spaced (Figure 5B). A similar cell and nucleoid morphology was observed in cells overexpressing wild-type MreB, indicating that chromosome segregation occurred despite the inhibition of cell division (Figure 5C). MreB proteins with the aspartic acid residue at +165 substituted with valine, glutamate or alanine expressed in a wild-type background produced elongated cells as described above. In these cells, however, nucleoid morphology was highly perturbed (Figure 5D–F). In all three cases, the nucleoids appeared as single diffuse and elongated entities without any clearly demarcated borders. We conclude that interference with MreB filament formation prevented nucleoid separation.
Fig. 5. Inhibition of nucleoid segregation by mutant forms of MreB. Nucleoid distribution in cells of MC1000 (mreBCD+) treated with cephalexin or MC1000 expressing native or mutated forms of mreB present in the expression vector pNDM220. Cells were grown at 30°C in AB glycerol medium supplemented with 0.025% casamino acids and stained with DAPI. MreB synthesis was induced with IPTG (100 µM) for five mass doublings. (A) MC1000 wild-type cells grown exponentially; (B) MC1000 wild-type cells were treated with 10 µg/ml cephalexin for three mass doublings prior to microscopy; (C) cells of MC1000/pTK505 (wild-type mreB); (D) MC1000/pTK509 (mreB165V); (E) MC1000/pTK510 (mreB165E); (F) MC1000/pTK511 (mreB165A).
The MreB filament defect causes abnormal localization of oriC and terC
Using the Gfp–ParB/parS system described above, we investigated the localization patterns of oriC and terC in cells overexpressing either wild-type or mutant forms of MreB in an otherwise wild-type background. MreB and Gfp–ParB syntheses were induced as described above for Figures 3 and 4, respectively. In cells overexpressing wild-type MreB, the origin- and terminus-proximal parS sites were observed to distribute as multiple, equally spaced foci along the long axis of the elongated cells (Figure 6B and G). A similar origin and terminus distribution was observed in cells treated with cephalexin (Figure 6A and F). These observations indicate that, in elongated cells, cell division was dispensable for the segregation and regular distribution of oriC and terC proximal regions.
In contrast, in the wild type/mreB165V, wild type/mreB165E and wild type/mreB165A strains, the origin-proximal parS sites were found to be severely dislocated (Figure 6C–E). Furthermore, the oriC proximal foci emitted a brighter fluorescent signal in these cells as compared with cells overexpressing wild-type MreB or cells treated with cephalexin, suggesting that at least some of the foci represented more than a single origin region. This result indicates a defect in the segregation of newly replicated copies of the oriC region.
The localization defect of the terminus region was even more dramatic (Figure 6H–J). Here, ectopic expression of the mutant MreBs caused the terminus regions to appear as single large and brightly fluorescent aggregates in the majority of the cells. Thus, interference with MreB function caused terminus cohesion.
Discussion
We show here that native MreB forms helical filaments that are required to maintain the non-spherical form of E.coli cells. Recently, a YFP–MreB fusion protein was shown to form helices similar to the ones described here (Shih et al., 2003). In B subtilis, MreB formed large, right-handed helices that were required to maintain cell shape (Jones et al., 2001). Mbl, a helix-forming MreB paralog in B.subtilis, was also required to maintain the shape of the cells (Jones et al., 2001; Carballido-Lopez and Errington, 2003) and was responsible for directing lateral cell wall synthesis in a helical pattern (Daniel and Errington, 2003). The MreB and Mbl filaments were responsible for width and linear axis control, respectively. We found here that expression of mutant forms of E.coli MreB resulted in altered width of the cells (Figures 4–6). Thus, MreB of E.coli also appears to control cell width. E.coli does not have MreB paralogs, suggesting that MreB of E.coli simultaneously controls cell width and linear axis. We conclude that the MreB family of proteins forms helical filaments pivotal in maintaining cell morphology in both Gram-negative and -positive bacteria.
It is not known how MreB filaments determine cell shape. The morphology of E.coli ΔmreB cells is very similar to that of cells lacking either penicillin-binding-protein-2 (PBP2) or RodA. In both cases, lateral cell wall synthesis was impaired (De Pedro et al., 2001). One attractive model is that MreB provides a scaffold that organizes the peptidoglycan synthesizing machinery (including PBP2) during cell elongation and thereby co-ordinates lateral cell wall growth with extension of the cell interior (Carballido-Lopez and Errington, 2003; Daniel and Errington, 2003).
MreB functions in chromosome segregation
The homology between MreB and the DNA segregation protein ParM of plasmid R1 inspired us to investigate whether MreB plays a role in chromosome segregation. The number of replication origins (oriC) in individual cells was measured by two independent methods, flow cytometry and fluorescence microscopy. Both methods showed that, in ΔmreB cells, the origin distribution was consistent with the proposal that origins segregated in pairs. Moreover, the oriC localization pattern was highly disordered in ΔmreB cells, especially at rapid growth conditions during which terC localization was disordered as well. A mini-cell producing strain, which also had an increased cell volume and number of chromosomes per cell, exhibited a normal oriC distribution pattern (Figure 2B). Thus, the disordered oriC distribution pattern in MreB-depleted cells was not a mere consequence of an increased chromosome number per cell.
In contrast, slowly growing ΔmreB cells had a terC focus number very similar to that of wild-type cells: small cells had one terC focus, while large, pre-divisional cells had two terC foci (Figure 3). Thus, in single cells, the number of oriC foci was increased without a concomitant increase in the number of terC foci. An increased chromosomal replication time of ΔmreB cells could explain the increased oriC/terC focus-ratio in these cells. Therefore, we measured the replication time in wild-type and ΔmreB cells by flow cytometry. However, the replication time of the chromosome was not significantly altered in ΔmreB cells (determined to be 70 ± 5 min for both strains; data not shown). Hence, we favor the alternative explanation that the increased oriC/terC focus ratio in ΔmreB cells reflects cohesion of the chromosomal termini regions. The cause of terminus cohesion is not known. However, terminus cohesion may be a consequence of a dysfunctional chromosome segregation machinery.
Overexpression of wild-type MreB inhibited cell division (Figures 4B and 5C). The MreB helix pattern in these elongated cells was indistinguishable from that seen in elongated cells obtained by cephalexin treatment (Figure 4A versus B). Even more importantly, the nucleoids were evenly distributed along the long axis of the elongated cells in both cases (Figure 5B and C). In stark contrast to this ‘wild-type’ nucleoid distribution pattern, overexpression of mutant MreB prevented nucleoid separation (Figure 5D–F) and at the same time perturbed the MreB filament pattern (Figure 4C–E). In cells expressing mutant MreB, bulk DNA was located in one large, coalesced nucleoid in the middle of the elongated cells. Concomitantly, oriC localization was highly disordered and the terC regions did not separate at all in these cells (Figure 6), indicating terminus cohesion. Thus, mutations in the phosphate-2 regions of the actin homologs ParM and MreB conferred DNA segregation defects in both cases—a striking coincidence. Taken together, our observations raise the possibility that MreB filaments participate directly or indirectly in chromosome segregation. However, we do not exclude the possibility that terminus cohesion reflects interference with the function of FtsK, a protein that couples separation of termini with completion of the divisional septum (Aussel et al., 2002).
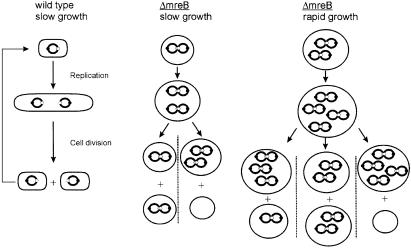
A model that explains all our observations regarding chromosome segregation in MreB depleted cells is shown in Figure 7. The model proposes that lack of MreB leads to sister chromosome cohesion, most likely via cohesion of the chromosome terminus regions. Newly replicated chromosomes stay together until a new round of replication leads to resolution of the pairs. However, the newly replicated chromosomes stay paired and the chromosomes will therefore, in all cases, segregate as paired structures. We find that our results strongly indicate that the paired chromosomes segregate randomly as depicted in Figure 7. This is because random segregation of paired chromosomes would lead to (spherical) cells containing two, four, six, etc., chromosomes. Thus, the model simultaneously explains the flow-cytometric data (Figure 2A), and the oriC and terC localization patterns (Figure 3B).
Fig. 7. Pairing explains the chromosome segregation pattern in cells lacking MreB. Chromosomes stay paired in ΔmreB cells (cohesion). The model assumes that replication allows separation of the old chromosomes and that the newly replicated sister chromosomes stay together as pairs. Pairing explains the even number of chromosomes in the ΔmreB strain seen at all growth rates. During division of ΔmreB cells, paired chromosomes segregate randomly, thus leading to anucleate cells and to cells with an even number of chromosomes. The lack of coordination of chromosome segregation also explains the broad copy-number distribution of chromosomes seen in the ΔmreB cells. The schematic does not show all possible combinations of the number of chromosomes in rapidly growing ΔmreB cells. Symbols: chromosomes are shown as large circles, origins of replication as closed dots and termini as open dots. For simplicity, chromosomes are shown as full circles and the figure does not take ongoing replication into account.
How do mutant MreB proteins interfere with MreB filament formation and function? MreB from T.maritima formed filaments in vitro (van den Ent et al., 2001), and it is reasonable to assume that the filamentous structures observed here (Figure 4) and by Jones et al. (2001) consist of MreB. We infer that the functionally inactive mutant MreB proteins retained the ability to interact with wild-type MreB and, doing so, interfered with its proper function. This interpretation is supported by the altered MreB filament morphology seen when mutant MreB proteins were expressed in wild-type cells (Figure 4C–E). In the case of the filament-forming ParM protein of plasmid R1, point mutations at the same position in the phosphate-2 region simultaneously inactivated its ATPase activity and its ability to sustain plasmid DNA segregation (Jensen and Gerdes, 1997; Moller-Jensen et al., 2002). Strikingly, the mutant ParM proteins also interfered with the function of wild-type ParM, indicating that wild-type and mutant ParM formed inactive complexes (Jensen and Gerdes, 1997).
What is the role of MreB in chromosome segregation? It is possible that the defect shape of ΔmreB cells affects chromosome segregation indirectly. However, the observation that rod-shaped cells expressing mutant MreBs do not segregate their chromosomes properly indicates that it is not the shape of the spherical cells per se that causes the chromosome segregation defect. Our results raise the possibility that MreB plays a more direct role in chromosome segregation analogous to what has been described for its plasmid-encoded homolog ParM (Moller-Jensen et al., 2002; van den Ent et al., 2002). One straightforward interpretation is that the actin-like MreB filaments actively move chromosomal DNA in opposite directions in co-ordination with lateral cell wall growth. Such a mechanism would require that MreB filaments are dynamic. This was shown recently to be the case for the Mbl filaments of B.subtilis (Carballido-Lopez and Errington, 2003).
Materials and methods
Bacterial strains
The standard E.coli K-12 strain MC1000 [araD139 Δ(ara, leu)7697 ΔlacX74 galU galK atrA] was used throughout (Casadaban and Cohen, 1980). MC1000ΔmreB contains an in-frame deletion in which all but the 3′ and 5′ 36 base pairs were deleted. The deletion was constructed by employing the ‘one-step inactivation of chromosomal genes’ procedure (Datsenko and Wanner, 2000). Briefly, a PCR fragment containing the cat gene flanked by two FLP recognition targets (FRT) was generated using the oligonucleotides KO-1 and KO-2 with pKD3 as template (sequences of DNA oligonucleotides are available upon request). This PCR product contains 36 nucleotide extensions that are homologous to the 3′ and 5′ 36 base pairs of mreB. The PCR fragment were introduced into BW25113/pKD46, resulting in insertion of the FRT cat FRT cassette in the mreB gene. The mreB disruption was transduced into MC1000 and the resistance gene deleted. The structure of the recombinants was confirmed by PCR analysis. The mini-cell producing strain PB114 (minB::aphA) was donated by Piet de Boer (de Boer et al., 1989).
Insertion of parS at oriC and terC
Using pKD3 (Datsenko and Wanner, 2000) as a template, the cat gene flanked by two FRT sites was PCR amplified using primers O1 and O2. The PCR fragment was cut with EcoRI and BamHI and inserted into pMG25 resulting in pTK532. Using P1 as a template, the parS sequence was PCR amplified using primers O3 and O4. This PCR fragment was digested with BamHI and HindIII and inserted into pTK532 resulting in pTK533. Plasmid pTK533 contains parS inserted downstream of the cat gene flanked by two FRT sites. OriC-1 and OriC-2 contain two flanking 36 nucleotide extensions homologous to sites in the bglF gene at 84 min (near oriC). TerC-1 and TerC-2 contains two flanking 36 nucleotide extensions homologous to sites in the relBE operon at 34 min (near terC). These sets of oligonucleotides were used to generate PCR fragments containing parS linked to the cat gene and the two FRT sites using pTK533 as a template. The PCR fragments were introduced into cells of BW25113. The bglF::parS and relBE::parS fusions were transduced to appropriate strains. The final strains were MC1000 bglF::parS, MC1000 relBE::parS, MC1000ΔmreB bglF::parS and MC1000ΔmreB relBE::parS.
Plasmids used and their construction
The plasmids used here are listed in Table I, and their construction is described in the Supplementary data, available at The EMBO Journal Online.
Table I. Plasmids used and constructed.
| Plasmid | Genotypes/plasmid propertiesa | Source or reference |
|---|---|---|
| pBAD33 | p15A cat araC pBAD | Guzman et al. (1995) |
| pEGFP | pUC plac::egfp bla | Clontech |
| pKD3 | cat, template plasmid | Datsenko and Wanner (2000) |
| pKD46 | bla, λ Red recombinase expression | Datsenko and Wanner (2000) |
| pNDM220 | mini-R1 bla lacIq pA1/O4/O3 | Gotfredsen and Gerdes (1998) |
| pMG25 | pUC bla lacIq pA1/O4/O3 | Laboratory collection |
| pTK500 | pMG25 pA1/O4/O3::his8::mreB bla | This work |
| pTK505 | pNDM220 pA1/O4/O3::mreB bla | This work |
| pTK509 | pTK505 pA1/O4/O3::mreB165V bla | This work |
| pTK510 | pTK505 pA1/O4/O3::mreB165E bla | This work |
| pTK511 | pTK505 pA1/O4/O3::mreB165A bla | This work |
| pTK512 | pMG25 pA1/O4/O3::mreBCD bla | This work |
| pTK514 | pTK512 pA1/O4/O3::mreB165VmreCD bla | This work |
| pTK515 | pTK512 pA1/O4/O3::mreB165EmreCD bla | This work |
| pTK516 | pTK512 pA1/O4/O3::mreB165AmreCD bla | This work |
| pTK521 | pNDM220 pA1/O4/O3::mreBCD bla | This work |
| pTK523 | pTK521 pA1/O4/O3::mreB165VmreCD bla | This work |
| pTK524 | pTK521 pA1/O4/O3::mreB165EmreCD bla | This work |
| pTK525 | pTK521 pA1/O4/O3::mreB165AmreCD bla | This work |
| pTK526 | pEGFP plac::gfp::parB bla | This work |
| pTK536 | pEGFP plac::SD-parM::gfp::parB bla | This work |
| pTK536 | pBAD33 pBAD::SD-parM::gfp::parB cat | This work |
acat denotes the chloramphenicol transacetylase gene; bla the β-lactamase gene.
Growth conditions and media
To express ParB–Gfp, cells containing pTK536 and the relevant plasmids were grown in AB minimal medium supplemented with 0.2% glycerol and 1 µg/ml thiamine or in AB minimal medium supplemented with 0.2% glycerol, 0.025% casamino acids and 1 µg/ml thiamine, as indicated. Expression of the ParB–Gfp fusion protein was induced by 0.2% arabinose. Induction for 180 min before microscopy yielded optimal results. For IFM, cells were grown in AB minimal medium.
Expression in MC1000 of native or mutant MreB from plasmids pTK505 (expresses native MreB), pTK509 (MreB165V), pTK510 (MreB165E) and pTK511 (MreB165A) was induced by adding 100 µM of IPTG. Cephalexin was added to a final concentration of 10 µg/ml.
Quantification of MreB by western blotting
For western blots, cells were grown in AB minimal medium with 0.2% glycerol, 0.025% casamino acids and 1 µg/ml thiamine to an OD450 of 0.3. Relative protein concentrations were determined by standard immunoblotting.
Fluorescence microscopy
Cells expressing the ParB–Gfp fusion protein were immobilized on microscope slides using a thin film of agarose (Glaser et al., 1997). Cells were observed with a Leica DMRA fluorescence and phase-contrast microscope with a Leica PL APO 100×/1.40 objective. Pictures were obtained with a Leica DC500 colour CCD camera and stored digitally using the Leica IM500 computer software.
For IFM, cells were grown in AB glycerol medium. Cells containing pTK505, pTK509, pTK510 and pTK511 were grown for four to five doublings with 100 µM IPTG. Cells were fixed and stained as described previously (Harry et al., 1995) with the following modifications: cells were fixed in phosphate-buffered saline (PBS) (pH 7.4) with a final concentration of 4% paraformaldehyde and 0.02% glutaraldehyde and immobilized on multi-well glass slides (VWR International) coated with poly-l-lysine. Permeabilization of cells was performed with a solution of freshly prepared 5 µg/ml lysozyme in PBS for 10 min. Affinity-purified anti-MreB antibodies and secondary Alexa488-conjugated goat anti-rabbit IgG antibodies (Molecular Probes) were used at 1:100 and 1:200 dilutions, respectively.
Flow cytometry
For the determination of numbers of origins per cell by flow cytometry, cells were grown in AB minimal medium supplemented with 0.2% glycerol or in LB medium as indicated. Prior to flow cytometry, cells were treated with 300 µg/ml rifampicin (to stop further replication initiations) and 3.6 µg/ml cephalexin (to stop further cell divisions). Flow cytometry was performed as described previously (Lobner-Olesen et al., 1989), using a Bryte HS instrument (Bio-Rad). Chromosomal replication time was measured by flow cytometry essentially as described previously (Bernander and Nordstrom, 1990).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank P.de Boer for the donation of PB103 and PB114. This work was supported by the Danish Biotechnology Instrument Centre (DABIC), The Danish Natural Research Council and The Novo Nordic Foundation.
References
- Aussel L., Barre,F.X., Aroyo,M., Stasiak,A., Stasiak,A.Z. and Sherratt,D. (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell, 108, 195–205. [DOI] [PubMed] [Google Scholar]
- Bernander R. and Nordstrom,K. (1990) Chromosome replication does not trigger cell division in E. coli. Cell, 60, 365–374. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander,C. and Valencia,A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R. and Errington,J. (2003) The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell, 4, 19–28. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen,S.N. (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol., 138, 179–207. [DOI] [PubMed] [Google Scholar]
- Daniel R.A. and Errington,J. (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell, 113, 767–776. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner,B.L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- De Pedro M.A., Donachie,W.D., Holtje,J.V. and Schwarz,H. (2001) Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol., 183, 4115–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Wachi,M., Ishino,F., Tomioka,S., Ito,M., Sakagami,Y., Suzuki,A. and Matsuhashi,M. (1988) Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J. Bacteriol., 170, 4619–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper G.C. and Gober,J.W. (2002) Bacterial chromosome segregation. Annu. Rev. Microbiol., 56, 567–597. [DOI] [PubMed] [Google Scholar]
- Dworkin J. and Losick,R. (2002) Does RNA polymerase help drive chromosome segregation in bacteria? Proc. Natl Acad. Sci. USA, 99, 14089–14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Moller-Jensen,J. and Jensen R.B (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol., 37, 455–466. [DOI] [PubMed] [Google Scholar]
- Glaser P., Sharpe,M.E., Raether,B., Perego,M., Ohlsen,K. and Errington,J. (1997) Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev., 11, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Gordon G.S. and Wright,A. (1998) DNA segregation: putting chromosomes in their place. Curr. Biol., 8, R925–R927. [DOI] [PubMed] [Google Scholar]
- Gordon G.S. and Wright,A. (2000) DNA segregation in bacteria. Annu. Rev. Microbiol., 54, 681–708. [DOI] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov,D., Webb,C.D., Teleman,A., Straight,A., Losick,R., Murray,A.W. and Wright,A. (1997) Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell, 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Gotfredsen M. and Gerdes,K. (1998) The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol. Microbiol., 29, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E.J., Pogliano,K. and Losick,R. (1995) Use of immuno fluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol., 177, 3386–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. (2000) The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol., 37, 528–541. [DOI] [PubMed] [Google Scholar]
- Jacob F., Brenner,S. and Cuzin,F. (1963) On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol., 23, 329–348. [Google Scholar]
- Jensen R.B. and Gerdes,K. (1997) Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR–parC complex. J. Mol. Biol., 269, 505–513. [DOI] [PubMed] [Google Scholar]
- Jensen R.B. and Gerdes,K. (1999) Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J., 18, 4076–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B. and Shapiro,L. (1999) Chromosome segregation during the prokaryotic cell division cycle. Curr. Opin. Cell Biol., 11, 726–731. [DOI] [PubMed] [Google Scholar]
- Jones L.J., Carballido-Lopez,R. and Errington,J. (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell, 104, 913–922. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2000) Movement of replicating DNA through a stationary replisome. Mol. Cell, 6, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev., 15, 2031–2041. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Errington,J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Li Y., Sergueev,K. and Austin,S. (2002) The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol., 46, 985–996. [DOI] [PubMed] [Google Scholar]
- Lin D.C. and Grossman,A.D. (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell, 92, 675–685. [DOI] [PubMed] [Google Scholar]
- Lobner-Olesen A., Skarstad,K., Hansen,F.G., von Meyenburg,K. and Boye,E. (1989) The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell, 57, 881–889. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J., Jensen,R.B., Lowe,J. and Gerdes,K. (2002) Prokaryotic DNA segregation by an actin-like filament. EMBO J., 21, 3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Jaffe,A., Imamura,R., Ogura,T. and Hiraga,S. (1991) The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J., 10, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi,Y. and Hiraga,S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Norris V. (1995) Hypothesis: chromosome separation in Escherichia coli involves autocatalytic gene expression, transertion and membrane-domain formation. Mol. Microbiol., 16, 1051–1057. [DOI] [PubMed] [Google Scholar]
- Rodionov O., Lobocka,M. and Yarmolinsky,M. (1999) Silencing of genes flanking the P1 plasmid centromere. Science, 283, 546–549. [DOI] [PubMed] [Google Scholar]
- Sawitzke J.A. and Austin,S. (2000) Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl Acad. Sci. USA, 97, 1671–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M.E. and Errington,J. (1999) Upheaval in the bacterial nucleoid. An active chromosome segregation mechanism. Trends Genet., 15, 70–74. [DOI] [PubMed] [Google Scholar]
- Shih Y.L., Le,T. and Rothfield,L. (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl Acad. Sci. USA, 100, 7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye,E. and Steen,H.B. (1986) Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J., 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A.A., Graumann,P.L., Lin,D.C.H., Grossman,A.D. and Losick,R. (1998) Chromosome arrangement within a bacterium. Curr. Biol., 8, 1102–1109. [DOI] [PubMed] [Google Scholar]
- van den Ent F., Amos,L.A. and Lowe,J. (2001) Prokaryotic origin of the actin cytoskeleton. Nature, 413, 39–44. [DOI] [PubMed] [Google Scholar]
- van den Ent F., Moller-Jensen,J., Amos,L.A., Gerdes,K. and Lowe,J. (2002) F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J., 21, 6935–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Helvoort J.M. and Woldringh,C.L. (1994) Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol., 13, 577–583. [DOI] [PubMed] [Google Scholar]
- Wachi M. and Matsuhashi,M. (1989) Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol, 171, 3123–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Doi,M., Tamaki,S., Park,W., Nakajima-Iijima,S. and Matsuhashi,M. (1987) Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol., 169, 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.D., Teleman,A., Gordon,S., Straight,A., Belmont,A., Lin,D.C., Grossman,A.D., Wright,A. and Losick,R. (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell, 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann,P.L., Kahana,J.A., Teleman,A.A., Silver,P.A. and Losick,R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Weitao T., Nordstrom,K. and Dasgupta,S. (1999) Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol., 34, 157–168. [DOI] [PubMed] [Google Scholar]
- Woldringh C.L. (2002) The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol., 45, 17–29. [DOI] [PubMed] [Google Scholar]
- Woldringh C.L., Zaritsky,A. and Grover,N.B. (1994) Nucleoid partitioning and the division plane in Escherichia coli. J. Bacteriol., 176, 6030–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J. and Errington,J. (2002) A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J., 21, 4001–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y. and Niki,H. (2000) Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 14656–14661. [DOI] [PMC free article] [PubMed] [Google Scholar]