Abstract
Abnormal gene expression patterns in somatic cell clones and their attrition in utero are commonly considered a consequence of errors in nuclear reprogramming. We observe that mouse clone blastocysts have less than half the normal cell number, and that higher cell number correlates with correct expression of Oct4, a gene essential for peri-implantation development and embryonic pluripotency. To increase the cell number, we aggregated genetically identical clones at the 4-cell stage. Clone–clone aggregates did not form more blastocysts, but the majority expressed Oct4 normally and had higher rates of fetal and postnatal development. Fertilized blastocysts with low cell numbers, induced by removal of two blastomeres at the 4-cell stage, did not exhibit abnormal Oct4 expression, indicating that improved gene expression and post-implantation development of clone–clone aggregates is not a consequence of increased cell number. Rather, we propose that complementation of non-cell-autonomous defects of genetically identical, but epigenetically different, embryos results in improved gene expression in clone–clone aggregates.
Keywords: aggregation/cloning/mouse chimera/nuclear transfer/pluripotency
Introduction
Reprogramming, in the context of cloning by transfer of a differentiated cell nucleus into an oocyte, can be defined as the transformation of a somatic cell nucleus into a functional embryonic nucleus capable of forming a viable organism. Correct expression of embryonic genes is a prerequisite for development and is indicative of nuclear reprogramming. We reported previously that only about one third of mouse clone blastocysts exhibit a normal expression pattern of Oct4, a gene essential for embryonic cell pluripotency and subsequent development (Boiani et al., 2002). Aberrant Oct4 expression, failure to activate genes that are expressed in pluripotent cells yet are silent in somatic donor cells (Bortvin et al., 2003) and incorrect expression of imprinted genes (Mann et al., 2003), are likely due to insufficient or aberrant reprogramming of the somatic cell nucleus within the ooplasm. As regulatory processes in pre-implantation-stage embryos are not only cell-autonomous but involve cell–cell interactions (Suzuki et al., 1995; Boni et al., 1999), it is conceivable that intercellular interactions also influence gene expression, and vice versa.
During pre-implantation development, somatic cell clones initially retain the metabolic preferences of the donor cell type, regarding the use of glucose as an energy substrate (Chung et al., 2002; Gao et al., 2003), indicating that some somatic cell characteristics are not eliminated in the metaphase II ooplasm, but change gradually. Gradual adaptation of the transplanted nucleus may lead to a lower rate of cell proliferation during pre-implantation development, resulting in a blastocyst comprised of fewer cells prior to implantation. Indeed, blastocyst-stage clones from several species are comprised of a lower number of cells than normal blastocysts (55% lower in mouse, Chung et al., 2002; 43% in rabbit, Chesne et al., 2002; 19% in porcine, Koo et al., 2000; and 9% in bovine, Koo et al., 2002). Mammalian blastocysts exhibit some plasticity in their ability to compensate for variations in the cell number (Buehr and McLaren, 1974; Lewis and Rossant, 1982; Conlon and Raff, 1999), but do not develop at a normal frequency when the cell number is substantially reduced. Several studies aimed at the production of identical offspring have effectively reduced cell number by dividing pre-implantation-stage embryos at various stages (bisection) or even isolating individual blastomeres. Although identical animals can be produced, the partitioned embryos exhibit lower rates of full-term development than controls (Willadsen, 1981; Willadsen and Polge, 1981; Ozil, 1983; Loskutoff et al., 1993). The low viability of blastocyst-stage clones may thus be related to both the failure to activate essential genes and failure to develop the sufficient cell numbers that allow early post-implantation survival. Insight into the contribution of each factor to clone development will increase our understanding of the capability of a somatic cell nucleus to undergo successful reprogramming.
In this study, we evaluated the cell number in somatic cell clones during the pre-implantation stage and correlated this variable to both the Oct4 gene expression and subsequent development. Our results indicate that low cell numbers are not directly related to Oct4 gene expression abnormalities in somatic cell clones. However, when clones at the 4-cell stage were combined with each other to generate blastocysts with a higher cell number, Oct4 expression was normal in most embryos and rates of post-implantation and full-term development markedly increased.
This indicates that either most single clones do not have a sufficient number of normal cells or/and that cell–cell interactions between blastomeres originating from different clones compensate for deficiencies and improve reprogramming.
Results
Low cell number but normal proportion of inner cell mass (ICM) cells in clones
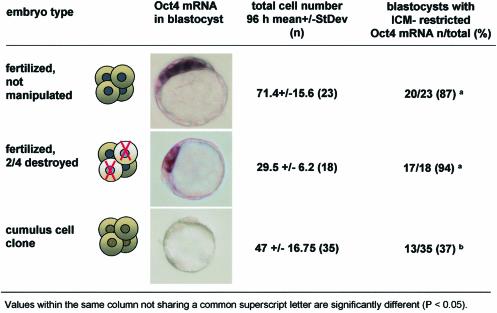
The ICM of mouse embryos must be comprised of at least three cells for subsequent development to take place (Markert and Petters, 1978). Disturbances in gene expression and low cell numbers observed in blastocyst-stage somatic cell clones may be indicative of failure to maintain an ICM and abnormal differentiation into trophectoderm (TE). We therefore analyzed the number of cells in the ICM and TE of mouse cumulus cell clones. At 96 h of development, the total cell number (ICM + TE) in somatic cell clones was less than half that of control embryos (Table I). Using differential labeling of ICM and TE cells, we found 10 cells or fewer present in the spatially defined ICM of a large proportion (50%) of clones versus 29 cells or more present in the ICM of most controls. The ratio of the number of cells in the ICM over the total number of cells in clones was normal compared with controls (Table I), and was independent of the total cell number. In contrast, a lower ratio was observed in fertilized embryos with a low cell number (<50 cells).
Table I. Developmental rate and cell number of clones and control embryos.
| Embryo | Total cell number | Differential cell count (mean ± SEM) (n) | ICM ratio | Blastocysts from 2 cells | ||
|---|---|---|---|---|---|---|
| 72 hmean ± SEM (n) | 96 hmean ± SEM (n) | ICM | TE | ICM/(ICM + TE) | % (n) | |
| Clone | 14 ± 0.7 (65) | 34 ± 1.8 (136)a | 13 ± 0.8 | 20 ± 0.8 (111) | 0.39a,c | 28 (4302) |
| Clone* | n.d. | 27 ± 3.3 (22)a | n.d. | n.d. | n.d. | 11 (174) |
| IVF | n.d. | 76 ± 5.7 (32)b | 38 ± 3.3 | 38 ± 3.8 (32) | 0.50c | 79 (932) |
| ICSI | 22 ± 0.8 (79) | 88 ± 2.4 (135)c | 25 ± 1.4 | 60 ± 2.1 (115) | 0.29a,b | 49 (1404) |
| In vivo | 19 ± 1.0 (36) | 103 ± 5.2 (58)d | 56 ± 5.0 | 64± 3.6 (30) | 0.47c | 100 (1000) |
*Clones transferred immediately into recipients; n.d., not determined.
Replicates in each embryo group ≥3. Values within the same column not sharing a common superscript letter are significantly different (P < 0.05).
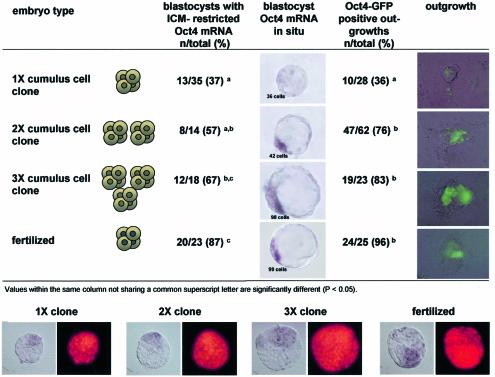
The cell number correlated with Oct4 transcript distribution in clone blastocysts. Blastocysts with ICM-restricted distribution of the Oct4 transcript had a higher average cell number (50 cells) than those lacking the Oct4 transcript (40 cells) or those exhibiting abnormal Oct4 distribution in both ICM and TE (48 cells). In clones with ICM-restricted Oct4 mRNA distribution (37%), Oct4-positive cells corresponded to the cells spatially defined as part of the ICM by differential staining. Based on the total and differential cell counts, we conclude that the lower cell number observed in cumulus cell clones is not associated with abnormal allocation of cells to the ICM and TE.
Low cell number in clones is due to a proliferation defect at the end of the morula/blastocyst transition
The low cell number observed in blastocyst-stage somatic cell clones could be caused by a number of events, including apoptotic cell death, delayed cell cycle progression or inhibition of cell proliferation due to metabolic restraints. We determined the incidence of apoptotic cell loss in clones using the TUNEL assay. On average, 4.0 cells per blastocyst-stage clone were apoptotic, compared with 3.1, 4.0 and 3.1 cells for in vitro fertilized (IVF), intra-cytoplasmic sperm injected (ICSI) and in vivo fertilized embryos, respectively. In clones and controls, apoptotic cells were only detected in blastocysts but never at the morula stage, consistent with the normal onset of apoptosis during mouse pre-implantation development (Pampfer and Donnay, 1999). Thus, apoptotic cell loss cannot account for the disparity in cell number between clones and controls prior to the blastocyst stage, and it affects an insufficient number of cells to explain the large cell number difference at the blastocyst stage.
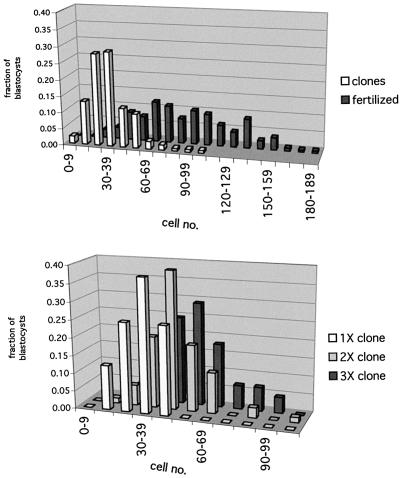
To identify the precise time point when the cell number of clones diverges from that of controls, we determined the total cell number of both groups at different times during pre-implantation development (Table I). Clones proliferated at a similar rate to controls up to the 8-cell stage, after which point the cell number was slightly lower compared with controls. At 96 h, the difference in cell number became significantly larger, indicating that clones as a population fail to enter the exponential proliferation phase that is observed in normal embryos subsequent to the morula stage. Clones cultured in vitro for an additional 24 h never attained the cell number of fertilized embryos at 96 h of development [61 ± 4.4 cells at 120 h in clones (n = 53) versus 103 ± 5.2 (n = 58) in fertilized embryos at 96 h]. As illustrated in Figure 1 (top), the distribution of the total number of cells in clones is consistent with one population of clones; two populations would have been expected if a subpopulation of clones with a higher cell number correlated with those undergoing post-implantation development. We conclude that the lower cell number of clones is primarily due to a non-lineage specific proliferation defect concomitant with the transition from the morula to blastocyst stage of development.
Fig. 1. Distribution of total cell number grouped into intervals of 10 cells for clone, fertilized and clone–clone aggregate embryos at 96 h of development. Top, distribution of total cell counts in clone and fertilized mouse embryos. Bars indicate the proportion (%) of embryos with counts that fall within each interval. The 50th percentile of the fertilized embryo distribution (80–89 total cells) coincides with the 98th percentile inclusive of the clone embryo distribution. Clones, n = 136; fertilized embryos, n = 225. Bottom, comparison of total cell counts of single clones and 2X and 3X clone–clone aggregates at 96 h. 1X clones, n = 16; 2X clones, n = 70; 3X clones, n = 45.
Increasing cell number in clones by aggregation
To address whether a larger cell number early in pre-implantation development influences gene expression and subsequent development, genetically identical clones were combined at the 4-cell stage to form double (2X) and triple (3X) clone aggregates (Figures 2 and 3). The 4-cell stage was chosen for aggregation as it precedes the onset of Oct4 expression and blastomere polarization at the morula stage. There was no net increase in the number of blastocysts that formed based on the initial number of 4-cell-stage clones used for aggregation (Table II). This may indicate that only those clones capable of forming blastocysts on their own did so in the aggregates. Compared with single clones, 2X and 3X clone blastocysts had higher total cell numbers at 96 h of development, but the average increase (1.2-fold for 2X and 1.5-fold for 3X; Table II) was lower than that observed in aggregates of fertilized embryos [1.75-fold, 172 versus 98 cells (n = 10) for 2X; and 1.9-fold, 191 cells (n = 10) for 3X] (Figure 1, bottom). It appears that one component of the clone–clone aggregate did not proliferate at the same rate as the other. The ratio of ICM to total cell number as determined by differential labeling did not differ between 1X clones and 2X aggregates (Table II), and was slightly higher in 3X aggregates.
Fig. 2. Spatial Oct4 expression in blastocysts and outgrowths of 1X, 2X and 3X clones and fertilized embryos. Top, Oct 4 mRNA distribution in blastocyst-stage cumulus cell clones, clone–clone aggregates and fertilized embryos was assessed by ISH. Outgrowths were analyzed for expression of Oct4–GFP. Bottom, analysis of Oct4 mRNA distribution and cell number in single and aggregate clone embryos. Subsequent to ISH and evaluation of Oct4 mRNA distribution, individual embryos were mounted on slides in a squash preparation (left) and stained with propidium iodide (right) for determination of the total cell number.
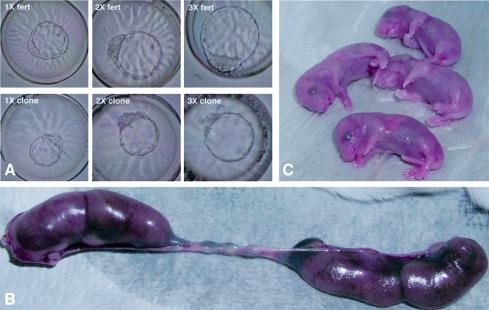
Fig. 3. Development of clone–clone aggregates to the blastocyst stage and full-term. (A) Blastocyst-stage fertilized (top) and clone–clone (bottom) 1X, 2X and 3X aggregation chimeras. (B) Development of 2X clone–clone aggregates to 19.5 d.p.c. Uterus from female receiving 29 blastocysts of 2X blastocyst-stage clones, with four fetuses. (C) Four 2X clone–clone chimeras after Caesarean section.
Table II. Development, cell number and ICM ratio of clone aggregates.
| Embryo | 4-cell (n) | Aggregates (n) | Blastocysts (n) (%) | Total cells (mean ± SEM) (n) | Cells range (n) | ICM cells (n) | ICM/(ICM + TE) |
|---|---|---|---|---|---|---|---|
| 1X clone | 262 | n.a. | 65 (25) | 42 ± 2.3 (51)a | 22–87 | 18 | 0.44 |
| 2X clone | 786 | 393 | 171 (44) | 50 ± 1.7 (84)b | 38–97 | 23 | 0.45 |
| 3X clone | 504 | 168 | 91 (54) | 62 ± 2.8 (62)c | 37–113 | 33 | 0.53 |
| Fertilized | 60 | n.a. | 57 (95) | 120 ± 5.2 (30)d | 33–171 | 56 | 0.47 |
Values within the same column not sharing a common superscript letter are significantly different (P < 0.05).
n.a., not applicable.
Aggregation of clones facilitates expression of Oct4
We evaluated Oct4 gene expression in single clones as well as in clone–clone aggregates at the blastocyst stage and in vitro outgrowths. The majority of 2X and 3X blastocyst-stage clones had a normal pattern of Oct4 distribution, which was restricted to the ICM (Figure 2). In contrast, the majority of single clones exhibited an abnormal Oct4 distribution. The frequency of in vitro outgrowth formation was similar in clone aggregates compared with 1X clones (74, 85 and 88% of blastocysts for 1X, 2X and 3X, respectively). However, the outgrowths from 1X clones were frequently small and often lacked an Oct4–GFP expressing cell mass. In contrast, the majority of outgrowths that formed from 2X and 3X aggregates were both larger and expressed Oct4–GFP (Figure 2).
Aggregation of clones improves development in vivo
The frequent formation of normal, Oct4-expressing in vitro outgrowths from 2X and 3X clones suggests that blastocysts resulting from clone–clone aggregates have improved developmental potential compared with single clones. We therefore assessed the ability for post-implantation development in both 1X (zona-free) and 2X clones by transfer to separate uterine horns of the same recipient. Analysis of fetal development at midgestation (E10.5) showed that 1X and 2X clone aggregates had a similar rate of implantation as determined by the number of decidua (Table III). The implantation rates of both groups were substantially higher than those of clones transferred with intact zonae pellucidae. Upon transfer of single clones into asynchronous recipients to extend the period in vivo during which embryos can develop, we observed a significant increase in implantation rates (15.9 versus 9.6%), although there was no improvement in fetal development at midgestation (1.3 versus 1.2%). Despite the high implantation rate, fetal development of zona-free 1X clones was as low as that observed for single, zona-intact clones. Removal of the zona pellucida was presumably responsible for the higher rate of implantation. In contrast to the low rate of fetus formation in single clones, clone–clone aggregates (2X) developed more frequently to midgestation, yielding 4-fold more fetuses than single clones (12 versus 3%; Table III). Aggregation also resulted in a better decidual response as determined by the size of the decidua, regardless of whether or not it contained a fetus. The size of the decidua in the largest cross-section was, on average, 30% larger in the aggregates than in single clones (33 513 area units, n = 18 1X clones; 43 658 area units, n = 38 2X clones). The improved developmental potential of aggregate clones was confirmed by the rate of full-term development. We observed an 8-fold increase in full-term development of 2X clone–clone chimeras compared with single clones (Table III).
Table III. Post-implantation development of single (1X) and aggregate (2X) clone blastocysts.
| Clone aggregate | Blastocysts transferred (n) | Pregnant uterine horns (n) | Decidua (n) (% of blastocysts) | 10.5 d.p.c. (n) (% of blastocysts) | Full-term (n) (% of blastocysts) |
|---|---|---|---|---|---|
| clone 1X | 109 | 14/16 | 62 (57)a | 2 (3.0)a | n.a. |
| clone 2X | 201 | 16/16 | 131 (65)a | 16 (12.0)b | n.a. |
| clone 1X | 391 | 11/23* | n.a. | n.a. | 4 (1.0)a |
| clone 2X | 98 | 5/7* | n.a. | n.a. | 8 (8.2)b |
n.a., not applicable; d.p.c., days post coitum. *The uterine horns were of separate recipients.
Replicates in each embryo group: 14 for midgestation analysis, >4 for full-term analysis. Values within the same column not sharing a common superscript letter are significantly different (P < 0.05).
Combination of genetically identical clones does not permit analysis of the contribution of the individual components. Therefore, in one experiment, we aggregated clones generated from nuclei of mouse strains with different coat colors, B6C3F1 (agouti) and B6D2F1 (black), and confirmed contribution of both clone components by coat color in postnatal animals.
Reduction in cell number does not impair Oct4 expression in fertilized embryos
To ascertain whether the improved Oct4 expression in aggregate clones was only a consequence of an increase in cell number, we determined the association between total cell number and Oct4 expression in fertilized mouse pre-implantation-stage embryos. Specifically, we tested whether an experimentally induced low cell number prior to the blastocyst stage had an influence on gene expression patterns. To produce fertilized embryos with a low cell number, we removed two of four blastomeres of fertilized B6C3F1 embryos at the 4-cell stage. The manipulated embryos developed to the blastocyst stage at a rate similar to that of controls (87 and 97% for manipulated and non-manipulated 4-cell-stage embryos, respectively), but had significantly reduced total cell numbers. On average, blastocysts that developed from half embryos had less than half the normal cell number of controls, similar to or less than observed in clones (Figure 4). In the large majority of manipulated blastocysts, however, Oct4 expression was restricted to the ICM (Figure 4) and signal intensity was comparable to controls, even though some of the embryos had only a few ICM cells. These observations show that in normal embryos, a lower cell number during pre-implantation stages does not result in absence or spatially abnormal Oct4 expression at the blastocyst stage.
Fig. 4. Total cell number and spatial Oct4 expression in clones, controls and bisected embryos. Embryos at 96 h of development were analyzed by ISH with an Oct4-mRNA-specific antisense riboprobe and subsequently stained with propidium iodide to determine cell counts in squashed preparations (see Figure 2, bottom).
Discussion
In the present study, we show that blastocyst-stage clones exhibit an abnormal gene expression pattern that is coincident with fewer cells than normal embryos. This raises the question whether a low cell number in clones is a consequence of reprogramming defects, or vice versa. Our results imply that a low cell number is not a direct cause for gene expression abnormalities in mouse embryos. Nevertheless, we observe that increasing the cell number in clones by aggregation of clones with one another dramatically improves spatial and temporal gene expression as well as developmental potential. Therefore, the developmental capacity of mouse clones can be rescued by events that are either related to an increase in the number of cells present at cleavage stages, or/and interaction between blastomeres from different clones.
Gap junctional intercellular communication between blastomeres is a prerequisite for the embryo to develop past the morula stage. In the mammalian embryo, gap junctions are first apparent at the 8-cell stage, and increase in number from compaction onwards (Lo and Gilula, 1979; Goodall and Johnson, 1984). The process of compaction involves establishment of an outer epithelial cell layer that will differentiate into the TE. A link between compaction, communication through gap junctions and development has been established for certain mouse strains (e.g. DDK); failure to maintain the compacted state at the morula stage due to defective gap junctional communication leads to blastocyst demise (Buehr et al., 1987). We observe that reduced proliferation and not apoptosis is responsible for the low cell number of clones at the blastocyst stage. The proliferation deficit is not evident at all pre-implantation stages, but becomes significant as the embryo transitions into compaction and morula formation. The more complex and integrated gene expression that is required during this period of morula compaction suggests that many clones fail because of poor coordination between cells, of the morula and blastocyst.
Clone–clone aggregates differ from single clones in two aspects: they have a higher cell number and are comprised of distinct, but genetically identical clones. Either one, or possibly both of these factors must confer the correct gene expression, and, as a consequence, the developmental potential of clones. In normal embryos, a drastic reduction in cell number, by removal of half the blastomeres at the 4-cell stage, had no effect on Oct4 expression in the embryo at the blastocyst stage. Therefore, the regulation of Oct4 expression in normal embryos is not affected by a 2-fold change in total cell number; and a cell number as low as that observed in clones is compatible with normal Oct4 expression. Therefore, the relatively small increase in cell number in aggregate clones compared with single clones is not likely to be the cause of the improved Oct4 gene expression that is observed in aggregates. Rather, the benefit of aggregation may be attributed to complementation of differences between clones. Variation in gene expression between and within somatic cell clones (e.g. mosaicism; Boiani et al., 2002; Park et al., 2002) demonstrates that despite being genetically identical, clones are epigenetically different. By combining two epigenetically different clones, non-cell-autonomous defects that are epigenetic in nature may be compensated for (Rideout et al., 2001). This could facilitate development of the whole embryo even if one of the two contributing embryos may not have been viable alone. Alternatively, if cells within one clone do not all have the same transcriptional activity or developmental potential due to genetic differences, aggregation of two may increase the number of normal cells with correct gene expression above a threshold required for further development. The presence of aneuploid cells within clones may explain why some, although having a similar number of cells to normal embryos, fail to develop. Precedent for aneuploidy in clones has been reported for several species (Pinto-Correia et al., 1993; Slimane-Bureau and King, 2002; Yin et al., 2002; Booth et al., 2003; Simerly et al., 2003) and therefore aggregation of clones containing aneuploid cells may produce a viable embryo as a consequence of combining a sufficient number of euploid cells.
Although allocation of cells to the inner and outer compartments of a large number of non-aggregated, single clones is normal, abnormal expression of Oct4 in blastocyst-stage single clones shows that the pluripotency of the ICM is disturbed. In contrast, blastocysts from clone–clone aggregates frequently exhibit normal Oct4 expression in the ICM, consistent with their improved post-implantation development. Therefore, expression of Oct4, other pluripotency-associated genes, or gene expression in general, is facilitated in aggregates versus single clones. Aggregation at the 4-cell stage precedes the onset of Oct4 expression, normally initiated between the 4- and 8-cell stages (Yeom et al., 1996). The normal spatial distribution of the Oct4 transcript in clone–clone aggregates at the blastocyst stage requires appropriate regulation of Oct4 gene expression in the ICM and TE of the whole embryo. This implies that there is synchronization or coordination of gene expression regulation between blastomeres of the different clone components of the aggregate. As Oct4 expression is improved in the clone–clone aggregates, the establishment and/or maintenance of its expression apparently involves a cell-non-autonomous component. It has been shown that physical contact with embryonic blastomeres can induce expression of Oct4 in somatic cells (Burnside and Collas, 2002), and that this is mediated by signaling through gap junctions. Intercellular junctions that can mediate cellular signaling between blastomeres of aggregation chimeras have been observed as early as the 2-cell stage. Mouse embryos from blocking strains that arrest at the 2-cell stage can resume development when aggregated with embryos from strains with a non-blocking genotype (Neganova et al., 2000). In these chimeras, E-cadherin/β-catenin-mediated signaling appeared to be required for developmental rescue. Therefore, it is conceivable that complementation via intercellular signaling between clone components can occur subsequent to aggregation, as early as the 4- and 8-cell stages, and prior to the blastocyst stage, at which point the effect becomes apparent.
In conclusion, this study shows that cells of defective clone embryos have more developmental potential than previously anticipated, as they can not only participate in normal development when combined with a normal embryo (Byrne et al., 2002), but they also can integrate or complement each other at pre-implantation stages. Our results provide a different perspective to the idea that reprogramming of gene expression is a major limiting factor in the development of clones. We propose that the regulation of gene expression and, possibly, reprogramming of nuclei in clones has a non-cell-autonomous component that can be complemented. The reprogramming process in the vast majority of clones may still be ongoing at the blastocyst stage and require assistance, which is provided in aggregates, to facilitate transition to successful post-implantation development. These issues can be addressed by evaluating the development of demi-embryos (from different clones) aggregated with each other, or clones aggregated with normal embryos. Clone–clone aggregation overcomes an early developmental obstacle. Although this procedure may help to reduce epigenetic disturbances, it remains to be determined whether the epigenetic defects observed in adult clones (Ogura et al., 2002; Tamashiro et al., 2002) would still persist.
Materials and methods
Recipient oocyte and donor nucleus collection
Eight- to ten-week-old C57Bl/6J X C3H/HeN female mice (referred to as B6C3F1; Taconic, Germantown, NY) were superovulated with 7.5 U of pregnant mare’s serum gonadotropin (PMSG, Calbiochem cat. no. 367222) followed 48 h later by 7.5 U of human chorionic gonadotropin (hCG; Calbiochem cat. no. 230734). Cumulus–oocyte complexes were collected 15 h post-hCG and cumulus cells removed by hyaluronidase at 50 U/ml in HEPES-buffered CZB medium at 27°C (ICN cat. no. 151271, activity >5000 IU/mg). The HEPES-buffered CZB medium (Chatot et al., 1989) was modified as follows: [5.56 mM glucose, 20 mM HEPES, 5 mM sodium bicarbonate, 0.1% w/v polyvinylpyrrolidone (PVP; 40 kDa, Calbiochem cat. no. 529504), no albumin] in the presence of protease inhibitors (Sigma cat. no. P1860; 0.2% v/v). After 15–20 min, the cumulus-free oocytes were washed three times in HEPES-buffered CZB medium and then placed in α-MEM culture medium (see Embryo culture). Donor cells from Oct4–GFP transgenic mice, subsequently referred to as OG2 (Szabò et al., 2002), were used to visualize expression of the transgene after nuclear transfer. Adult cumulus cells were isolated from the cumulus–oocyte complexes of superovulated C57Bl/6J X OG2 females. The cumulus cells were left in the hyaluronidase drop and stored at 4°C after adding an equal volume of α-MEM.
Animals were maintained and used for experimentation according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania.
Microsurgery and activation of reconstructed oocytes
Removal of metaphase chromosomes (′enucleation′) and nuclear transfer were carried out as described previously (Boiani et al., 2002) with the following modifications. Recipient oocytes were enucleated in HEPES-buffered CZB medium (PVP 0.1% w/v; albumin-free) in the presence of cytochalasin B (ICN cat. no.195119, 0.25 µg/ml) and nocodazole (Sigma cat. no. M1404, 0.5 µg/ml). After enucleation, the oocytes were washed and left to recover for 1–2 h in drops of α-MEM medium at 37°C. Nuclear transplantation was carried out in hypertonic (110%) HEPES-buffered CZB medium (as above, except with PVP 1% w/v). Oocytes were processed in batches of 20 within a 10 min time span, using piezo-driven (PMM 150 FU, PrimeTech, Japan) borosilicate needles (Clark Instruments, UK) and DIC optics (Nikon, Japan) at 27°C. After micromanipulation, the oocytes were left to recover in 1:1 part mixture of HEPES-buffered CZB and α-MEM medium for 1 h at 27°C prior to incubation in α-MEM medium at 37°C. One to 2 h later, they were activated for 6 h in modified (Ca-free, 10 mM SrCl2) M16 medium supplemented with bovine serum albumin (0.4% w/v, ICN cat. no. 103700), vitamins (1X, Sigma cat. no. R7256) in the presence of cytochalasin B (ICN cat. no. 195119, 5 µg/ml; prepared in DMSO as a 200× stock solution). The presence of cytochalasin B was required to prevent extrusion of a pseudo polar body.
Embryo culture
Embryos were cultured in groups of 50–100 in 20 µl drops of α-MEM medium (Wun et al., 1994) in 35 mm dishes (Corning cat. no. 430588). α-MEM was supplemented with bovine serum albumin (0.4% w/v, ICN cat. no. 103700) and filtered on a cellulose acetate membrane (0.22 µm, Fisher cat no. 09–719A; preconditioned with medium) prior to use. Culture drops were overlaid with silicon oil (5 centistokes, Sigma cat. no. DMPS-V) and incubated under 5% CO2 at 37°C. After 48 h of culture, the microdrop was supplemented with an additional 10 µl of α-MEM. Embryos were assessed for morula and blastocyst formation at 72 and 96 h post-activation, respectively, as well as for the expression of GFP from the OG2 transgene.
IVF and ICSI
Sperm was isolated in Whittingham medium (3% w/v bovine serum albumin, fraction V, Sigma cat. no. A3311) by swim-up from the cauda epididymis of mature OG2 (Oct4–GFP+/+) males. Sperm at a concentration of 2 × 106/ml were capacitated in Whittingham’s medium for 1.5 h prior to addition to the oocyte insemination drop or microinjection (ICSI). ICSI was performed by piezo-driven injection into oocytes of acrosome-reacted sperm heads without a tail (Kimura and Yanagimachi, 1995). Successful fertilization was measured by the extrusion of the second polar body, which usually occurred within 2 h (IVF) or 3 h (ICSI). In vivo fertilized zygotes were also obtained by mating B6C3F1 females with OG2 males.
Aggregation of embryos
The zonae pellucidae of 4-cell-stage embryos were removed by treatment with pronase (ICN cat. no. 537088, 1% w/v in HEPES-buffered CZB at 27°C for 3 min). Zona-free embryos were washed in α-MEM containing 3% albumin and protease inhibitors (1% v/v; see Recipient oocytes for nuclear transfer). Clusters of depressions in the bottom of a culture dish, generated by gentle pressure with a darning needle (cat. no. DN-09, BLS, Hungary; Nagy and Rossant, 1993), were covered with microdrops (30 µl volume) of α-MEM, overlaid with oil, and incubated for at least 1 h prior to use. Aggregation was accomplished by placing the zona-free embryos in groups of two or three within each microwell. Single clones (1X) and double (2X) or triple (3X) clone aggregates, as well as aggregates of fertilized embryos, were cultured in parallel in separate drops within the same dish.
Production of blastocysts with low cell number
Two-cell embryos were collected 40–42 h post-hCG from the oviduct of B6C3F1 females mated with ICR males. For manipulation at the 4-cell stage, embryos were cultured in bicarbonate-buffered CZB to 63–65 h post-hCG. Using a piezo-driven needle (∼15 µm), two blastomeres of each embryo were removed. Embryos were cultured in bicarbonate-buffered CZB culture medium until 95–97 h post-hCG, and were fixed in 4% paraformaldehyde, 0.1% glutaraldehyde in PBS for in situ gene expression analysis and cell count.
In vivo development of embryos
Two- and 4-cell-stage clone embryos were transferred into the oviducts of ICR females (Taconic, Germantown, NY) that were pseudopregnant, as ascertained by presence of a copulatory plug subsequent to mating with vasectomized ICR males, on the day of the plug detection [0.5 days post coitum (d.p.c.)]. For analysis of total cell number at the blastocyst stage, clones were transferred to the oviduct (0.5 d.p.c.) at 6 h post-activation, and were collected from the uterus 66 h later and cultured in vitro for an additional 24 h. Blastocyst-stage clones were transferred into the uterine horns of 2.5 d.p.c. pseudopregnant recipients. At 10.5 d.p.c., females with a weight increase consistent with a pregnancy were sacrificed. Fetuses were genotyped from cells of the amniotic sac for presence of the Oct4–GFP transgene as described previously (Boiani et al., 2002). Estimates of the largest cross-section (LCS) of the decidua were obtained by the following measurement. The decidua were positioned so as to lie on their flatter side. The area projected in a digital picture under the same magnification was presented as a LCS, and scanned by image software (Adobe Photoshop). The area units of the LCS are pixels.
Embryonic outgrowths
Zona-free single and aggregate clones were placed on a feeder layer of mitomycin C-inactivated confluent STO neo LIF (SNL) cells in a 4-well plate (Nunc cat. no. 176740). Culture of feeder cells and embryos (outgrowths) was in DMEM (Specialty Media SLM-220B, 4.5 g/L glucose supplemented with 0.1 mM non-essential amino acids, 2 mM l-glutamine, 0.5 mM β-mercaptoethanol, 14% HyClone fetal bovine serum, 50 U/ml penicillin–streptomycin). Outgrowth formation was defined by the spreading of trophoblast cells from the attached blastocyst.
Whole-mount in situ hybridization (ISH) of mouse embryos and outgrowths
Digoxigenin-labeled riboprobes (Oct4 cDNA, sense and antisense) were generated by in vitro transcription with T3 and T7 polymerases from linearized pBluescript containing a full-length Oct4 cDNA insert (Schöler et al., 1990) using Dig-labeling components (Roche Bioscience), according to the manufacturer’s protocol. Control and cloned embryos were cultured to the blastocyst stage. Embryos and outgrowths were fixed in PBS containing 4% paraformaldehyde and 0.1% glutaraldehyde for 30 min at room temperature. ISH was performed as described previously (Oblin and Clarke, 1997), with the following modifications. Hybridization was at 58°C overnight, in 5× SSC pH 5, 50% formamide, 50 µg/ml heparin, 0.1% Tween-20, 100 µg/ml tRNA, 100 µg/ml denatured sheared salmon sperm DNA. Post-hybridization washes were in 2× SSC pH 4.5, 50% formamide, 0.1% Tween-20, twice at room temperature and three times for 30 min at 59°C. Embryos were mounted in microdrops and positioned to localize the ICM.
Differential labeling of the ICM and TE
The nuclei of blastocyst ICM and TE cells were differentially labeled with polynucleotide-specific fluorochromes following a modification of the immunodissection procedure of Handyside and Hunter (1984). Subsequent to removal of the zonae pellucidae in acidic Tyrode’s solution, blastocysts were washed in HEPES-buffered CZB medium containing 0.4% albumin and incubated in 10% rabbit anti-mouse serum (ICN cat. no. 8650141) at 27°C in the presence of bis-benzimide (Hoechst 33342, 10 µg/ml). After 30 min, blastocysts were washed in HEPES-buffered CZB medium and incubated in a 1:10 dilution of guinea pig complement (ICN cat. no. 8642831) in HEPES-buffered CZB containing 1 µg/ml propidium iodide for 30 min. Blastocysts were washed in PBS and placed on microscope slides where they were partially flattened under a coverslip. Excitation with green light (excitation 535 nm, dichroic 565 nm, emission 590 nm) revealed the red stain of nuclei of the TE cells that had been lysed by complement. Under UV light (excitation 360 nm, dichroic 400 nm, emission 420 nm), nuclei of all cells were visible, labeled blue by the membrane permeable Hoechst 33342 stain.
DNA labeling for apoptosis (TUNEL)
Embryos were fixed for 30 min in 3% paraformaldehyde in PBS and washed in PBS containing 0.4% BSA. They were processed in microdrops with intervening PBS washes through 0.1% Triton X-100 for 15 min, equilibration buffer, terminal transferase reaction (incorporating digoxigenin-labeled deoxynucleotides) and finally fluorescein-labeled anti-digoxigenin antibody reaction (all reagents from ApoTag in situ apoptosis detection kit, Intergen). As positive controls, embryos were treated with 0.5 µg/ml DNAse I after the permeabilization step. The embryos washed in PBS containing 1 µg/ml Hoechst 33342, then transferred on microscope slides to be squashed under a coverslip. The slides were viewed under UV and blue light (excitation 470 nm, dichroic 495 nm, emission 515 nm) using a fluorescence microscope.
Statistical analysis
Values expressed as proportions were compared by a simple two-tailed z-test not requiring the square root and arc sin transformation. Values expressed as counts were analyzed in cross tabs through χ2 and the Fisher’s exact test. Tests were performed as described previously (Glantz, 1992).
Acknowledgments
Acknowledgements
The authors would like to thank Davor Solter and Jeffrey R.Mann for helpful comments on the manuscript, as well as Marianne Friez, Jaclyn O’Shea and Janet Turpin for technical and animal husbandry assistance. We also thank Areti Malapetsa for editing the manuscript. This work was supported, in part, by the Marion Dilley and David George Jones Funds and the Commonwealth and General Assembly of Pennsylvania (K.J.M., S.E., N.A.L., M.B., H.S.), grant NIH 1RO1HD42011–01 (M.B., H.S.) and University of Pennsylvania Research Foundation and U.S.D.A. (K.J.M).
References
- Boiani M., Eckardt,S., Schöler,H.R. and McLaughlin,K.J. (2002) Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev., 16, 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni R., Tosti,E., Roviello,S. and Dale,B. (1999) Intercellular communication in in vivo- and in vitro-produced bovine embryos. Biol. Reprod., 61, 1050–1055. [DOI] [PubMed] [Google Scholar]
- Booth P.J., Viuff,D., Tan,S., Holm,P., Greve,T. and Callesen,H. (2003) Numerical chromosome errors in day 7 somatic nuclear transfer bovine blastocysts. Biol. Reprod., 68, 922–928. [DOI] [PubMed] [Google Scholar]
- Bortvin A., Eggan,K., Skaletsky,H., Akutsu,H., Berry,D.L., Yanagimachi,R., Page,D.C. and Jaenisch,R. (2003) Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development, 130, 1673–1680. [DOI] [PubMed] [Google Scholar]
- Buehr M. and McLaren,A. (1974) Size regulation in chimaeric mouse embryos. J. Embryol. Exp. Morphol., 31, 229–234. [PubMed] [Google Scholar]
- Buehr M., Lee,S., McLaren,A. and Warner,A. (1987) Reduced gap junctional communication is associated with the lethal condition characteristic of DDK mouse eggs fertilized by foreign sperm. Development, 101, 449–459. [DOI] [PubMed] [Google Scholar]
- Burnside A.S. and Collas,P. (2002) Induction of Oct-3/4 expression in somatic cells by gap junction-mediated cAMP signaling from blastomeres. Eur. J. Cell Biol., 81, 585–591. [DOI] [PubMed] [Google Scholar]
- Byrne J.A., Simonsson,S. and Gurdon,J.B. (2002) From intestine to muscle: nuclear reprogramming through defective cloned embryos. Proc. Natl Acad. Sci. USA, 99, 6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatot C.L., Ziomek,C.A., Bavister,B.D., Lewis,J.L. and Torres,I. (1989) An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil., 86, 679–688. [DOI] [PubMed] [Google Scholar]
- Chesne P., Adenot,P.G., Viglietta,C., Baratte,M., Boulanger,L. and Renard,J.P. (2002) Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol., 20, 366–369. [DOI] [PubMed] [Google Scholar]
- Chung Y.G., Mann,M.R., Bartolomei,M.S. and Latham,K.E. (2002) Nuclear-cytoplasmic ‘tug of war’ during cloning: effects of somatic cell nuclei on culture medium preferences of preimplantation cloned mouse embryos. Biol. Reprod., 66, 1178–1184. [DOI] [PubMed] [Google Scholar]
- Conlon I. and Raff,M. (1999) Size control in animal development. Cell, 96, 235–244. [DOI] [PubMed] [Google Scholar]
- Gao S., Chung,Y.G., Williams,J.W., Riley,J., Moley,K. and Latham,K.E. (2003) Somatic cell-like features of cloned mouse embryos prepared with cultured myoblast nuclei. Biol. Reprod., 69, 48–56. [DOI] [PubMed] [Google Scholar]
- Glantz S.A. (1992) Primer of Biostatistic. McGraw Hill Inc., New York, NY. [Google Scholar]
- Goodall H. and Johnson,M.H. (1984) The nature of intercellular coupling within the preimplantation moue embryo. J. Embryol. Exp. Morphol., 79, 53–76. [PubMed] [Google Scholar]
- Handyside A.H. and Hunter,S. (1984) A rapid procedure for visualising the inner cell mass and trophectoderm nuclei of mouse blastocysts in situ using polynucleotide-specific fluorochromes. J. Exp. Zool., 231, 429–434. [DOI] [PubMed] [Google Scholar]
- Kimura Y. and Yanagimachi,R. (1995) Intracytoplasmic sperm injection in the mouse. Biol. Reprod., 52, 709–720. [DOI] [PubMed] [Google Scholar]
- Koo D.B. et al. (2000) In vitro development of reconstructed porcine oocytes after somatic cell nuclear transfer. Biol. Reprod., 63, 986–992. [DOI] [PubMed] [Google Scholar]
- Koo D.B. et al. (2002) Aberrant allocations of inner cell mass and trophectoderm cells in bovine nuclear transfer blastocysts. Biol. Reprod., 67, 487–492. [DOI] [PubMed] [Google Scholar]
- Lewis N.E. and Rossant,J. (1982) Mechanism of size regulation in mouse embryo aggregates. J. Embryol. Exp. Morphol., 72, 169–181. [PubMed] [Google Scholar]
- Lo C.W. and Gilula,N.B. (1979) Gap junctional communication in the preimplantation mouse embryo. Cell, 18, 399–409. [DOI] [PubMed] [Google Scholar]
- Loskutoff N.M., Johnson,W.H. and Betteridge,K.J. (1993) The developmental competence of bovine embryos with reduced cell numbers. Theriogenology, 39, 95–107. [Google Scholar]
- Mann M.R., Chung,Y.G., Nolen,L.D., Verona,R.I., Latham,K.E. and Bartolomei,M.S. (2003) Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod., May 14 [epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Markert C.L. and Petters,R.M. (1978) Manufactured hexaparental mice show that adults are derived from three embyronic cells. Science, 202, 56–58. [DOI] [PubMed] [Google Scholar]
- Nagy A. and Rossant,J. (1993) Production of completely ES cell-derived fetuses. In Joyner,A. (ed.), Gene targeting: a practical approach. IRL Press, Oxford, UK. [Google Scholar]
- Neganova I.E., Sekirina,G.G. and Eichenlaub-Ritter,U. (2000) Surface-expressed E-cadherin, and mitochondrial and microtubule distribution in rescue of mouse embryos from 2-cell block by aggregation. Mol. Hum. Reprod., 6, 454–464. [DOI] [PubMed] [Google Scholar]
- Oblin C. and Clarke,H. (1997) Rapid whole-mount in situ hybridization protocol for mammalian oocytes and preimplantation embryos. Elsevier Trends Journals Technical Tips Online TTO, 1, 5. [Google Scholar]
- Ogura A., Inoue,K., Ogonuki,N., Lee,J., Kohda,T. and Ishino,F. (2002) Phenotypic effects of somatic cell cloning in the mouse. Cloning Stem Cells, 4, 397–405. [DOI] [PubMed] [Google Scholar]
- Ozil J.P. (1983) Production of identical twins by bisection of blastocysts in the cow. J. Reprod. Fertil., 69, 463–468. [DOI] [PubMed] [Google Scholar]
- Pampfer S. and Donnay,I. (1999) Apoptosis at the time of embryo implantation in mouse and rat. Cell Death Differ., 6, 533–545. [DOI] [PubMed] [Google Scholar]
- Park K.W. et al. (2002) Mosaic gene expression in nuclear transfer-derived embryos and the production of cloned transgenic pigs from ear-derived fibroblasts. Biol. Reprod., 66, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Pinto-Correia C., Collas,P., Ponce de Leon,F.A. and Robl,J.M. (1993) Chromatin and microtubule organization in the first cell cycle in rabbit parthenotes and nuclear transplant embryos. Mol. Reprod. Dev., 34, 33–42. [DOI] [PubMed] [Google Scholar]
- Rideout W.M. III, Eggan,K. and Jaenisch,R. (2001) Nuclear cloning and epigenetic reprogramming of the genome. Science, 293, 1093–1098. [DOI] [PubMed] [Google Scholar]
- Schöler H.R., Ruppert,S., Suzuki,N., Chowdhury,K. and Gruss,P. (1990) New type of POU domain in germ line-specific protein Oct-4. Nature, 344, 435–439. [DOI] [PubMed] [Google Scholar]
- Simerly C. et al. (2003) Molecular correlates of primate nuclear transfer failures. Science, 300, 297. [DOI] [PubMed] [Google Scholar]
- Slimane-Bureau W.C. and King,W.A. (2002) Chromosomal abnormalities: a potential quality issue for cloned cattle embryos. Cloning Stem Cells, 4, 319–329. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Togashi,M., Adachi,J. and Toyoda,Y. (1995) Developmental ability of zona-free mouse embryos is influenced by cell association at the 4-cell stage. Biol. Reprod., 53, 78–83. [DOI] [PubMed] [Google Scholar]
- Szabò P.E., Hübner,K., Schöler,H. and Mann,J.R. (2002) Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev., 115, 157–160. [DOI] [PubMed] [Google Scholar]
- Tamashiro K.L. et al. (2002) Cloned mice have an obese phenotype not transmitted to their offspring. Nat. Med., 8, 262–267. [DOI] [PubMed] [Google Scholar]
- Willadsen S.M. (1981) The development capacity of blastomeres from 4- and 8-cell sheep embryos. J. Embryol. Exp. Morphol., 65, 165–172. [PubMed] [Google Scholar]
- Willadsen S.M. and Polge,C. (1981) Attempts to produce monozygotic quadruplets in cattle by blastomere separation. Vet. Rec., 108, 211–213. [DOI] [PubMed] [Google Scholar]
- Wun W.S., Wun,C.C. and Grunert,G.M. (1994) Minimum essential medium α (MEM) enhances assisted reproductive technology results. I. Mouse embryo study. J. Assist. Reprod. Genet., 11, 303–307. [DOI] [PubMed] [Google Scholar]
- Yeom Y.I. et al. (1996) Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development, 122, 881–894. [DOI] [PubMed] [Google Scholar]
- Yin X.J., Kato,Y. and Tsunoda,Y. (2002) Effect of delayed enucleation on the developmental potential of nuclear-transferred oocytes receiving adult and fetal fibroblast cells. Zygote, 10, 217–222. [DOI] [PubMed] [Google Scholar]