Abstract
The programme of gene expression induced by RelA/NF-κB transcription factors is critical to the control of cell survival. Ligation of ‘death receptors’ such as tumor necrosis factor receptor 1 (TNF-R1) triggers apoptosis, as well as NF-κB, which counteracts this process by activating the transcription of anti-apoptotic genes. In addition to activating caspases, TNF-R1 stimulation causes the release of cathepsins, most notably cathepsin B, from the lysosome into the cytoplasm where they induce apoptosis. Here we report a mechanism by which NF-κB protects cells against TNF-α-induced apoptosis: inhibition of the lysosomal pathway of apoptosis. NF-κB can protect cells from death after TNF-R1 stimulation, by extinguishing cathepsin B activity in the cytosol. This activity of NF-κB is mediated, at least in part, by the upregulation of Serine protease inhibitor 2A (Spi2A), a potent inhibitor of cathepsin B. Indeed, Spi2A can substitute for NF-κB in suppressing the induction of cathepsin B activity in the cytosol. Thus, inhibition of cathepsin B by Spi2A is a mechanism by which NF-κB protects cells from lysosome-mediated apoptosis.
Keywords: apoptosis/cathepsin/lysosome/NF-κB/serpin
Introduction
During immune and inflammatory responses, NF-κB/Rel transcription factors control cell survival (Ghosh et al., 1998). Normally, NF-κB heterodimers of p50-Rel A (p65) are sequestered in the cytoplasm by binding to inhibitory κB proteins (IκB), and can be activated by signals that induce sequential phosphorylation and proteolysis of IκBs. The analysis of RelA-deficient mice first revealed a role for NF-κB in suppressing apoptosis. RelA–/– mice die before birth due to liver apoptosis (Beg et al., 1995), and RelA–/– murine embryonic fibroblasts (MEFs) are sensitive to apoptosis caused by TNF-α. This is in contrast to wild-type MEFs and cells, which are only sensitive to TNF-α if the induction of protective factors by NF-κB is blocked by inhibitors of protein synthesis (Beg and Baltimore, 1996). The protective proteins upregulated by NF-κB act in concert to antagonize the release of mitochondrial proteins into the cytoplasm and the caspase protease cascade that leads to apoptosis (Budihardjo et al., 1999).
Recent evidence has revealed that, in addition to the mitochondrion, the lysosome also participates in cell death (Ferri and Kroemer, 2001). Importantly, studies with knockout mice have revealed a role for lysosomal cathepsins, notably cathepsin B, as a potent inducer of apoptosis (Guicciardi et al., 2000; Ferri and Kroemer, 2001; Foghsgaard et al., 2001). Ligation of TNF-R1 has been shown to result in the activation of the lysosomal enzymes acid sphingomyelinase and ceramidase (Schutze et al., 1999), and the production of the lysosomotropic detergent sphingosine which induces lysosomal breakdown (Kagedal et al., 2001; Werneburg et al., 2002). Cathepsin B is released into the cytoplasm where it activates caspase-dependent and caspase-independent pathways of cell death (Guicciardi et al., 2000; Ferri and Kroemer, 2001; Foghsgaard et al., 2001). Although the means by which cathepsins induce apoptosis are not fully elucidated, cleavage of the pro-apoptotic member of the Bcl-2 family (Bid) is a possible mechanism by which cathepsin B activates the mitochondrial pathway of apoptosis (Budihardjo et al., 1999; Stoka et al., 2001). The ability of NF-κB to block apoptosis completely implies that the transcription factor activates genes that are able to block the lysosomal pathway. However, these genes have not been identified.
Members of the superfamily of serine protease inhibitors (serpins) can modulate apoptosis through the inhibition of cytosolic executioner proteases. For example, the ovalbumin (ova) like serpins cytokine response modifier A (Crm A), from cowpox, and proteinase inhibitor 9 (PI9), from humans, are potent inhibitors of the serine protease granzyme B and so protect cells from lysis by cytotoxic lymphocytes (Quan et al., 1995; Sun et al., 1996). In addition, both proteins can also inhibit cysteine proteases such as caspases and so can protect cells from apoptosis triggered by the death receptors Fas and TNF-R1 (Komiyama et al., 1994; Tewari and Dixit, 1995; Zhou et al., 1997; Annand et al., 1999). Thus, given the ability of NF-κB to block TNF-α-mediated apoptosis completely, we investigated whether the transcription factor activated anti-apoptotic serpins.
The transcription of the mouse gene encoding the anti-chymotrypsin-like serpin, Serine protease inhibitor 2A (Spi2A), is induced by inflammatory stimulation and, uniquely for a serpin gene, depends on NF-κ-binding (Inglis et al., 1991; Hampson et al., 1997, 2001). The predicted reactive center domain of Spi2A, which is thought to interact with a target protease, is unusual for a member of the anti-chymotryspin family. Indeed, the cytosolic location of Spi2A (Morris et al., 2003) and the presence of cysteines at the critical P1 and P1′ positions of the reactive center are reminiscent of anti-apoptotic ova-serpins such as CrmA and PI9 (Quan et al., 1995; Sun et al., 1996). We found that complementation of RelA–/– MEFs with Rel A abrogates the induction of cytosolic cathepsin B after stimulation of TNF-R1, and protects against apoptosis. Purification of Spi2A revealed that it is an inhibitor of several cysteine cathepsins including cathepsin B. Importantly, we observed that Spi2A can substitute for RelA as a physiologically relevant inhibitor of cytosolic cathepsin B and a cytoprotective factor. Our findings describe a mechanism by which NF-κB suppresses the lysosomal pathway of cell death.
Results
NF-κB antagonizes the lysosomal pathway of cell death
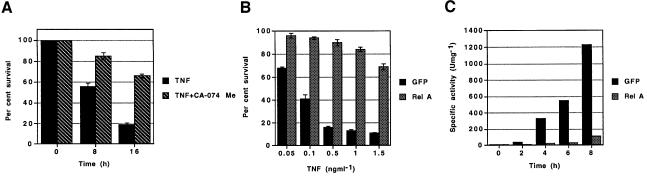
NF-κB protects cells from TNF-α-mediated death through the upregulation of protective genes which inhibit the apoptotic cascade at several different points. A role for cathepsin B has been demonstrated in the TNF-R1-induced death of several types of tumor cells using specific inhibitors of cathepsin B, such as CA-074 Me (Foghsgaard et al., 2001). The complete inhibition of cathepsin B activity by CA-074 Me (30 µM) protected RelA–/– MEFs from TNF-α-induced death (Figure 1A). Therefore, cathepsin B activity contributes to the susceptibility of RelA–/– MEFs to TNF-α-induced apoptosis.
Fig. 1. NF-κB antagonizes the lysosomal pathway of cell death (A) Percentage survival of RelA–/– MEFs treatment with TNF-α (0.5 ng/ml) and CHX (0.1 µg/ml) in the presence (TNF + CA-074 Me) or absence (TNF) of CA-074 Me (30 µM). The recovery of cells was compared with those incubated with CHX alone (100% recovery) to determine the percentage of recovery. (B) Percentage survival of RelA–/– MEFs transduced by retrovirus encoding GFP alone or Rel A. The recovery of cells after 16 h was compared with those incubated with CHX alone (0.1 µg/ml) to determine the percentage of recovery (100% recovery). (C) Cathepsin B activity in crude cytoplasmic extracts from RelA–/– MEFs transduced by retrovirus encoding GFP alone or RelA after treatment with TNF-α (0.2 ng/ml) and CHX (0.1 µg/ml). This experiment is representative of two independent experiments.
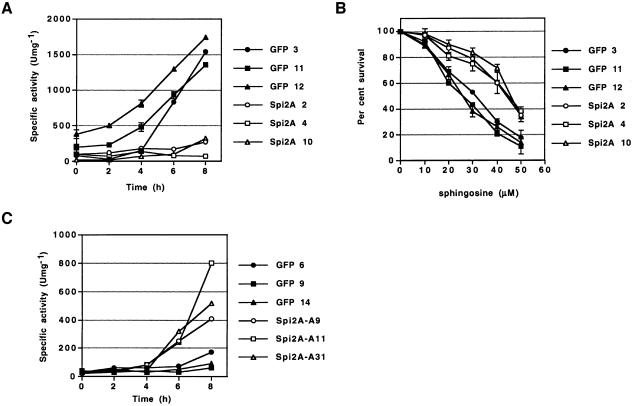
Studies in primary and tumor cells have demonstrated that activation of TNF-R1 results in the release of cathepsin B from the lysosome into the cytoplasm where it triggers apoptosis (Guicciardi et al., 2000; Foghsgaard et al., 2001; Werneburg et al., 2002). Using RelA–/– MEFs, we examined the effect of RelA complementation on the induction of cytosolic cathepsin B activity after TNF-α treatment. RelA–/– MEFs were transduced with retrovirus encoding RelA on a polycistronic mRNA encoding GFP (Zhang and Ren, 1998). As has been shown before, expression of RelA in RelA–/– MEFs (see Supplementary figure 1 available at The EMBO Journal Online) restored NF-κB function and gave complete protection from TNF-α-cytotoxicity (Figure 1B) (Beg and Baltimore, 1996). We next examined the influence of NF-κB/RelA on the induction of cathepsin B activity in the cytosol after treatment with TNF-α. We observed an increase in cathepsin B activity of cytosolic extracts from control RelA–/– MEFs as early as 2 h after treatment with TNF-α, which then increased with time (Figure 1C) . In contrast, transduction with RelA extinguished cathepsin B activity in the cytoplasm of RelA–/– MEFs for as long as 8 h after treatment with TNF-α (Figure 1C). Thus NF-κB may upregulate genes that inhibit cathepsin B activity in the cytosol.
Induction of Spi2A by NF-κB protects from TNF-α-mediated cell death
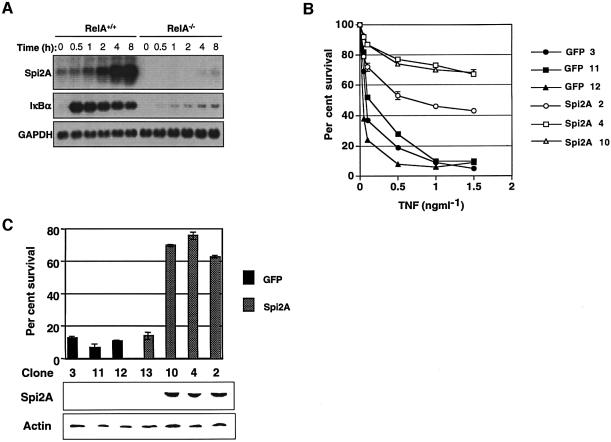
The transcription of Spi2A is induced by inflammatory stimulation and depends on NF-κ binding (Hampson et al., 1997, 2001; Inglis et al., 1991). Initially we examined whether Spi2A was a physiologic target of NF-κB. Spi2A mRNA (2.3 kb) was strongly induced by TNF-α in RelA+/+ MEFs, but this induction was completely abolished in NF-κB/RelA–/– MEFs (Beg and Baltimore, 1996) (Figure 2A). While dramatic, the induction of Spi2A expression occurred with slower kinetics than the expression of ikbα, a known target of NF-κB (De Smaele et al., 2001). We conclude that Spi2A is a physiological target of NF-κB.
Fig. 2. Induction of Spi2A by NF-κB protects from TNF-α-mediated death. (A) Northern blots of mRNA from MEFs treated with TNF-α (0.2 ng/ml) and CHX (0.1 µg/ml) (B) Percentage survival of RelA–/– MEFs transduced by retrovirus encoding GFP alone or Spi2A. The recovery of cells after 16 h was compared with those incubated with CHX alone (100% recovery) to determine the percentage of recovery. (C) Western blot detection of Spi2A from GFP and Spi2A clones of RelA–/– MEF cells and correlation with survival after treatment with TNF-α (1 ng/ml) and CHX (0.1 µg/ml).
The control of cell survival is critically dependent on the induction of protective genes by NF-κB transcription factors (Karin and Lin, 2002). We examined whether Spi2A can protect RelA–/– MEFs from TNF-α–induced death. RelA–/– MEFs were transduced with retrovirus encoding Spi2A on a polycistronic mRNA with the GFP gene (Zhang and Ren, 1998). Cells from stable clones transduced with Spi2A (Spi2A cells) exhibited markedly improved survival against TNF-α, whereas cloned cells transduced with vector alone (GFP cells) did not (Figure 2B). Protection of RelA–/– MEFs from TNF-α correlated with the expression of Spi2A protein (Figure 2C). At low concentrations of TNF-α protection by Spi2A was virtually complete (Figure 2B; see 0.5 ng/ml TNF-α) and was dramatic even after 16 h at high concentrations, indicating that Spi2A can temporarily substitute for NF-κB complexes in inhibiting TNF-α-induced apoptosis.
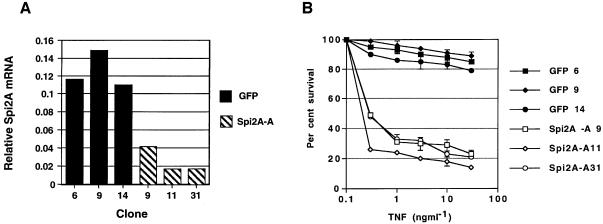
To verify that cytoprotection mediated by Spi2A was not due to overexpression, we generated wild-type (RelA+/+) MEFs expressing Spi2A in an antisense orientation (Spi2A-A cells). After treatment with TNF-α, analysis by real-time PCR revealed that the upregulation of endogenous Spi2A mRNA was abrogated in stable clones of Spi2A-A cells (Medhurst et al., 2000) (Figure 3A). Despite their ability to activate NF-κB (Supplementary figure 2), Spi2A-A cells exhibited a marked susceptibility to TNF-α-induced cell death (Figure 3B). The sensitivity of Spi2-A cells to TNF-α was also observed in the absence of cyclohexamide (CHX), indicating that TNF-α cytotoxicity was not due to an inhibition of protein synthesis in Rel A+/+ MEFs (Supplementary figure 3). Thus Spi2A is required to antagonize TNF-α-induced apoptosis, and protection from death is a physiological function of Spi2A.
Fig. 3. Spi2A is required for the protection of wild-type MEFs from TNF-α-induced death. (A) Quantitation of endogenous Spi2A mRNA levels by real-time PCR in cloned RelA+/+ MEFs transduced by retrovirus encoding GFP alone or antisense Spi2A (Spi2A-A) 4 h after treatment with TNF-α (10 ng/ml) and CHX(10 µg/ml). (B) Percentage survival of GFP clones and Spi2A-A clones of RelA+/+ MEFs 16 h after treatment with TNF-α and CHX (10 µg/ml).
Spi2A protects from apoptosis
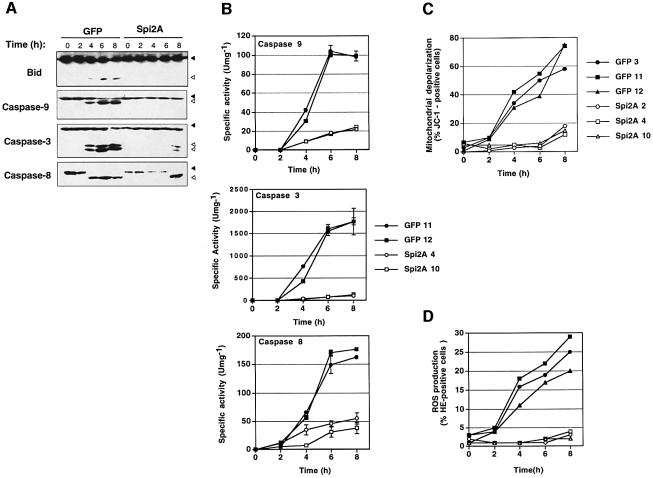
NF-κB protects cells from death induced by TNF-α by upregulating the expression of genes which antagonize the mitochondrial pathway of apoptosis (Beg and Baltimore, 1996; Baldwin, 2001). Given the ability of Spi2A to substitute for NF-κB complexes in protecting from TNF-α, we determined whether Spi2A could inhibit the mitochondrial pathway of apoptosis. In RelA–/– MEFs, TNF-α activation of caspases 3, 8 and 9, and the pro-apoptotic Bcl-2 family member Bid, was assessed by western blots (Figure 4A) and in vitro enzyme assays (Budihardjo et al., 1999; Stegh et al., 2000) (Figure 4B). Remarkably, the activation of both apical and executioner caspases, as well as Bid, was suppressed in RelA–/– MEFs that expressed high levels of Spi2A. In these cells mitochondrial depolarization, a key indicator of apoptosis, was virtually abrogated by Spi2A (Budihardjo et al., 1999) (Figure 4C). Importantly, Spi2A also suppressed the production of reactive oxygen species (ROS) which mediate TNF-α cytotoxicity (Goossens et al., 1995) (Figure 4D). Thus Spi2A abrogates TNF-α-induced caspase activation, mitochondrial depolarization and ROS production in NF-κB null cells, thereby recapitulating the effects of the transcription factor on apoptosis (Wang et al., 1998).
Fig. 4. Spi2A inhibits apoptosis induced by TNF-α. (A) Western blots showing the proteolytic activation of effector molecules from RelA–/– MEFs [GFP (clone 11) or Spi2A (clone 4)] after treatment with TNF-α (0.2 ng/ml) and CHX (0.1 µg/ml). Filled arrows indicate inactive pro-form and open arrows indicate active form of each protein. RelA–/– MEFs [GFP (clone 11) or Spi2A (clone 4)] were treated with TNF-α and CHX as above and the following were measured: (B) caspase activity, (C) mitochondrial depolarization and (D) ROS.
Spi2A inhibits lysosomal cysteine cathepsins
To determine the mechanism by which Spi2A antagonized apoptosis we examined the protease specificity of Spi2A in vitro. Spi2A was purified from RelA–/– MEFs transduced with retrovirus encoding epitope-tagged Spi2A (Cooley et al., 2001) (Figure 5A). Spi2A inhibited both serine and cysteine proteases, similar to the serpin SQN-5 (Al-Khunaizi et al., 2002). Spi2A inhibited the chymotrypsin-like serine protease cathepsin G, but not elastase or either granzyme B or granzyme A (Figure 5B). The specificity of Spi2A for cysteine proteases extended to all of the lysosomal papain-like proteases that were examined: cathepsins B, V, L, K and H. Spi2A inhibited cathepsin B with a rate constant k >106 M–1 s–1 (Supplementary data, part 4), and so is likely to be a physiologically relevant inhibitor in vivo (Silverman et al., 2001). However, the inhibitory effects of Spi2A did not extend to any of the caspases tested (caspases 3, 8 and 9) (Figure 5B). Thus, Spi2A is a cross-class specific inhibitor of both serine proteases and lysosomal cysteine cathepsins.
Fig. 5. The protease specificity of Spi2A. (A) SDS–PAGE showing Spi2A (lane P, 53 kDa) purified from lysates (lane L) of RelA–/– MEFs transduced with retrovirus encoding Spi2A-3xFLAG. (B) Inhibition of proteases by Spi2A. The activity of protease after preincubation with Spi2A was compared with activity from protease incubated alone (0% inhibition) and is expressed as mean ± SEM of three or four independent experiments with assays performed in duplicate.
Spi2A localizes to the cytoplasm and nucleus
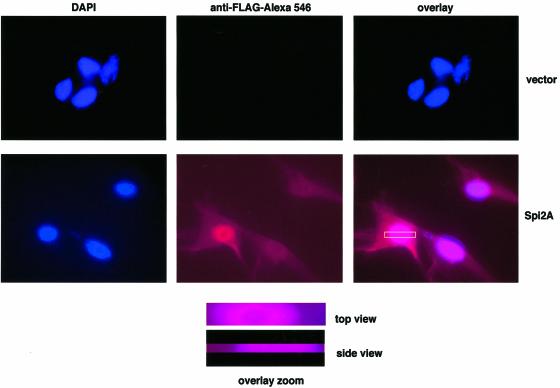
Spi2A is an unusual member of the chymotrypsin-like family of serpins in that it lacks a secretory signal sequence and so is likely to be located intracellularly (Hampson et al., 1997). To examine further the effect of Spi2A in protecting from TNF-α-induced apoptosis we first determined the intracellular location of FLAG-tagged Spi2A in stably transduced Rel A–/– MEFs (Figure 2B). Immunofluorescence studies revealed staining with anti-FLAG antibodies in the cytoplasm and nucleus (Figure 6). Z-section analysis confirmed uniform distribution of anti-FLAG staining throughout the cytoplasm rather than in the plasma membrane. We conclude that Spi2A resides in the cytoplasm and nucleus. The nucleocytoplasmic localization of Spi2A revealed by our studies is concordant with findings of others with macrophage cell lines and COS cells using Spi2A antisera in immunofluorescence studies (Morris et al., 2003). Localization in the cytoplasm raises the possibility that Spi2A may protect from apoptosis through the inhibition of cathepsin activity after release from the lysosome (Figure 1C).
Fig. 6. Intracellular localization of Spi2A. Vector (GFP clone 11, top) or Spi2A transfected (Spi2A clone 4, bottom) Rel A–/– MEFs were stained for FLAG (middle panel) or DAPI (left panel); the right panel shows the overlay of the two channels. Overlay zoom, top and side views. In both views, the FLAG label appears to be uniformly distributed through the cytoplasm.
Spi2A antagonizes the lysosomal pathway of cell death
The upregulation of Spi2A by NF-κB protects cells from apoptosis following ligation of TNF-R1 (Figure 2). Spi2A can inhibit cathepsin B in vitro (Figure 5B), and is located in the cytosol (Figure 6). Therefore the induction of Spi2A and inhibition of cathepsin B after it is released into the cytoplasm may be a mechanism by which NF-κB antagonizes the lysosomal pathway of cell death (Figure 1).
As was observed with Rel A complementation (Figure 1B), Spi2A inhibited the induction of cytosolic cathepsin B activity after treatment of Rel A–/– MEFs with TNF-α (Figure 7A). Direct treatment of cells with sphingosine causes the release of cathepsin B from the lysosome and the induction of apoptosis (Foghsgaard et al., 2001; Kagedal et al., 2001; Werneburg et al., 2002). Consistent with a role in protecting from lysosome-mediated apoptosis, Spi2A could protect Rel A–/– MEFs from death after treatment with sphingosine (Figure 7B). Overall, we conclude that Spi2A abrogates TNF-α-induced activation of cytoplasmic cathepsin B in NF-κB null cells, thereby recapitulating the effects of the transcription factor on the lysosomal pathway of apoptosis.
Fig. 7. Spi2A antagonizes the lysosomal pathway of cell death. (A) Cathepsin B activity in crude cytoplasmic extracts from cloned RelA–/– MEFs transduced by retrovirus encoding GFP alone or Spi2A after treatment with TNF-α and CHX as described before. (B) Percentage survival of GFP and Spi2A clones of RelA–/– MEFs 2 h after treatment with sphingosine. (C) Cathepsin B activity in crude cytoplasmic extracts from cloned RelA+/+ MEFs transduced by retrovirus encoding GFP alone or antisense Spi2A (Spi2A-A) after treatment with TNF-α (10 ng/ml) and CHX (10 µg/ml).
Importantly, the inhibition of endogenous Spi2A mRNA expression by antisense Spi2A resulted in the induction of cytoplasmic cathepsin B activity after treatment of RelA+/+ MEFs with TNF-α (Figure 7C). We conclude that the inhibition of cathepsin B activity in the cytosol by Spi2A is a physiologically relevant mechanism by which NF-κB protects cells from the lysosomal pathway of apoptosis.
Discussion
The program of gene expression induced by RelA/NF-κB transcription factors is critical in controlling cell survival in response to a variety of apoptotic stimuli (Baldwin, 2001). NF-κB activates multiple target genes whose products can block the apoptotic program triggered by death receptors or the mitochondrial pathway. NF-κB-inducible anti-apoptotic factors include those that inhibit caspase function, such as the cellular inhibitor of apoptosis proteins (c-IAPs), X-chromosome-linked IAP (XIAP) and caspase-8-c-FLIP (FLICE inhibitory protein), those that inhibit NF-κB signaling after TNF-R1 stimulation, such as TNF-R-associated factors 1 and 2 (TRAF1 and TRAF2), those that preserve mitochondrial function, such as Bcl-xL and A1/Bf1, and those that inhibit pro-apoptotic signaling by the c-Jun kinase (JNK) pathway, such as Gadd45β and XIAP (Karin and Lin, 2002). Stimulation of TNF-R1 induces the breakdown of the lysosome and the induction of apoptosis by cathepsins released into the cytoplasm (Guicciardi et al., 2000; Ferri and Kroemer, 2001; Foghsgaard et al., 2001). We report a novel mechanism by which NF-κB protects cells against TNF-α-induced apoptosis: inhibition of the lysosomal pathway of apoptosis. The upregulation of Spi2A and inhibition of cytoplasmic cathepsin B, after release from lysosomes, represents a physiologically relevant mechanism by which NF-κB blocks the lysosomal pathway of cell death in MEFs. The wider physiological importance of cytoprotection from TNF-α by Spi2A is suggested by the finding that the Spi2A gene is located in the ‘TNF protection locus’ on chromosome 12 in mice (Libert et al., 1999).
Several observations lead us to conclude that the induction of Spi2A by NF-κB protects cells from TNF-α by inhibiting the lysosomal pathway of apoptosis. Inhibition of cathepsin B by CA-074 Me protects RelA–/– from TNF-α-induced apoptosis, confirming the observations that cathepsin B plays a direct role in apoptosis in other cell types (Guicciardi et al., 2000; Foghsgaard et al., 2001). Treatment of RelA–/– MEFs with TNF-α leads to the induction of cytosolic cathepsin B activity after only 2 h (Figures 1C and 7A). This precedes the onset of apoptosis, as measured by caspase or Bid activation (Figure 4A and B), mitochondrial depolarization (Figure 4C) and ROS production (Figure 4D), by 2 h. Therefore, in our system, the release of cathepsin B into the cytoplasm does not seem to be a consequence of apoptosis but rather precedes it. We demonstrate that Spi2A is a broad specific inhibitor of lysosomal cathepsins, including cathepsin B, and is located in the cytoplasm (Figures 5B and 6). Therefore the suppression of cathepsin B activity by Spi2A is at least one mechanism by which NF-κB antagonizes the lysosomal pathway of apoptosis.
We observed that Spi2A can partially inhibit the increase in the pH of lysosomes that occurs after TNF-α treatment, as assessed by the appearance of acridine orange (AO) low cells (Kagedal et al., 2001) (Supplementary figure 4). This could mean that, in addition to inhibiting cathepsin B activity in the cytoplasm, Spi2A may also prevent lysosomal breakdown. In the light of a recent study that suggests a direct role for cathepsin B in lysosomal breakdown (Werneburg et al., 2002), there is a possibility that Spi2A acts to preserve lysosomal integrity. Spi2A is located in the cytosol, but it is not clear whether lysosomal or cytoplasmic cathepsin B catalyzes lysosomal breakdown. Direct measurement of cathepsin traffic out of the lysosome in Spi2A-expressing Rel A–/–MEFs will be required to test this possibility when suitable reagents become available.
The relationship between the lysosomal and caspase pathways of apoptosis is not fully elucidated and seems to depend on the nature of the apoptotic signal and cell type in question (Ferri and Kroemer, 2001; Foghsgaard et al., 2001). After cleavage by caspase 8, Bid activates the mitochondrial pathway of apoptosis (Budihardjo et al., 1999). Importantly, cathepsin B can cleave Bid but not pro-caspases in vitro, and so Bid may also be a possible link between the lysosomal and mitochondrial pathways of apoptosis (Stoka et al., 2001). Spi2A cannot directly inhibit caspases 3, 8 or 9, and yet caspase activity in RelA–/– MEFs treated with TNF-α is abrogated by Spi2A expression (Figures 4A and B, and 5B). After ligation of TNF-R1, Spi2A inhibits cytoplasmic cathepsin B activity and so may prevent apoptosis by inhibiting the cleavage of Bid by cathepsin B and subsequent caspase activation by proteins released from mitochondria (Scaffidi et al., 1998). The interconnectedness of the lysosomal and mitochondrial pathways of apoptosis and the ability of damaged mitochondria to feedback and activate caspase 8 (Scaffidi et al., 1998) likely account for the ability of Spi2A to restore so robustly the NF-κB-phenotype of resistance to TNF-α to Rel A–/– MEFs. However, compared with other protective genes induced by NF-κB, Spi2A is certainly not unique in this regard (De Smaele et al., 2001).
The inhibition of papain-like cysteine cathepsins has been observed not only for Spi2A (Figure 5B) but also for other serpins, such as squamous cell carcinoma antigen 1 (SCCA1) from humans (Schick et al., 1998) and SQN-5 from mice (Al-Khunaizi et al., 2002). Unlike other inhibitors of executioner protease, such as c-IAPs which inhibit caspases (Deveraux et al., 1998) and cystatins which inhibit papain-like cysteine cathepsins (Turk and Bode, 1991), serpins characteristically act as ‘suicide substrates’ and inactivate proteases through the formation of a 1:1 covalent complex (Silverman et al., 2001). The mechanism by which Spi2A inhibits cathepsins may be by acting as a suicide substrate because Spi2A can be cleaved by cathepsin G, cathepsin B and cathepsin K in vitro (Morris et al., 2003; S.M.Raja, C.J.Froelich and P.G.Ashton-Rickardt, unpublished data). However, whether inhibition of cysteine cathepsins by Spi2A is mediated by the formation of covalent complexes, as with some but not all cross-class specific serpins, remains to be determined (Komiyama et al., 1994; Annand et al., 1999; Al-Khunaizi et al., 2002).
Despite the fact that the mechanism by which Spi2A inhibits pro-apoptotic cathepsins is not fully elucidated, antisense experiments show that Spi2A is a physiologically relevant inhibitor of cytoplasmic cathepsin B (Figure 7C). Furthermore, we demonstrate that cathepsin B is a mediator of TNF-α-induced apoptosis in RelA–/– MEFs (Figure 1A). Spi2A inhibited every cysteine cathepsin we tested, and so it is possible that the protective function of Spi2A may extend to the inhibition of other pro-apoptotic cathepsins. Indeed, the inhibition of cathepsins by other serpins seems to protect cells from TNF-α-induced apoptosis. For example, the overexpression of the human serpin SCCA 2 can inhibit TNF-α-induced apoptosis of HeLa cells possibly through inhibition of cathepsin G (McGettrick et al., 2001). Although our studies support an anti-apoptotic mechanism by which Spi2A inhibits cathepsin B activity in the cytoplasm, Spi2A in the nucleus may also protect from apoptosis. Cathepsin B has been localized to the nuclear membrane in human tumor cells and so, like cystatin B, Spi2A may protect cells by inhibiting the activity of papain-like cathepsins in the nucleus (Speiss et al., 1994; Riccio et al., 2001).
The loss of lysosome integrity and the release of cathepsins and other digestive enzymes is a critical event in the induction not only of apoptosis but also of coagulative necrosis (Wyllie et al., 1981; Ferri and Kroemer, 2001). Therefore one could predict that induction of Spi2A and NF-κB may provide protection from necrosis as well as apoptosis. Further investigation of the potential role of NF-κB and Spi2A in affording protection from necrosis during vertebrate development (Chautan et al., 1999) and the pathogenesis of disease (Wyllie et al., 1981), using in vivo models, will shed light on this potentially exciting new aspect of NF-κB biology.
Materials and methods
Spi2A mRNA expression
Total RNA (4 µg) was extracted from MEFs after treatment with TNF-α (0.2 ng/ml) (R&D) and cyclohexamide (CHX) (0.1 µg/ml) using Trizol Reagent according to the manufacturer’s instructions (Invitrogen) and northern blots were prepared using standard procedures (Sambrook et al., 1989). Blots were probed with an [α-32P]dCTP-hexamer-labeled cDNA probe encoding Spi2A (Hampson et al., 1997). Blots were stripped and reprobed with similarly labeled control probes encoding IκBα or GAPDH (De Smaele et al., 2001).
In Spi2A-antisense experiments, the level of Spi2A mRNA was quantitated by real-time PCR using primers and probes specific for Spi2A (forward primer 5′-AACCAGAGACCCTGAGGAAGTG-3′; reverse primer 5′-AACTTGGGCAGGCGCAG-3′; probe 5′-AAGAACTC TCTGAAGCCCAGGATGATACATGA-3′) (Inglis et al., 1991) and the cyclophilin A housekeeping control gene (Medhurst et al., 2000) (forward primer 5′-CCATCAAACCATTCCTTCTGTAGC-3′; reverse primer 5′-AGCAGAGATTACAGGACATTGCG-3′; probe 5′-CAGGAGAGCGT GCCTACCCCATCTG-3′) (Megabases Inc). Probes were labeled with the fluorescent reporter dye FAM. Four hours after treatment with CHX (10 µg/ml) and TNF-α (10 ng/ml), RNA was extracted from RelA+/+MEFs using Trizol Reagent (Invitrogen) and then cDNA was generated using Superscript First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time PCR reactions were carried out using TaqMan Universal PCR Master Mix according to the manufacturer’s recommended protocol (PE Applied Biosystems) and analyzed on an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems). Data were captured and analyzed using Sequence Detector software (PE Applied Biosystems). The slope of the standard curve describes the efficiency of the real-time PCR, which allowed us to ensure that the real-time PCR reactions consistently ran at >90% efficiency. The relative concentration of Spi2A RNA was calculated by dividing the concentration of Spi2A RNA by that of the cyclophilin A control gene (Hasel and Sutcliffe, 1990).
Retroviral transduction of MEFs
The cDNA for human RelA (p65) was subcloned into the Hpa 1 restriction site of the MIGR1 retroviral vector in the forward orientation (Franzoso et al., 1996; Zhang and Ren, 1998). The Spi2A open reading frame (ORF) was amplified by PCR from cDNA prepared from purified T cells using a forward primer (5′-AGAATTCGCCACCATGGCTG GTGTCTCCCCTG-3′) and a reverse primer (5′-TGTGGATCCTCCC TGTCAAATCAGGCAGCATAGCGGAT-3′). These primers introduced 5′ BamHI and 3′ EcoR1 restriction sites and mutated the stop codon of Spi2A ORF to facilitate the production of an inframe fusion protein between Spi2A and C-terminal 3xFLAG (22 amino acids) after cloning into the 3xFLAG-CMV-14 expression vector (Sigma-Aldrich). Using the same forward primer and a reverse primer specific for 3xFLAG DNA that introduced an EcoR1 restriction site (5′-GTGAATTCATCACTAC TTGTCATCGT-3′), the Spi2A-3xFLAG ORF was amplified by PCR and then subcloned into the EcoR1 site of the MIGR1 retroviral vector in the forward or reverse orientations. The MIGR1 retroviral vector directed the expression of RelA, Spi2A-3xFLAG or Spi2A-3xFLAG antisense mRNA as a biscistronic mRNA encoding GFP.
Retrovirus was produced as described previously (Burns et al., 1993). Briefly, cells of the 293 GP packaging line (4 × 106) were transiently transfected with MIGR1-Spi2A-3xFLAG DNA (6 µg) and DNA-encoding vesticular stomatis virus (VSV) glycoprotein (6 µg) using Lipofectamine PLUS reagent according to manufacturer’s instructions (Invitrogen). After 48 and 72 h supernatant containing retrovirus was harvested, filtered and stored at –80°C until needed. MEFs (1–2 × 105) were seeded in six-well plates and transduced with 4 ml of retroviral supernatant containing polybrene (8 µg/ml) by centrifugation at 1000 g for 1 h at room temperature, followed by incubation at 37°C for 24 h. After 48 h, the transduction efficiency was determined by measuring the percentage of GFP-positive MEFs by FACS, which was routinely 96–98%. Transduced MEFs that were in the top 5% of GFP expression were purified by FACS and cloned.
Fluorescence microscopy
RelA–/– MEFs were transduced with retrovirus encoding Spi2A-3xFLAG or empty vector and plated in a Chamber-Slide (Lab-Tek, Nalge Nunc) overnight at 10 000 cells per chamber in 10% fetal calf serum (FCS) containing Dulbecco′s modified Eagle′s medium (DMEM). Immunofluorescence localization for Spi2A was performed using the anti-FLAG antibody (Sigma). Briefly, the cells were washed three times with chilled phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde (PFA)–PBS for 15 min at room temperature (RT), permeabilized using 0.5% Triton X-100 (15 min at RT) followed by five washes with chilled PBS. The slides were blocked with 2% normal mouse serum (NMS) in PBS (45 min at RT) followed by incubation (90 min at RT) with the biotinylated anti-FLAG antibody (10 µg/ml). After washing off the unbound antibody, the slides were incubated (60 min at RT) with Streptavidin (SA)–Alexa 546 (1 µg/ml; Molecular Probes) followed by washes with chilled PBS. Finally the cells were mounted in Vectashield mounting medium (Vector Labs) containing DAPI as the nuclear stain. The cells were observed and imaged on a Leica DMIRE2 inverted microscope equipped with a Photometrics CoolSNAP HQ digital camera (Roper Scientific). Fluorescence images of each of the fluorophores were acquired sequentially using the following Chroma filter cubes: DAPI, cube 31000 (Ex. 340–380 nm, Em. 435–485 nm); FLAG, cube 41004 (Ex. 535–585, Em. 610–680 nm). Images were acquired and then overlaid using Meta Imaging Series software version 4.6.5 (Universal Imaging Corporation). To determine whether FLAG was distributed throughout the cytoplasm (rather than being bound to the plasma membrane), Z-series of individual cells were captured and deconvolved using MetaMorph. A three-dimensional model was reconstructed from the Z-series, and then sliced and rotated (again using MetaMorph) to obtain a side view.
Protein expression
Antiserum specific to Spi2A peptides [peptide 1 (amino acids 406–423), NH2-(C) NPERSTNFPNGEGASSQR-COOH; peptide 2 (amino acids 278–294), NH2-(C) SLQPETLRKWKNSLKPR-COOH ] was raised in rabbits using standard procedures (Coligan et al., 1995). Briefly, two rabbits were immunized with each peptide conjugated to KLH and then boosted twice with immunogen over a period of 3 months. Anti-Spi2A antibodies were affinity purified on columns of immunizing peptide, eluted in 3M KSCN and then dialyzed against PBS.
Detergent extracts from RelA–/– MEFs transduced with control or retrovirus encoding Spi2A were resolved by SDS–PAGE and then immunoblotted (25 µg per lane) and probed with either peptide-1- or peptide-2-specific anti-Spi2A antibodies (10 µg/ml) using standard protocols (Coligan et al., 1995). Spi2A was detected after probing with goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (Sigma-Aldrich) at 2 µg/ml and chemiluminescence (ECL-kit, Amersham). Antibodies purified from the two rabbits immunized with either peptide 1 or peptide 2 detected Spi2A as a 52 kDa protein in extracts from Spi2A cells but not GFP cells. To control for equal loading, blots were stripped and reprobed for actin (42 kDa) with anti-actin monoclonal antibody clone ACTN05 ( RDI Research Diagnostics Inc) at 0.5 µg/ml and anti-mouse IgG-HRP (Sigma-Aldrich) at 2 µg/ml.
Survival assays
Although RelA–/– MEFs are markedly more sensitive to TNF-α than RelA+/+ MEFs, we still used low levels of CHX (0.1 µg/ml) in our survival assays. This was to suppress any protective activity of constitutively active non-RelA NF-κB molecules that are present in RelA–/– MEFs (Figure 4). Thus RelA–/– MEFs were treated with CHX and TNF-α (R&D) and the number of live GFP-positive adherent cells was counted by flow cytometry after 16 h, if not indicated otherwise. Live cells were defined as those that excluded propidium iodide (PI-negative) and had the appropriate size, as defined by forward- and side-scatter characteristics (Coligan et al., 1995). For RelA+/+ MEFs, TNF-α cytotoxicity was determined after 16 h with CHX (10 µg/ml), if not indicated otherwise. Cathepsin B activity was inhibited by a 1 h pretreatment of MEFs with CA-074 Me (30 µM) (Peptide Institute). Complete inhibition of cathepsin B activity was verified by enzyme assay. RelA–/– MEFs were treated with 10–50 µM sphingosine (Calbiochem) and the number of live GFP-positive adherent cells was counted by flow cytometry after 2 h.
Death effector assays
Death effector pathways were induced in RelA–/– MEFs by treatment with TNF-α (0.2 ng/ml) and CHX (0.1 µg/ml). Assays for executioner proteases (caspases and cathepsin B) were performed on crude cytoplasmic extracts (Stegh et al., 2000). Briefly, MEFs (106) were lysed in 10 mM Tris–Cl pH 7.5, 100 mM NaCl, 1 mM EDTA and 0.01% Triton X-100 (50 µl) for 30 min on ice, and then centrifuged at 15 000 g for 30 min at 4°C and the supernatant recovered. Protein concentration was determined by Lowry assay (DC-protein assay kit, Bio-Rad). Western immunoblots were performed on crude cytosolic extracts (50 µg per lane) using standard protocols and probed with the following antibodies: goat anti-mouse Bid antiserum (1 µg/ml) (R&D systems), rabbit anti-human caspase 9 antiserum (2 µg/ml) (Cell Signaling Technology), rabbit anti-human caspase 3 antiserum (2 µg/ml) (Cell Signaling Technology) and mouse anti-human caspase 8 monoclonal antibody clone 12F5 (1 µg/ml) (Axxora). The following secondary antibodies were used: anti-goat IgG HRP (0.5 µg/ml) (Santa Cruz Technology), anti-rabbit IgG HRP (0.5 µg/ml) (Amersham), anti-mouse IgG HRP (0.5 µg/ml) (Santa Cruz Technology). Specific proteins were visualized using chemiluminesence (ECL-kit, Amersham).
Colorimetric assays for caspases were performed in reaction buffer [50 mM HEPES pH 7.4, 100 mM NaCl, 10 mM dithiothreitol (DTT), 1 mM EDTA, 10% glycerol, 0.1% CHAPS] at 37°C on crude cytoplasmic extracts using p-nitroaniline (pNA)-labeled substrates (Calbiochem) specific for caspases 3 and 7 (Ac-DEVD-pNA), caspase 8 (Ac-IETD-pNA) and caspase 9 (Ac-LEHD-pNA), each at 0.2 mM. Specific activity was determined by subtracting the apparent activity detected after preincubation of extract for 1 h with the pan-caspase inhibitor Z-VAD.fmk (50 µM) (ICN Biomedicals Inc.) and then normalizing for the amount of protein. Assays for cathepsin B activity in crude cytoplasmic extracts was performed in reaction buffer (100 mM KHPO4 pH 6.1, 2 mM DTT, 1 mM EDTA) at 37°C using the Z-RR-pNA substrate (Calbiochem), which is specific for cathepsin B but not for other cysteine cathepsins from the lysosome (Barrett and Kirschke, 1981), at 0.4 mM. Specific activity was determined by subtracting the apparent activity detected after 30 min preincubation of extract at 37°C with the cathepsin B inhibitor CA-074 Me (Peptide Institute) at 30 µM and then normalizing for the amount of protein.
Mitochondrial membrane potential and ROS production were measured using the fluorescent dyes JC-1 (3 µg/ml) and dihydroethidium (HE) (5 µM) (Molecular Probes), respectively, and flow cytometry according to the manufacturer’s instructions.
Protease specificity of Spi2A
RelA–/– MEFs were transduced with retrovirus encoding Spi2A with a C-terminal 3 xFLAG epitope tag and Spi2A-3xFLAG purified using the method described previously (Cooley et al., 1998). Briefly, cells (3 × 109) were lysed and Spi2A-3xFLAG (75 µg) was purified by batch Q-Fast Flow ion-exchange chromatography (Pharmacia Biotech) after elution at 160–220 mM NaCl followed by anti-FLAG antibody columns, performed according to the manufacturer’s instructions (Sigma-Aldrich). Spi2A-3xFLAG was dialyzed into PBS and stored as aliquots at –80°C until needed.
Proteases were purchased from the manufacturers (Calbiochem or Athens Research and Technology) except for granzymes A and B which were purified as described (Hanna et al., 1993) and cathepsins V and K which were purified as described (Linnevers et al., 1997; Bromme et al., 1999). Proteases (20 nM) were incubated in the appropriate assay buffer with Spi2A at 200 nM (at least 10-fold excess of inhibitor to maintain pseudo-first-order conditions) for 1 h at 37°C. Control samples included only the enzyme, without the inhibitor. After 1 h protease activity was assayed. For serine proteases the following substrates (Calbiochem) were used at 1 mM (Cooley et al., 2001; Al-Khunaizi et al., 2002): human cathepsin G, Suc-AAPF-pNA; human elastase, MeOSuc-AAPV-pNA in assay buffer (20 mM Tris–HCl pH 7.4, 500 mM NaCl, 0.1% PEG); human granzyme B, IETD-pNA; human granzyme A, BLT-pNA. For cysteine cathepsins the following substrates (Molecular Probes) were used at 5 µM: human cathepsins B, L, K and V, (Z-FR)2-R110; human cathepsin H, (Z-PR)2-R110 in assay buffer (50 mM NaAc pH 5.4, 4 mM DTT,1 mM EDTA) (Al-Khunaizi et al., 2002). Substrate hydrolysis was measured in a fluorescence microtiter plate reader (Spectramax Gemini XS, Molecular Devices). Percentage inhibition was calculated from the residual enzyme activity compared with no Spi2A controls. Incubation with alkaline phosphatase tagged with C-terminal 3 x FLAG (Sigma-Aldrich) under the same conditions had no effect on protease activity.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.Crispino for the MIGRI vector, M.Nishimura for the 293 GP packaging line and A.Murmann for laser confocal microscopy. We thank C.-R.Wang and A.H.Wyllie for useful comments. This work was supported by NIH grant AI45108 to P.G.A.-R. Requests for material should be addressed to P.G.A.-R.
References
- Al-Khunaizi M. et al. (2002) The serpin SQN-5 is a dual mechanistic-class inhibitor of serine and cysteine proteinases. Biochemistry, 41, 3189–3199. [DOI] [PubMed] [Google Scholar]
- Annand R.R., Dahlen,J.R., Sprecher,C.A., de Drue,P., Foster,D.C., Mankovich,J.A., Talanian,R.V., Kisiel,W. and Giegel,D.A. (1999) Caspase-1 (interleukin-1β-coverting enzyme) is inhibited by the human serpin proteinase inhibitor 9. Biochem. J., 342, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Baldwin A.S. (2001) Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J.Clin.Invest., 107, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A.J. and Kirschke,H. (1981) Cathepsin B, Cathepsin H and Cathepsin L. Methods Enzymol., 80, 535–559. [DOI] [PubMed] [Google Scholar]
- Beg A.A. and Baltimore,D. (1996) An essential role for NF-κB in preventing TNF-a-induced cell death. Science, 274, 782–787. [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha,W.C., Bronson,R.T., Ghost,S. and Baltimore,D. (1995) Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature, 376, 167–169. [DOI] [PubMed] [Google Scholar]
- Bromme D., Li,Z., Barnes,M. and Mehler,E. (1999) Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization and chromosomal localization. Biochemistry, 38, 2377–2385. [DOI] [PubMed] [Google Scholar]
- Budihardjo I., Oliver,H., Lutter,M., Luo,X. and Wang,X. (1999) Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell. Dev. Biol., 15, 269–290. [DOI] [PubMed] [Google Scholar]
- Burns J.C., Friedmann,T., Driever,W., Burrascano,M. and Yee,J.-K. (1993) Vesticular stomatitis viruse G glycoprotein pseudotyped retrovirus vectors: concentration to very high titer and efficient gene transfer into mammalian and non-mammalian cells. Proc. Natl Acad. Sci. USA, 90, 8033–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautan M., Chazal,G., Cecconi,F., Gruss,P. and Golstein,P. (1999) Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr. Biol, 9, 967–970. [DOI] [PubMed] [Google Scholar]
- Coligan J.E., Kruisbeek,A.M., Margulis,D.H., Shevach,E.M. and Strober,W. (1995) Current Protocols in Immunology. John Wiley, New York, NY. [Google Scholar]
- Cooley J., Mathieu,B., Remold-O’Donnell,E. and Mandle,R.J. (1998) Production of recombinant human monocyte/neutrophil elastase inhibitor (rM/NEI). Protein Expr. Purif., 14, 38–44. [DOI] [PubMed] [Google Scholar]
- Cooley J., Takayama,T.K., Shapiro,S.D., Schechter,N.M. and Remold-O’Donnell,E. (2001) The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through effecient reactions at two reactive sites. Biochemistry, 40, 15762–15770. [DOI] [PubMed] [Google Scholar]
- De Smaele E., Zazzeroni,F., Papa,S., Nguyen,D.U., Jin,R., Jones,J., Cong,R. and Franzoso,G. (2001) Induction of gadd45β by NF-κB downregulates pro-apoptotic JNK signalling. Nature, 414, 308–313. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Roy,N., Stennicke,H.R., Van Arsdale,T., Zhou,Q., Srinivasula,S.M., Alnemri,E.S., Salvesen,G.S. and Reed,J.C. (1998) IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J., 17, 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri K.F. and Kroemer,G. (2001) Organelle-specific initiation of cell death pathways. Nat. Cell. Biol., 3, E255–E263. [DOI] [PubMed] [Google Scholar]
- Foghsgaard L., Wissing,D., Mauch,D., Lademann,U., Bastholm,L., Boes,M., Elling,F., Leist,M. and Jaattela,M. (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol., 153, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G., Carlson,L., Brown,K., Daucher,M.B., Bressler,P. and Siebenlist,U. (1996) Activation of the serum response factor by p65/NF-κB. EMBO J., 3403–3411. [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 255–260. [DOI] [PubMed] [Google Scholar]
- Goossens V., Grooten,J., De Vos,K. and Fiers,W. (1995) Direct evidence for tumor necrosis factor-induced mitochondrial oxygen intermediates and their involvement in cytotoxicity. Proc. Natl Acad. Sci. USA, 92, 8115–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi M.E., Deussing,J., Miyoshi,H., Bronk,S.F., Svingen,P.A., Peters,C., Kaufmann,S.H. and Gores,G.J. (2000) Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest., 106, 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson I.N., Hampson,L., Pinkoski,M., Cross,C.M., Heyworth,C.M., Bleackley,R.C., Atkinson,E. and Dexter,T.M. (1997) Identification of a serpin specifically expressed in multipotent and bipotent hematopoietic progenitor cell and in activated T cells. Blood, 89, 108–118. [PubMed] [Google Scholar]
- Hampson L., Hammond,G.L., Babichuk,C.K., Cotter,L., Bleackley,R.C., Dexter,T.M. and Cross,C.M. (2001) A minimal serpin promoter with high activity in haematopoietic progenitors and activated T cells. Hematol. J., 2, 150–160. [DOI] [PubMed] [Google Scholar]
- Hanna W.L., Zhang,H., Turbov,J., Winkler,U., Hudig,D. and Froelich,C.J. (1993) Rapid purification of cationic granule proteases: application to human granzymes. Protein Expr. Purif., 4, 398–404. [DOI] [PubMed] [Google Scholar]
- Hasel K.W. and Sutcliffe,J.G. (1990) Nucleotide sequence of a cDNA coding for mouse cyclophilin. Nucleic Acids Res., 18, 4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis J.D., Lee,M., Davidson,D.R. and Hill,R.E. (1991) Isolation of two cDNAs encoding novel a1-antichymotrypsin-like proteins in a murine chondrocytic cell line. Gene, 106, 213–220. [DOI] [PubMed] [Google Scholar]
- Kagedal K., Zhao,M. and Brunk,U.T. (2001) Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem. J., 359, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. and Lin,A. (2002) NF-κB at the crossroads of life and death. Nat. Immunol., 3, 221–227. [DOI] [PubMed] [Google Scholar]
- Komiyama T., Ray,C.A., Pickup D.J., Howard,A.D., Thornberry,N.A., Peterson,E.P. and Salversen,G. (1994) Inhibition of interleukin-1β coverting enzyme by the cowpox virus serpin Crm A. An example of cross-class inhibition. J. Biol. Chem., 269, 19331–19337. [PubMed] [Google Scholar]
- Libert C., Wielockx,B., Hammond,G.L., Brouckaert,P., Fiers,W. and Elliott,R.W. (1999) Identification of a locus on distal mouse chromosome 12 that controls resistance to tumor necrosis factor-induced lethal shock. Genomics, 55, 284–289. [DOI] [PubMed] [Google Scholar]
- Linnevers C.J., McGrath,M.E., Armstrong,R., Misdtry,F.R., Barnes,M.G., Klaus,J.L., Palmer,J.T., Katz,B.A. and Bromme,D. (1997) Expression of human cathepsin K in Pichia pastoris and preliminary studies of an inhibitor complex. Protein Sci., 6, 919–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettrick A.F., Barnes,R.C. and Worrall,M. (2001) SCCA2 inhibits TNF-mediated apoptosis in transfected HeLa cells. Eur. J. Biochem., 268, 5868–5875. [DOI] [PubMed] [Google Scholar]
- Medhurst A., Harrison,D.C., Read,S.J., Campbell,C.A., Robbins,M.J. and Pangalos,M.N. (2000) The use of TaqMan RT-PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J. Neurosci. Methods, 98, 9–20. [DOI] [PubMed] [Google Scholar]
- Morris E.C. et al. (2003) Murine serpin 2A is a redox sensitive intracellular protein. Biochem. J., 371, 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan L.T., Caputo,A., Bleackley,R.C., Pickup,D.J. and Salvesen,G.S. (1995) Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem., 270, 10377–10379. [DOI] [PubMed] [Google Scholar]
- Riccio M., Di Giamo,R., Pianetti,S., Palmieri,P.P., Melli,M. and Santi,S. (2001) Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPMI). Exp. Cell Res., 262, 84–94. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Scaffidi C., Fulda,S., Srinivasan,A., Friesen,C., Li,F., Tomaselli,K.J., Debatin,K.-M., Krammer,P.H. and Peter,M.E. (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J., 17, 1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick C. et al. (1998) Cross-class inhibition of the cysteine proteinase cathepsins K, L and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry, 37, 5258–5266. [DOI] [PubMed] [Google Scholar]
- Schutze S., Machliedt,T., Adam,D., Schwander,R., Weigmann,K., Kruse,M.-L., Heinrich,M., Wickel,M. and Kronke,M. (1999) Inhibition of receptor internalization by monodansylcadaverine selectively p55 tumor necrosis receptor death domain signaling. J. Biol. Chem., 274, 10203–10212. [DOI] [PubMed] [Google Scholar]
- Silverman G.A. et al. (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. J. Biol. Chem., 276, 33293–33296. [DOI] [PubMed] [Google Scholar]
- Speiss E., Bruning,A., Gack,S., Ulbricht,B., Spring,H., Trefz,G. and Ebert,W. (1994) Cathepsin B activity in human lung tumor cell lines: ultrastructural localization, pH sensitivity and inhibitor status at the cellular level. J. Histochem. Cytochem., 42, 917–929. [DOI] [PubMed] [Google Scholar]
- Stegh A.H., Herrmann,H., Lampel,S., Weisenberger,D. Andra,K., Seper,M., Wiche,G., Krammer,P.E. and Peter,M.E. (2000) Identification of the cytolinker plectin as a major early in vivo substrate for caspase 8 during CD95- and tumor necrosis factor receptor-mediated apoptosis. Mol. Cell Biol., 20, 5665–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoka V. et al. (2001) Lysosomal protease pathways to apoptosis: cleavage of Bid, not pro-caspases is the most likely route. J. Biol. Chem., 276, 3149–3157. [DOI] [PubMed] [Google Scholar]
- Sun J., Bird,C.H., Sutton,V., McDonald,L., Coughlin,P.B., De Jong,T.A., Trapani,J.A. and Bird,P.I. (1996) A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J. Biol. Chem., 271, 27802–27809. [DOI] [PubMed] [Google Scholar]
- Tewari M. and Dixit,V.M. (1995) Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J. Biol. Chem., 270, 3255–3260. [DOI] [PubMed] [Google Scholar]
- Turk V. and Bode,W. (1991) The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett., 285, 213–219. [DOI] [PubMed] [Google Scholar]
- Wang C.-Y., Mayo,M.W., Korneluk,R.G., Goeddel,D.V. and Baldwin,A.S., Jr. (1998) Induction of TRAF 1 and TRAF 2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science, 281, 1680–1683. [DOI] [PubMed] [Google Scholar]
- Werneburg N.W., Guicciardi,M.E., Bronk,S.F. and Gores,G.J. (2002) Tumor necrosis factor-α-associated lysosomal permeabilization is cathepsin B dependent. Am. J. Physiol., 283, G947–G956. [DOI] [PubMed] [Google Scholar]
- Wyllie A.H., Kerr,J.F.R. and Currie,A.R. (1981) Cell death: the significance of apoptosis. Int. Rev. Cytol., 68, 251–305. [DOI] [PubMed] [Google Scholar]
- Zhang X. and Ren,R. (1998) Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin 3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model of chronic myelogenous leukemia. Blood, 92, 3829–3840. [PubMed] [Google Scholar]
- Zhou Q., Snipas,S., Orth,K., Muzio,M., Dixit,V.M. and Salvesen,G.S. (1997) Target protease specificity of the viral serpin CrmA: analysis of five caspases. J. Biol. Chem., 272, 7797–7800. [DOI] [PubMed] [Google Scholar]