Abstract
The clathrin triskelion self-assembles into a polyhedral coat surrounding membrane vesicles that sort receptor cargo to the endocytic pathway. A triskelion comprises three clathrin heavy chains joined at their C-termini, extending into proximal and distal leg segments ending in a globular N-terminal domain. In the clathrin coat, leg segments entwine into parallel and anti-parallel interactions. Here we define the contributions of segmental interactions to the clathrin assembly reaction and measure the strength of their interactions. Proximal and distal leg segments were found to lack sufficient affinity to form stable homo- or heterodimers under assembly conditions. However, chimeric constructs of proximal or distal leg segments, trimerized by replacement of the clathrin trimerization domain with that of the invariant chain protein, were able to self-assemble in reversible reactions. Thus clathrin assembly occurs because weak leg segment affinities are coordinated through trimerization, sharing a dependence on multiple weak interactions with other biopolymers. Such polymerization is sensitive to small environmental changes and is therefore compatible with cellular regulation of assembly, disassembly and curvature during formation of clathrin-coated vesicles.
Keywords: assembly/clathrin/cooperativity/curvature/trimerization
Introduction
Clathrin self-assembly creates a basket-like polyhedral protein lattice that coats intracellular membranes involved in receptor-mediated endocytosis, organelle biogenesis from the trans-Golgi network, and lysosomal targeting (Brodsky et al., 2001). The clathrin lattice organizes associated adaptor molecules to sequester receptors for selective sorting into clathrin-coated membrane domains or transport vesicles. The self-assembling unit is a clathrin triskelion comprising three heavy chains, non-covalently trimerized at their C-termini, each bound by a light chain subunit that associates irreversibly during biosynthesis. While purified clathrin can spontaneously and reversibly form closed polyhedral baskets, polymerization in the cell is highly regulated by accessory proteins. Clathrin light chains prevent random heavy chain assembly. Clathrin association with adaptor proteins and/or assembly co-factors overcomes this inhibition, directing polymerization onto membranes (Brodsky et al., 2001; Mishra et al., 2002). Adaptors and their partners, such as epsin (Ford et al., 2002), contribute to the transformation of a hexagonal clathrin lattice into a curved lattice, ultimately containing the twelve pentagons needed for polyhedron closure. The heat shock family protein Hsc70 and its co-factor auxilin promote disassembly (Lemmon, 2001). To resolve the apparent thermodynamic paradox of how spontaneous clathrin polymerization can be regulated with sensitivity and reversibility in the cell, we analyzed the interactions between individual domains of the clathrin triskelion legs. In this study we define their binding properties and establish how these properties facilitate cellular regulation of clathrin self-assembly.
The structure and morphology of assembled clathrin is well characterized (Brodsky et al., 2001). Clathrin heavy chains are comprised of an N-terminal globular domain (1–390), a helical linker (391–493), a distal leg segment (494–1073), a proximal leg segment (1074–1522), and a C-terminal trimerization/vertex domain (1523–1675). In the assembled clathrin basket, each polygonal side is composed of two anti-parallel proximal leg segments and two anti-parallel distal leg segments contributed by four separate triskelia (Figure 1A and B) (Smith et al., 1998). The clathrin heavy chain proximal leg segment is a super-helix of α-helices ∼160 Å long with an oval cross section of 24 × 28 Å and comprises three clathrin heavy chain repeat (CHCR) domains (Ybe et al., 1999). These repeats are predicted to extend through the distal leg, ending at the linker region (Ybe et al., 1999). High resolution cryo-electron microscopy images of assembled clathrin baskets (Smith et al., 1998) revealed close intertwining of the clathrin proximal and distal leg segments (Musacchio et al., 1999) (Figure 1C). This analysis in conjunction with biochemical evidence suggests that interactions between leg segments are the driving force for clathrin assembly. Monoclonal antibodies against the proximal leg segment block assembly (Blank and Brodsky, 1986) and recombinant ‘Hub’ fragments (1074–1675), which contain three proximal leg segments and the trimerization domain, reversibly assemble into lattice-like structures (Liu et al., 1995). In addition, recombinant Hub and terminal-distal (1–1074) fragments of clathrin reconstitute assembly of baskets when stimulated with adaptor fragments (Greene et al., 2000), demonstrating that distal leg segments have a significant role in orienting the triskelia into a closed basket shape.
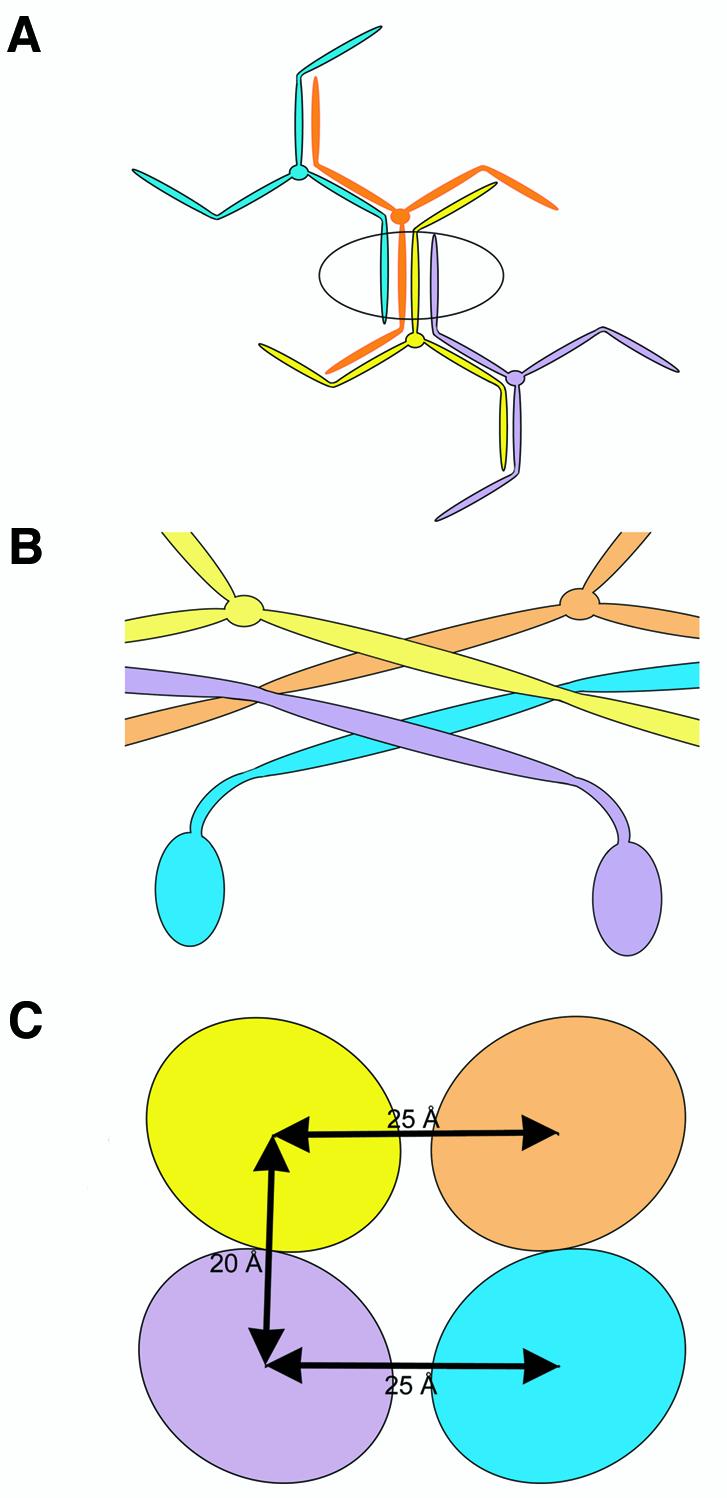
Fig. 1. Clathrin polymerization. (A) Four triskelia together form one edge of a clathrin polyhedron. The completed edge (circled) is composed of four leg segments: two anti-parallel distal leg segments (lavender and cyan) and two anti-parallel proximal leg segments (yellow and orange). (B) This predicted arrangement of triskelia in one edge of the clathrin polyhedron, viewed from the side, shows two adjacent proximal leg segments (yellow, orange) coming from a vertex downward and crossing at an angle on the outer surface of the basket. Two crossing distal leg segments (lavender, cyan) lie underneath, with the globular N-terminal domain protruding down toward the inside of the basket. Based on the 21 Å structure of a clathrin basket (Smith et al., 1998). (C) A cross-section view of one edge of the clathrin polyhedron with two proximal leg segments on top (yellow, orange) and two distal leg segments beneath (lavender, cyan) indicates the center-to-center distance separation between leg segments. Based on the 21 Å structure of a clathrin basket (Smith et al., 1998).
To understand how assembly is regulated, the coordination and affinities of leg segment binding interactions during clathrin polymerization needs to be established. In a typical protein polymerization reaction, the entropic cost of ordering molecules during assembly is overcome primarily by entropy gain from exclusion of water from hydrophobic subunit interfaces in the assembled structure (Cantor and Schimmel, 1980a). Entropy-driven reactions such as the polymerization of actin or tobacco mosaic virus are typically maintained by extensive weak hydrophobic van der Waals interactions supplemented by a few electrostatic hydrogen bonds. Shape complementarity and electrostatic interactions provide more specificity than affinity to such interactions (Fersht et al., 1985; Prevelige, 1998). Experiments focusing on individual aspects of clathrin assembly have suggested that while electrostatic enthalpic interactions are involved (Ybe et al., 1998), entropic hydrophobic effects also make important contributions (Nandi and Edelhoch, 1984). In addition, theoretical models of the energetics of clathrin assembly (Nossal, 2001) have suggested that the binding energy between assembled triskelia is quite low, on the order of thermal energetic fluctuations, suggesting assembly may indeed be driven by weak van der Waals interactions. Resolution of this latter issue is critical for establishing whether curvature of a clathrin lattice can occur by rearrangement of a hexagonal lattice, which would involve local disruption of binding interactions, or is limited to addition of new triskelia at the lattice edge.
To establish the relative contributions of clathrin leg segment interactions to the overall assembly reaction, we studied the dimerization of recombinant clathrin proximal and distal leg segments, the assembly of chimeric trimers of leg segments, and measured the binding kinetics between isolated leg segments, Hubs, or chimeras. Our data demonstrate that proximal–proximal, proximal–distal, and distal–distal interactions all contribute affinity to the clathrin polymerization process, but individually their dimerization interactions in solution are too weak to detect. Thus clathrin assembly is consistent with the classical model of protein polymerization driven by multiple weak hydrophobic interactions. These data account for the intrinsic ability of clathrin assembly to be regulated in the cell with respect to curvature changes and disassembly.
Results
Three complementary approaches were used to address how segments of clathrin mediate the affinity for self-interaction. In the first approach, we tested for possible strong interactions between leg segments by analyzing whether dimerization of individual leg segments occurs under conditions favorable for clathrin assembly. In the second approach, we tested cooperative interactions between leg segments by engineering protein chimeras in which leg segments were trimerized using the trimerization domain from the invariant chain protein. In the third approach, we measured the strength of interactions between isolated or trimerized leg segments using surface plasmon resonance assays in which high concentrations of protein were immobilized to mimic assembly induction on the surface of a membrane.
Individual leg segments are monomeric under assembly conditions
In order to characterize contacts between clathrin leg domains, isolated leg fragments were tested for their ability to dimerize under conditions where clathrin Hubs (1074–1675) assemble into lattices. Initially, size-exclusion chromatography was used to assess leg segment dimerization. Proximal (1074–1522) and terminal-distal (1–1074) leg domains were expressed and purified as recombinant polyhistidine-tagged protein fragments (Ybe et al., 1999; Greene et al., 2000). The proximal leg segment was chromatographed on a calibrated high resolution size-exclusion column, and eluted as a monomer (Figure 2A) under conditions (pH 6.2) where clathrin Hubs form a lattice (Liu et al., 1995). In a separate experiment using a lower resolution size-exclusion column, recombinant terminal-distal domain (TDD) eluted as a monomer significantly smaller than Hub at pH 6.7, also recapitulating conditions favorable for Hub assembly (Figure 2B). Mixtures of TDD and proximal leg segments contained the two component peaks, but no new peak in the eluate corresponding to a complex (Figure 2B). A mixture of the proximal leg with clathrin light chain LCb at pH 6.2 eluted as an isolated heterodimeric complex (Figure 2C), indicating that LC binding did not promote proximal leg homodimeric interactions to detectable levels. The presence of both subunits in the eluate was confirmed by gel analysis (unpublished data) and demonstrated that the folding of the proximal leg segment was representative of the native triskelion structure, in which the clathrin heavy and light chains form a stable complex on a sizing column (Ungewickell and Branton, 1981). Over all, these results suggest that clathrin leg segment interactions in the presence or absence of light chain are too weak to detect using chromatography, and are thus significantly weaker than the heavy chain–light chain interaction.
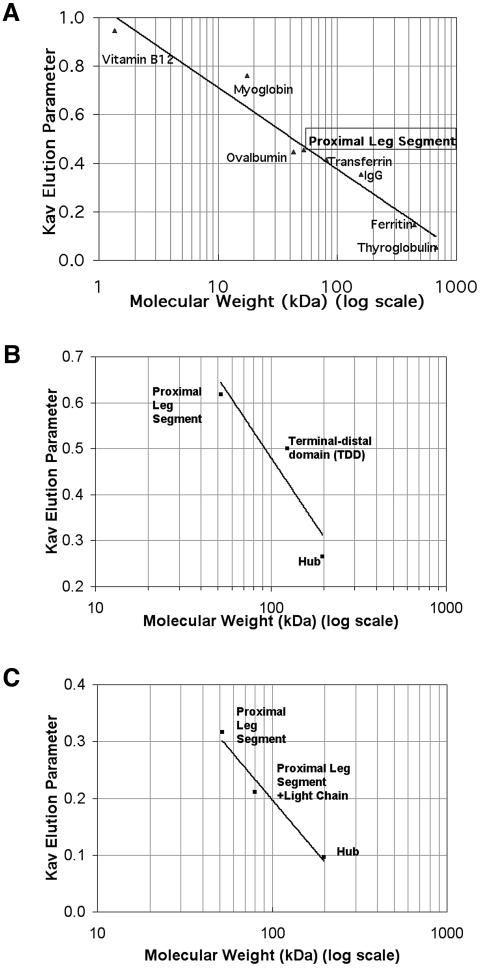
Fig. 2. Leg segments are monomeric by FPLC. Fragments representing the clathrin proximal leg and terminal–distal domain were expressed as polyhistidine-tagged recombinant proteins and analyzed by gel permeation chromatography. Kav versus molecular weight for sample and standards were plotted. Proximal leg segments are monomeric on a calibrated FPLC column. (A) Proximal leg fragment and calibration standards were independently chromatographed on a Superdex 200 HR 10/30 column in Bis–TRIS pH 6.2 and elution times were noted. Proximal leg segment Kav was calculated as 0.454, indicating a size for clathrin proximal leg segment consistent with a monomeric 52 kDa species. (B) Isolated proximal and distal leg segments are monomeric and do not form a complex when combined. Proximal and terminal-distal domains were chromatographed on a Superose 6 prep grade column in MES/TRIS pH 6.7 and elution times were compared with elution of Hub or leg segments at pH 7.3. Kav was calculated. Mixtures of proximal and distal domain segments eluted as two separate populations; no larger peak corresponding to a complex was observed. (C) Proximal leg segments bind light chains normally but do not dimerize in the presence of light chain. Proximal leg segments with and without purified light chains and Hub were chromatographed on a Superose 6 prep grade column in Bis–TRIS pH 6.2 and elution times were noted for calculation of Kav. While the mixture of proximal leg with light chain eluted as a complex, indicating proper folding of the recombinant protein fragments, the Kav of the complex is consistent with each complex containing only one light chain and one proximal leg. Thus the presence of light chain did not induce homodimerization of the proximal leg.
More sensitive methods were then chosen to assay the dimerization of clathrin proximal leg segments. Analytical ultracentrifugation by sedimentation velocity yielded data fitting a single population of molecules in solution at pH 6.2 (Figure 3A) or pH 6.7 (unpublished data), with an apparent sedimentation coefficient of S = 2.3. These data indicate a monomer with a frictional ratio of 1.8 (proportional to the axial ratio of the protein). The prolate ellipsoid shape of the proximal leg segment would be expected to have a frictional ratio larger than that typical for globular proteins (1.2) (Cantor and Schimmel, 1980b). In order to confirm that this elongated rod represented a monomer, the proximal leg segment was further analyzed by equilibrium sedimentation at pH 6.2. Because this assay relies on the distribution of the species after equilibrium has been reached, it is insensitive to non-spherical shapes of proteins. Equilibrium sedimentation data at multiple speeds and concentrations were fitted simultaneously to a single set of parameters and were consistent with the proximal leg segment behaving as a monomer (Figure 4). The corresponding Kd for interaction of proximal leg segments determined from data that was fitted artificially to dimer behavior is at best on the order of 115 µM. This value is weaker than the nanomolar range affinity of enthalpy-driven self-assembly reactions (Mejean et al., 2001; Parker et al., 2001) and is more consistent with the order of magnitude expected for an entropy-driven network of weak hydrophobic subunit interactions (Cantor and Schimmel, 1980a). When complexes of proximal leg with LCb were tested under similar conditions, the equilibrium sedimentation data were also characteristic of behavior as an isolated heterodimeric complex, consistent with the chromatography experiments above (unpublished data).
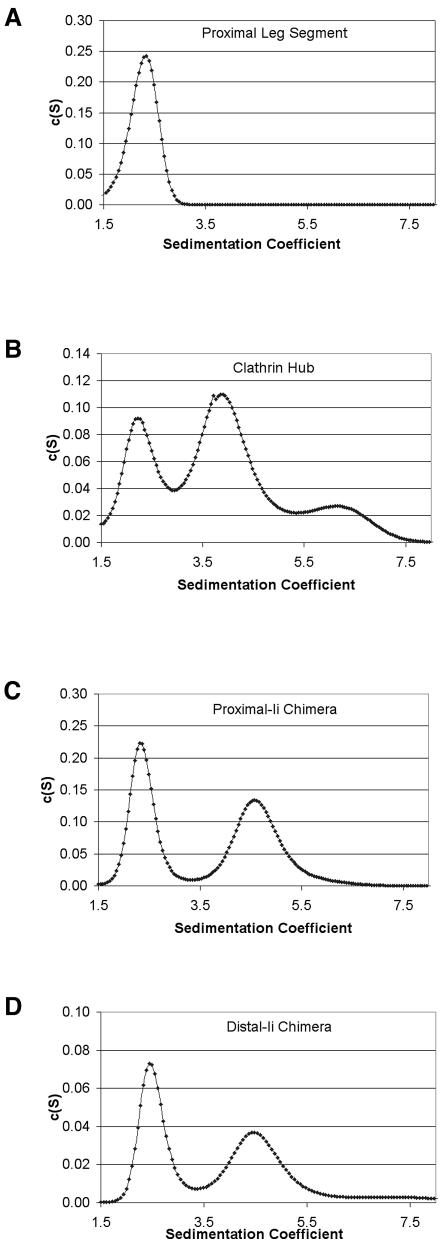
Fig. 3. Sedimentation velocity assays. Polyhistidine-tagged constructs were expressed and purified and their interaction assessed using sedimentation velocity by analytical ultracentrifugation in a Beckman XL-I at 40K r.p.m., 4°C. SedFit was used for a continuous c(S) distribution analysis of the data. For each fit, the root mean square deviation (r.m.s.d.) for the fit of the curves to the data, the number of data points (n), sum of squared residuals (SSR) and frictional ratio (F, indicating shape deviations from a sphere) were noted. The confidence level was 0.95. (A) Proximal leg segment at pH 6.2 is composed of a single species with sedimentation coefficient S = 2.3 (r.m.s.d. = 0.006503, n = 8372, SSR = 0.354059, F = 1.8). This species was confirmed as a monomer by equilibrium sedimentation (Figure 4), and components of trimeric constructs with S = 2.3 were thus assigned as monomeric species in plots (B–D). (B) Hub at pH 7.9 is composed of three species in solution: the first two are at S = 2.3 and S = 3.9 and correspond to monomer (35.2%) and trimer (64.8%), respectively, and the third is a minor larger species at S = 6.1 (r.m.s.d. = 0.007666, n = 14198, SSR = 0.834386, F = 1.6). (C) Proximal-Ii chimera at pH 7.9 is composed of two species in solution, at S = 2.4 and S = 4.7 (r.m.s.d. = 0.013468, n = 16365, SSR = 2.968339, F = 1.3), which correspond to monomer (46.8%) and trimer (53.2%), respectively. (D) Distal-Ii chimera at pH 7.9 is composed of two species in solution, at S = 2.5 and S = 4.6 (r.m.s.d. = 0.0.008542, n = 18212, SSR = 1.328910, F = 1.2), which correspond to monomer (49.5%) and trimer (50.5%), respectively.
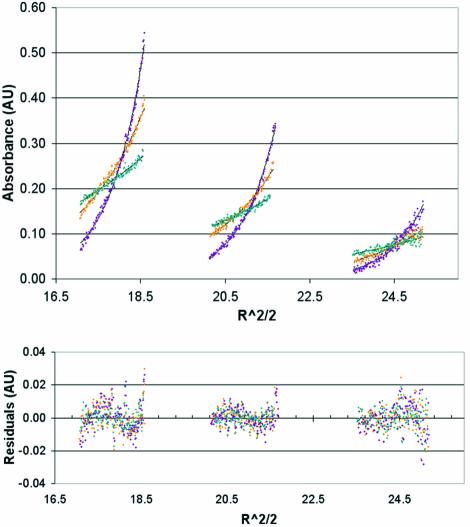
Fig. 4. Proximal leg segment is a monomer at pH 6.2 in an equilibrium sedimentation assay. Proximal leg was expressed and purified and its self-interaction was assessed by equilibrium sedimentation in a Beckman XL-I analytical ultracentrifuge at 40K r.p.m., 4°C, pH 6.2. WinNonLin was used to simultaneously fit data points from samples of 0.4 (left), 0.2 (center), and 0.1 mg/ml (right) at 7K (green), 10K (orange), and 14K (purple) to the lines shown in black. (Radial distance)2/2 (R^2/2) is plotted against A280 (AU). A plot of the residuals (lower panel) indicates the systematic error in the fit between the lines and the raw data points. The fit gave values for parameter σ = 0.623 (0.547–0.696) where for the expected monomer σ = 0.667. The fit is consistent with a proximal leg segment fitted to an N-mer of N = 0.94, which represents monomer. Variance of fit was 4.6 × 10–5 with 1234 degrees of freedom.
A yeast two-hybrid assay was employed to examine further the possibility of weak leg segment interactions under more physiological conditions. Clathrin fragments were cloned into two-hybrid bait and prey vectors to express them as fusion proteins attached to GAL4 DNA binding or activation domains (Table I). As a positive control, a strong interaction between the proximal leg segment and bovine clathrin light chain LCb, cloned into the same prey, was demonstrated. In the same assay, it was demonstrated that yeast light chains do not bind to bovine proximal leg (unpublished data). Low levels of Hub assembly occur at physiological pH (in the absence of bound clathrin light chains) (Liu et al., 1995) and correspondingly, a weak interaction between Hub constructs (1073–1675) was detected. However, the proximal leg segment (1073–1522) and its fragments showed no detectable interaction with one another or with Hub fragments. To rule out the possibility that the cloning orientation inhibited interaction, the proximal leg segment was also engineered upstream of the GAL4 activation domain in the bait vector. In this configuration the GAL4 domains would be adjacent during an anti-parallel interaction, but this construction also failed to show any interactions in the assay (unpublished data).
Table I. Proximal leg segments do not interact in a yeast-two-hybrid assaya.
| BD |
|||
|---|---|---|---|
| 1073–1675 | 1073–1522 | 1204–1522 | |
| AD | Hub | Proximal leg segment | |
| 1073–1675 Hub | 4 | ||
| 1073–1522 | – | – | – |
| 1204–1522 | – | – | |
| 1271–1414 | – | – | |
| LCb (control) | 65 | 65 | 200 |
aInteractions between binding domain (BD) and activation domain (AD) are noted in β-galactosidase units. Numbers refer to encoded clathrin heavy chain residues attached to the AD and BD.
– indicates interaction was tested but could not be detected.
The failure to detect interaction of proximal leg segments using assays dependent on dimerization indicates that isolated clathrin leg segment interactions have extremely weak affinity. These observations explain our previously published finding that an excess of proximal leg segments did not act as dominant-negative inhibitors of clathrin assembly, because they could not bind Hub efficiently (Greene et al., 2000).
Chimeric constructs assemble using cooperative leg segment interactions
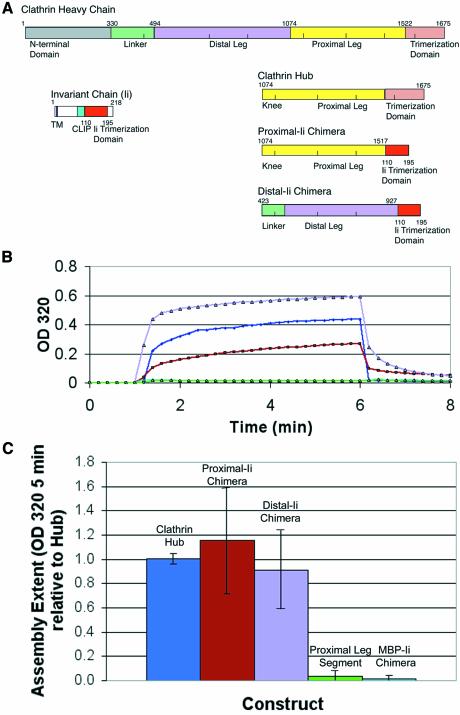
Given the lack of detectable dimerization between leg segments, clathrin or Hub assembly could either rely on weak cooperative interactions, or on interactions involving domains missing from the leg constructs. The only additional domains in the Hub fragments (Liu et al., 1995) are the sequences mediating trimerization and extending to the vertex. These could potentially bind the ‘knees’ between proximal and distal leg segments, which are localized beneath the vertices in the assembled clathrin lattice and are present in the Hub constructs. If these interactions are primarily responsible for assembly, then inhibition by antibodies to the proximal leg segment (Blank and Brodsky, 1986) would have to be ascribed to steric hindrance. While a role for these non-leg interactions is certainly possible, the effects of the proximal leg-specific antibodies on assembly are also consistent with a model in which assembly occurs due to weak leg–leg interactions that are coordinated by a trimeric configuration. We therefore investigated whether trimerization of leg segments with a non-clathrin trimerization domain would allow cooperative assembly between leg segments without the involvement of the trimerization domain itself. To this end, chimeric constructs were created with proximal or distal leg segments attached to the C-terminal trimerization domain of the invariant chain (Ii) protein (Figure 5A).
Fig. 5. Chimeric trimerized clathrin proximal or distal leg segments assemble. (A) Chimeric constructs distal-Ii and proximal-Ii were created with clathrin distal or proximal leg segments, respectively, attached to the trimerization domain of the invariant chain protein. The bar diagrams indicate the protein domains of each donor protein from which chimeras were constructed and the domains in the resulting chimeras. Numbers indicate the residues flanking protein domains and the junctions where fragments were recombined. (B) Hub (blue) and purified chimeric proteins proximal-Ii (brown) and distal-Ii (lavender) at pH 7.9 were induced to assemble by a drop in pH 6.7 through the addition of 1/25 volume of 1 M MES pH 6.7, and turbidity (change in OD at 320 nm) was monitored for 5 min. Disassembly was induced by increasing the pH with an addition of 1/25 volume of 1 M TRIS pH 9 and monitoring OD320 for an additional 2 min as turbidity returned to baseline levels, demonstrating reversibility of the assembly process. Monomeric proximal leg segment (green) and trimeric MBP-Ii (cyan) served as controls (overlapping lines near the baseline). (C) The chimeras had a similar overall extent of assembly to purified clathrin Hub in the pH 6.7 turbidity assay. Samples were induced to assemble as above and the extent of assembly (OD320 after 5 min) was recorded. Extent of assembly was normalized to average hub assembly over several preparations and averaged for this plot.
In an antigen-presenting cell, trimerized Ii proteins occupy the peptide binding sites of three nascent MHC Class II heterodimers, forming a nonamer, to stabilize their structure until they encounter antigen in a peptide loading compartment (Cresswell, 1996). The structure of the Ii trimerization domain reveals a cluster of helices connected by short loops (Jasanoff et al., 1998). Like the clathrin trimerization domain, the three helical subunits enter the C-terminal trimerization domain at a slightly puckered 120° angle. Chimeric Ii trimerization domain constructs were engineered in which the Ii trimerization domain was joined to either the triskelion proximal leg (proximal-Ii) or distal leg (distal-Ii) segment by connecting loop segments between helices, creating a non-clathrin trimerization domain for the clathrin leg segments.
In order to confirm that the chimeric proteins were trimers, sedimentation velocity experiments were performed under conditions where clathrin and Hub are disassembled (pH 7.9). The purified proximal-Ii or distal-Ii chimeras each had a sedimentation coefficient distribution very similar to Hub (Figure 3B–D). Each sample was composed of two species, the first of which (S ∼2.4) had the same apparent sedimentation coefficient as monomeric proximal leg (Figure 3A). For both Hubs and chimeras, the distribution indicated a predominance of the second, trimeric peak (S = 4–5) over monomer. The Hub and two chimeras are oblate ellipsoids and the data fitted to an apparent frictional ratio of 1.6 for Hub (Figure 3B), indicating perhaps a slightly flatter disc shape than the two chimeras with apparent frictional ratios of 1.2 and 1.3 (Figure 3C and D). Thus, for both engineered constructs, the invariant chain trimerization domain trimerized a significant proportion of chimeric leg segments in the expressed protein.
The chimeric proteins were then assessed for their ability to assemble under conditions that induce Hub assembly (Liu et al., 1995), using a turbidity assay. In this assay, self-assembly is distinguishable from protein aggregation in that it requires proper folding of the protein and is reversible (Kentsis et al., 2002). Spectrophotometric data indicated that both the proximal-Ii and distal-Ii constructs were able to assemble at either pH 6.2 or 6.7. Assembly of the chimeric constructs proceeded at a rate similar to that of Hub assembly (Figure 5B) and to a similar extent, as revealed by the endpoint spectrophotometric measurements (Figure 5C). The assembly reactions of the chimeras were reversible by raising the pH to 7.9, which causes assembled Hub to disassemble, thus demonstrating specificity and reversibility of their assembly (Figure 5B). Analysis of assembled chimeric constructs by electron microscopy revealed pseudo-lattices similar to those previously observed for assembled Hub (Liu et al., 1995; Greene et al., 2000) in which assembly proceeds in multiple planes, so that lattice morphology is poorly stained (unpublished data). In control reactions, the proximal leg segment did not produce a spectrophotometric signal upon lowering the pH to 6.2 or 6.7, indicating no formation of larger aggregates under these conditions, consistent with their lack of dimerization in other assays and providing a specificity control for assembly of the chimeras. A control construct of trimerized maltose binding proteins in a chimera with invariant chain trimerization domains (MBP-Ii) did not assemble (Figure 5B and C). Thus interactions between clathrin leg chimeras were due to clathrin segments and not self interactions of the invariant chain trimerization domains. Chimera assembly showing distal–distal interactions in a trimeric configuration complements previous data regarding the role of distal leg segments in clathrin assembly (Greene et al., 2000). In that study, distal leg segments were extended to include the terminal domain and multimerized by adaptors into a counter-Hub, which influenced Hub assembly to form a curved structure. The distal–distal interactions demonstrated here must have contributed to the counter-Hub effect along with the implicated proximal–distal interactions.
Transient interactions between isolated leg segments quantitated using SPR
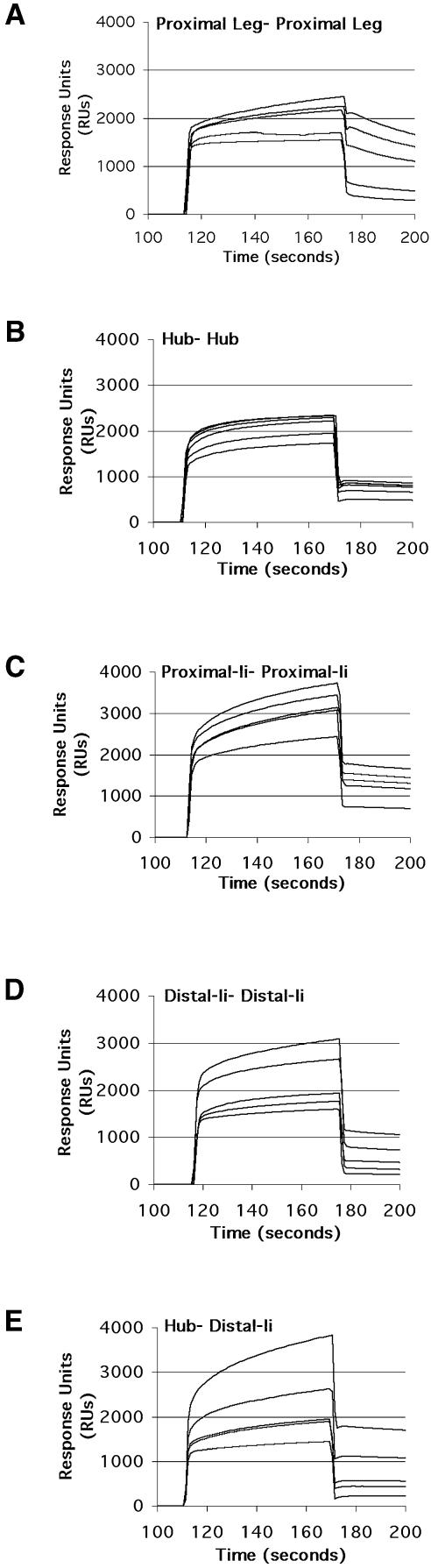
Surface plasmon resonance (SPR) was used to assay for the transient proximal leg segment interactions implied by the data on leg interactions obtained so far. In this assay proximal leg segment, Hub, or chimeras were densely immobilized on the surface of a chip. Solutions of proximal leg segments, Hubs, or chimeras were flowed over the chip surface in the chamber of a BIAcore Processing Unit. Changes in the refractive index of the immobilized protein on the chip revealed transient binding interactions. When tested under conditions favoring clathrin assembly, interactions were detected between isolated proximal leg segments, Hubs, and chimeras (Figure 6). The dissociation rate constant for proximal leg segment self interactions was five times greater than that for the trimerized Hub self-interaction (Table II), indicating lower stability of interactions between isolated leg segments. In addition, the concentration threshold necessary to observe interaction was almost an order of magnitude lower for trimerized leg segments than for isolated proximal legs, demonstrating a greater overall binding affinity when leg segments were arranged in a trimeric conformation in Hubs or chimeras. Proximal-Ii self interactions were slightly less stable than Hubs but more stable than interactions between isolated proximal legs. Interactions between trimerized distal leg segments (distal-Ii) were similar to those measured for proximal-Ii self interaction. Distal-Ii interactions with Hub had higher stability than Hub–Hub interactions. These latter measurements confirm the contributions of distal leg interactions to clathrin assembly through both distal–distal and distal–proximal contacts. The latter appears to be the strongest contact in basket formation.
Fig. 6. Surface plasmon resonance detects interaction between proximal leg segments. Clathrin constructs were expressed, purified and tested for interactions using a BIAcore Processing Unit. Proximal leg segments, Hubs, or chimeras were immobilized to a CM5 chip. Mobile solutions of clathrin fragments or chimeras were flowed over the surface at pH 6.7 and changes in refractive index monitored. Lower curves represent lower concentrations of mobile species. (A) Immobilized proximal leg segment (5029 RU) interacts with proximal leg segments at 2.5, 5.0, 7.5, 10.0 and 12.5 µM. (B) Immobilized Hub (6144 RU) interacts with Hub at 0.5, 1.0, 1.5, 2.0, 2.5 and 3.0 µM. (C) Immobilized proximal-Ii chimera (5731 RU) interacts with proximal-Ii chimera at 1.0, 2.0, 2.5, 3.0, 4.0 and 5.0 µM. (D) Immobilized distal-Ii chimera (5115 RU) interacts with distal-Ii chimera at 0.25, 0.50, 0.75, 2.5 and 5.0 µM. (E) Immobilized Hub (6144 RU) interacts with distal-Ii chimera at 0.25, 0.50, 0.75, 2.5 and 5.0 µM.
Table II. Kinetic parameters for clathrin interactions determined by SPR.
| Immobilized protein/mobile protein | Concentration of immobilized species | Concentration threshold for binding | kdiss |
|---|---|---|---|
| Proximal leg/proximal leg | 0.92 mM | 1.0–2.5 µM | 0.012 |
| Hub/Hub | 0.28 mM | 100–500 nM | 0.0026 |
| Proximal-Ii/proximal-Ii | 0.30 mM | 250–500 nM | 0.0051 |
| Distal-Ii/distal-Ii | 0.24 mM | 100–250 nM | 0.0048 |
| Hub/distal-Ii | 0.28 mM | 100–250 nM | 0.0018 |
The concentration of the immobilized species in each of these assays was above the critical concentration to initiate assembly and was often higher than the concentration in the fluxed solution. The fact that the mobile species was exposed to an effective high local concentration on the immobilized surface likely accounts for ability to detect binding in this assay even when the fluxed solution was significantly below the critical concentration for assembly. Substantial rebinding evident in the slopes of the dissociation curves demonstrates that at such a high density of immobilized protein (1 mM for isolated proximal leg segments) the interactions observed are more representative of bulk self-assembly than dimerization. Overall, this real-time SPR binding data verifies our conclusion that clathrin self-assembly involves multiple weak interactions between leg segments that must be directed by their connections to one another, in order to stabilize and strengthen the clathrin cage.
Discussion
Here we define the nature of domain interactions contributing to clathrin assembly. We demonstrate that individual leg segment affinities are weak and rely on trimerization to allow coordinated contributions to the formation of a clathrin lattice. In addition, we confirm that clathrin light chain binding is not required for trimerized proximal leg segment interactions and we demonstrate that light chain binding does not promote interaction of individual proximal leg segments. Finally, we show that distal–distal leg segment interaction contributes to clathrin assembly, in addition to proximal–proximal and proximal–distal interactions that have been previously implicated (Blank and Brodsky, 1986; Smith et al., 1998; Greene et al., 2000). Measurements of leg dissociation constants suggest a hierarchy with proximal–distal being the strongest, and homotypic interactions of the leg segments being about equal contributors to assembly. We cannot rule out supplementary roles for vertex–knee interactions in assembly from the data reported here, but the chimeric constructs and SPR data demonstrate that leg segment interactions when trimerized are sufficient to sustain assembly without these interactions. From this analysis of clathrin assembly, we can infer important features of the thermodynamics of clathrin assembly that provide insight into the regulation of clathrin assembly and curvature in the cell.
Thermodynamic properties of clathrin assembly
Multiple weak, simultaneous interactions have been proposed as the mechanism for the assembly of clathrin terminal domains with adaptor complexes and beta-arrestin (Kirchhausen, 2000). Here we demonstrate that similar principles govern clathrin self-assembly. Thus, as hypothesized over twenty years ago (Crowther and Pearse, 1981), we now have evidence that the process of clathrin coat formation joins actin, amyloid, tubulin, and others in the large and diverse group of entropy-driven condensation reactions, for which the tobacco mosaic virus rod-forming polymerization process is a model (Lauffer, 1975). Based on what is known about these reactions, we must consider that the primary driving force in clathrin assembly is hydrophobic, rather than electrostatic. Typically, electrostatically mediated assembly would involve early formation of weak long-range interactions, followed by the docking of proteins into a higher affinity configuration requiring strong hydrophobic interactions (Schreiber and Fersht, 1996). The predicted electrostatic interactions between clathrin domains (Ybe et al., 1998) would be unlikely to act attractively at long range due to electrostatic repulsion between predominantly acidic leg segments, especially if acidic clathrin light chains are bound. Furthermore, the absence of strong binding affinity between isolated leg segments suggests that strong short-range salt bridges are also absent in this interface. However, electrostatic repulsion can function as a specificity determinant to block misalignment of leg segments, as has been seen for other self-assembling systems (Coleman et al., 1999). Thus, the weakness of the interaction between leg segments demonstrates a possible role for repulsive electrostatic specificity-determining interactions which are balanced out by hydrophobic attractive interactions, as previously suggested for clathrin assembly (Nandi and Edelhoch, 1984). In fact, the pH and salt sensitivity observed for clathrin assembly is common among entropy-driven polymerizations (Lauffer, 1975) and may result from a simple reduction of the electrostatic repulsion between acidic heavy chains near the isoelectric point of the leg segments. We have proposed that ionization of surface-accessible histidines could influence clathrin assembly (Ybe et al., 1998). In light of the weak individual interactions in assembly, we now suggest those histidines could be involved in pH-dependent electrostatic hydrogen bonds contributing to an electrostatic effect, as has been found in other proteins (Mejean et al., 2001).
Based on the density map derived from images of assembled baskets (Smith et al., 1998), we used GRASP (Nicholls et al., 1991) to predict that the interface between two proximal leg segments has a buried surface area on the order of 1500 Å2. Affinity interactions over this area would be expected to include van der Waals associations, several charged and uncharged hydrogen bonds, and possibly a small contribution from one or more salt bridges (Prevelige, 1998). Taking all of these properties into consideration, along with the data reported here, we propose that clathrin self-assembly is driven primarily by the entropic gain of the hydrophobic effect when the assembled leg segments exclude water from their interface.
Cellular regulation of clathrin assembly and curvature
The unstable and weak interaction between isolated leg segments raises the question of how nucleation of clathrin assembly occurs in the cell. Clathrin polymerization appears to proceed through an intermediate structure with 4–7 triskelia (Schwarz et al., 1989). With our measurement of weak interactions between isolated leg segments, we expect that intermediates smaller than this would be unstable. In the cell, adaptors and accessory co-factors AP180 (Ford et al., 2001) and epsin (Ford et al., 2002) are therefore required to play a key role in initiating assembly through recruitment and stabilization of clathrin polymerization on the membrane. Based on the thermodynamic considerations above, assembly could also be readily stimulated by locally lowered pH due to proton pump activity (Heuser, 1989) or recruitment to acidic membrane lipids, which may have a similar effect on clathrin assembly (Maezawa et al., 1989) as they do on laminin assembly (Freire and Coelho-Sampaio, 2000). Clathrin propagation, when recruited to the membrane in the vicinity of a stable clathrin-coated pit locus (Gaidarov et al., 1999) and stimulated to undergo a coordinated assembly, would occur more readily than nucleation because it would by nature involve multiple segment interactions joining an existing lattice.
A second issue relevant to our data is how curvature can be introduced into a clathrin lattice. There are three factors that contribute to lattice curvature. The first is the geometric requirement for introduction of twelve pentagons into a hexagonal lattice in order to achieve a closed sphere or vesicle coat (Kaneseki and Kadota, 1969). The second factor is the shape of the triskelion itself and its inherent flexibility. In an assembled clathrin basket, a triskelion has increased pucker at the vertex as well as greater curvature of the legs than it displays in solution (Musacchio et al., 1999). It has been suggested that the knee region between proximal and distal leg segments, which has little intrinsic curvature in solution (Yoshimura et al., 1991) and no inherent additional elasticity compared with the proximal or distal leg segments (Kocsis et al., 1991; Jin and Nossal, 2000), is responsible for most of the curvature in the assembled basket (Kirchhausen et al., 1986; Musacchio et al., 1999). We have further shown that ultimate flexibility at the knee by dissociation of the distal from the proximal leg domain is compatible with introduction of curvature into a clathrin lattice (Greene et al., 2000). In addition, theoretical analyses have indicated that triskelia may form pentagons as well as hexagons with some torsional strain but little overall free energy cost (Jin and Nossal, 1993), particularly in the presence of tetrameric adaptors (Nossal, 2001). Thus changes in triskelion conformation can readily occur and contribute to lattice curvature without posing a significant energy barrier. The third factor regulating curvature of a clathrin lattice at the cellular membrane is the participation of adaptors and accessory proteins in this process. Tetrameric adaptor proteins AP1 (at the TGN) and AP2 (at the plasma membrane) work with monomeric adaptors such as GGAs (Robinson and Bonifacino, 2001) and Dab2 (Mishra et al., 2002) to induce invaginations in the clathrin lattice. Epsin, which binds lipid, tetrameric adaptors and clathrin, is able to accomplish this curvature even without the aid of tetrameric adaptors (Ford et al., 2002). In this case, epsin’s mechanism of action relies on deformation of the membrane by insertion of its hydrophobic domain and its ability to manipulate clathrin to form pentagons through binding to clathrin. Whether epsin and other adaptors can perform clathrin manipulations within a preformed hexagonal array or only at the edge of such a lattice by interaction with newly recruited triskelia depends on the energetics of the interaction of individual clathrin leg domains.
Here we show that the energetics of clathrin self-assembly are compatible with lattice-modifying proteins producing a bud from the interior of an existing clathrin lattice, essentially to regulate rearrangement of a lattice. Evidence presented here for weak interactions between clathrin leg segments supports earlier theoretical free energy calculations for the assembly process (Nossal, 2001), which estimated that the free energy gain on the addition of an individual triskelion to a partially formed lattice is quite small, on the order of thermal fluctuations. Such a low free energy threshold permits rearrangement in the center of a lattice by cytosolic exchange of triskelia (Brown and Petersen, 1999; Wu et al., 2001) and is compatible with removal of a triskelion for rearrangement of a hexagon into a pentagon (Kaneseki and Kadota, 1969; Nossal, 2001). While our data support such a model for lattice rearrangement, they do not absolutely contradict the suggestion that such pentagonal arrangements are limited to the edges of the lattice (Harrison and Kirchhausen, 1983; Kirchhausen, 2000). In either case, we would suggest that the presence of lattice-modifying proteins in a budding segment of membrane allows rearranged or new triskelia to be oriented so that they interact with more than one leg segment of the partially or fully completed lattice at the same time, stabilizing the interaction when compared with isolated leg segment interactions. Thus, a delicate balance between individually weak, repulsive and attractive electrostatic, hydrogen bonding, and hydrophobic interactions can be exploited by regulatory proteins. This allows clathrin self-assembly to be spontaneous, readily reversible and exquisitely regulated in the cell.
Materials and methods
Construct production and purification
Production of the plasmids for polyhistidine-tagged distal leg segment (1–1074) in pET 23d (Greene et al., 2000), and for the proximal leg segment (1074–1522), and Hub (1074–1675) in pET15b (Liu et al., 1995) and the protein purification protocols using nickel-affinity chromatography have been previously described. Light chain LCb was expressed and purified as above along with Hub, and then isolated by boiling the Hub-LCb complex to release light chains. Fresh proximal legs were then incubated with excess light chain on ice to reoccupy light chain binding sites, as previously described (Liu et al., 1995).
Chimeric constructs were created using the Seamless Cloning Kit (Stratagene). Seamless primers were used to amplify cDNA for the invariant chain trimerization domain (110–195) from previously described plasmids (Liu et al., 1998) and to ligate that together with the cDNA for bovine clathrin proximal leg segments (1074–1517) or distal leg segments (423–925) within pET15b (Novagen) according to Seamless Protocol. Purification protocol was identical to previously published Hub purification. Control chimera MBP-Ii was created by PCR amplification of the invariant chain trimerization domain cloned into the BamHI site of pMALc2X (New England Biolabs) to trimerize maltose binding protein, and purified using Amylose Resin (New England Biolabs). Trimerization was confirmed by size-exclusion chromatography.
Size-exclusion chromatography
Size exclusion columns were run by fast protein liquid chromatography (FPLC) and the distribution of proteins in eluate was confirmed by gel electrophoresis. A Superdex HR200 10/30 column (Amersham) in pH 6.2 50 mM Bis–TRIS buffer was calibrated at 0.5 ml/min by injection of individual standards noted in Figure 2A (Amersham Gel Filtration Calibration Kits), along with Acetone and Dextran Blue. Kav = (Ve – Vo)/(Vt – Vo) of proximal leg segments and calibration standards were plotted against the log molecular weight to determine molecular size of proximal leg segments, following the Amersham Instruction Manual.
Additionally, a 60 ml XK16/40 Superose 6 column (Amersham) in pH 6.7 MES 40 mM/TRIS 10 mM was used to determine the size of proximal leg segments, terminal/distal domain, or mixtures of both. These were compared with the size of Hub and terminal/distal domain at pH 7.3 on the same column.
This 60 ml XK16/40 Superose 6 column was also used to look at light chain–proximal leg segment complexes, isolated proximal leg segments, and Hub at a flow rate of 1 mg/ml with a 10 mM Bis–TRIS pH 6.2 mobile phase. Hub at pH 6.2 assembled into a large aggregate that was centrifuged and removed, leaving small amounts of unassembled Hub in the supernatant for this analysis.
Analytical ultracentrifugation
A Beckman XL-A or XL-I was used in absorbance mode at 280 nm for both sedimentation velocity and equilibrium assays. For equilibrium assays, proximal leg segments at 0.1, 0.2, and 0.4 mg/ml in 10 mM TRIS/40 mM MES pH 6.2 were equilibrated at 4°C in a six-channel centerpiece with data collected at 7000 r.p.m., 10 000 r.p.m. and 14 000 r.p.m. These nine data files were simultaneously fitted using WinNonLin (Johnson et al., 1981) to find σ and compare it to a calculated σ for the monomer under those conditions. A forced fit to a dimer was used to estimate lnK2. Under similar conditions, Hub at pH 7.4 was fitted to an N-mer of N = 2.9 as a control (unpublished data). The partial specific volume of proximal leg and trimeric hubs was calculated as 0.732 g/ml using Ultrascan (Demeler and Saber, 1998) and solvent density was estimated at 1.00 g/ml. For velocity experiments, a 3 mg/ml sample of proximal leg segment was used at pH 6.2 and a 0.3 mg/ml sample at pH 6.7. Data collection proceeded in a two-channel centerpiece at 4°C at 40K at pH 6.2 or 8°C at 50K for pH 6.7. Each data set was fitted using continuous c(S) distribution analysis in SedFit (Schuck, 2000). The proximal-Ii and distal-Ii chimeras and Hubs were tested at 40K, 8°C at pH 7.9 10 mM TRIS at 0.3 mg/ml.
Yeast two-hybrid assays
Clathrin heavy chain fragments were PCR amplified from bovine brain cDNAs (Liu et al., 1995) and cloned into BamHI–SalI sites of pGBT9 or pGAD424 (Clontech). Yeast transformation, qualitative liquid β-galactosidase [using o-nitrophenyl β-d-galactopyranoside (ONPG) as substrate], filter assays were performed using Clontech yeast protocols handbook, as previously described (Chen et al., 2002).
Spectrophotometric turbidity assay for assembly
Hub or chimera assembly was assayed as increased OD320 in pH-induced assembly as described previously (Liu et al., 1995). Extent was recorded after 5 min assembly, at which point pH was increased to reverse the assembly reaction. All preparations were scaled relative to Hub assembly assayed on the same day and were averaged over multiple purifications (Hub n = 8, 4 batches; proximal-Ii and distal-Ii, n = 4, 2 batches). Proximal leg segment (n = 3, 2 batches) and MBP-Ii (n = 3, 1 batch) at 0.3 mg/ml were used as controls.
Surface plasmon resonance
A BIAcore Processing Unit was used to quantify real-time interactions. Twelve microliters of clathrin Hubs, proximal leg segments, distal-Ii chimeras, or proximal-Ii chimeras at 50 µg/ml in 10 mM acetate pH 4.0 were immobilized to a carboxymethylated dextran matrix (CM5) chip at 5 µl/min in HBS-EP buffer, using standard amine coupling conditions. The resulting effective concentration of protein immobilized was calculated assuming a 100 nm dextran layer, where 1 response unit (RU)= 1 pg/mm2.
Solutions of clathrin fragments in 10 mM MES pH 6.7, 9 mM HEPES were flowed over the surface of the immobilized proximal leg for 60 s at 15 µl/min in 10 mM MES pH 6.7. From a range of concentrations, a minimum concentration threshold was determined to bring the local concentration of mobile protein near the chip surface above the critical concentration for assembly. After each dissociation, residual bound protein was removed by regeneration with 10 mM sodium tetraborate pH 8.5, 1 M NaCl.
The dissociation rate constant kdiss was determined using BIAEvaluate software, at the highest concentration of interacting solute over a 10 s time frame beginning ∼5 s after the injection was completed (175–185 s). The apparent binding rate constant ks (and thus Kd) could not be determined due to the highly cooperative nature of the self-assembly reaction, wherein the rate saturated so readily above the threshold that the on rate was independent of solute concentration.
Acknowledgments
Acknowledgements
The authors are grateful to R.Tsai and J.Marks for generously allowing our access and training for the BIAcore instrument, G.Knudsen and D.Mullins for helpful suggestions on AUC data collection and analysis, and to M.Reese for helpful comments on the manuscript. This work was supported by NIH grants GM55143 and GM38093 to F.M.B., an ARCS (Achievement Rewards for College Scientists) Fellowship and NIH grant CA-09043 to D.E.W., a postdoctoral fellowship from the Arthritis Foundation to C.-Y.C. and grant F97-SF-031 from the California Universitywide AIDS Research Program to B.G.
References
- Blank G.S. and Brodsky,F.M. (1986) Site-specific disruption of clathrin assembly produces novel structures. EMBO J., 5, 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky F.M., Chen,C.Y., Knuehl,C., Towler,M.C. and Wakeham,D.E. (2001) Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu. Rev. Cell. Dev. Biol., 17, 517–568. [DOI] [PubMed] [Google Scholar]
- Brown C.M. and Petersen,N.O. (1999) Free clathrin triskelions are required for the stability of clathrin-associated adaptor protein (AP-2) coated pit nucleation sites. Biochem. Cell Biol., 77, 439–448. [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel,P.R. (1980a) The conformation of biological macromolecules. W.H. Freeman and Company, San Francisco, CA, pp. 144–145. [Google Scholar]
- Cantor C.R. and Schimmel,P.R. (1980b) Techniques for the study of biological structure and function. W.H. Freeman and Company, San Francisco, CA, pp. 560–570. [Google Scholar]
- Chen C.Y., Reese,M.L., Hwang,P.K., Ota,N., Agard,D. and Brodsky,F.M. (2002) Clathrin light and heavy chain interface: alpha-helix binding superhelix loops via critical tryptophans. EMBO J., 21, 6072–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J., Eaton,S., Merkel,G., Skalka,A.M. and Laue,T. (1999) Characterization of the self association of Avian sarcoma virus integrase by analytical ultracentrifugation. J. Biol. Chem., 274, 32842–32846. [DOI] [PubMed] [Google Scholar]
- Cresswell P. (1996) Invariant chain structure and MHC class II function. Cell, 84, 505–507. [DOI] [PubMed] [Google Scholar]
- Crowther R.A. and Pearse,B.M.F. (1981) Assembly and packing of clathrin into coats. J. Cell Biol., 91, 790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B. and Saber,H. (1998) Determination of molecular parameters by fitting sedimentation data to finite-element solutions of the Lamm equation. Biophys. J., 74, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A.R. et al. (1985) Hydrogen bonding and biological specificity analysed by protein engineering. Nature, 314, 235–238. [DOI] [PubMed] [Google Scholar]
- Ford M.G., Mills,I.G., Peter,B.J., Vallis,Y., Praefcke,G.J., Evans,P.R. and McMahon,H.T. (2002) Curvature of clathrin-coated pits driven by epsin. Nature, 419, 361–366. [DOI] [PubMed] [Google Scholar]
- Ford M.G., Pearse,B.M., Higgins,M.K., Vallis,Y., Owen,D.J., Gibson,A., Hopkins,C.R., Evans,P.R. and McMahon,H.T. (2001) Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science, 291, 1051–1055. [DOI] [PubMed] [Google Scholar]
- Freire E. and Coelho-Sampaio,T. (2000) Self-assembly of laminin induced by acidic pH. J. Biol. Chem., 275, 817–822. [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Santini,F., Warren,R.A. and Keen,J.H. (1999) Spatial control of coated-pit dynamics in living cells. Nat. Cell Biol., 1, 1–7. [DOI] [PubMed] [Google Scholar]
- Greene B., Liu,S.-H., Wilde,A. and Brodsky,F.M. (2000) Complete reconstitution of clathrin basket formation with recombinant protein fragments: Adaptor control of clathrin self-assembly. Traffic, 1, 69–75. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. and Kirchhausen,T. (1983) Clathrin, cages and coated vesicles. Cell, 33, 650–652. [DOI] [PubMed] [Google Scholar]
- Heuser J. (1989) Effects of cytoplasmic acidification on clathrin lattice morphology. J. Cell Biol., 108, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasanoff A., Wagner,G. and Wiley,D.C. (1998) Structure of a trimeric domain of the MHC class II-associated chaperonin and targeting protein Ii. EMBO J., 17, 6812–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A.J. and Nossal,R. (1993) Topological mechanisms involved in the formation of clathrin-coated vesicles. Biophys. J., 65, 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A.J. and Nossal,R. (2000) Rigidity of triskelion arms and clathrin nets. Biophys. J., 78, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.L., Correia,J.J., Yphantis,D.A. and Halvorson,H.R. (1981) Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J., 36, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneseki T. and Kadota,K. (1969) The ‘vesicle in a basket.’ A morphological study of the coated vesicle isolated from the nerve endings of the guinea pig brain, with special reference to the mechanism of membrane movements. J. Cell Biol., 42, 202–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A., Gordon,R.E. and Borden,K.L. (2002) Self-assembly properties of a model RING domain. Proc. Natl Acad. Sci. USA, 99, 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. (2000) Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol., 1, 187–198. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Harrison,S.C. and Heuser,J. (1986) Configuration of clathrin trimers: Evidence from electron microscopy. J. Ultrastruct. Mol. Struct. Res., 94, 199–208. [DOI] [PubMed] [Google Scholar]
- Kocsis E., Trus,B.L., Steer,C.J., Bisher,M.E. and Steven,A.C. (1991) Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J. Struct. Biol., 107, 6–14. [DOI] [PubMed] [Google Scholar]
- Lauffer M.A. (1975) Entropy-driven processes in biology. Mol. Biol. Biochem. Biophys., 20, 1–264. [DOI] [PubMed] [Google Scholar]
- Lemmon S.K. (2001) Clathrin uncoating: Auxilin comes to life. Curr. Biol., 11, R49–R52. [DOI] [PubMed] [Google Scholar]
- Liu S.-H., Wong,M.L., Craik,C.S. and Brodsky,F.M. (1995) Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell, 83, 257–267. [DOI] [PubMed] [Google Scholar]
- Liu S.H., Marks,M.S. and Brodsky,F.M. (1998) A dominant-negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II major histocompatibility complex (MHC) pathway. J. Cell Biol., 140, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa S., Yoshimura,T., Hong,K., Düzgünes,N. and Papahadjopoulos,D. (1989) Mechanism of protein-induced membrane fusion: fusion of phospholipid vesicles by clathrin associated with its membrane binding and conformational change. Biochemistry, 28, 1422–1428. [DOI] [PubMed] [Google Scholar]
- Mejean A., Bodenreider,C., Schuerer,K. and Goldberg,M.E. (2001) Kinetic characterization of the pH-dependent oligomerization of R67 dihydrofolate reductase. Biochemistry, 40, 8169–8179. [DOI] [PubMed] [Google Scholar]
- Mishra S.K., Keyel,P.A., Hawryluk,M.J., Agostinelli,N.R., Watkins,S.C. and Traub,L.M. (2002) Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J., 21, 4915–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Smith,C.J., Roseman,A.M., Harrison,S.C., Kirchhausen,T. and Pearse,B.M.F. (1999) Functional organization of clathrin in coats: Combining electron cryomicroscopy and X-ray crstallography. Mol. Cell, 3, 761–770. [DOI] [PubMed] [Google Scholar]
- Nandi P.K. and Edelhoch,H. (1984) The effects of lyotropic (Hofmeister) salts on the stability of clathrin coat structure in coated vesicles and baskets. J. Biol. Chem., 259, 11290–11296. [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nossal R. (2001) Energetics of clathrin basket assembly. Traffic, 2, 138–147. [DOI] [PubMed] [Google Scholar]
- Parker M.H., Brouillette,C.G. and Prevelige,P.E.,Jr (2001) Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry, 40, 8962–8970. [DOI] [PubMed] [Google Scholar]
- Prevelige P.E. Jr (1998) Inhibiting virus-capsid assembly by altering the polymerisation pathway. Trends Biotechnol., 16, 61–65. [DOI] [PubMed] [Google Scholar]
- Robinson M.S. and Bonifacino,J.S. (2001) Adaptor-related proteins. Curr. Opin. Cell Biol., 13, 444–453. [DOI] [PubMed] [Google Scholar]
- Schreiber G. and Fersht,A.R. (1996) Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol., 3, 427–431. [DOI] [PubMed] [Google Scholar]
- Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J., 78, 1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F.P., Steer,C.J. and Kirchhoff,W.H. (1989) A differential scanning calorimetric study of brain clathrin. Arch. Biochem. Biophys., 273, 433–439. [DOI] [PubMed] [Google Scholar]
- Smith C.J., Grigorieff,N. and Pearse,B.M.F. (1998) Clathrin coats at 21 Å resolution: A cellular assembly designed to recycle multiple membrane receptors. EMBO J., 17, 4943–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E. and Branton,D. (1981) Assembly units of clathrin coats. Nature, 289, 420–422. [DOI] [PubMed] [Google Scholar]
- Wu X., Zhao,X., Baylor,L., Kaushal,S., Eisenberg,E. and Greene,L.E. (2001) Clathrin exchange during clathrin-mediated endocytosis. J. Cell Biol., 155, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe J.A., Brodsky,F.M., Hofmann,K., Lin,K., Liu,S.H., Chen,L., Earnest,T.N., Fletterick,R.J. and Hwang,P.K. (1999) Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature, 399, 371–375. [DOI] [PubMed] [Google Scholar]
- Ybe J.A., Greene,B., Liu,S.H., Pley,U., Parham,P. and Brodsky,F.M. (1998) Clathrin self-assembly is regulated by three light chain residues controlling the formation of critical salt bridges. EMBO J., 17, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T., Kameyama,K., Maezawa,S. and Takagi,T. (1991) Skeletal structure of clathrin triskelion in solution: experimental and theoretical approaches. Biochemistry, 30, 4528–4534. [DOI] [PubMed] [Google Scholar]