Abstract
Intracellular replication of the bacterial pathogen Salmonella enterica occurs in membrane-bound compartments called Salmonella-containing vacuoles (SCVs). Maturation of the SCV has been shown to occur by selective interactions with the endocytic pathway. We show here that after invasion of epithelial cells and migration to a perinuclear location, the majority of SCVs become surrounded by membranes of the Golgi network. This process is dependent on the Salmonella pathogenicity island 2 type III secretion system effector SseG. In infected cells, SseG was associated with the SCV and peripheral punctate structures. Only bacterial cells closely associated with the Golgi network were able to multiply; furthermore, mutation of sseG or disruption of the Golgi network inhibited intracellular bacterial growth. When expressed in epithelial cells, SseG co-localized extensively with markers of the trans-Golgi network. We identify a Golgi-targeting domain within SseG, and other regions of the protein that are required for localization of bacteria to the Golgi network. Therefore, replication of Salmonella in epithelial cells is dependent on simultaneous and selective interactions with both endocytic and secretory pathways.
Keywords: Golgi/Salmonella/SseG/trafficking/type III secretion
Introduction
Salmonella enterica serovar Typhimurium (S.typhi murium) is a bacterial pathogen that invades and replicates within a membrane-bound compartment (Salmonella-containing vacuole; SCV) inside a variety of host cell types. The biogenesis and maturation of the SCV has been studied in detail in both macrophages and epithelial cells. Within a few minutes of bacterial entry, components of the early endocytic pathway, such as EEA1 and the transferrin receptor, are recruited to the SCV (early SCV; Méresse et al., 1999; Steele-Mortimer et al., 1999). These proteins are then gradually lost, and interactions between the SCV and late endosomal compartments lead to the progressive acquisition of the vacuolar ATPase and several lysosomal membrane proteins (lgps), such as LAMP1 (intermediate SCV; Garcia-del Portillo and Finlay, 1995; Rathman et al., 1997; Méresse et al., 1999; Steele-Mortimer et al., 1999). The late SCV represents the third stage of maturation which begins 3–4 h after invasion. It requires the intracellular synthesis of bacterial proteins and is characterized by the further recruitment of lgps and the initiation of Sif formation. Sifs are tubular structures enriched in lgps and which appear to be contiguous with the SCV (Garcia-del Portillo et al., 1993). However, the SCV does not accumulate significant amounts of the lysosomal enzymes that are normally delivered to a maturing phagolysosome by the cation-independent mannose-6-phosphate receptor (Garcia-del Portillo and Finlay, 1995; Rathman et al., 1997). Interactions between the SCV and the endocytic pathway are therefore selective, and this unique trafficking pathway creates an environment permissive for bacterial cell division, which begins after a lag period of 3–4 h after invasion (Garcia-del Portillo et al., 1993).
Intracellular replication of S.typhimurium involves a large number of virulence proteins, including the type III secretion system (TTSS), encoded by the SPI-2 pathogenicity island (Ochman et al., 1996; Cirillo et al., 1998; Hensel et al., 1998). This TTSS translocates effector proteins into the vacuolar membrane and cytosol of the host cell. Several effectors have been identified in recent years, encoded both within and outside the pathogenicity island. SpiC is a SPI-2-encoded protein that prevents interactions between SCVs and endocytic compartments in macrophages (Uchiya et al., 1999). SifA is encoded outside SPI-2, and is required for maintenance of the vacuolar membrane, most likely by facilitating the recruitment and fusion of vesicles with the SCV (Beuzón et al., 2000). SifA is also required for the formation of Sifs (Stein et al., 1996). SCV membrane dynamics are also influenced by the action of another effector, the predicted acyl transferase SseJ (Ruiz-Albert et al., 2002). Several other effectors have been described, but the functions of most of them are not understood (Waterman and Holden, 2003).
While interactions between the SCV and markers of the endocytic pathway have been studied in depth, little is known about the spatial distribution of SCVs within infected cells and the potential relevance of this to bacterial multiplication. We show here that the majority of SCVs become surrounded by Golgi membranes 4 h after invasion of epithelial cells, a process that is dependent on the SPI-2 TTSS effector SseG. Mutational analysis of this protein identified a Golgi-targeting domain and defined further regions that are required for recruitment of SCVs to the Golgi network. The physiological relevance of Golgi targeting by Salmonella is demonstrated by the requirement of both SseG and an intact Golgi network for Salmonella replication. Therefore, SCV–Golgi interactions represent a crucial stage in the intracellular life cycle of Salmonella.
Results
Salmonella localizes to the Golgi network in epithelial cells
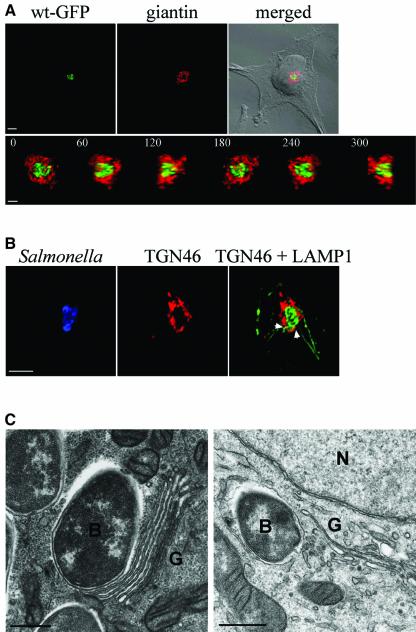
To investigate the intracellular localization of Salmonella, HeLa epithelial cells were infected with GFP-expressing S.typhimurium and analysed by confocal immunofluorescence microscopy. Developing microcolonies usually comprised tight clusters of bacteria, positioned close to the nucleus (Figures 1A and 2A) and were often found closely associated with the Golgi network. Confocal X/Z reconstructions revealed that these microcolonies were partially or completely enveloped by Golgi membranes (Figure 1A; Supplementary figure S1 available at The EMBO Journal Online). This phenomenon was observed using antibodies against a variety of Golgi proteins, including giantin and Golgi matrix protein 130 (GM130) (Figure 1A and data not shown). Similar results were obtained using an antibody against TGN46, a glycoprotein localized primarily in the trans-Golgi network (TGN) (Figure 1B). In contrast, calreticulin, an endoplasmic reticulum (ER) luminal protein, was not associated with microcolonies. As expected (Steele-Mortimer et al., 1999), there was no association between bacteria and early or recycling endosomal markers, including EEA1 and the transferrin receptor (data not shown). At 8 h after invasion, 84 ± 6% of microcolonies of GFP-expressing wild-type bacteria were associated with Golgi membranes. The labelled Golgi proteins were concentrated in the area around the developing microcolony, rarely between individual SCVs.
Fig. 1. Salmonella associates with the Golgi network in HeLa cells. (A) Upper panel, confocal immunofluorescence micrograph showing the subcellular localization of GFP-expressing wild-type S.typhimurium (wt-GFP, green) in relation to the cis-Golgi protein giantin (red), and the host cell (DIC in merged image), 8 h after invasion. Scale bar corresponds to 5 µm. Lower panel shows a 360° rotation on the y-axis of a 3D reconstruction obtained from a z-stack of the cell shown in the upper panel. Scale bar corresponds to 2 µm. (B) Infected cells were labelled for TGN46 (red), Salmonella (blue) and LAMP1, a marker of the SCV membrane (green). Points of co-localization between LAMP1 and TGN46 are indicated by arrowheads. Scale bar corresponds to 5 µm. (C) Transmission electron micrographs of representative HeLa cells showing wild-type Salmonella (B) in close proximity to Golgi cisternae (G). The nucleus is marked as (N). Scale bars correspond to 500 nm.
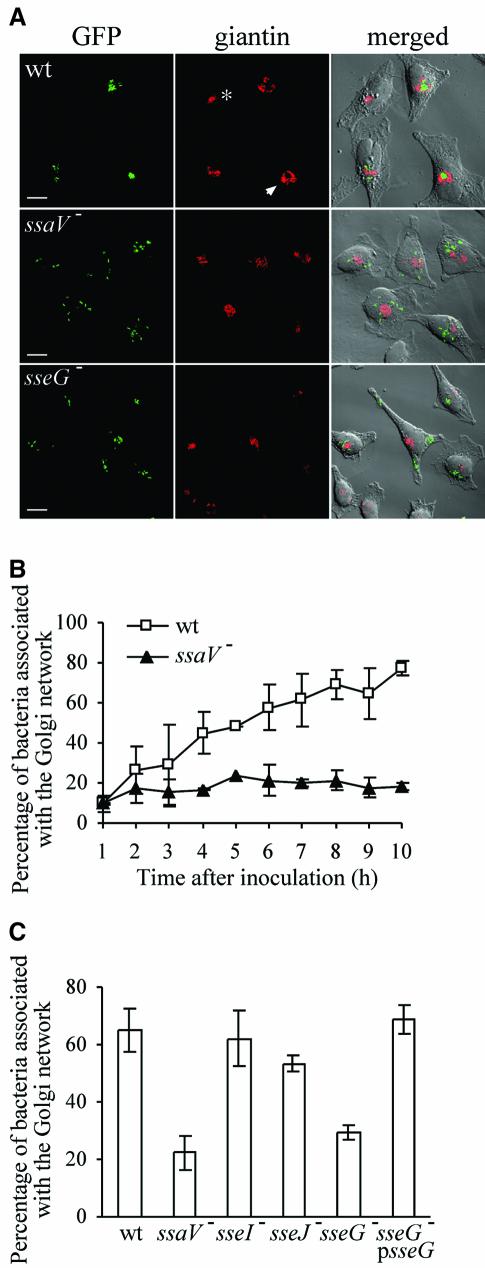
Fig. 2. Salmonella–Golgi association requires the SPI-2 TTSS effector protein SseG. (A) Intracellular distribution of GFP-expressing wild-type (wt), ssaV or sseG mutant strains in relation to giantin, 8 h after invasion of HeLa cells. Arrowhead indicates a distorted Golgi structure associated with a bacterial microcolony. Asterisk indicates compact Golgi network in an uninfected cell. Scale bars correspond to 10 µm. (B) Time course showing the increased association of wild-type S.typhimurium with the Golgi network (revealed by giantin labelling) in contrast to the ssaV mutant strain. Standard deviations from the mean are shown; results correspond to three independent experiments. (C) Association of SPI-2 effector mutant strains with the Golgi, 8 h after invasion of HeLa cells. Standard deviations from the mean are shown; results correspond to three independent experiments.
To determine if Golgi proteins co-localized with LAMP1, a marker of the SCV membrane (Méresse et al., 1999; Beuzón et al., 2002), infected cells were labelled with antibodies against LAMP1 and either giantin or TGN46. Neither Golgi protein was found to co-localize extensively with LAMP1 present on the vacuolar membrane, but frequent discrete points of co-localization were observed with both markers (arrowheads, Figure 1B). Neither COPI nor COPII coat proteins, which mediate vesicle formation within the secretory pathway, were recruited to the SCV membrane (data not shown). To determine whether SCV membranes were fused with Golgi membranes, infected HeLa cells were examined by transmission electron microscopy. SCVs were frequently found within 50–100 nm of Golgi cisternae (Figure 1C), and in some cases less than 50 nm. However, there was no evidence of fusion between SCV and Golgi membranes in over 50 infected cells analysed.
In uninfected HeLa cells, the Golgi network has a relatively compact appearance (asterisk, Figure 2A). In contrast, by 8 h after infection, the morphology of the network was frequently distorted, with the bacterial microcolony in the centre, suggesting that Golgi membranes are displaced by the presence of the growing microcolony (Figure 1A and arrowhead, Figure 2A). Association of S.typhimurium with the Golgi network was also observed in the human small intestinal epithelial cell line INT 407, but not in RAW 264.7 or elicited peritoneal murine macrophages, where replicating bacteria were frequently perinuclear, but neither associated in tight clusters nor surrounded by Golgi membranes (data not shown).
Association with the Golgi requires the SPI-2 TTSS effector SseG
We next investigated whether bacterial virulence proteins that are released intracellularly are involved in Salmonella–Golgi interactions. HeLa cells were infected with different mutant strains, then fixed and analysed by confocal microscopy for bacterial association with the Golgi network. SCVs were considered Golgi-associated if they were at least partially surrounded by Golgi membranes. The percentage of wild-type bacteria associated with the Golgi steadily increased to ∼80% over a 10 h period. However, a strain carrying a mutation in ssaV, which is defective for secretion of all SPI-2 TTSS effector proteins, had a significantly reduced level of association with the Golgi network; <20% of bacteria were associated with the Golgi at any time during the experiment (Figure 2A and B). Although ssaV mutant bacteria remained predominantly perinuclear, in contrast to the wild-type strain they displayed a scattered distribution (Figure 2A). No differences were observed between the wild-type and strains carrying mutations in phoP or spvB (data not shown), two virulence loci that are involved in growth of Salmonella in macrophages (Holden, 2002). To identify the SPI-2 effector responsible for this phenotype, strains carrying mutations in genes encoding different SPI-2-translocated proteins were examined. Whereas sseJ and sseI mutant strains were indistinguishable from the wild-type, an sseG mutant strain had a phenotype similar to that of the ssaV mutant (Figure 2A and C). The Golgi-targeting defect of the sseG mutant could be attributed to a lack of SseG because a plasmid containing the wild-type sseG allele restored association with the Golgi to the level of the wild-type strain (Figure 2C). Inactivation of SseG leads to a replication defect in macrophages and moderate virulence attenuation in the mouse model of systemic salmonellosis (Hensel et al., 1998). SseG is predicted to have at least two transmembrane domains; its primary amino acid sequence displays no significant similarities to protein database entries, apart from a 31% similarity to SseF, which is encoded immediately downstream of sseG (Hensel et al., 1998).
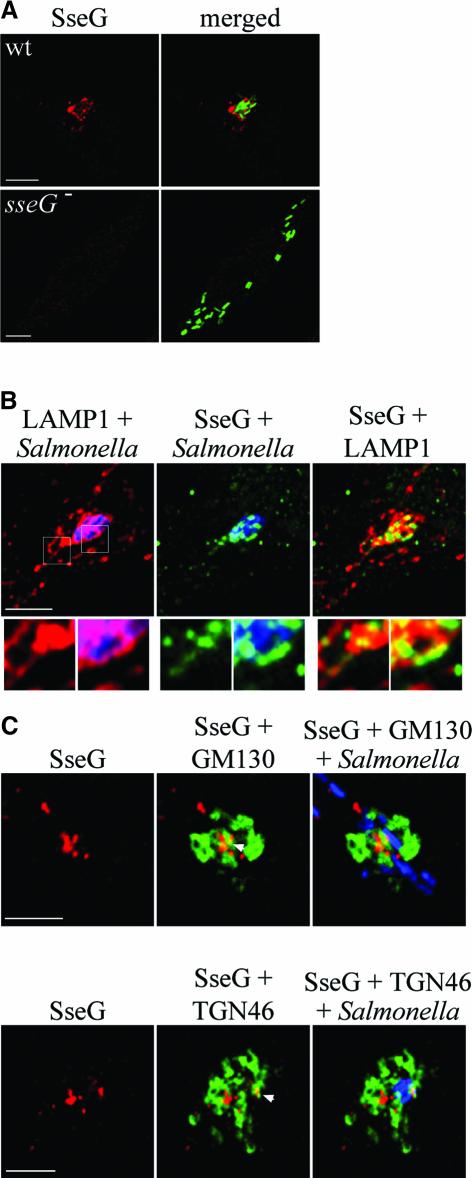
An antibody against a C-terminal peptide of SseG was used to examine the intracellular distribution of the protein. Intense labelling was typically observed outlining part of the bacterial microcolony, and also in peripheral punctate structures (Figure 3A and B). SseG co-localized with LAMP1 in the SCV membrane and on Sifs (Figure 3B). This observation is consistent with the localization of an M45 peptide-tagged version of SseG (Kuhle and Hensel, 2002). However, the peripheral SseG-positive, punctate structures did not co-localize with LAMP1. They were found mostly within the immediate vicinity of the Golgi network and occasionally co-localized with both cis-Golgi and TGN markers (arrowheads, Figure 3C).
Fig. 3. Intracellular distribution of SseG in HeLa cells. (A) Confocal immunofluorescence microscopy of HeLa cells infected for 10 h by GFP-expressing wild-type strain or the sseG mutant as a control. Cells were labelled with the anti-SseG antibody (red). Scale bars correspond to 5 µm. (B) Confocal immunofluorescence microscopy of HeLa cells infected for 10 h by wild-type S.typhimurium. Boxed areas shown in higher magnification below show co-localization between SseG (green) and LAMP1 (red) on a Sif and SCV membrane. Salmonella typhimurium was labelled with an anti-Salmonella antibody (blue). Punctate labelling of SseG can also be seen in the vicinity of the microcolony. Scale bar corresponds to 5 µm. (C) Distribution of SseG in relation to the Golgi network in HeLa cells infected for 10 h with wild-type bacteria (blue in merged image). Arrowheads indicate points of co-localization between SseG (red) and either GM130 (green, upper panel) or TGN46 (green, lower panel). Scale bars correspond to 5 µm.
SseG and an intact Golgi network are required for replication of Salmonella in epithelial cells
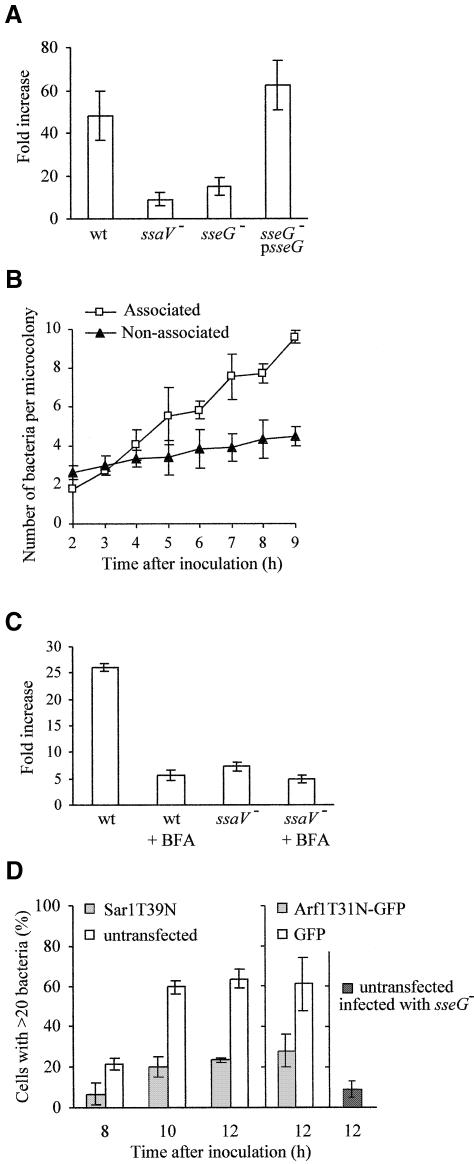
To establish if SseG contributes to growth of S.typhimurium in epithelial cells, intracellular replication assays were carried out. The numbers of wild-type and complemented sseG mutant strains underwent an ∼50-fold increase over a 14 h period. In contrast, the sseG mutant numbers increased by <10-fold over the same time period, a replication defect similar to that of the ssaV mutant (Figure 4A). Since SseG is required for both Golgi localization of bacteria and intracellular replication, we hypothesized that the Golgi network might be exploited by Salmonella for its replication. Therefore, the number of wild-type bacteria in Golgi-associated microcolonies was compared over time with those located elsewhere in the cell by microscopic examination. There was no significant increase in numbers of bacteria not associated with the Golgi network, whereas the numbers of Golgi-associated bacteria underwent a significant increase over a 9 h period (Figure 4B). This result suggests that bacterial replication is restricted to those associated with the Golgi.
Fig. 4. Interaction of Salmonella with the Golgi network is required for intracellular replication in HeLa cells. (A) Intracellular replication of S.typhimurium strains in HeLa cells. The values shown are representative of three independent experiments and indicate the fold increase calculated as a ratio of the intracellular bacteria between 16 and 2 h after invasion. Each infection was performed in triplicate and the standard errors from the mean are shown. (B) Analysis of the number of bacteria per microcolony in HeLa cells during a 9 h time course. Microcolonies associated with the Golgi network are compared with those found elsewhere in the cell. Values represent the mean of three independent experiments and the standard deviations from the mean are shown. (C) Intracellular replication of S.typhimurium strains in HeLa cells treated with BFA. The assay was carried out as for (A) above, except that BFA (5 µg/ml) was added at 15 min after invasion. There was no statistical difference between the numbers of internalized bacteria in treated or untreated cells at the 2 h time point. (D) Effect of expression of SarT39N or ArfT31N on intracellular replication of wild-type S.typhimurium. HeLa cells were transfected for 14 h, then infected for a further 8–12 h. Values represent the percentage of host cells containing more than 20 bacteria at different time points after invasion. The percentage of untransfected cells with more than 20 sseG mutant bacteria is shown on the right. Values represent the means of three independent experiments and the standard deviations from the mean are shown.
To test the requirement for an intact Golgi network in intracellular growth of Salmonella, cells were treated 15 min after bacterial invasion with Brefeldin A (BFA). BFA inhibits nucleotide exchange on the small GTPase ADP-ribosylating factor (Arf1), which is required for formation of COPI coated vesicles (Chardin and McCormick, 1999). As a result, the Golgi network rapidly redistributes into the ER (Lippincott-Schwartz et al., 1989). BFA strongly inhibited intracellular growth of wild-type S.typhimurium, but had little effect on numbers of ssaV mutant bacteria (Figure 4C). However, BFA had no noticeable effect on markers of the maturation and trafficking of the SCV, such as LAMP1 recruitment and cathepsin D exclusion (data not shown). Therefore, the effects of BFA appear to be independent of the selective interactions between SCVs and the late endocytic pathway. As a further test for the requirement of a functional Golgi network in S.typhimurium replication, bacteria were examined in HeLa cells expressing Arf1T31N-GFP, a dominant interfering variant of Arf1 (Dascher and Balch, 1994). A significant reduction in intracellular bacterial numbers was observed at 12 h after invasion, compared with bacterial replication in cells expressing GFP alone (Figure 4D). Sar1 is another small GTPase, involved in COPII-mediated vesicle formation, and its inhibition by constitutively inactive Sar1T39N prevents the formation of early secretory vesicles at ER exit sites (Ward et al., 2001). Bacterial replication was also severely inhibited in cells expressing this mutant protein (Figure 4D). Expression of Arf1T31N-GFP or Sar1T39N had no effect on bacterial internalization (Supplementary table S1). Together, these experiments establish that an intact and functional Golgi network is required for intracellular replication of S.typhimurium.
SseG is a Golgi-targeting protein
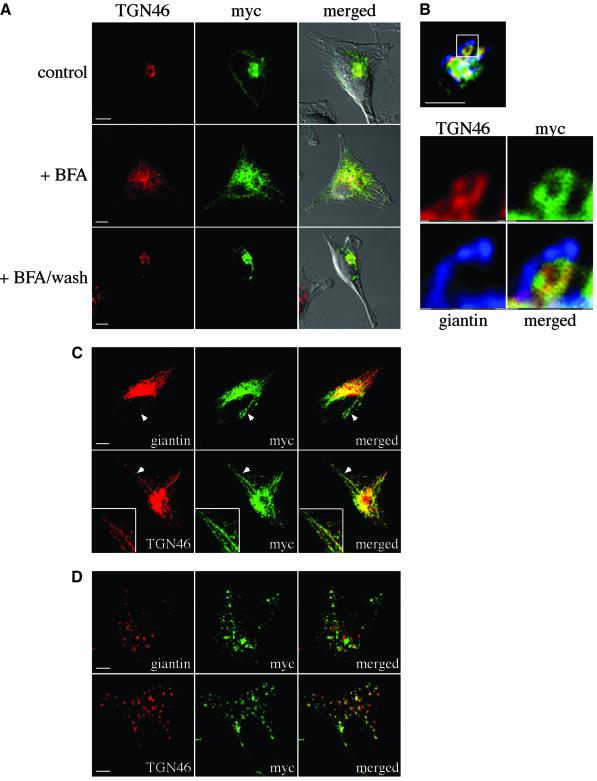
To gain further insight into the function of SseG, an epitope-tagged version of the protein (myc::SseG) was expressed in HeLa cells. The protein was concentrated in a perinuclear region, where it co-localized extensively with Golgi markers including giantin (data not shown) and TGN46 (Figure 5A). To verify that myc::SseG was localized to the Golgi network, transfected cells were incubated with BFA for 1 h, and either fixed immediately or washed in medium lacking BFA, and incubated for a further 1 h. In all transfected cells, BFA caused extensive redistribution of myc::SseG throughout the cell, where most of it co-localized with dispersed TGN46 (Figure 5A). Following BFA wash-out, TGN46 moved back to its original perinuclear position and was accompanied by myc::SseG (Figure 5A). Subcellular fractionation of transfected cells and immunoblotting showed that SseG localized to the membrane fraction, as expected for a predicted integral membrane protein (data not shown). In transfected cells that strongly overexpressed myc::SseG, the Golgi network was disrupted. However, expression of other Salmonella SPI-2 secreted proteins, including two with predicted membrane-spanning domains (myc::SseC and myc::SseD), did not result in localization to the Golgi or perturbation of its structure (data not shown).
Fig. 5. Expression of myc::SseG in HeLa cells results in its localization to the Golgi network. (A) Cells expressing a myc-tagged version of SseG (green) were co-labelled with an antibody against TGN46 (red), and examined by confocal microscopy. The majority of myc::SseG co-localizes with TGN46 (upper panel) and its redistribution follows that of the TGN marker upon BFA treatment (middle panel). BFA washout leads to recovery of a compact Golgi structure, and the re-localization of myc::SseG (lower panel). Scale bars correspond to 5 µm. (B) Cells expressing myc::SseG (green) were triple labelled with anti-myc (green), anti-TGN46 (red) and anti-giantin (blue) antibodies. (C) Cells expressing myc::SseG (green) were treated with BFA for 10 min, fixed and labelled with either anti-giantin or anti-TGN46 antibodies. In the upper panel the arrowheads indicate tubules containing myc::SseG (green) which does not co-localize with giantin (red). Scale bar corresponds to 5 µm. In the lower panel, myc::SseG co-localizes with TGN46 in tubules. Arrowheads indicate tubulating regions shown at a higher magnification in insets. (D) Distribution of myc::SseG and Golgi markers after exposure of transfected cells to nocodazole. Cells were labelled with anti-myc (green) and either anti-giantin (red, upper panel) or anti-TGN46 (red, lower panel) antibodies. Scale bars correspond to 5 µm.
The distribution of myc::SseG within the Golgi network was further analysed by confocal microscopy using Golgi markers. Cells expressing myc::SseG also showed some co-localization with the cis-Golgi protein giantin. However, triple labelling with both cis-Golgi and TGN markers showed that there was a greater degree of co-localization between myc::SseG and TGN46 than with giantin (Figure 5B). Expression of myc::SseG and p58-GFP, a protein of the ER-Golgi intermediate compartment, did not result in their co-localization (data not shown).
Golgi membrane tubulation induced by BFA results in rapid redistribution of the cis-Golgi network into the ER (Lippincott-Schwartz et al., 1990). Although the TGN also undergoes tubulation, these tubules do not interact with the ER but instead fuse with compartments of the early endosomal network (Lippincott-Schwartz et al., 1991; Wood and Brown, 1992). To confirm that myc::SseG localizes preferentially to the TGN, the kinetics of redistribution of myc::SseG were analysed after treatment with BFA. HeLa cells expressing myc::SseG were incubated with BFA and fixed for immunofluorescence microscopy at short time intervals thereafter. At 2 and 4 min after BFA treatment no significant tubulation of the cis-Golgi was observed. However, at the same time points, myc::SseG was found on tubulating membranes that also contained TGN46 (data not shown). From 6 to 10 min after BFA treatment, myc::SseG was still detected in tubular structures containing TGN46 but did not co-localize significantly with giantin-containing tubules (Figure 5C). As expected, a significant proportion of SseG also co-localized with the early endosomal marker EEA1 (Supplementary figure S2). By 14 min, the majority of giantin was redistributed in the ER whereas myc::SseG remained on TGN46-containing tubules (data not shown). Although the two proteins did not co-localize completely on tubules (arrowheads, Figure 5C), the kinetics of tubulation of compartments containing myc::SseG and TGN46 were indistinguishable.
Depolymerization of microtubules with nocodazole causes slow dispersal of the Golgi complex (Cole et al., 1996). Short stacked cisternae assemble slowly at peripheral sites to form small structures that closely resemble intact Golgi stacks (Thyberg and Moskalewski, 1999). Cells expressing myc::SseG were treated with nocodazole for 5 h and then analysed by labelling myc::SseG with either giantin or TGN46. There was little co-localization between myc::SseG and giantin, but a significant degree of co-localization of myc::SseG and TGN46 was observed in many of the scattered Golgi structures (Figure 5D). Triple labelling with anti-myc, anti-giantin and anti-TGN46 antibodies revealed that some of the peripheral structures containing myc::SseG were negative for both the cis-Golgi and TGN markers (data not shown). Collectively, these experiments show that a significant proportion of SseG localizes specifically to the TGN when expressed ectopically in HeLa cells.
The Golgi-targeting domain of SseG
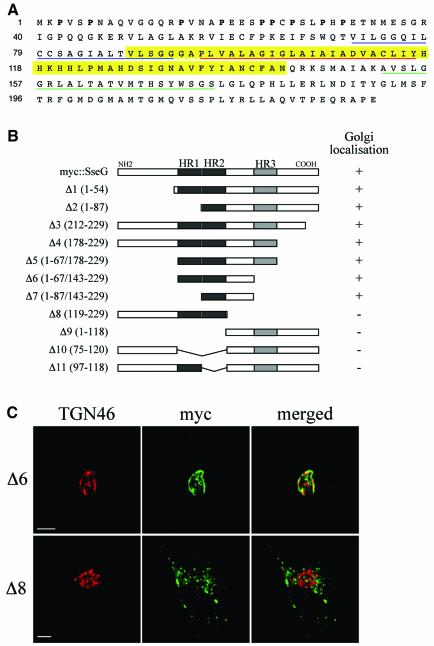
SseG has two strongly predicted transmembrane domains (TMDs), designated HR1 and HR2, and a third hydrophobic region (HR3) that could also constitute a TMD (Figure 6A). In addition, the N-terminal region is unusual in that 9 of the first 31 amino acids are prolines. To identify the region(s) of SseG that confer its localization to the Golgi, a series of myc-tagged truncated versions was constructed and expressed in HeLa cells after transfection. Progressive deletions from the N- and C-termini (Δ1, Δ2, Δ3, Δ4, Δ5, Δ6, Δ7) identified a 55 amino acid region including HR2 that is necessary and sufficient for Golgi targeting of SseG (Figure 6B). Removal of either HR2 (Δ9, Δ10, Δ11) or the region between HR2 and HR3 (Δ8) resulted in a complete loss of Golgi targeting and the mutant proteins were instead redistributed in a punctate manner throughout the cell (Figure 6B and C). Therefore, information in the second TMD and its carboxy-flanking region are required for Golgi targeting. Sequence comparisons between this domain and known eukaryotic and viral Golgi-targeting sequences have failed to reveal any significant similarities.
Fig. 6. Identification of Golgi-targeting regions of SseG. (A) Amino acid sequence of SseG. Hydrophobic regions HR1, HR2 and HR3 are underlined in blue, red and green, respectively. The Golgi targeting region is highlighted in yellow. Prolines in the N-terminal 31 amino acids are in bold. (B) Schematic representation of truncated polypeptides derived from myc::SseG. The extent of each deletion is shown in parentheses to the left of each construct. The ability of each polypeptide to associate with the Golgi is indicated to the right of each construct. (C) Representative images of Δ6 (upper panel) showing extensive co-localization with TGN46 and Δ8 (lower panel) showing a scattered distribution, with no co-localization with the Golgi marker. Scale bars correspond to 5 µm.
Topology of SseG and regions required for bacterial association with the Golgi
The results described above show that SseG is required for the association between SCVs and the Golgi network, and that this protein has a novel Golgi-targeting signal. Therefore, one can predict that additional information must be present within SseG that localizes SCVs to the Golgi network.
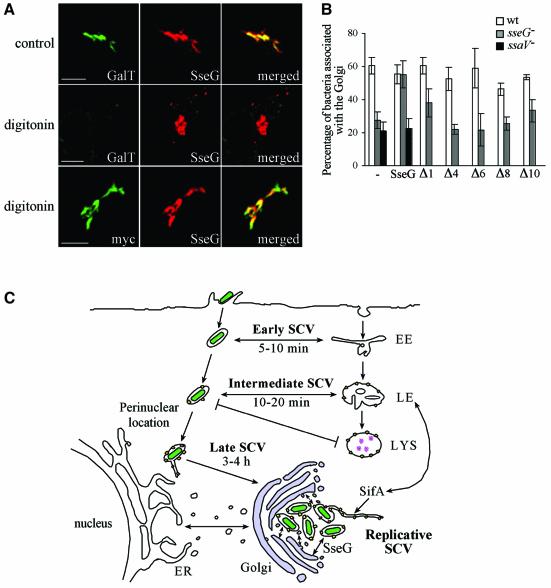
To identify regions of SseG that are exposed on the cytoplasmic face of Golgi membranes, we determined the membrane topology of SseG. HeLa cells expressing myc-tagged SseG were first incubated with digitonin (which renders the plasma membrane permeable to large molecules but which does not alter the integrity of secretory pathway compartments; Plutner et al., 1992), then labelled with anti-myc or anti-SseG antibodies in the absence of detergent. The myc epitope serves as a marker for the N-terminus of SseG and the anti-SseG antibody recognizes the C-terminus of the protein. An antibody against a luminal epitope of galactosyltransferase (GalT) was used as a control for the integrity of Golgi membranes. Following exposure of cells to digitonin, Golgi-associated SseG was readily detected by the anti-myc and anti-SseG antibodies. However, there was no labelling of GalT unless the cells were exposed to Triton X-100, which is a non-specific membrane permeabilizing detergent. Co-labelling of GalT and SseG revealed extensive co-localization of the two proteins (Figure 7A).
Fig. 7. Topology of SseG and functional activity of mutant polypeptides. (A) HeLa cells expressing myc::SseG were permeabilized with Triton X-100 (control) or digitonin and labelled with antibodies against myc, SseG or a luminal epitope of human galactosyltransferase (GalT). Scale bars correspond to 5 µm. (B) HeLa cells expressing wild-type or mutant myc::SseG polypeptides were infected for 10 h with GFP-expressing wild-type, ssaV or sseG mutant S.typhimurium. Cells were labelled with anti-myc and anti-giantin antibodies, and the level of bacterial association with the Golgi network was determined in both transfected and untransfected cells. Values represent means of three independent experiments and the standard deviations from the mean are shown. (C) Model of SCV trafficking in relation to localization within epithelial cells. Following host cell invasion, development of the early and intermediate SCV proceeds by progressive and selective interactions with compartments of the endocytic pathway (LAMP1 and cathepsin D are shown as yellow and pink symbols, respectively), as bacteria (green) migrate to a perinuclear location. This leads to the formation of the late SCV. The translocation of the SPI-2 TTSS effector SifA is required for recruitment of lgp-containing vesicles and the formation of Sifs, while SseG localizes SCVs to the Golgi network. As a consequence of this, the replicative phase of development can begin through acquisition of nutrients that enable bacterial replication, and membrane which encloses the growing population of intracellular bacteria. EE, early endosome; LE, late endosome; LYS, lysosome.
From this experiment we conclude that both N- and C-terminal domains of Golgi-associated SseG are exposed on the cytoplasmic face of the membrane. It follows that SseG contains only two TMDs, which are most likely HR1 and HR2. Therefore, either or both N- and C-terminal regions of the protein might be involved in localizing SCVs to the Golgi network. To test this, variants of SseG were expressed in HeLa cells and assayed for their ability to restore association of the sseG mutant strain with the Golgi network. First, cells were transfected with a construct expressing full-length myc::SseG, then infected with GFP-expressing wild-type or sseG mutant bacteria for 10 h, and examined for bacterial localization. Expression of myc::SseG or mutant versions did not affect the ability of wild-type bacteria to localize efficiently to the Golgi network (Figure 7B), indicating that these proteins did not exert a dominant-negative effect on Golgi targeting by wild-type bacteria. Ectopically expressed myc::SseG restored Golgi localization of the sseG mutant strain to the level seen with the wild-type strain (Figure 7B). Expression of mutants Δ8 and Δ10, which were not Golgi associated, did not rescue Golgi localization to the sseG mutant. However, mutants Δ1, Δ4 and Δ6, which were efficiently Golgi-localized (Figure 6B), also failed to rescue the sseG mutant phenotype (Figure 7B). These results demonstrate that in addition to a requirement for the Golgi-targeting region of SseG, information in both the C- and N-terminal regions of the protein is required for the interaction between SCVs and the Golgi network. To determine if other SPI-2 effectors might be involved in Golgi-targeting, myc::SseG was tested for its ability to restore Golgi localization of ssaV (SPI-2 null mutant) bacteria. In contrast to the sseG mutant, Golgi localization of the ssaV mutant could not be rescued by ectopic expression of SseG (Figure 7B). This result suggests that other SPI-2 translocated effectors contribute to the process of Salmonella localization to the Golgi network.
Discussion
SCV maturation and association with the Golgi
After invasion of epithelial cells by Salmonella, the SCV begins to migrate to the perinuclear region, and undergoes three stages of maturation, which reflect selective interactions with the endocytic pathway of the host cell (Steele-Mortimer et al., 1999). In this work we have shown that these events are followed by a further, crucial phase of development, in which SCVs interact with the Golgi network through the action of SseG. Since this stage involves bacterial replication, we refer to it as the replicative SCV. Therefore, maturation of the SCV proceeds not only by progressive and selective interactions with the endocytic pathway but also in terms of the spatial distribution of SCVs in the host cell, and their interaction with the secretory pathway. A model of SCV trafficking in relation to its intracellular localization is shown in Figure 7C.
Physical continuities between SCV and Golgi membranes were not observed at the ultrastructural level, but it seems likely that transient interactions must occur, since intracellular bacterial replication can be inhibited either by perturbation of Golgi function, or by mutation of sseG, which prevents SCVs from localizing to the Golgi network. The close apposition of SCVs and Golgi membranes, and an associated membrane fusion machinery, could enable transient fusion events between Golgi-derived vesicles and SCVs, resulting in the acquisition of molecules required for Salmonella replication. Since expression of SseG in HeLa cells led to its concentration in the TGN, it seems likely that these vesicles are derived from late Golgi compartments.
In HeLa cells, vacuoles containing Chlamydia spp. (inclusion bodies) become fusogenic with sphingomyelin-containing exocytic vesicles that traffic between the TGN and the plasma membrane. Incorporation of Golgi-derived lipids is thought to provide a mechanism for growth and maintenance of the Chlamydia inclusion membrane (Hackstadt et al., 1997). Therefore it is possible that SseG could mediate a similar function in acquiring lipids for the SCV membrane. BFA treatment of Chlamydia-infected cells leads to smaller, morphologically abnormal inclusions, but does not affect bacterial multiplication (Hackstadt et al., 1996). In contrast, BFA had no effect on the integrity or morphology of the SCV membrane, as judged by LAMP1 labelling (data not shown), but strongly inhibited Salmonella replication. Furthermore, Chlamydia inclusions do not interact with endocytic organelles (Wyrick, 2000), whereas the SPI-2 effector SifA has a major role in the recruitment and probably fusion of lgp-containing vesicles to the SCV membrane (Ruiz-Albert et al., 2002). Indeed, mutation of sifA results in a strain that cannot maintain the physical integrity of its vacuolar membrane (Beuzón et al., 2000). Therefore Salmonella appears to exploit late endocytic organelles as a source of membrane that is required to enclose replicating bacteria, and it seems more likely that interactions between SCVs and the Golgi satisfy a nutritional requirement, for example by providing carbon and/or nitrogen sources which are necessary for bacterial replication.
Role of SseG in bacterial association with the Golgi
Two lines of evidence show that the SPI-2 TTSS effector protein SseG and its Golgi-targeting region are necessary for S.typhimurium association with the Golgi network. First, sseG mutant bacteria fail to localize to the Golgi network of infected cells. Second, this defect can be rescued efficiently by ectopic expression of full-length SseG in infected cells, but not by a version lacking the Golgi-targeting domain. In infected cells, SseG was found associated with SCVs and in peripheral punctate structures, which occasionally co-localized with Golgi markers. How then does translocated SseG account for bacterial association with the Golgi? Apart from its Golgi-targeting region, both the proline-rich N-terminal domain and information in the C-terminal 51 amino acids contribute to the localization of SCVs to the Golgi network. Furthermore, localization of sseG mutant but not SPI-2 null mutant bacteria to the Golgi can be rescued by the ectopic expression of SseG in HeLa cells. These results suggest the existence of another SPI-2 effector that functions together with SseG to mediate Golgi localization of SCVs. Alternatively the null mutant may be defective in earlier unrelated processes of vacuole maturation, on which SseG function is dependent. Translocated SseG might interact with its target membrane via the Golgi-targeting domain, and to another effector through its N- or C-terminal regions. It is possible that SseG is a component of a multiprotein complex that mediates interactions between SCVs and Golgi membranes.
A variety of proteins are targeted to Golgi membranes by their TMDs. These include integral membrane glycosyltransferases and several viral proteins. For example, the M protein of the coronavirus mouse hepatitis virus, which targets the TGN, has one signal in its TMD and a second signal in the cytoplasmic tail (Gleeson, 1998). In the M protein of the avian coronavirus infectious bronchitis virus, the first of three TMDs contains the signal that targets the protein to the Golgi network (Gleeson, 1998). However, no similarity between SseG and either eukaryotic or prokaryotic Golgi targeting sequences has been found. Therefore the Golgi-targeting signal present within amino acids 88 and 142 of SseG appears to be novel.
Interestingly, SseG has also been reported to be present on, and required for the formation of, Sifs (Guy et al., 2000; Kuhle and Hensel, 2002). However, this process is likely to be independent of its interactions with the Golgi network, as BFA treatment does not prevent Sif formation (Brumell et al., 2001; data not shown). Therefore, SseG might perform more than one function in the maturation of SCVs.
Role of SseG in Salmonella virulence
Although it is clear that the Salmonella SPI-2 TTSS plays an important role in intracellular replication of bacteria in epithelial cells (Cirillo et al., 1998; Bispham et al., 2001; Beuzón et al., 2002; Paesold et al., 2002; this study) the functions of SPI-2 have been mainly studied in macrophages, which are colonized during the systemic phase of the disease in mice (Holden, 2002). Microscopic examination of S.typhimurium in primary and RAW 264.7 macrophages failed to reveal significant association of SCVs with the Golgi network. However, other evidence indicates that S.typhimurium–Golgi interactions might be relevant in macrophages. First, the virulence of the sseG mutant strain is attenuated in the murine model of infection (Hensel et al., 1998). Second, the sseG mutant strain has a replication defect in RAW 264.7 macrophages (Hensel et al., 1998), and in primary peritoneal macrophages (data not shown). Third, treatment of infected macrophages with BFA significantly inhibited S.typhimurium replication (Supplementary figure S3). Therefore, it seems probable that S.typhimurium does interact with the Golgi network in macrophages, but in a way that does not involve clustering of bacterial microcolonies within Golgi membranes.
It is well established that in epithelial cells, maturation of the SCV is dependent on a series of selective interactions with the endocytic pathway. The major conclusion to emerge from the work described here is that while these interactions are necessary for bacterial survival, they are not sufficient for intracellular replication: this requires SCV interactions with the secretory pathway. The identification of SseG as a Golgi-targeting virulence protein provides the basis for further investigation into the mechanisms by which this pathogen replicates in host cells, could provide insights into the normal functioning of the Golgi network and might ultimately offer novel approaches to inhibit bacterial growth in host cells.
Materials and methods
Bacterial strains, plasmids and growth conditions
The S.typhimurium strains used in this study were the wild-type 12023s, and its isogenic mutant derivatives: ssaV::aphT (Deiwick et al., 1998), sseG::aphT (Hensel et al., 1998) sseI::pGP704 (Ruiz-Albert et al., 2002) and ΔsseJ (Ruiz-Albert et al., 2002). Bacteria were grown in Luria Bertani (LB) medium supplemented with ampicillin (50 µg/ml), kanamycin (50 µg/ml) or chloramphenicol (35 µg/ml) for plasmid-containing strains, as appropriate.
Plasmid pFVP25.1, carrying gfpmut3A under the control of the rpsM constitutive promoter (Valdivia and Falkow, 1997), was introduced into bacterial strains for fluorescence visualization where indicated. The plasmids pCLXSN-GFP and pCLXSN-Arf1T39N-GFP (Kagan and Roy, 2002) were provided by Dr C. Roy. The Sar1T39N plasmid (Ward et al., 2001) was obtained from Dr J. Lippincott-Schwartz. Construction of the complementing plasmid psseG and the expression plasmid pmyc::sseG are described in the Supplementary data. The computer program TMpred was used to identify membrane-spanning domains within SseG (Persson and Argos, 1994). Construction of plasmids used for expression of truncated versions of SseG are described in the Supplementary data.
Antibodies and reagents
The mouse anti-LAMP1 antibody H4A3 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, maintained by the University of Iowa and was used at a dilution of 1:1000. Anti-Salmonella goat antibody CSA-1 (Kirkegaard and Perry Laboratories) was used at a dilution of 1:400. The rabbit antibodies anti-GM130 and anti-giantin were obtained from Dr A.De Matteis and used at a dilution of 1:1000 and 1:2000, respectively. The mouse anti-GM130 antibody (Transduction Laboratories) was used at a dilution of 1:1000. The sheep anti-human TGN46 antibody (Serotec) was used at a dilution of 1:100. A rabbit antibody against human TGN46 (Ponnambalam et al., 1996) was obtained from Dr G.Banting and was used at the dilution of 1:500. The mouse anti-c-myc antibody 9E10 (Santa Cruz Biotechnology) was used at a dilution of 1:100. An antibody to galactosyltransferase was obtained from Dr E.Berger and was used at a dilution of 1:4 (Berger et al., 1986). Rhodamine Red X (RRX)-, Cyanine 2-, Cyanine 5 (Cy5)- and aminomethylcoumarin acetate-conjugated donkey anti-mouse, anti-rabbit and anti-goat antibodies (Jackson Immunoresearch Laboratories) were used at a dilution of 1:400. RRX- and Cy5-conjugated donkey anti-sheep antibodies (Jackson Immunoresearch Laboratories) were used at dilutions of 1:100 and 1:600, respectively. BFA (Sigma) was used at 5 µg/ml. At this concentration BFA was not cytotoxic to HeLa cells and did not affect S.typhimurium growth in LB medium over 16 h. Nocodazole (Calbiochem) was used at a final concentration of 10 µg/ml.
Affinity purification of the anti-SseG antibody
A peptide with the sequence SSPLYRLLAQVTPEQRAPE corresponding to the last 19 amino acids of SseG was conjugated to keyhole limpet haemocyanin and this was used to immunize rabbits. Polyclonal rabbit serum was affinity purified using Affi-Gel 15 (BioRad), to which the peptide had been coupled, following the manufacturer’s procedure.
Cell culture
HeLa (clone HtTA1) cells were kindly provided by Dr S.Méresse. The INT 407 cell line U510 was a gift from Dr A.L.Servin. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and 2 mM glutamine at 37°C in 5% CO2.
Bacterial infection of cultured cells and replication assays
HeLa cells were infected with equivalent numbers of exponential phase S.typhimurium strains, as described previously (Beuzón et al., 2000). In order to follow a synchronized population of bacteria, HeLa cells were washed after 15 min of exposure to S.typhimurium and subsequently incubated in medium containing gentamicin to kill extracellular bacteria. For enumeration of intracellular bacteria, HeLa cells were washed three times with phosphate-buffered saline, lysed with 0.1% Triton X-100 for 10 min and a dilution series was plated onto LB agar.
Microscopy
For electron microscopy, samples were prepared as described previously for HeLa cells (Beuzón et al., 2002). For immunofluorescence, cell monolayers were fixed and labelled as described previously (Beuzón et al., 2002). For the affinity purified anti-SseG antibody, coverslips were permeabilized for 10 min with 0.1% Triton X-100 and then labelled as described above, in the absence of saponin. Samples were analysed using a confocal laser scanning microscope (LSM510; Zeiss).
Transfection of HeLa cells
HeLa cells were transiently transfected by the calcium phosphate DNA precipitation method as described previously (Ruiz-Albert et al., 2002). Bacterial infection of transfected HeLa cells was performed as described above, 14 h after DNA addition, and cells were fixed 24 h after DNA addition (10 h after infection). To determine the topology of myc::SseG, HeLa cells were transfected for 24 h with 2 µg of DNA using Polyfect (Quiagen) and then permeabilized with 25 µg/ml digitonin (Calbiochem) for 5 min on ice (Plutner et al., 1992). Cells were then fixed with PFA and labelled with antibodies in the absence of detergent.
Quantification of Salmonella association with the Golgi network
Quantification of the number of bacteria associated with the Golgi network was performed by confocal microscopy. Only bacteria that were either completely or at least 50% surrounded by the labelled Golgi marker were counted as being Golgi associated. Bacterial clusters that were found adjacent to the Golgi but did not fulfil the above criteria were counted as non-associated. At least 50 host cells, corresponding to more than 100 bacteria, were scored blind in each experiment and all experiments were repeated three times.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are very grateful to Gustavo Egea, Paul Freemont and Gareth Griffiths for helpful suggestions, and to Jennifer Lippincott-Schwartz, Antonietta De Matteis, George Banting, Craig Roy, Eric Berger, Stephane Méresse and Alain Servin for providing reagents. We thank Dave Goulding for electron microscopy, Sarah Gilliland for technical assistance and Javier Ruiz-Albert for technical advice. We are grateful to Christoph Tang, Emmanuelle Caron and members of the Holden laboratory for critical review. S.P.S. is supported by a fellowship from the Portuguese Foundation for Science and Technology. This work was also supported by a grant from the Medical Research Council, UK.
References
- Berger E.G., Aegerter,E., Mandel,T. and Hauri,H.P. (1986) Monoclonal antibodies to soluble, human milk galactosyltransferase (lactose synthase A protein). Carbohydr. Res., 149, 23–33. [DOI] [PubMed] [Google Scholar]
- Beuzón C.R., Méresse,S., Unsworth,K.E., Ruiz-Albert,J., Garvis,S., Waterman,S.R., Ryder,T.A., Boucrot,E. and Holden,D.W. (2000) Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J., 19, 3235–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón C.R., Salcedo,S.P. and Holden,D.W. (2002) Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology, 148, 2705–2715. [DOI] [PubMed] [Google Scholar]
- Bispham J., Tripathi,B.N., Watson,P.R. and Wallis,T.S. (2001) Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun., 69, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell J.H., Tang,P., Mills,S.D. and Finlay,B.B. (2001) Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic, 2, 643–653. [DOI] [PubMed] [Google Scholar]
- Chardin P. and McCormick,F. (1999) Brefeldin A: the advantage of being uncompetitive. Cell, 97, 153–155. [DOI] [PubMed] [Google Scholar]
- Cirillo D.M., Valdivia,R.H., Monack,D.M. and Falkow,S. (1998) Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol., 30, 175–188. [DOI] [PubMed] [Google Scholar]
- Cole N.B., Sciaky,N., Marotta,A., Song,J. and Lippincott-Schwartz,J. (1996) Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell, 7, 631–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C. and Balch,W.E. (1994) Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem., 269, 1437–1448. [PubMed] [Google Scholar]
- Deiwick J., Nikolaus,T., Shea,J.E., Gleeson,C., Holden,D.W. and Hensel,M. (1998) Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol., 180, 4775–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F. and Finlay,B.B. (1995) Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol., 129, 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F., Zwick,M.B., Leung,K.Y. and Finlay,B.B. (1993) Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl Acad. Sci. USA, 90, 10544–10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson P.A. (1998) Targeting of proteins to the Golgi apparatus. Histochem. Cell. Biol., 109, 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy R.L., Gonias,L.A. and Stein,M.A. (2000) Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol., 37, 1417–1435. [DOI] [PubMed] [Google Scholar]
- Hackstadt T., Rockey,D.D., Heinzen,R.A. and Scidmore,M.A. (1996) Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J., 15, 964–977. [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T., Fischer,E.R., Scidmore,M.A., Rockey,D.D. and Heinzen,R.A. (1997) Origins and functions of the chlamydial inclusion. Trends Microbiol., 5, 288–293. [DOI] [PubMed] [Google Scholar]
- Hensel M., Shea,J.E., Waterman,S.R., Mundy,R., Nikolaus,T., Banks,G., Vazquez-Torres,A., Gleeson,C., Fang,F.C. and Holden,D.W. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol., 30, 163–174. [DOI] [PubMed] [Google Scholar]
- Holden D.W. (2002) Trafficking of the Salmonella vacuole in macrophages. Traffic, 3, 161–169. [DOI] [PubMed] [Google Scholar]
- Kagan J.C. and Roy,C.R. (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol., 4, 945–954. [DOI] [PubMed] [Google Scholar]
- Kuhle V. and Hensel,M. (2002) SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol., 4, 813–824. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan,L.C., Bonifacino,J.S. and Klausner,R.D. (1989) Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell, 56, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Donaldson,J.G., Schweizer,A., Berger,E.G., Hauri,H.P., Yuan,L.C. and Klausner,R.D. (1990) Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell, 60, 821–836. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan,L., Tipper,C., Amherdt,M., Orci,L. and Klausner,R.D. (1991) Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell, 67, 601–616. [DOI] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer,O., Finlay,B.B. and Gorvel,J.P. (1999) The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J., 18, 4394–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Soncini,F.C., Solomon,F. and Groisman,E.A. (1996) Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl Acad. Sci. USA, 93, 7800–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paesold G., Guiney,D.G., Eckmann,L. and Kagnoff,M.F. (2002) Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella-induced apoptosis in intestinal epithelial cells. Cell. Microbiol., 4, 771–781. [DOI] [PubMed] [Google Scholar]
- Persson B. and Argos,P. (1994) Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol., 237, 182–192. [DOI] [PubMed] [Google Scholar]
- Plutner H., Davidson,H.W., Saraste,J. and Balch,W.E. (1992) Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol., 119, 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Girotti,M., Yaspo,M.L., Owen,C.E., Perry,A.C., Suganuma,T., Nilsson,T., Fried,M., Banting,G. and Warren,G. (1996) Primate homologues of rat TGN38: primary structure, expression and functional implications. J. Cell Sci., 109, 675–685. [DOI] [PubMed] [Google Scholar]
- Rathman M., Barker,L.P. and Falkow,S. (1997) The unique trafficking pattern of Salmonella typhimurium-containing phagosomes in murine macrophages is independent of the mechanism of bacterial entry. Infect. Immun., 65, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Albert J., Yu,X.J., Beuzón,C.R., Blakey,A.N., Galyov,E.E. and Holden,D.W. (2002) Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol., 44, 645–661. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O., Méresse,S., Gorvel,J.P., Toh,B.H. and Finlay,B.B. (1999) Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol., 1, 33–49. [DOI] [PubMed] [Google Scholar]
- Stein M.A., Leung,K.Y., Zwick,M., García-del Portillo,F. and Finlay,B.B. (1996) Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol., 20, 151–164. [DOI] [PubMed] [Google Scholar]
- Thyberg J. and Moskalewski,S. (1999) Role of microtubules in the organization of the Golgi complex. Exp. Cell Res., 246, 263–279. [DOI] [PubMed] [Google Scholar]
- Uchiya K., Barbieri,M.A., Funato,K., Shah,A.H., Stahl,P.D. and Groisman,E.A. (1999) A Salmonella virulence protein that inhibits cellular trafficking. EMBO J., 18, 3924–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R.H. and Falkow,S. (1997) Fluorescence-based isolation of bacterial genes expressed within host cells. Science, 277, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Ward T.H., Polishchuk,R.S., Caplan,S., Hirschberg,K. and Lippincott-Schwartz,J. (2001) Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol., 155, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman S.R. and Holden,D.W. (2003) Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol., 5, 501–511. [DOI] [PubMed] [Google Scholar]
- Wood S.A. and Brown,W.J. (1992) The morphology but not the function of endosomes and lysosomes is altered by brefeldin A. J. Cell Biol., 119, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick P.B. (2000) Intracellular survival by Chlamydia. Cell. Microbiol., 2, 275–282. [DOI] [PubMed] [Google Scholar]