Abstract
The morphology of influenza virions was found to depend on cellular determinants. Influenza viral filaments up to 30 μm in length were observed to form at high levels on surfaces of various polarized epithelial cell types infected with the A/Udorn/72 virus. In contrast, virions produced by nonpolarized cell types infected with this virus were almost exclusively of spherical morphology. Disruption of the actin microfilament array by cytochalasin D treatment of polarized MDCK cells had a profound effect on viral morphology. Although virus titers and release of spherical particles were not reduced in the presence of cytochalasin D, we observed a 15-fold reduction in the release of filamentous particles. In contrast, the ratio of filaments to spheres produced by infected MDCK cells was not altered by the microtubule-disrupting agent nocodazole. These observations indicate that the polarized cell phenotype and the integrity of the actin microfilament network are important cellular determinants of the morphology of a filamentous influenza virus.
Influenza virions are pleomorphic, with sizes ranging from small spherical particles with a mean diameter of approximately 100–150 nm to long filamentous particles, several μm in length. Filamentous particles were reported to exhibit a higher specific infectivity and a higher RNA content than spherical virions (1, 2). The viral morphology is a genetic trait, which depends on the amino acid sequences of viral structural proteins (2, 7). Early egg passages of original human isolates are predominantly filamentous, whereas most laboratory-adapted strains are predominantly spherical because of a more rapid growth rate of the small spherical variants (1, 8, 9). The molecular mechanisms responsible for filamentous particle formation and the role of such particles in viral transmission and pathogenicity are still not well defined.
The cellular cytoskeleton may play a role in release of some enveloped viruses (10, 14). Actin microfilaments have been implicated in the cell-to-cell spread of vaccinia virus (14). Actin and actin-binding proteins of the ezrin–radixin–moesin family have been isolated from purified enveloped virions, including rabies virus, HIV, and influenza virus (15, 18). Although disruption of actin microfilaments does not prevent influenza virus replication (19, 20), disruption of the microtubule network has been reported to cause redirection of a fraction of influenza viral proteins from the apical to basolateral plasma membrane domains in polarized epithelial cells (21) .
Because of their extreme length, influenza viral filaments can be readily visualized by immunofluorescence and light microscopy. In the present study, we report the surprising observation that the host cell type and the integrity of the cytoskeletal network are important determinants of the morphology of influenza virions.
MATERIALS AND METHODS
Cells and Viruses.
Madin–Darby canine kidney (MDCK), Madin–Darby bovine kidney (MDBK), human endometrial (HEC-1), monkey kidney Vero C1008 and Vero 76, and baby hamster kidney (BHK-21) cells were grown by described procedures (22, 23). MDCK-derived stock of influenza A/Udorn/72 (H3N2) was described previously (7). Influenza A/Memphis/1/96 virus was kindly provided by J. Katz (Centers for Disease Control and Prevention, Atlanta, GA). Stocks of influenza A/WSN and A/PR8 were prepared in MDBK cells (24).
Virus Titers.
Infectivity titers were determined by plaque assay in MDCK cells (25). Hemagglutination titers were determined as described previously (26).
Virus Infection.
Cells were grown to 80% confluency on 12-mm glass coverslips for immunofluorescence studies or plastic dishes for viral yield studies and were incubated with 3 plaque-forming units per cell of influenza virus stocks. After adsorption for 1 hr at 37°C, the inoculum was removed and the cells were incubated in medium supplemented with 2% fetal bovine serum (FBS).
Cytoskeletal Inhibitors.
Stocks of cytochalasin D (5 mg/ml in dimethyl sulfoxide, DMSO), or nocodazole (33 mM in DMSO) were diluted in medium just prior to use, added at 3 hours post infection (h.p.i.), and maintained throughout the infection period. In recovery experiments, cytochalasin D was removed at 6 h.p.i., the cells were washed twice in medium without inhibitor, and the infection was continued for 3 h. In experiments using nocodazole, the cells at 2.5 h.p.i. were incubated at 4°C for 30 min in the presence of the inhibitor and then shifted to 37°C for the duration of the experiment. Control cells were incubated with the respective concentration of DMSO.
Immunofluorescence.
Cell surface staining was performed at 4°C on either live unfixed cells or paraformaldehyde-fixed monolayers as described previously (7). Cells were incubated with goat antisera specific for M1, HA (hemagglutinin), NA, or NP proteins (Influenza Virus Repository, National Institute of Allergy and Infectious Diseases; Bethesda, MD) or monoclonal antibodies to tubulin, vimentin, or cytokeratins (Sigma), followed by fluorescein isothiocyanate (FITC)- or tetramethylrhodamine isothiocyanate (TRITC)-conjugated rabbit anti-goat or mouse Ig. Actin was visualized by staining with FITC- or TRITC-conjugated phalloidin (Sigma).
Sucrose Gradient Centrifugation.
[35S]Methionine/cysteine-labeled virus was separated by sucrose gradient centrifugation as described previously (7). The gradients were fractionated from the bottom into 500-μl fractions and aliquots were analyzed by scintillation counting.
Electron Microscopy.
Thin-section electron microscopy of A/Udorn-infected MDCK cells and negative staining of virus released into the medium were performed as described previously (7, 27). The percentage of filamentous and spherical virus particles was determined by counting a minimum of 600 particles in low-magnification micrographs of several different preparations. Filamentous particles were defined as particles with a length greater than 500 nm.
RESULTS
Visualization of Viral Filament Formation.
In contrast to most laboratory-adapted strains of influenza virus, which are found to produce virions of roughly spherical morphology and 100- to 150-nm diameter, the A/Udorn/72 virus produces a large number of filamentous particles (7, 27). When MDCK cells infected with the A/Udorn virus were examined at 9 h.p.i., dramatic changes were observed at the cell surface. Long filamentous particles were observed in large numbers by immunofluorescence microscopy (Fig. 1a), and the formation of viral filaments could also be observed by phase-contrast microscopy (Fig. 1b). To confirm that the structures seen by light microscopy correspond to filamentous virions, we examined sections of infected MDCK cells (Fig. 1c) as well as negatively stained virus particles released into the medium (Fig. 1 d and e). The results support the conclusion that the filamentous structures observed by light microscopy correspond to filamentous virions, which exhibit the characteristic spikes covering the surface and have lengths ranging from 500 nm to at least 15 μm. By immunofluorescence the filamentous particles were found to be stained by antibodies specific for each of the major viral structural proteins, including HA, NA, M1, M2, and NP (data not shown). We also found a high level of production of filamentous particles in MDCK cells infected with the A/Memphis/1/96 virus, a recent human isolate that has been passaged only four times in MDCK cells (not shown).
Figure 1.
Visualization of filamentous particle formation in MDCK cells. (a and b) Immunofluorescence (a) and phase-contrast microscopy (b) at 9 h.p.i., revealing the presence of numerous influenza virus filaments emanating from the cell surface. The immunofluorescence shows double staining with HA-specific antibody (red) and antibody to vimentin (green). The latter did not colocalize to viral filaments. (c) Low-magnification electron micrograph showing the presence of many long viral filaments and filament bundles projecting from the surface of A/Udorn-infected MDCK cells. (d and e) Electron micrographs of negatively stained filamentous particles released into the medium. The viral filaments depicted in d are 5–7 μm in length. The characteristic viral spike-like surface projections are readily visible at higher magnification (e). (a and b = ×1,000; c = ×15,000; d = ×10,000; e = ×85,000.)
Host Cell Dependence of Viral Morphology.
We observed extensive filament formation in various polarized epithelial cell types, including MDCK, MDBK, HEC-1, and Vero C1008 cells, but not in the nonpolarized Vero 76 and BHK-21 cell types. The Vero C1008 cell line is a polarized derivative of the parental Vero 76 cell line. Filament formation was observed by surface fluorescence in the polarized Vero C1008 cells at 9 h.p.i. (Fig. 2b) but was not observed in Vero 76 cells, even though intense surface antigen expression was evident (Fig. 2a). The virus released from A/Udorn-infected Vero 76 or BHK-21 cells was spherical or slightly elongated in morphology (Fig. 2c). In contrast, virus released from any of the polarized epithelial cell types was composed predominantly of long filamentous particles (Fig. 2d). About 15% of the particle number released from MDCK cells were filaments. Because the mass of a typical filamentous particle is likely to be at least 10-fold greater than that of a spherical particle, more than 60% of the viral mass is in the form of filaments.
Figure 2.
(a and b) Immunofluorescence micrographs of cell surfaces of Vero C1008 and Vero 76 cells stained with anti-Udorn antisera and FITC-conjugated rabbit anti-guinea pig Ig. Note the intense cell surface staining of Vero 76 cells but lack of filamentous particles (a), whereas filamentous particles (arrows) are budding from infected Vero C1008 cells (b). (c and d) Electron micrographs of negatively stained virus released from BHK-21 cells or HEC-1 cells. Filamentous particles were frequently observed in the virus released from HEC-1 cells (d) but spherical or slightly elongated particles were produced by BHK-21 cells (c). (a and b = ×500; c and d = ×3,800.)
To further compare virus assembly in different cell types, we separated spherical and filamentous A/Udorn particles by sucrose gradient centrifugation (Fig. 3). Filamentous particles sediment as a faster-migrating species, whereas the slower-migrating species consist of spherical particles (7). Virus derived from BHK-21 cells was found to sediment predominantly in fractions 11–13, similar to the spherical A/WSN virus used as a control. In contrast, MDCK cells show a rapidly sedimenting fraction consisting of filamentous particles (fractions 8–10). We also examined the synthesis of viral proteins at 5, 7, and 9 h.p.i. and did not observe any differences in relative amounts of viral proteins expressed between the different cell lines (data not shown). Taken together, these observations indicate that viral filament formation appears to require some cellular feature that is present in polarized epithelial cells but lacking in the nonpolarized cells we examined.
Figure 3.
Sucrose gradient separation of spherical and filamentous virus particles. Spherical particles derived from A/Udorn-infected MDCK cells sediment in fractions 11–13, whereas the faster-sedimenting filamentous particles are found in fractions 8–10 (7). The laboratory-adapted strain A/WSN (WSN), which produces strictly spherical particles, sediments in fractions 11–13. Virus derived from infected BHK-21 or from MDCK cells treated with cytochalasin D (10 μg/ml) also predominantly sediments in fractions 11–13.
An Intact Actin Microfilament Network Is Required for Viral Filament Formation.
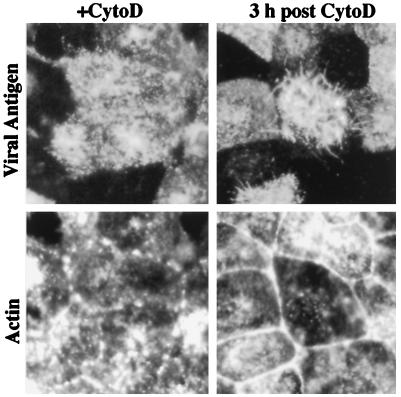
Previous studies demonstrated that disruption of actin microfilaments by cytochalasin D treatment did not impair influenza virus release (19, 20). These studies utilized laboratory-adapted strains, which are predominantly spherical in morphology, and the possible role of the cytoskeleton in the assembly of filamentous influenza virus particles has not been investigated. At concentrations of 3–10 μg/ml cytochalasin D, we observed that viral filament formation was almost completely suppressed (Fig. 4). It is evident from the continued surface fluorescence, however, that viral antigens continue to be transported to the cell surface in the presence of cytochalasin D (Fig. 4). Viral filament formation was not observed at either 6 or 9 h.p.i. in the presence of inhibitor, indicating that the inhibitory effect is not simply a result of a delay in viral filament production. By double labeling with phalloidin-FITC, it can be seen that the actin microfilament network has been disrupted into aggregates upon cytochalasin D treatment (Fig. 4). To determine the specificity of the inhibitory effect, we also examined whether disruption of the microtubule network by nocodazole could affect formation of viral filaments. Viral filament formation was found to continue at high levels in infected cells in the presence of 33 μM nocodazole (data not shown).
Figure 4.
Inhibition of filament formation and recovery from cytochalasin D treatment. Cells were incubated in the presence of cytochalasin D (CytoD) for 6 hr, the drug was removed, and the infection was allowed to continue at 37°C. At 0 or 3 hr after removal of the inhibitor, cells were doubly stained with guinea pig anti-Udorn antisera and rabbit anti-guinea pig-TRITC (viral antigen) and with FITC-phalloidin (actin). In cytochalasin D-treated cells at 6 h.p.i., intense cell surface staining of viral antigen was observed, but filament formation was not observed. Phalloidin staining confirmed that actin microfilaments were disrupted into aggregates. At 3 h after drug removal, short viral filaments were evident, concomitant with reformation of the actin microfilament array. (×550.)
Because cytochalasin D disruption of actin microfilaments is reversible, we determined whether the production of viral filaments would resume if actin microfilaments were allowed to reassemble. At 3 h after drug removal, the actin microfilament network had reassembled as evident by the dissociation of aggregates and reformation of the cortical actin ring at sites of cell to cell contact (Fig. 4). Concomitant with F-actin reassembly after drug removal, viral filaments were also observed emanating from the cells, albeit at lower levels than at 9 h.p.i. in untreated cells (see Fig. 1a). Furthermore, the viral filaments were significantly shorter (maximum length approximately 5 μm) than those observed in untreated control cells (compare with Fig. 1).
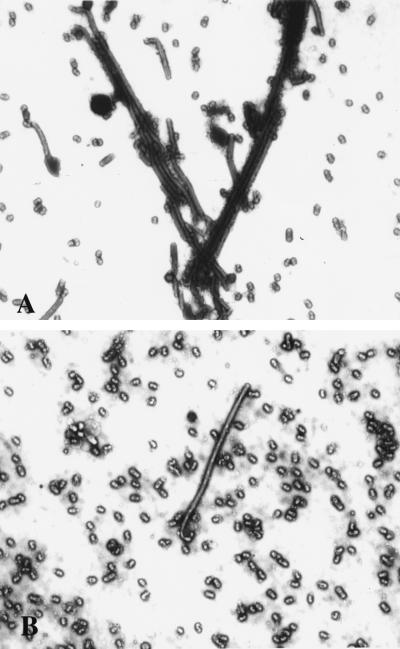
To investigate whether the inhibition of viral filament formation by cytochalasin D reflected a failure of virions to assemble or bud at the cell surface, virus titers released from infected cells were determined (Table 1). We observed no reduction in viral yields in the presence of cytochalasin D, a result that is similar to previous results indicating that cytochalasin D does not impair influenza virus release in a spherical strain (19). In fact, we observed enhancement of viral release in the presence of cytochalasin D in some cell types (Table 1). About 15% of the particles produced by untreated cells were long, filamentous virions (Fig. 5A). In contrast, in cytochalasin D-treated cells, the released particles were predominantly spherical (less than 1% filaments) (Fig. 5B). Virus produced by MDCK cells treated with cytochalasin D also exhibited a sedimentation profile similar to that observed for spherical strains of influenza (A/WSN) or for A/Udorn virus derived from BHK-21 cells (Fig. 3). Thus, disruption of actin microfilaments by cytochalasin D selectively inhibits viral filament formation, and it results in preferential assembly and release of spherical particles. In MDCK cells infected with the A/Memphis/1/96 virus, we also observed that filamentous virus formation was inhibited by cytochalasin D (data not shown), demonstrating that the observed effect is not specific to the A/Udorn virus. We therefore conclude that the actin microfilament network plays an important role in the assembly of filamentous influenza virus particles.
Table 1.
Cytochalasin D does not reduce virus yields
| Cell type | Virus yield, pfu × 10−5/ml
|
|||
|---|---|---|---|---|
| 9 h.p.i.
|
24 h.p.i.
|
|||
| −CytoD | +CytoD | −CytoD | +CytoD | |
| MDCK | 68 | 87 | 110 | 96 |
| MDBK | 7.1 | 7.8 | 15 | 21 |
| Vero 76 | 3.3 | 2.1 | 11 | 29 |
| Vero C1008 | 0.7 | 2.1 | 1.4 | 4.5 |
| BHK-21 | 4.8 | 3.2 | 13 | 17 |
Infection was carried out either in the absence of cytochalasin D or in the presence of cytochalasin D at 10 μg/ml. pfu, Plaque-forming units.
Figure 5.
Effects of cytochalasin D on virus morphology. Negatively stained preparations of A/Udorn virus produced in untreated control MDCK cells (A) and cytochalasin D-treated MDCK cells (B) were examined by electron microscopy. Note the reduction of filamentous particles and abundance of spherical forms present in the cytochalasin D-treated sample. (×8,900.)
Specificity of Viral Filament Assembly.
Although viral morphology is largely genetically determined (2, 7), some reports indicated that alteration of the host cell membrane by exogenous surfactants such as vitamin A alcohol could alter viral morphology, resulting in the production of filamentous virus particles by influenza virus strains that normally produce spherical particles (28). We did not observe the production of filament-like particles in vitamin A alcohol-treated MDCK cells infected with spherical strains (A/WSN/34, A/PR8) either by cell surface immunofluorescence or by negative staining of released particles (data not shown). Thus, at least in the cell culture system we used, assembly of filamentous influenza virus particles does not result from the nonspecific effect of a surfactant on cellular membranes.
DISCUSSION
We have presented evidence that host cell factors are important determinants of influenza virus morphology. The extensive production of filamentous virions by the influenza A/Udorn/72 strain was observed in various epithelial cells with polarized phenotypes. Viral filament production occurred in cells of human, monkey, canine, or bovine origin that were derived from various tissues such as kidney or cervical or endometrial epithelium. In contrast, viral filament production was severely restricted in BHK-21 fibroblasts or the nonpolarized Vero 76 cell type. It is known that the cytoskeleton plays an important role in establishment and maintenance of epithelial cell polarity. Actin and actin-binding proteins have been implicated in restriction of basolateral proteins to this domain (29). Actin association with enveloped virus particles has been well documented for several paramyxoviruses (10, 30, 31). The presence of actin within virions and the projection of actin-like microfilaments into developing particles at the cell membrane have been visualized during measles virus infection (10), and actin-binding proteins have been isolated from purified virions of several enveloped viruses, including influenza A virus (16, 17). During vaccinia virus infection, it was recently demonstrated that intracellular vaccinia virions utilize host actin microfilaments to project themselves out of the cytoplasm and into neighboring cells (14). Studies with cytochalasin D have also supported actin involvement in release of intracellular enveloped vaccinia virions (14, 32). During HIV infection, monocyte–epithelial interaction triggers a rearrangement of F-actin in the HIV-infected monocyte to form a pseudopod (12), and virus is preferentially released onto the epithelial cell surface from this site. It is evident from our data, and previous reports, that F-actin microfilaments are not required for spherical influenza virion assembly and release (15, 19, 20), since disruption of the microfilament network by cytochalasin D treatment actually enhances spherical virion release (Table 1 and ref. 19). Cytochalasin D enhancement of release or spread of virus has also been observed in some other viral systems, such as Newcastle disease virus (33) and rotavirus (34). In the former study, the enhancement was attributed to faster particle release. Although cytochalasin D treatment did not inhibit spherical particle assembly and release, we found that formation of filamentous influenza virus particles was drastically inhibited, indicating that assembly of viral filaments requires an intact cellular microfilament network. This role of actin as a determinant of influenza viral morphology differs from the role described for HIV and vaccinia (12, 14), in that no obvious structural rearrangement of F-actin occurs during influenza virus infection and no actin-dependence of virion morphology has been reported for other viruses.
It has been suggested that viral budding may be regulated by the concerted actions of proteins exerting opposing forces at the plasma membrane, namely a pulling out of the membrane by transmembrane-oriented viral glycoproteins and a concomitant thrusting out from within by the viral matrix proteins (35, 37). While such a mechanism may apply for spherical particles, the mechanism of filamentous particle elongation may be more complex. We observed that viral surface glycoproteins frequently appear to be concentrated at regions of filament growth. This was not observed in cells producing spherical influenza virions, suggesting that filament formation requires the concentration of viral glycoproteins at specific membrane domains. We suggest a model of filamentous particle formation in which the push–pull effect exerted by the viral matrix and glycoproteins is enhanced by forces exerted by the actin microfilament array. Both the viral matrix and nucleoproteins have been reported to interact with the cytoskeleton (38, 39). Incorporation of the matrix protein M1 into extending regions of virus filament growth may provide the force that extends the developing particle and may selectively dissociate or laterally displace membrane-associated microfilaments and actin-binding proteins. The M2 protein via its long cytoplasmic tail may aid this process by binding transiently to the actin network or by directing M1 molecules to regions of virus elongation.
Vitamin A alcohol was reported to induce formation of filamentous particles and distended, balloon-like particles in embryonated eggs infected with the spherical A/PR8 virus (28, 40). Subsequent studies indicated that in the absence of surfactants, pleomorphic forms of virus resembling the distended, balloon-like structures observed with vitamin A alcohol (28) are also produced in eggs infected with a filamentous influenza A virus strain (3). We did not observe an alteration in the morphology of influenza A virions by the addition of vitamin A alcohol to infected MDCK cells. Thus, the present results support previous conclusions (3, 4, 6, 7, 37) that the formation of filamentous influenza virions reflects genetically determined features of the viral structural proteins. However, we have provided new evidence that in addition, the nature of the host cell type and the integrity of the actin cytoskeleton network also play a role in influenza virus assembly that was previously unrecognized.
The site of human infection by influenza viruses is the respiratory epithelium, and on the basis of our results it is anticipated that such epithelial cells would support the production of filamentous virions. The predominance of filamentous particles in early passages of human isolates also strongly suggests a role for such particles in human infections. Our results (7) and earlier studies (1, 2) indicate that filamentous particles have a higher specific infectivity than do spherical particles. The filamentous morphology may be advantageous in cell-to-cell transmission of the virus in the respiratory mucosa. In the upper respiratory tract, the virus must traverse the mucus layers lining the mucosal surfaces. It is presumed that the viral neuraminidase functions to decrease the viscosity of the sialic-acid rich mucus, thereby facilitating virus access to the underlying mucosal epithelial cells. Filamentous virions, by virtue of their elongated morphology and greater number of neuraminidase molecules, may represent an infectious agent that can penetrate the protective mucus layers more readily. Filaments may also facilitate transmission of virus to neighboring cells, because their length may enable them to infect adjacent cells prior to release. Such a transmission process would be more resistant to clearance by upward mucocilliary removal of fluids by the respiratory tract, and possibly to humoral and cellular immune defense mechanisms. Spherical virions, which are also produced by epithelial cells, would be expected to be incorporated more readily into aerosols and may thus play the major role in person-to-person transmission. It will therefore be of interest to obtain further information on the role of the different forms of virus in the pathogenesis of infection.
Acknowledgments
We thank Lawrence Melsen for expert assistance with photography and electron microscopy, and Tanya Cassingham for assistance in preparing the manuscript. We thank Miroslav Novak for influenza A/Udorn virus and antisera. This study was supported by National Institutes of Health Grant AI 12680.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: h.p.i., hours post infection; FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine isothiocyanate.
References
- 1.Ada G L, Perry B T, Abbot A. J Gen Microbiol. 1958;19:23–39. doi: 10.1099/00221287-19-1-23. [DOI] [PubMed] [Google Scholar]
- 2.Smirnov Y A, Kuznetsova M A, Kaverin N V. Arch Virol. 1991;118:279–284. doi: 10.1007/BF01314038. [DOI] [PubMed] [Google Scholar]
- 3.Choppin P W. Virology. 1963;21:342–352. doi: 10.1016/0042-6822(63)90195-8. [DOI] [PubMed] [Google Scholar]
- 4.Kilbourne E D, Murphy J S. J Exp Med. 1960;111:387–405. doi: 10.1084/jem.111.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilbourne E D. Prog Med Virol. 1963;5:79–126. [Google Scholar]
- 6.Mitnaul L J, Castrucci M R, Murti K G, Kawaoka Y. J Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts P C, Lamb R A, Compans R W. Virology. 1998;240:127–138. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 8.Choppin P W, Murphy J S, Tamm I. J Exp Med. 1960;112:945–952. doi: 10.1084/jem.112.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu C M, Dawson I M, Elford W J. Lancet. 1949;i:602–603. doi: 10.1016/s0140-6736(49)91699-2. [DOI] [PubMed] [Google Scholar]
- 10.Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. Virology. 1986;149:91–106. doi: 10.1016/0042-6822(86)90090-5. [DOI] [PubMed] [Google Scholar]
- 11.Tashiro M, Seto J T, Klenk H-D, Rott R. J Virol. 1993;67:5902–5910. doi: 10.1128/jvi.67.10.5902-5910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce-Pratt R, Malamud D, Phillips D M. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki H, Nakamura M, Ohno T, Matusuda Y, Yuda Y, Nonomura Y. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cudmore S, Cossart P, Griffiths G, Way M. Nature (London) 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 15.Griffin J A, Compans R W. J Exp Med. 1979;150:379–391. doi: 10.1084/jem.150.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder R C, Chertova E N, Arthur L O, Henderson L E. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagara J, Tsukita S, Yonemura S, Tsukita S, Kawai A. Virology. 1995;206:485–494. doi: 10.1016/s0042-6822(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 18.Wang E, Wolf B A, Lamb R A, Choppin P W, Goldberg A R. In: Cell Motility. Goldman R, Pollard T, Rosenbaum F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 589–599. [Google Scholar]
- 19.Griffin J A, Basak S, Compans R W. Virology. 1983;125:324–334. doi: 10.1016/0042-6822(83)90205-2. [DOI] [PubMed] [Google Scholar]
- 20.Salas P J I, Misek D E, Vega-Salas D E, Gundersen D, Cereijido M, Rodriguez-Boulan E. J Cell Biol. 1986;102:1853–1867. doi: 10.1083/jcb.102.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rindler M J, Ivanov I E, Sabatini D D. J Cell Biol. 1987;104:231–241. doi: 10.1083/jcb.104.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ball J M, Moldoveanu Z, Melsen L R, Kowalski P, Jackson S, Mulligan M, Mestecky J, Compans R W. In Vitro. 1995;31:197–207. doi: 10.1007/BF02639434. [DOI] [PubMed] [Google Scholar]
- 23.Blau D M, Compans R W. Virology. 1995;210:91–99. doi: 10.1006/viro.1995.1320. [DOI] [PubMed] [Google Scholar]
- 24.Choppin P W. Virology. 1969;39:130–134. doi: 10.1016/0042-6822(69)90354-7. [DOI] [PubMed] [Google Scholar]
- 25.Tobita K, Sugiura A, Enomoto C, Furuyama M. Med Microbiol Immunol. 1975;162:9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- 26.Compans R W, Dimmock N J, Meier-Ewert H. J Virol. 1969;4:528–534. doi: 10.1128/jvi.4.4.528-534.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughey P G, Roberts P C, Holsinger L J, Zebedee S L, Lamb R A, Compans R W. Virology. 1995;212:411–421. doi: 10.1006/viro.1995.1498. [DOI] [PubMed] [Google Scholar]
- 28.Blough H A. Virology. 1963;19:349–358. doi: 10.1016/0042-6822(63)90074-6. [DOI] [PubMed] [Google Scholar]
- 29.Nelson W J, Hammerton R W. J Cell Biology. 1989;108:893–903. doi: 10.1083/jcb.108.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyrrell D L J, Norrby E. J Gen Virol. 1978;39:219–229. doi: 10.1099/0022-1317-39-2-219. [DOI] [PubMed] [Google Scholar]
- 31.Giuffre R M, Tovell D R, Kay C M, Tyrrell D L J. J Virol. 1982;42:963–968. doi: 10.1128/jvi.42.3.963-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne L G, Kristensson K. Arch Virol. 1982;74:11–20. doi: 10.1007/BF01320778. [DOI] [PubMed] [Google Scholar]
- 33.Morrison T G, McGinnes L J. Virus Research. 1985;4:93–106. doi: 10.1016/0168-1702(85)90023-1. [DOI] [PubMed] [Google Scholar]
- 34.Bass D M, Baylor M, Chen C, Upadhyayula U. Virology. 1995;212:429–437. doi: 10.1006/viro.1995.1500. [DOI] [PubMed] [Google Scholar]
- 35.Simons K, Garoff H. J Gen Virol. 1980;50:1–21. doi: 10.1099/0022-1317-50-1-1. [DOI] [PubMed] [Google Scholar]
- 36.Mebatsion T, Konig M, Conzelmann K-K. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 37.Jin H, Leser G P, Zhang J, Lamb R A. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avalos R T, Yu Z, Nayak D P. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Lamb R A. Virology. 1996;225:255–261. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
- 40.Blough H A. Virology. 1963;19:112–114. doi: 10.1016/0042-6822(63)90033-3. [DOI] [PubMed] [Google Scholar]