Abstract
Rad54, a member of the Swi2/Snf2 protein family, works in concert with the RecA-like recombinase Rad51 during the early and late stages of homologous recombination. Rad51 markedly enhances the activities of Rad54, including the induction of topological changes in DNA and the remodeling of chromatin structure. Reciprocally, Rad54 promotes Rad51-mediated DNA strand invasion with either naked or chromatinized DNA. Here, using various Saccharomyces cerevisiae rad51 and rad54 mutant proteins, mechanistic aspects of Rad54/Rad51-mediated chromatin remodeling are defined. Disruption of the Rad51-Rad54 complex leads to a marked attenuation of chromatin remodeling activity. Moreover, we present evidence that assembly of the Rad51 presynaptic filament represents an obligatory step in the enhancement of the chromatin remodeling reaction. Interestingly, we find a specific interaction of the N-terminal tail of histone H3 with Rad54 and show that the H3 tail interaction domain resides within the amino terminus of Rad54. These results suggest that Rad54-mediated chromatin remodeling coincides with DNA homology search by the Rad51 presynaptic filament and that this process is facilitated by an interaction of Rad54 with histone H3.
Keywords: Rad51, Rad54, Chromatin Remodeling, D-loop, Homologous Recombination
1. Introduction
The Saccharomyces cerevisiae RAD54 gene encodes an evolutionarily conserved protein that plays a central role in homologous recombination (HR) and DNA repair reactions. Accordingly, deletion of RAD54 in yeast and mammalian cells engenders hypersensitivity to DNA damaging agents and leads to chromosome fragility [1-3]. Mutations in highly conserved residues of the human Rad54 protein are found in primary tumors [4]. Studies involving the tumor-associated mutations in the yeast Rad54 protein have shown that these mutations compromise Rad54 function [5], providing support for the premise that Rad54 contributes to the suppression of the tumor phenotype.
Structurally and functionally, Rad54 belongs to the Swi2/Snf2 protein family, whose members are known to be involved in chromatin remodeling and the removal of protein targets from DNA. Rad54 has a potent ATPase activity that is activated by DNA, duplex DNA in particular [6,7]. Biochemical studies, scanning force microscopy, and a recent single-molecule study together have provided compelling evidence for an ATP hydrolysis-fueled DNA translocase activity in Rad54 [8-11]. As a result of DNA translocation, Rad54 induces dynamic topological changes and transient strand opening in the DNA [9,10]. Like many members of the Swi2/Snf2 protein family, Rad54 remodels chromatin structure, and this chromatin remodeling activity requires ATP hydrolysis [12-14]. Moreover, Rad54 utilizes the free energy from ATP hydrolysis to accelerate the DNA strand transfer reaction [15, 16] and to process branched DNA structures [16], including the Holliday junction [16], that are expected to be generated during certain HR reactions [17].
In the initial stage of HR, a single-stranded DNA molecule, such as that derived from the nucleolytic processing of a DNA double-stranded break, is paired with a homologous duplex partner to form a DNA joint called D-loop. D-loop formation is catalyzed by a recombinase enzyme, either Rad51 or Dmc1 [18], in a reaction termed homologous DNA pairing [17,19,20]. Like their prokaryotic counterparts E. coli RecA and T4 UvsX, Rad51 and Dmc1 form a filamentous structure, referred to as the presynaptic filament, on ssDNA [18,19]. Assembly of the presynaptic filament requires ATP binding by the recombinase protein [19,20], and ATP hydrolysis by the DNA-bound recombinase promotes the turnover of the presynaptic filament [21-24]. All the subsequent biochemical steps, including the capture of the duplex DNA partner and the search for homology in the engaged ssDNA and dsDNA molecules, that lead to D-loop formation occur within the confines of the presynaptic filament [17,19,20]. Thus, the presynaptic filament represents the key catalytic recombinase-DNA complex critical for the successful initiation and completion of the HR reaction.
Rad54 protein physically interacts with Rad51 [7,25,26] and greatly enhances the ability of Rad51 to form DNA joints [7]. The promotion of Rad51-mediated homologous DNA pairing and strand exchange by Rad54 requires the ATPase activity of the latter and physical interaction between these two HR factors [27-29]. Interestingly, Rad54 dissociates Rad51 from duplex DNA via its DNA translocase activity, a function believed to be important for the intracellular recycling of Rad51 and possibly for freeing the primer end in the D-loop structure to initiate the DNA synthesis reaction [30]. Rad54 facilitates the assembly and maintenance of the Rad51 presynaptic filament as well [31,32], and the interactions of Rad54 and Rad51 are germane for their mutual targeting to a site-specific DSB in cells [31,33,34]. The Rad51 presynaptic filament stabilization role of Rad54 does not require its ATPase activity [32,34]. Thus, Rad54 plays a major role in regulating the activities of Rad51. Reciprocally, the enzymatic activities of Rad54 - ATP hydrolysis, DNA supercoiling, DNA strand opening, DNA branch migration, and chromatin remodeling - are enhanced by Rad51 [9,12-14,16,35]. In strains deleted for RAD54, ssDNA associated with a site-specific DSB at the MAT locus can synapse with its homologous donor sequence, but DNA strand invasion appears to be impaired [33,36]. This latter observation provides in vivo evidence that successful execution of the early stage of HR - DNA strand invasion and/or the initiation of repair DNA synthesis - is reliant on Rad54.
Much remains to be learned about the functional synergy between Rad51 and Rad54. Here, we attempt to define the mechanistic basis for the functional interactions between Rad54 and Rad51 in chromatin remodeling. Using specific rad51 and rad54 mutants, we demonstrate that Rad54 must physically interact with Rad51 in the context of the presynaptic filament for efficient chromatin remodeling to occur. We also show that Rad54 binds the N-terminal tail of histone H3 and that the N-terminal region of Rad54 mediates this interaction. These results should be valuable for guiding the ongoing dissection of the mechanisms that underlie the actions of Rad51 and Rad54 in HR.
2. Experimental Procedures
2.1. Recombination proteins
The expression and purification of Rad54 and N-terminally truncated forms (Δ113 and Δ129) of Rad54 have been described [28]. For the expression of the rad51 G103E protein, the mutant rad51 gene was placed under the PGK promoter, as described for the wild type protein [37]. Rad51, rad51 K191R, rad51 K191A, and rad51 G103E were purified from the rad51Δ strain (LSY411) transformed with protein expression plasmids, as described [37].
2.2. Histones and histone tails
Core histones were purified from the nuclei of chicken erythrocytes (Lampire Biological Laboratories, Pipersville, PA), as described [38]. Plasmids that express glutathione-S-transferase (GST) fusion proteins harboring amino-terminal residues 1-46, 1-25, or 21-46 of yeast histone H3 or residues 1-35 of yeast histone H2A were introduced into E. coli BL21 (DE3). Overnight cultures of bacteria were diluted with 2× LB medium and protein expression was induced by 0.5 mM IPTG at 37 °C for 3 h. Cell lysate was prepared from 7 g of bacterial cell paste using a sonicator in 100 ml cell breakage buffer (50 mM Tris-HCl, pH 7.5, 10% sucrose, 10 mM EDTA, 150 mM KCl, 1 mM phenylmehtyl sulfonyl fluoride and 5 μg/ml of each of the following protease inhibitors: aprotinin, chymostatin, leupeptin, pepstatin A). The lysate was clarified by centrifugation (100,000 × g, 90 min) and the supernatant was mixed with 4 ml of glutathione Sepharose 4 Fast Flow matrix (GE Healthcare) for 3 h at 4 °C. The matrix was washed with 100 ml of the cell breakage buffer and the bound GST-fusion protein was eluted from the matrix with 10 mM reduced glutathione in buffer P (10 mM KH2PO4, pH 7.4, 150 mM KCl, 1 mM DTT and 0.01% Igepal CA-630 (Sigma)) and the above-mentioned protease inhibitors. Glutathione in the elution factions was removed by dialysis in buffer P and the GST-fusion proteins were concentrated using a Centricon-30 microconcentrator to 5-10 mg/ml. The concentrated protein preparations were stored in small portions at −80 °C.
2.3. Chromatinized template
Chromatin was assembled with the purified chicken histone octamer and pBluescript SK dsDNA by the salt dialysis method as described [12,39]. Chromatin assembly was monitored by EMSA, micrococcal nuclease digestion, and accessibility to the restriction enzyme HaeIII [14,40].
2.4. MNase digestion of chromatinized DNA
Naked or chromatinized DNA (1 μg) was incubated with Micrococcal nuclease (MNase) in 10 μl of buffer M (10 mM Tris-HCl, pH 7.5, 1 mM CaCl2) at 23 °C for 1 min. The reactions were stopped by adding SDS and EDTA to final concentrations of 0.2% and 20 mM, respectively. DNA was deproteinized with proteinase K (1 mg/ml) at 37 °C for 5 min and resolved in a 1.5% agarose gel in TAE buffer (40 mM Tris acetate, pH 7.4, 0.5 mM EDTA) at 25 °C.
2.5. D-loop assay
D-loop reactions involving the use of 5′ end-labeled 90-mer oligonucleotide D1 and naked or chromatinized pBluscript SK dsDNA were performed as described previously [5]. Briefly, Rad51 (0.9 μM or as indicated) was incubated with oligonucleotide D1 (2.4 μM nucleotides) at 37°C in buffer D (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 100 μg/ml BSA, 50 mM KCl, 5 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate, and 30 μg/ml creatine kinase) for 5 min to assemble the presynaptic filament, which was incubated with Rad54 (120 nM) for 1 min at 30 °C, followed by the addition of free or chromatinized pBluescript DNA (35 μM base pairs) and a further incubation at 30 °C for 2.5 min. The reaction mixtures (12.5 μl) were mixed with an equal volume of 1% SDS and proteinase K (1 mg/ml) and then incubated at 37 °C for 5 min. The deproteinized samples were resolved in 0.9% agarose gels in TAE buffer at 25 °C. The gels were dried onto DE81 paper (Whatman) and the radiolabeled DNA species were visualized and quantified by phosphorimaging analysis.
2.6. Chromatin accessibility assay
The accessibility of the chromatinzed DNA template was tested by treatment with the restriction enzyme HaeIII [14]. Briefly, the chromatinized DNA template (35 μM base pairs) was incubated with Rad54 or rad54 Δ129 (120 nM) with or without the Rad51 or RecA presynaptic filament assembled on oligonucleotide D1 (consisting of 0.9 μM Rad51 or RecA and 2.4 μM nucleotides of DNA) and the restriction enzyme HaeIII (0.8 unit/μl). Buffer D was used for these reactions and the final reaction volume was 12.5 μl. After a 60 min incubation at 30 °C, the reaction mixtures were treated with SDS and proteinase K as described above. The deproteinized reaction mixtures were fractionated in 1.5% agarose gels and the DNA species were visualized by staining with ethidium bromide. In reactions that contained Rad51, the omission of the oligonucleotide did not result in a significant drop in remodeling efficiency. This is likely due to the fact that the linker DNA situated in-between nucleosomes served as the template for presynaptic filament assembly. Alternatively, or in addition, ssDNA region resulting from the DNA supercoiling activity of Rad54 [9, 11] provided the template for presynaptic filament assembly.
2.7. ATPase assay
The ATPase assay was performed essentially as described [28]. Briefly, Rad54 or rad54 Δ129 (30 nM in Fig. 3B and 20 nM in Fig. 4A) and Rad51 or mutants (300 nM or as indicated) were incubated on ice for 20 min in 24.5 μl of buffer A (30 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM DTT, and 100 μg/ml BSA) with 1.5 mM [γ-32P] ATP. The reaction was initiated by the addition of relaxed ϕX174 replicative form I DNA (30 μM base pairs) at 30 °C. At the indicated times, a 1.8 μl of the reaction was mixed with 2 μl of 0.5 M EDTA before being analyzed by thin layer chromatography in a polyethyleneimine plate (J. T. Baker), as described previously [7]. The level of ATP hydrolysis was determined by phosphorimaging analysis of the chromatography plate.
Fig. 3. Enhancement of Rad54 ATPase activity by rad51 K191A.
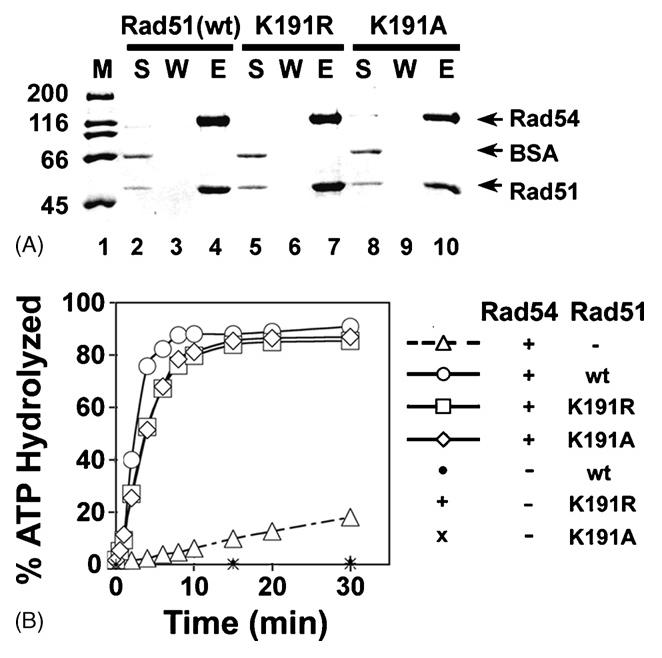
(A) Rad51, rad51 K191A (K191A), and rad51 K191R (K191R) were incubated with His-tagged Rad54. Protein complexes were captured on nickel NTA agarose beads and then eluted with SDS. The supernatant (S), wash (W), and SDS-eluate (E) were analyzed by SDS-PAGE.
(B) ATP hydrolysis by Rad54 was examined with or without Rad51, rad51 K191A, and rad51 K191R.
Fig. 4. Examination of the rad51 G103E mutant protein for interactions with Rad54.
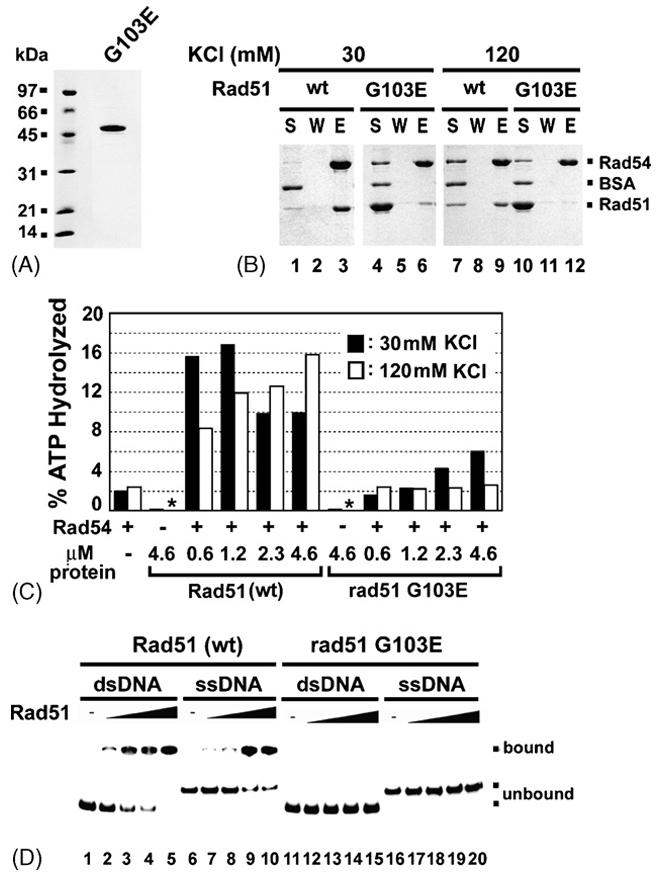
(A) Purified rad51 G103E mutant protein (2 μg) was analyzed by SDS-PAGE.
(B) Affinity pulldown to examine complex formation of rad51 G103E with Rad54, conducted with 30 or 120 mM KCl, as described for Fig. 3A.
(C) The ability of rad51 G103E and Rad51 to stimulate the Rad54 ATPase activity was examined at either 30 mM (black bar) or 120 mM KCl (white bar). ATP hydrolysis was carried out with the indicated amounts of Rad51 or rad51 G103E at 30 °C for 5 min. An asterisk indicates that ATP hydrolysis was not detectable.
(D) Binding of ssDNA and dsDNA by Rad51 (lanes 1-10) or rad51-G103E (lanes 11-20) was examined.
2.8. Affinity pulldown assays
To test for complex formation, Rad51 (5 μg) or mutant rad51 (5 μg of rad51 K191A or rad51 K191R and 12 μg of rad51 G103E) was incubated with (His)6-tagged Rad54 (7 μg) for 30 min on ice in 20 μl of buffer B (20 mM KH2PO4, pH 7.4, 150 mM KCl, 0.01% Igepal CA-630, 10 mM imidazole, and 1 mM 2-mercaptoethanol). The reaction was mixed gently with 10 μl of nickel NTA-agarose beads (Qiagen) at 4 °C for 1 h. The beads were collected by centrifugation, washed three times with 20 μl buffer B, and treated with 20 μl of 2% SDS to elute the bound proteins. The supernatant (S), the third wash (W), and the SDS-eluate (E), 5 μl each, were analyzed by 10% SDS-PAGE and Coomassie Blue staining.
To test for the association of Rad54 with histone tails, GST fusion proteins (15 μg) containing the N-terminus of H2A (residues 1-35) and various portions of the N-terminus of H3 (residues 1-46, 1-25 or 21-46) were mixed with 6 μl of glutathione-Sepharose-4B beads (GE Healthcare) in 30 μl buffer P containing protease inhibitors for 30 min at 4 °C. The beads were washed twice with 100 μl buffer P and then mixed with either 5 μg of Rad54 or its deletion mutants or Rad51 in 30 μl Buffer P for 30 min at 4 °C. The beads were collected by centrifugation, washed twice with 30 μl buffer P, and treated with 30 μl 2% SDS to elute the bound proteins. The supernatant (S), second wash (W), and SDS eluate (E), 10 μl each, were analyzed by SDS-PAGE with Coomassie Blue staining.
2.9. DNA Mobility Shift Assay
To examine DNA binding by Rad51 and rad51 G103E, these proteins (0.3, 0.6, 0.9, and 1.2 μM) were incubated with 32P-labeled oligonucleotide D1 (3 μM nucleotides) or dsDNA (3 μM base pairs; consisting of the radiolabeled D1 being hybridized to its exact complement) for 10 min at 37°C in 10 μl of buffer E (30 mM Tris-HCl, pH 7.5, 2.5 mM ATP, 5 mM MgCl2, 1 mM DTT, and 50 mM KCl). The reaction mixtures were resolved in a 10% polyacrylamide gel in TAE buffer at 4°C. The gel was dried and subject to phosphorimaging analysis.
3. Results
3.1. Dependence of Rad51/Rad54-mediated chromatin remodeling on protein complex formation
The amino-terminal region of Rad54 makes a major contribution to the interaction with Rad51, and a rad54 deletion mutant, rad54 Δ129, lacking the amino-terminal 129 residues, is compromised for Rad51 interaction, especially when the salt concentration is raised [28]. Thus, a moderately high concentration of KCl (60 to 100 mM KCl) nearly ablates the cooperative activities - e.g, hydrolysis of ATP by Rad54, DNA supercoiling, and D-loop formation – of the Rad51-rad54 Δ129 protein pair but has little impact on the same activities of the Rad51-Rad54 complex [28].
We wished to determine whether the efficiency of chromatin remodeling and D-loop formation with a chromatinized template mediated by Rad51 and rad54 Δ129 is also sensitive to the salt concentration. To examine chromatin remodeling, we employed an assay similar to that previously used to characterize the Drosophila melanogaster Rad51 and Rad54 proteins [14]. The chromatinized template was assembled using a plasmid DNA molecule and purified chicken histone octamers using the salt dialysis method. The template was subject to micrococcal nuclease digestion (Fig 1A). Unlike free DNA (Fig. 1A, lanes 2 and 3), the chromatinized template (Fig. 1A, lanes 5 to 7) displayed discrete bands, indicating the presence of arrays of nucleosomes. The chromatinized DNA template was notably resistant to digestion by the restriction enzyme HaeIII (Fig.1B, compare lanes 3 and 5), and an elevation of the sensitivity of the chromatinized template to HaeIII served as the indication of a chromatin remodeling activity in the protein factors under testing.
Fig. 1. Salt-sensitivity of Rad51-rad54 Δ129 in chromatin remodeling.
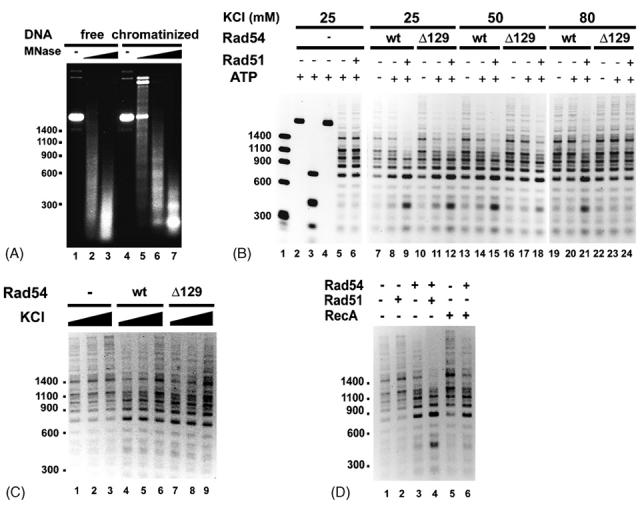
(A) MNase (5 units/ml in lanes 2 and 5, 10 units/ml in lanes 3 and 6, and 15 units/ml in lane 7) analysis of free (lanes 1-3) and chromatinized (lanes 4-7) DNA.
(B) The ability of Rad54 (wt) or rad54 Δ129 (Δ129) to render the chromatinized DNA template sensitive to the restriction enzyme HaeIII was examined as a function of the KCl concentration. The analysis was conducted with or without Rad51, as indicated. Naked DNA was used in lanes 2 and 3 and chromatinized DNA was used in lanes 4 to 24. Size markers were run in lane 1. HaeIII was added to all the other reactions except those in lanes 2 and 4.
(C) Chromatin remodeling by Rad54 or rad54 Δ129 at various KCl concentrations (25, 50, and 80 mM).
(D) Chromatin remodeling by Rad54 with or without Rad51 or RecA, as indicated.
As shown in Figure 1B, incubation of the chromatinized template with Rad54 protein rendered it more sensitive to HaeIII, and, consistent with previously published data [12-14], the chromatin remodeling reaction was greatly enhanced upon the inclusion of Rad51 (lanes 7 to 9). As expected, Rad51 on its own did not exhibit any chromatin remodeling activity (Fig. 1B, lanes 5 and 6). At 25 mM KCl, chromatin remodeling by either Rad54 or rad54 Δ129 was enhanced by Rad51 to the same degree (Fig. 1B, lanes 8, 9, 11, and 12). Importantly, with rad54 Δ129, the stimulatory effect of Rad51 was greatly diminished at 50 mM KCl (Fig. 1B, lane 18) and was no longer evident at 80 mM KCl (Fig. 1B, lane 24). In sharp contrast, increasing the KCl concentration from 25 mM to either 50 mM or 80 mM had very little or no impact on the ability of Rad51 to stimulate the chromatin remodeling activity of full length Rad54 (Fig. 1B, lanes 15 and 21). To verify that the lack of functional synergy between Rad51 and rad54 Δ129 at 50 and 80 mM KCl is not due to a differential sensitivity of the truncated rad54 protein to KCl, we examined chromatin remodeling by the rad54 Δ129 alongside full length Rad54 at 25, 50, and 80 mM KCl. As shown in Figure 1C, the chromatin remodeling activity of both proteins was unaffected by 50 mM KCl, whereas a very slight decrease in reaction efficiency was noted for both proteins at 80 mM KCl. These results clearly indicate that the chromatin remodeling activity of rad54 Δ129 is just as resistant to KCl as full length Rad54. We note that the DNA-dependent ATPase activity of rad54 Δ129 protein is also no more sensitive to KCl than the full length protein [28]. Based on these observations, we concluded that the salt sensitivity of the chromatin remodeling activity of the Rad51-rad54 Δ129 protein pair stems from a weakened interaction between the two proteins. Our conclusion that complex formation between Rad51-Rad54 is critical for functional synergy in chromatin remodeling is further bolstered by the observation that E. coli RecA protein has no effect on this Rad54 activity (Fig. 1D).
3.2. Salt sensitivity of D-loop formation by Rad51 and rad54 Δ129 with chromatinized DNA
The ability of Rad54 to promote Rad51-dependent D-loop formation has been well documented [20]. More recent studies that utilized highly purified S. cerevisiae and D. melanogaster protein factors have shown that Rad54 enables Rad51 to make D-loop with a chromatinized DNA template as well [12,14]. We showed previously [28] and have confirmed here (Fig. 2C) that while rad54 Δ129 is just as effective as full length Rad54 in promoting the Rad51-mediated D-loop reaction with a naked DNA template at low ionic strength, raising the salt concentration results in progressive inhibition of this activity, such that little or no D-loop was seen at 110 mM KCl (Fig. 2, C, D, and E). In contrast, the D-loop reaction mediated by Rad51 and full length Rad54 is impervious to 110 mM KCl ([28]; Fig. 2, B, D, and E). As shown in Figure 2, with the chromatinized DNA template also, rad54 Δ129 was just as adept as full length Rad54 in promoting Rad51-dependent D-loop formation at 25 mM KCl, but the ability of the rad54 mutant to enhance homologous pairing diminished greatly upon elevating the KCl concentration. Thus, the D-loop reaction that involves the rad54 Δ129 mutant protein is highly salt sensitive, regardless of whether a naked or chromatinized duplex template is used.
Fig. 2. Salt-sensitivity of Rad51-rad54 Δ129 in D-loop formation with a chromatinized template.
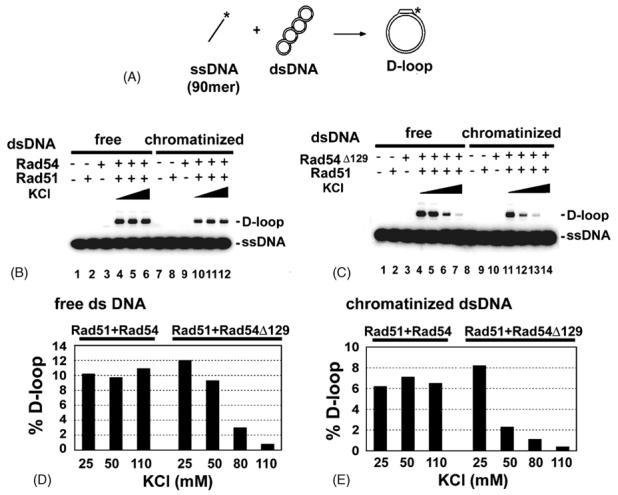
(A) Schematic of the D-loop reaction.
(B) D-loop reactions were carried out with Rad51 and Rad54 in the presence of an increasing concentration of KCl (25, 50, and 110 mM). Either free or chromatinized duplex DNA template was used, as indicated.
(C) D-loop reactions were carried out with Rad51 and rad54 Δ129 with an increasing concentration of KCl (25, 50, 80, and 110 mM). Either free or chromatinized duplex DNA template was used, as indicated.
(D) and (E) The results from (B) and (C) are presented in the histograms.
3.3. The DNA binding defective rad51 K191A mutant associates with Rad54 and stimulates the Rad54 ATPase activity
Our biochemical and EM studies have found that assembly of the ScRad51 presynaptic filament requires ATP or a non-hydrolyzable ATP analogue [41]. In an effort to further define the role of ATP binding and hydrolysis in Rad51 function, we previously constructed two rad51 mutants that harbor a change of the highly conserved lysine (K191) residue in the Walker A motif to Ala (K191A) or Arg (K191R). The rad51 K191A mutant does not bind ATP or DNA [9]. This mutant was included in our present study so as to address the role of the presynaptic filament in chromatin remodeling. Although being defective in ATP hydrolysis [37], the rad51 K191R mutant binds ATP and is capable of forming a functional presynaptic filament ([9,37], Fig 6), so it was included in this study for comparison. We wished to verify that both of the rad51 mutant proteins retain the ability to interact with Rad54. To do this, the rad51 mutant proteins were individually mixed with (His)6-tagged Rad54, and protein complexes that formed were captured on nickel NTA agarose, which recognizes the (His)6-tag on the latter. The results in Figure 3A showed that both rad51 K191A and rad51 K191R are able to bind Rad54 (lanes 4, 7, and 10). Because Rad51 and the rad51 K191R mutant proteins both enhance the ATPase activity of Rad54 [9], it was of interest to determine whether rad51 K191A is capable of the same. Consistent with the published results, Rad51 and rad51 K191R markedly stimulated (∼8-fold) the ATPase activity of Rad54 (Fig. 3B). Importantly, we observed a comparable level of Rad54 ATPase stimulation by rad51 K191A (Fig. 3B). Taken together, the above data indicate that the rad51 K191A and K191R mutations do not affect Rad51-Rad54 complex formation, and that the Rad51 presynaptic filament is not needed (as in the case of rad51 K191A) for Rad54 ATPase enhancement.
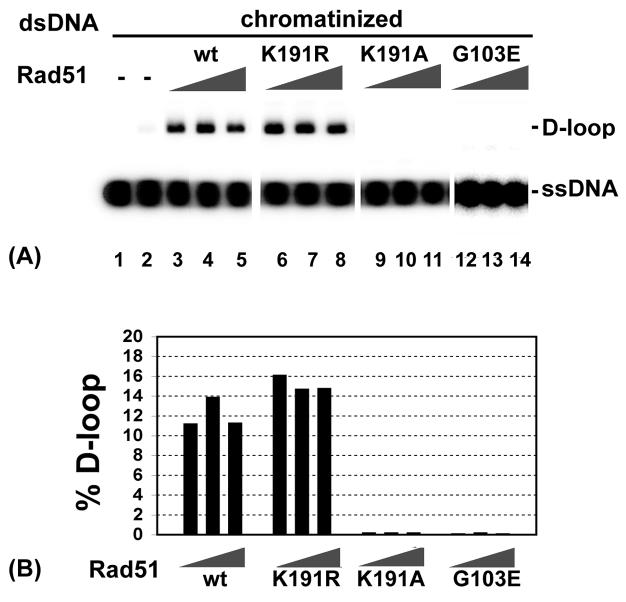
Fig. 6. D-loop formation involving rad51 mutant proteins and a chromatinized template.
(A) D-loop reactions were carried out with Rad54 and increasing amounts (0.6, 0.8, and 1.2 μM) of Rad51 or mutant proteins (rad51 K191A K191R, or G103E) and chromatinzed dsDNA as described for Fig. 2. (B) The results in (A) were quantified and shown in the histogram.
3.4. The rad51 G103E mutant is defective in Rad54 interaction
The rad51 G103E protein was found to be defective in interaction with Rad54 in the yeast two-hybrid system [42]. However, it was recently reported that purified rad51 G103E binds GST-tagged Rad54 as strongly as wild type Rad51 [43], although it appears to be deficient in DNA binding and DNA-activated ATPase activity [43,44]. We wished to examine the functional interactions of the rad51 G103E mutant protein with Rad54 in the context of chromatin remodeling and D-loop formation involving a chromatinized template. For this purpose, we overexpressed the rad51 G103E-encoded protein in rad51Δ yeast cells by using the PGK promoter in a multi-copy plasmid. The rad51 G103E protein is soluble and can be purified to near homogeneity (Fig. 4A) using the procedure that we previously developed for wild type Rad51 [37]. We first asked whether rad51 G103E would form a complex with (His)6-tagged Rad54 by affinity pulldown using nickel NTA-agarose beads, as described earlier (Fig. 3A). To determine the strength of the protein-protein interaction, we carried out the pulldown assay under low (30 mM) and high (120 mM) KCl concentrations. As shown in Figure 4B, the rad51 G103E mutant protein interacted weakly with Rad54 at 30 mM KCl (lane 6), and little, if any, association with Rad54 occurred at 120 mM KCl (Fig. 4B, lane 12). In contrast, the interaction of wild type Rad51 with Rad54 remained robust at 120 mM KCl (Fig. 4B, lane 9).
Consistent with the premise that Rad54 ATPase enhancement requires a specific interaction with Rad51, little or no stimulation of Rad54-mediated ATP hydrolysis was seen over a wide range of rad51 G103E concentration at 120 mM KCl (Fig. 4C). At 30 mM KCl, where we observed weak interaction between the two proteins (Fig. 4B, lane 6), there was a slight stimulation of the Rad54 ATPase activity by rad51 G103E. As was observed by Zhang et al [43], we found the rad51 G103E mutant protein to be greatly deficient in DNA binding activity (Fig. 4D).
3.5. Functional linkage of chromatin remodeling to the Rad51 presynaptic filament
The results obtained with the rad51 K191A and G103E mutant proteins have provided evidence that Rad54 ATPase enhancement requires a specific interaction with Rad51 but can occur outside of the context of the presynaptic filament. We next asked whether the various rad51 mutant proteins (i.e. K191A, K191R, and G103E) are capable of functional synergy with Rad54 in chromatin remodeling, by testing these proteins with Rad54 in the chromatin remodeling assay. Rad51 was included in this series of experiments for comparison. The rad51 K191R protein was just as effective as wild type Rad51 in enhancing the chromatin remodeling activity of Rad54 (Fig. 5A, lanes 17 and 19). Interestingly, no stimulation was seen with rad51 K191A (lanes 21 and 23). Since rad51 K191A is proficient in complex formation with Rad54 and in the enhancement of the Rad54 ATPase activity (Fig. 3, A and B), the lack of functional synergy in the chromatin remodeling reactions likely stems from the inability of the mutant protein to form the presynaptic filament. Consistent with this premise, rad51 G103E also failed to stimulate the chromatin remodeling activity of Rad54 (Fig. 5B, lanes 6 to 8).
Fig. 5. Functional linkage of chromatin remodeling to the Rad51 presynaptic filament.
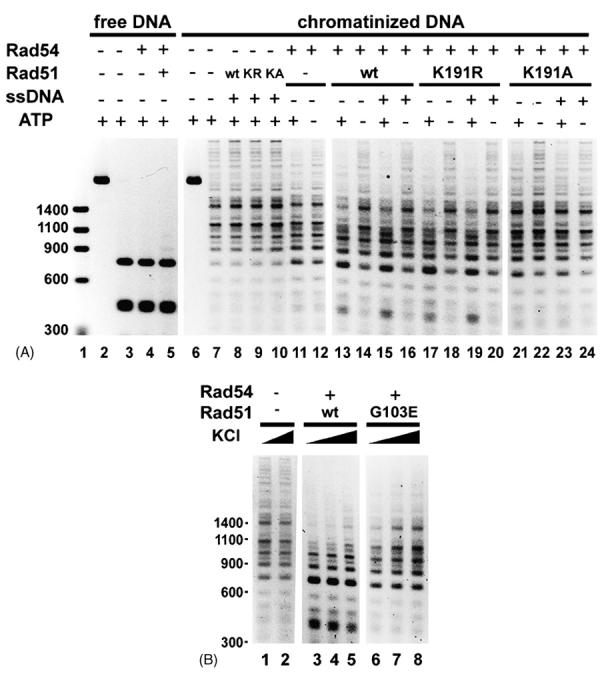
(A) The ability of Rad51, rad51 K191A, and rad51 K191R to enhance the chromatin remodeling activity of Rad54 was examined as described for Fig. 1B. DNA size markers were run in lane 1. HaeIII was added to all the other reactions except those in lanes 2 and 6.
(B) Chromatin remodeling by Rad54-Rad51 (lanes 3 to 5) and Rad54-rad51 G103E (lanes 6 to 8) was examined with increasing KCl (30, 50, 80 mM) concentration. Lanes 1 and 2 are reaction blanks that contain 30 or 80 mM KCl, respectively.
We wished to verify whether or not the rad51 K191A, K191R, and G103E mutant proteins are capable of making D-loop in conjunction with Rad54. As shown in Figure 6, while rad51 K191R was just as active as Rad51 in the D-loop reaction with the chromatinized template, neither rad51 K191A nor rad51 G103E showed any D-loop forming ability.
3.6. Physical interaction of Rad54 with Histone H3 tail
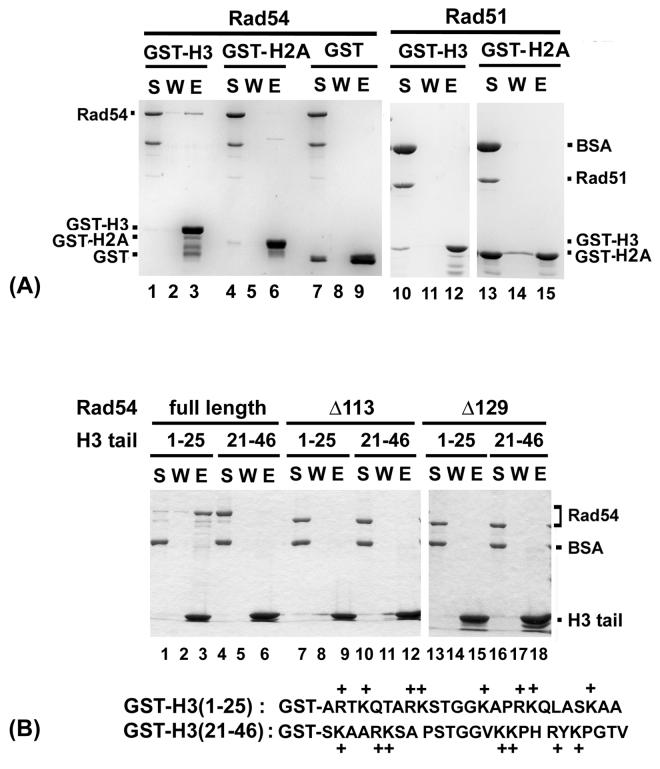
Interactions of various chromatin remodeling factors with the histone components of chromatin are believed to facilitate their action in the chromatin remodeling reaction [45]. We have investigated whether Rad54 interacts with the amino-terminal tail of either histone H2A or H3 by an in vitro pull-down assay with purified proteins. Interestingly, Rad54 was retained on glutathione Sepharose beads coated with GST-histone H3 tail (Fig. 7A, lane 3). The association of Rad54 with the H3 tail is specific, as we did not see any binding of Rad54 to glutathione Sepharose beads decorated with either the N-terminal tail of H2A (lane 6) or with GST (lane 9), and Rad51 failed to associate with glutathione beads containing either of the two N-terminal histone tail species (Fig. 7A, lanes 12 and 15)
Fig. 7. Interaction of Rad54 with yeast histone H3 tail.
(A) Rad54 (lanes 1-9) or Rad51 (lanes 10-15) was mixed with glutathione-Sepharose beads decorated with GST-H2A tail, GST-H3 tail, or GST. Proteins were eluted from the beads with SDS. The supernatant (S), wash (W), and SDS eluate from the reactions were analyzed.
(B) Glutathione Sepharose beads decorated with a GST fusion protein harboring either the proximal (residues 1 to 25) or distal (residues 21 to 46) portion of the H3 tail were incubated with Rad54, rad54 Δ113, or rad54 Δ129. Analysis for protein complex formation followed the procedure in (A).
The GST-H3 species used in the above pulldown assays harbors the entire N-terminal tail from residues 1 through 46. In the study of Hecht et al [46], the chromatin-silencing factor Sir3 was found to bind the proximal segment of the H3 tail spanning residues 1-25, but little or no interaction was noted between Sir3 and the distal segment of the histone tail encompassing residues 21-46. We asked whether Rad54 would bind either of the aforementioned H3 N-terminal tail segments. As shown in Fig. 7B, avid association of Rad54 with GST-H3 (1-25) was seen (lane 3), whereas no binding to GST-H3 (21-46) occurred. Since the net charges of the two H3 tail segments are quite similar (Fig. 7B) and the intact H3 tail was not bound more avidly than was H3 (1-25) by Rad54, the association of Rad54 with the H3 tail very likely involves more than ionic interactions. As expected, Rad51 failed to associate with either of the H3 tail pieces (data not shown).
3.7. The H3 tail binding domain resides within the Rad54 amino-terminus
We have shown that deletion of the amino-terminal 113 residues of Rad54, as in the rad54 Δ113 mutant, has only a minimal effect on the ability to interact with Rad51 [28], but the deletion of an additional 16 residues, as in rad54 Δ129, greatly impairs complex formation with Rad51 [28]. We were interested to know whether these rad54 N-terminal truncation mutants retain the ability to interact with the H3 tail or not. We used the same pulldown assay with GST-H3 (1-25) to answer this question, and the results showed that both the Δ113 and Δ129 rad54 mutant variants are devoid of the ability to associate with the H3 tail fragment (Fig. 7B, lanes 9 and 15).
4. Discussion
Genetic studies have found an involvement of Rad54 protein in homologous recombination and the DNA homology-directed elimination of DNA breaks and other chromosome lesions [3,17]. Consistent with the genetic data, studies with purified proteins have shown synergistic interactions of Rad54 with Rad51, in a manner that (1) greatly improves the ability of Rad51 to mediate the DNA strand invasion reaction with both naked and chromatinized DNA templates, and (2) enhances the ATPase, DNA supercoiling, DNA strand opening, DNA branch migration, and chromatin remodeling functions of Rad54 [9,10,12-14,16]. Here, we have examined whether efficient chromatin remodeling by the combination of Rad54 and Rad51 is contingent upon complex formation between the two HR factors, and have in addition addressed the functional significance of the Rad51 presynaptic filament in the chromatin remodeling reaction. To achieve these goals, we employed (1) a rad54 mutant (Δ129) that has a conditional defect (induced by elevating the ionic strength) in Rad51 interaction, (2) rad51 K191R that is differentially inactivated for ATP hydrolysis but retains the ability to bind ATP and for presynaptic filament assembly, (3) rad51 K191A that does not bind ATP or DNA, (4) rad51 G103E that is greatly attenuated for DNA binding and ablated for interaction with Rad54, and (5) the heterologous recombinase RecA. Altogether, the results help establish that complex formation between Rad54 and Rad51 is critically important for ATPase activation, chromatin remodeling, and for DNA strand invasion involving a chromatinized template. Moreover, the experiments involving rad51 K191A have yielded the first evidence that while Rad54 ATPase enhancement by Rad51 can occur without the presynaptic filament being assembled, optimal chromatin remodeling is somehow functionally linked to the presynaptic filament. Alternatively, or in addition, the chromatin remodeling reaction is dependent on the ATP-bound form of Rad51 within the context of the Rad51-Rad54 complex. Future studies involving other appropriate mutants of Rad51 will be necessary to distinguish between these two models.
Whereas Zhang et al [43] observed that rad51 G103E has the same affinity for Rad54 as wild type Rad51, in our hands, this mutant protein is greatly attenuated for the ability to bind Rad54. We do not currently have a satisfactory explanation for this discrepancy between the two studies, but note that the same mutation clearly abolishes the ability of Rad51 to interact with Rad54 in the two-hybrid system [42] and ablates the synergistic effect of Rad51 and Rad54 in ATP hydrolysis and chromatin remodeling reactions ([43]; this work).
In the structure of the nucleosome core particle, the histone amino-termini are seen extending away from the DNA gyres [47]. These histone tails represent major targets for post-translational modifications, and a number of biochemical studies have demonstrated that they are bound by and acted on by a variety of chromatin modifying proteins and enzymes [48-53]. In addition to demonstrating a strong reliance of the chromatin remodeling efficacy on Rad51-Rad54 complex formation and the Rad51 presynaptic filament, we have also shown that Rad54 specifically interacts with the amino-terminal tail of histone H3, but that no such interaction of the H3 tail with Rad51 is evident. Furthermore, the interaction of Rad54 with the H3 tail likely involves more than ionic interactions, because Rad54 binds the full length H3 tail (encompassing residues 1-46) no better than the proximal portion (encompassing residues 1-25) of the tail, and the distal portion (encompassing residues 21-46) of the H3 tail, despite it having a very similar charge density as the proximal portion, is not bound by Rad54 at all. It will be interesting to test whether the Rad54 counterpart from other organisms also interacts specifically with the amino-terminal tail of its cognate histone H3. We note that Rad54 may not recognize the N-terminal tail of chicken histone H3, which could explain why little or no difference in reaction efficiency between full length Rad54 and the rad54 Δ129 mutant could be revealed in the chromatin remodeling reaction that used nucleosomal arrays assembled with chicken histones. Alternatively, or in addition, another set of reaction conditions are needed in order to reveal such a difference between the full length and truncated Rad54 proteins.
We showed previously that the N-terminus of Rad54 is particularly prone to proteolytic attack, providing evidence that this region is structurally separate from the core catalytic domain that imparts ATPase, DNA binding, and DNA translocase activities [28]. Importantly, we and others have presented evidence that the flexible N-terminus of Rad54 harbors the Rad51 interaction domain [28,29]. We have shown here that the histone H3 tail binding domain likewise resides within the N-terminus of Rad54. Since the rad54 Δ113 mutant protein binds Rad51 [28] but not the H3 tail, distinct sub-domains in the N-terminus of Rad54 likely mediate the interactions with Rad51 and histone H3.
In addition to promoting the Rad51-mediated D-loop reaction, Rad54 also (1) stabilizes the Rad51 presynaptic filament [32] and seems to have a role in nucleating Rad51 onto ssDNA in cells [31] and (2) removes Rad51 protein from duplex DNA [30]. The former activity (i.e. Rad51 loading and presynaptic filament stabilization) could be germane for the assembly and maintenance of the Rad51 presynaptic filament in cells and the latter function (i.e. Rad51 removal from dsDNA) is likely important for the intracellular recycling of Rad51 and for exposing the primer-template junction in the newly made D-loop to initiate DNA repair synthesis. It remains to be determined whether complex formation with Rad51 is critical for these Rad54 functions. Likewise, it will be important to test whether complex formation with Rad51 is critical for up-regulating the DNA branch migration activity of Rad54 [16].
Acknowledgements
We are grateful to Craig Peterson for providing the plasmids that express the GST-histone tails and to members of our laboratory for reading the manuscript. This study was supported by National Institutes of Health research grants RO1GM053738, RO1GM57814, RO1CA110415 and by the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE) grant RTI04-03-06.
Abbreviations
- HR
homologous recombination
- ss
single-stranded
- ds
double-stranded
- NTA
nitrilotriacetic acid
- BSA
bovine serum albumin
- GST
glutathione S-transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde JM. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 2.Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 3.Schmuckli-Maurer J, Rolfsmeier M, Nguyen H, Heyer WD. Genome instability in rad54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1013–1023. doi: 10.1093/nar/gkg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuda M, Miyagawa K, Takahashi M, Fukuda T, Kataoka T, Asahara T, Inui H, Watatani M, Yasutomi M, Kamada N, Dohi K, Kamiya K. Mutations in the RAD54 recombination gene in primary cancers. Oncogene. 1999;18:3427–3430. doi: 10.1038/sj.onc.1202692. [DOI] [PubMed] [Google Scholar]
- 5.Smirnova M, Van Komen S, Sung P, Klein HL. Effects of tumor-associated mutations on Rad54 functions. J. Biol. Chem. 2004;279:24081–24088. doi: 10.1074/jbc.M402719200. [DOI] [PubMed] [Google Scholar]
- 6.Swagemakers SM, Essers J, de Wit J, Hoeijmakers JH, Kanaar R. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 7.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 8.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 10.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 12.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 13.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remo deling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 14.Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solinger JA, Heyer WD. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 17.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell. Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 19.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 20.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 21.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Cox MM. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 24.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 26.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer WD. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 28.Raschle M, Van Komen S, Chi P, Ellenberger T, Sung P. Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J. Biol. Chem. 2004;279:51973–51980. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- 29.Alexiadis V, Lusser A, Kadonaga JT. A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. J. Biol. Chem. 2004;279:27824–27829. doi: 10.1074/jbc.M402648200. [DOI] [PubMed] [Google Scholar]
- 30.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 31.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 32.Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 33.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 34.Wolner B, Peterson CL. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- 35.Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 36.Houston PL, Broach JR. The dynamics of homologous pairing during mating type interconversion in budding yeast. PLoS Genet. 2006;2:e98. doi: 10.1371/journal.pgen.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 38.Stein A. Reconstitution of chromatin from purified components. Methods Enzymol. 1989;170:585–603. doi: 10.1016/0076-6879(89)70066-5. [DOI] [PubMed] [Google Scholar]
- 39.Jeong SW, Lauderdale JD, Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J. Mol. Biol. 1991;222:1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- 40.Lusser A, Kadonaga JT. Strategies for the reconstitution of chromatin. Nat. Methods. 2004;1:19–26. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 41.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 42.Krejci L, Damborsky J, Thomsen B, Duno M, Bendixen C. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol. 2001;21:966–976. doi: 10.1128/MCB.21.3.966-976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Zhang XP, Lee KI, Solinger JA, Kiianitsa K, Heyer WD. Gly-103 in the N-terminal domain of Saccharomyces cerevisiae Rad51 protein is critical for DNA binding. J. Biol. Chem. 2005;280:26303–26311. doi: 10.1074/jbc.M503244200. [DOI] [PubMed] [Google Scholar]
- 44.Galkin VE, Wu Y, Zhang XP, Qian X, He Y, Yu X, Heyer WD, Luo Y, Egelman EH. The Rad51/RadA N-terminal domain activates nucleoprotein filament ATPase activity. Structure. 2006;14:983–992. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 46.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 47.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 48.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell. Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 49.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol. Cell. Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Georgel PT, Palacios DeBeer MA, Pietz G, Fox CA, Hansen JC. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8584–8589. doi: 10.1073/pnas.151258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]