SUMMARY
Mutations in the amyloid precursor protein (APP) cause early-onset Alzheimer's disease (AD), but the only genetic risk factor for late-onset AD is the ε4 allele of apolipoprotein E (apoE), a major cholesterol carrier. Using Cre-lox conditional knockout mice, we demonstrate that lipoprotein receptor LRP1 expression regulates apoE and cholesterol levels within the CNS. We also found that deletion of APP and its homologue APLP2, or components of the γ-secretase complex, significantly enhanced the expression and function of LRP1, which was reversed by forced expression of the APP intracellular domain (AICD). We further show that AICD, together with Fe65 and Tip60, interacts with the LRP1 promoter and suppresses its transcription. Together, our findings support that the γ-secretase cleavage of APP plays a central role in regulating apoE and cholesterol metabolism in the CNS via LRP1 and establish a biological linkage between APP and apoE, the two major genetic determinants of AD.
INTRODUCTION
Mounting genetic and biochemical evidence strongly supports the hypothesis that amyloid β-peptide (Aβ) accumulation in the brain is an early and toxic event in the pathogenesis of Alzheimer's disease (Hardy and Selkoe, 2002). Accordingly, reducing brain Aβ production and/or increasing its clearance have become attractive targets for AD drug development (Hardy and Selkoe, 2002). Aβ is derived from sequential proteolytic processing of amyloid precursor protein (APP), a ubiquitously expressed type I transmembrane protein that undergoes two distinct processing pathways (Selkoe and Kopan, 2003; Zheng and Koo, 2006). In the non-amyloidogenic pathway, APP undergoes ectodomain shedding by α-secretase, identified as members of the ADAM metalloprotease family (Zheng and Koo, 2006). Subsequent cleavage of the APP C-terminal membrane-associated stub by γ-secretase (Selkoe and Kopan, 2003) generates a non-toxic p3 peptide, as well as the APP intracellular domain (AICD) (Selkoe and Kopan, 2003). In the amyloidogenic pathway, APP is first cleaved by the β-secretase BACE1 (β-site APP cleaving enzyme 1) (Vassar et al., 1999) and then by γ-secretase to generate Aβ and AICD. Mutations associated with early-onset familial forms of AD (FAD) are found in the APP gene itself or in the genes of presenilin 1 (PS1) and PS2, whose products, along with nicastrin, Pen-2 and Aph-1, are obligate components of a multi-protein complex that gives rise to γ-secretase activity (Selkoe and Kopan, 2003). A common feature of all FAD mutations is that they increase the generation of Aβ peptides, or the proportion of the longer Aβ42 form, considered to be more amyloidogenic and pathogenic than the shorter Aβ40 due to its highly aggregative nature (Hardy and Selkoe, 2002).
Although FAD genetics and mouse models have generated tremendous insights into AD pathogenesis, the vast majority of AD cases are sporadic with late-onset. A major genetic risk factor that was initially discovered in 1993 and has since been validated in numerous genetic studies is the presence of the ε4 allele of the apolipoprotein E (APOE) gene (Corder et al., 1993). ApoE is a major apolipoprotein in the brain, and exists in three isoforms in humans (apoE2, apoE3, apoE4); each differing by a single amino acid (Mahley, 1988). In the brain, apoE/lipoprotein particles are produced primarily by astrocytes and are believed to deliver cholesterol and other lipids to neurons via lipoprotein receptors, namely members of the low-density lipoprotein receptor (LDLR) family (Herz and Bock, 2002; Herz and Chen, 2006). Although the mechanisms underlying the pathogenic nature of apoE4 in sporadic AD are still poorly understood, several models have been proposed and supported by in vitro and in vivo studies. First, apoE interacts with Aβ, and apoE4 likely possesses greater ability to promote Aβ fibrillogenesis and amyloid plaque formation (Holtzman et al., 2000). Second, apoE facilitates Aβ clearance via apoE receptors expressed either in neurons (Zerbinatti and Bu, 2005) or in the blood brain barrier (Zlokovic, 2005). ApoE4 is less functional in Aβ clearance owing to its weaker affinity to Aβ (LaDu et al., 1994). Third, apoE4 may be toxic to neurons independently of Aβ aggregation and clearance (Huang, 2006). It is possible that multiple pathways contribute to the pathogenic nature of apoE4 in AD.
Cholesterol is an essential component of membranes and myelin sheathes and is crucial for synaptic integrity and neuronal functions (Pfrieger, 2003). Interestingly, an association between brain cholesterol metabolism and the risk of AD has been proposed (Shobab et al., 2005). Early studies indicated that the use of statins, which inhibit cholesterol synthesis, is associated with a significant decrease in AD prevalence; however, several recent prospective studies do not support such a conclusion (Shobab et al., 2005). Additionally, the effect of cholesterol on the amyloidogenic processing of APP to Aβ remains controversial (Ledesma and Dotti, 2006). Intriguingly, apoE4 knock-in mice exhibit decreased brain cholesterol levels even though the peripheral cholesterol levels are increased (Hamanaka et al., 2000). A reduction of brain cholesterol levels is also observed in AD brains (Ledesma and Dotti, 2006). These disparate findings raise the need for an understanding of brain cholesterol metabolism and its potential dysregulation in AD.
In this manuscript we present a novel linkage between APP processing and apoE/cholesterol metabolism. Specifically, we found that lack of either APP/APLP2 or PS1/PS2 leads to increased brain apoE/cholesterol catabolism. We show that the APP processing product, AICD, suppressed expression of the major apoE/lipoprotein receptor LRP1 by binding directly to its promoter following association with the adaptor protein Fe65. Defective apoE/cholesterol catabolism in APP/APLP2 and PS1/PS2 knockout cells was restored by a forced expression of AICD. Together, our results reveal a novel biological function of APP in the regulation of brain apoE and cholesterol metabolism, offering new alternatives for the design of therapeutic strategies to treat AD.
RESULTS
APP and APLP2 Regulate Brain ApoE and Cholesterol Metabolism
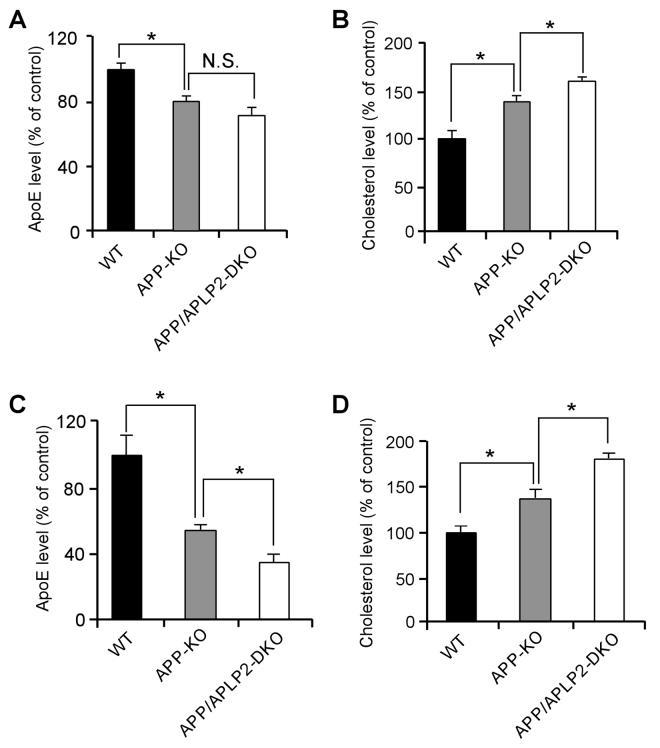
APP processing and apoE/cholesterol metabolism are two important events in the pathogenesis of AD. To examine whether these two pathways are functionally related, we analyzed cellular apoE and cholesterol levels in mouse embryonic fibroblasts (MEFs) of WT, APP-KO, or APP and APLP2 double knockout (APP/APLP2-DKO). To minimize potential clonal effects when MEF cells were established, APP-KO MEF cells stably retransfected with APP695 cDNA were used as WT controls. The APP-KO and APP/APLP2-DKO cells displayed a significant decrease in apoE levels and a concomitant increase in cholesterol levels compared to WT controls (Figures 1A and 1B). The apoE and cholesterol levels we measured likely reflect a combination of those derived from both serum and cells. One possible explanation to these findings is that lack of APP or APP/APLP2 lead to increased catabolism of apoE/lipoprotein resulting in increased intracellular cholesterol levels (Mahley, 1988). To assess whether these changes also occur in vivo, we compared brain apoE and cholesterol levels in WT, APP-KO and APP/APLP2-DKO mouse brain. ApoE levels were decreased by ∼50% in APP-KO and further decreased in APP/APLP2-DKO when compared to WT littermate controls (Figure 1C). A corresponding increase in cholesterol levels was also observed in APP-KO and APP/APLP2-DKO mouse brain when compared to WT controls (Figure 1D). Together, these results raise the possibility that apoE and cholesterol levels are modulated, either directly or indirectly, by APP and APLP2 and/or their processing products.
Figure 1. APP Regulates ApoE and Cholesterol Metabolism.
(A and B) ApoE levels were decreased and cholesterol content was increased in the absence of APP or APP/APLP2. ApoE and cholesterol levels were measured in triplicates in lysates of WT, APP-KO and APP/APLP2-DKO MEF cells, normalized against total protein and plotted as a percentage of WT controls. (C and D) ApoE levels were also increased and cholesterol content decreased in brain lysates of APP-KO and APP/APLP2-DKO mice. ApoE and cholesterol levels were measured in brain lysates of WT, APP-KO and APP/APLP2-DKO mice (n=4), normalized against total protein and plotted as a percentage of WT controls. For this and subsequent figures, data represent mean ± SEM; N.S., not significant; *, P<0.05; **, P<0.01.
APP and APLP2 Regulate LRP1 Expression and Function
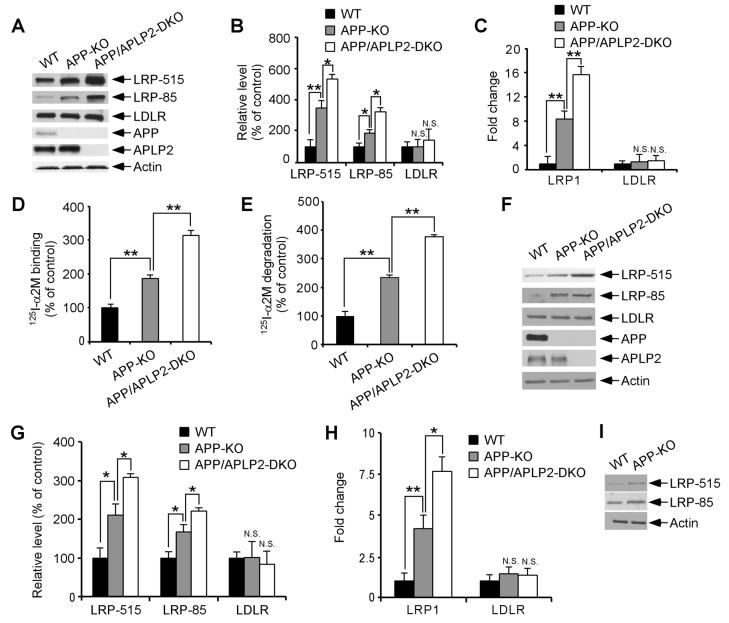
The increased catabolism of apoE/lipoprotein by APP-KO and APP/APLP2-DKO cells lead us to investigate the possibility that apoE receptor levels maybe up-regulated in these cells. To test this possibility, we compared expression levels of two major brain apoE receptors LRP1 and LDLR (Herz and Bock, 2002) in WT, APP-KO and APP/APLP2-DKO MEF cells. Western blotting using two different antibodies to LRP1 showed that LRP1 levels were significantly increased in APP-KO MEF cells and in APP/APLP2-DKO MEF cells compared to WT MEF cells (Figures 2A and 2B). The expression of the LDLR was not altered by APP/APLP2 deletion (Figures 2A and 2B). To analyze whether changes in LRP1 expression were at the transcriptional or post-transcriptional level, we compared LRP1 mRNA levels by real-time PCR. As shown in Figure 2C, LRP1 mRNA levels were significantly increased in both APP-KO and APP/APLP2-DKO MEF cells when compared to WT control cells. To investigate whether changes in LRP1 expression also correlate with changes in LRP1 function, we analyzed binding and endocytosis of α2-macroglobulin (α2M), a high affinity ligand for LRP1 (Herz and Bock, 2002). Ligand binding assays using 125I-α2M demonstrated increased binding capacity in APP-KO and APP/APLP2-DKO MEF cells when compared to WT control cells (Figure 2D). Similar increase was seen when 125I-α2M uptake and degradation were analyzed (Figure 2E).
Figure 2. APP/APLP2 Regulates LRP1 Expression and Function.
(A) LRP1 and LDLR expression levels were compared between WT, APP-KO and APP/APLP2-DKO MEF cells by Western blot. Equal amount of protein in this and subsequent figures was loaded to each lane. (B) Densitometric analyses of Western blots from triplicate samples demonstrate a significant increase in the expression of LRP1, but not LDLR, in the absence of APP/APLP2. (C) LRP1 and LDLR mRNA levels were quantified in WT, APP-KO and APP/APLP2-DKO MEF cells by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of APP/APLP2. (D) 125I-α2M (1 nM) binding to WT, APP-KO and APP/APLP2-DKO MEF cells was performed at 4°C for 1 h in the absence or presence of RAP (500 nM). RAP-inhibitable 125I-α2M binding was normalized against total cellular protein and plotted as a percentage of WT controls. (E) 125I-α2M (1 nM) uptake and degradation assays were performed at 37°C for 4 h in the absence or presence of RAP (500 nM). RAP-inhibitable 125I-α2M degradation was normalized against total cellular protein and plotted as a percentage of WT controls. (F) LRP1 and LDLR expression levels in the brain were compared between WT, APP-KO and APP/APLP2-DKO newborn mice by Western blot. (G) Densitometric analyses of Western blots (n=3) indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of APP or APP/APLP2. (H) LRP1 and LDLR mRNA levels were quantified in the brain of WT, APP-KO and APP/APLP2-DKO newborn mice (n=3) by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of APP/APLP2. (I) LRP1 expression in the brain was compared by Western blot between WT and APP-KO mice at 4 months of age. Similar increase in LRP1 expression was observed in adult APP-KO mouse brain.
To examine whether our in vitro findings were reproduced in vivo, we compared LRP1 expression in the brains of newborn APP-KO, APP/APLP2-DKO and littermate control mice. LRP1 expression, but not LDLR expression, was significantly increased in APP-KO and APP/APLP2-DKO mouse brains when compared to WT controls both at the protein (Figures 2F and 2G) and mRNA levels (Figure 2H). Similar increase in LRP1 expression was also observed in the brain of APP-KO mice at 4 months of age. These results demonstrate that LRP1 expression and function are modulated by APP/APLP2 or their processing products and suggest that APP/APLP2 likely modulate brain apoE and cholesterol metabolism via regulation of LRP1 levels.
LRP1 Modulates Brain ApoE/Lipoprotein Metabolism
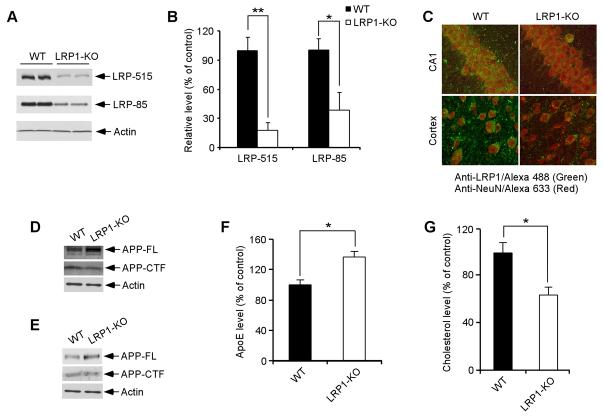
LRP1 is abundantly expressed in the brain, but in vivo evidence that LRP1 regulates brain apoE and cholesterol metabolism is lacking. Because conventional knockout of LRP1 is early embryonic lethal (Herz et al., 1992), we generated conditional LRP1 forebrain knockout mice by crossing LRP1 floxP mice (Rohlmann et al., 1998) with αCamKII-Cre mice (Tsien et al., 1996). LRP1 expression was significantly decreased in the forebrain as determined by immunobloting using antibodies directed against either the 515-kDa subunit or the 85-kDa subunit (Figures 3A and 3B). Remaining LRP1 expression detected by Western blotting likely represents expression in glial cells (Moestrup et al., 1992). By double immunofluorescence staining using LRP1-specific antibody and NeuN antibody we confirmed that LRP1 expression was nearly abolished in CA1 neurons of the hippocampus and in pyramidal neurons of the frontal cortex (Figure 3C). To examine how an LRP1 deletion affects APP levels and processing, we compare the steady-state levels of the full-length APP and APP C-terminal fragment (CTF). We found that deletion of LRP1 slightly increased the steady-state levels of full-length APP while decreased the levels of APP-CTF both in LRP1-KO MEF cells (Figure 3D) and in mouse brain (Figure 3E); likely reflecting a stabilization of APP at the cell surface due to reduced endocytosis (Cam et al., 2005).
Figure 3. LRP1 Is Essential for Brain ApoE and Cholesterol Metabolism.
(A) LRP1 expression in the forebrain was evaluated in LRP1 forebrain knockout (LRP1-KO) and WT littermate control mice by Western blot using two distinct LRP1 antibodies (against 515-kDa subunit and 85-kDa subunit, respectively). Equal amount of protein in this and subsequent figures was loaded to each lane. (B) Densitometric quantification of LRP1 expression was performed as described in Materials and Methods from two independent experiments (n=4). (C) Double immunofluorescence staining using an LRP1 antibody (detected with Alexa 488, green) and neuronal marker NeuN antibody (detected with Alexa 633, red). Shown are representative staining in CA1 neurons and pyramidal neurons of frontal cortex. Note that LRP1 expression was almost absent in these forebrain neurons. (D) APP-FL and APP-CTF levels were compared by Western blot between WT and LRP1-KO MEF cells. (E) APP-FL and APP-CTF levels were compared by Western blot between WT and LRP1-KO mouse brains. (F and G) ApoE and cholesterol levels in the brain were compared between LRP1-KO and WT controls (n=4). ApoE levels were significantly higher (F) and cholesterol levels were significantly lower (G) in LRP1-KO mice.
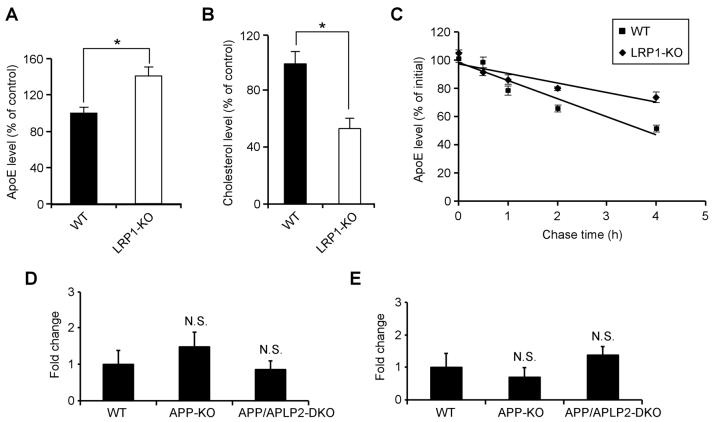
To evaluate the impact of LRP1 deletion on apoE and cholesterol metabolism, we compared apoE and cholesterol levels in the forebrain of LRP1-KO and WT littermate controls. We found that while apoE levels were significantly increased (Figure 3F), cholesterol levels were decreased (Figure 3G) in the LRP1-KO mice, suggesting impaired catabolism of apoE/lipoprotein particles. The changes in apoE and cholesterol levels in the absence of LRP1 were also observed in LRP1-KO MEF cells (Figures 4A and 4B). Interestingly, the half-life of apoE is significantly increased in LRP1-KO MEF cells (Figure 4C), suggesting that in the absence of LRP1, apoE catabolism is decreased. ApoE mRNA levels were not changed between WT and LRP1-KO MEF cells (Figure 4D) or brain tissues (Figure 4E). Together, these results demonstrate that LRP1 is a bona fide apoE/lipoprotein receptor which modulates apoE and cholesterol metabolism in the brain.
Figure 4. Absence of LRP1 Expression Increases ApoE Half-life.
(A and B) ApoE (A) and cholesterol (B) levels were measured in triplicates in lysates of WT and LRP1-KO MEF cells, normalized against total protein and plotted as a percentage of WT controls. (C) WT and LRP1-KO MEF cells were incubated with serum-free medium for 2 h and chased in the presence of protein synthesis inhibitor cycloheximide for 0, 1, 2, or 4 h. ApoE levels under each condition were measured by ELISA and plotted against chase time. Note apoE half-life is significantly increased in LRP1-KO MEF cells. (D and E) ApoE mRNA was quantified by real-time PCR in WT, APP-KO, APP/APLP2-DKO MEF cells (D) or brain tissues (E). Note apoE mRNA levels were not significantly affected by APP/APLP2 deletion.
Aβ Is Not Required for LRP1 Regulation
A recent study demonstrated a role for Aβ in regulating the biosynthesis of cholesterol and sphingomyelin (Grimm et al., 2005). Therefore we evaluated the possibility that Aβ may be the processing product of APP which regulates LRP1 expression levels. Because β-secretase BACE1 is necessary for Aβ production (Luo et al., 2001), we evaluated the effects of BACE1 knockout on LRP1 expression. Deficiency of BACE1 in either MEF cells or in mouse brain did not alter LRP1 expression or function (Supplementary Figure 1). Furthermore, no difference in LRP1 expression was found in MEF cells over-expressing a human APP bearing the Aβ-overproducing Swedish mutation (APPsw) in either the WT background (high Aβ levels) or BACE1-KO background (no Aβ) (Supplementary Figure 1). These results indicate that Aβ production is not required for LRP1 regulation.
γ-Secretase Activity Regulates LRP1 Expression and Function
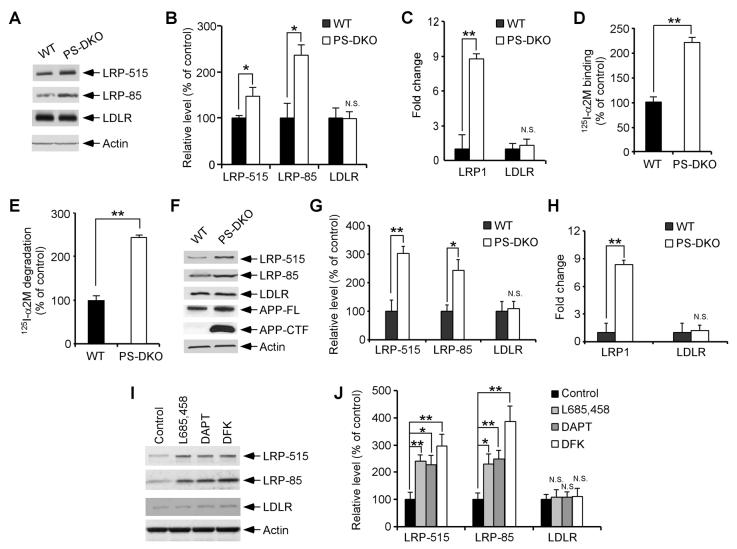
Following α- or β-cleavage, the C-terminal stub of APP is processed by γ-secretase to generate AICD, which has been previously reported to translocate to the nucleus and regulate gene transcription (Baek et al., 2002; Cao and Sudhof, 2001; Pardossi-Piquard et al., 2005). To examine whether γ-secretase cleavage is required for APP-mediated regulation of LRP1 expression, we first compared LRP1 expression in WT MEF cells to those lacking presenilin (PS), an essential component of the γ-secretase complex. Western blotting showed that LRP1 expression was significantly increased in PS1/2 double knockout (PS-DKO) MEF cells (Figures 5A and 5B) when compared to WT MEF cells, while the expression of the LDLR was not affected. A similar increase in LRP1 expression was seen in PS1-KO MEF cells (Supplementary Figure 2). Real-time PCR analysis confirmed increase in LRP1 expression at the mRNA levels (Figure 5C, Supplementary Figure 2). PS-DKO MEF cells also showed increased binding and degradation of the LRP1 ligand 125I-α2M when compared to WT MEF cells (Figures 5D, 5E and Supplementary Figure 2). To test whether LRP1 expression was altered in the absence of the γ-secretase function in vivo, brain tissues from PS-DKO, PS1-KO and their littermate controls were analyzed for LRP1 expression by Western blotting and real-time PCR. The loss of γ-secretase function in PS-DKO mouse brains was demonstrated by an accumulation of APP-CTF, a substrate for γ-secretase (Figure 5F). We found that expression of LRP1 was significantly increased in PS-DKO (Figures 5F-5H) and PS1-KO (Supplementary Figure 2) mouse brains.
Figure 5. γ-Secretase Regulates LRP1 Expression.
(A) LRP1 and LDLR expression levels were compared between WT and PS-DKO MEF cells by Western blot. (B) Densitometric analyses of Western blots from quadruplicate samples indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of PS. (C) LRP1 and LDLR mRNA levels were quantified in WT and PS-DKO MEF cells by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of PS. (D and E) 125I-α2M binding (D) and degradation assays (E) were performed in WT and PS-DKO MEF cells as described in Figure 2. (F) LRP1 and LDLR expression levels were also analyzed in WT and PS-DKO mouse brains by Western blot. Note the significant accumulation of APP CTF in PS-DKO mouse brains. (G) Densitometric analyses (n=3) indicate a significant increase in the expression of LRP1, but not LDLR, in the absence of PS. (H) LRP1 and LDLR mRNA levels were quantified in WT and PS-DKO mouse brain (n=3) by real-time PCR. LRP1 mRNA, but not LDLR mRNA, was significantly increased in the absence of PS. (I) WT MEF cells were treated with vehicle control or γ-secretase inhibitors DAPT (2 μM), DFK (100 μM), or L685,458 (1 μM) for 48 h. Expression levels of LRP1 and LDLR were measured by Western blot. (J) Densitometric analyses of Western blots from triplicate samples indicate an increase in the expression of LRP1, but not LDLR, upon γ-secretase inhibitor treatments.
The requirement of γ-secretase activity in LRP1 regulation was further verified by two alternative approaches. MEF cells lacking nicastrin, another essential component of the γ-secretase complex (Selkoe and Kopan, 2003), also showed increased LRP1 expression and function compared to WT control MEF cells (Supplementary Figure 3). Furthermore, treatment with three distinct γ-secretase inhibitors (L685,458, DAPT, DFK) enhanced LRP1 expression in WT MEF cells (Figures 5I and 5J). Together, our results demonstrate that γ-secretase activity regulates LRP1 expression and function both in vitro and in vivo.
AICD Rescues LRP1 Expression in the Absence of APP/APLP2 or PS Expression
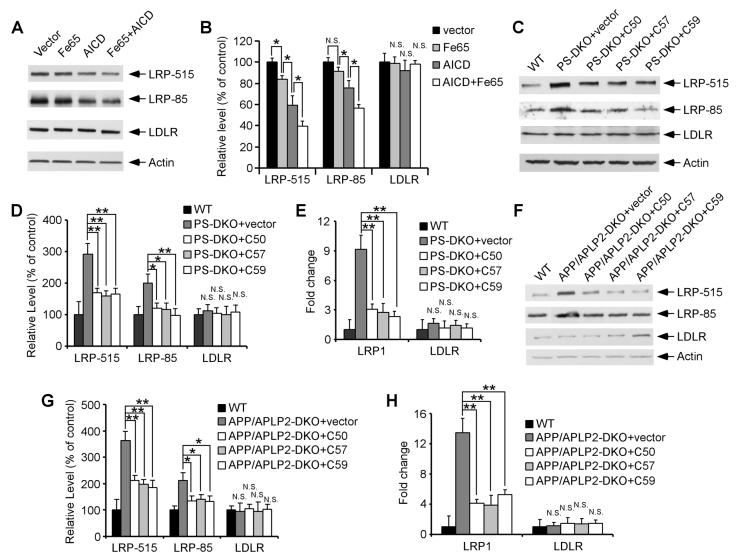
To directly address whether the γ-secretase product, AICD, is involved in LRP1 regulation, we first analyzed the effects of AICD forced expression on LRP1 expression. Transient transfection of AICD into U87 cells significantly suppressed LRP1 expression without affecting LDLR levels (Figures 6A and 6B). The adaptor protein Fe65 has been previously shown to modulate AICD stability and potentiate its subsequent nuclear translocation (Cao and Sudhof, 2001). When overexpressed in U87 cells, Fe65 alone slightly suppressed LRP1 expression; however, co-expression of Fe65 with AICD further enhanced the AICD-mediated suppression of LRP1 expression (Figures 6A and 6B).
Figure 6. AICD Rescues LRP1 Expression in the Absence of APP/APLP2 or PS1/2.
(A) U87 cells were transiently transfected with plasmid constructs as indicated. LRP1 and LDLR expression levels were analyzed by Western blot. (B) Densitometric analyses of Western blots from triplicate samples indicate that a forced expression of AICD and Fe65 suppressed the expression of LRP1, but not LDLR. (C) PS-DKO MEF cells were transiently infected with retroviral vector control or AICD C50, C57, or C59 cDNA. LRP1 and LDLR expression levels were then analyzed by Western blot and compared with those of WT MEF cells. (D) Densitometric analyses of Western blot from triplicate samples indicate that AICD expression suppressed LRP1 expression close to the levels seen in WT MEF cells. (E) The rescuing effect of AICD on LRP1 expression was also observed at the mRNA levels as measured by real-time PCR. (F-H) Similar experiments to those shown in (C-E) were performed with APP/APLP2-DKO MEF cells.
BACE1 cleavage of APP followed by γ-secretase cleavage generates Aβ40 or Aβ42 with concomitant production of AICD consisting of either 59 or 57 amino acids respectively (referred to as AICD C57 and C59). An additional γ-secretase cleavage, referred to as ε cleavage, occurs several amino acids downstream and releases a 50 amino acid fragment (termed AICD C50). To test whether AICD can rescue LRP1 expression in cells deficient for either PS or APP, we cloned AICD C50, C57 and C59 into a retroviral vector. Infection with retrovirus expressing AICD C50, C57, or C59 significantly suppressed LRP1 expression in MEF cells deficient for either PS1/2 or APP/APLP2 (Figures 6C, 6D, 6F and 6G). A reduction in LRP1 mRNA levels was also observed when analyzed by real-time PCR (Figures 6E and 6H). Our data indicate that AICD is capable of rescuing the defective LRP1 expression observed in APP/APLP2-DKO and PS-DKO cells and strongly suggest that an AICD-dependent signaling pathway is crucial for the regulation of LRP1 cellular levels.
AICD Nuclear Signaling Inhibits LRP1 Promoter Activation
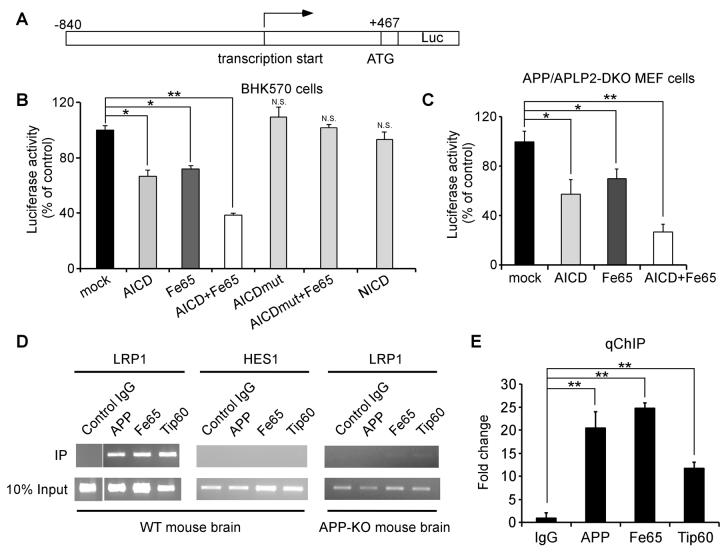
Since AICD down-regulates LRP1 expression at the transcriptional level and AICD has been shown to modulate promoter function, we next tested whether LRP1 promoter activity is repressed by AICD. The LRP1 promoter was cloned into a luciferase reporter vector pGL3 (Figure 7A) and its activity was measured in BHK570 cells and APP/APLP2-DKO MEF cells after transfection of AICD, Fe65, or both. We found that AICD reduced LRP1 promoter activity and Fe65 further potentiated this effect in both cell types (Figures 7B and 7C). An AICD mutant bearing a functional mutation in the 682YxNPxY motif (Y682G, see Borg et al., 1996) lost the ability to regulate LRP1 promoter, and Notch intracellular domain (NICD) did not change LRP1 promoter function in this assay (Figure 7B). To examine whether AICD binds directly to the LRP1 promoter, we performed chromatin immunoprecipitation (ChIP) assay. Immunoprecipitation of mouse brain lysates with an antibody that recognizes the APP C-terminal domain showed that AICD associates with the LRP1 promoter (Figures 7D and 7E). Because biochemical and functional interactions among AICD, Fe65 and Tip60 have been demonstrated (Baek et al., 2002; Cao and Sudhof, 2001), we also analyzed the ability of antibodies to Fe65 and Tip60 to immunoprecipitate the LRP1 promoter. Indeed, we found that both Fe65 and Tip60 antibodies immunoprecipitated the LRP1 promoter (Figures 7D and 7E). The association of AICD, Fe65 and Tip60 with the LRP1 promoter was specific because normal rabbit IgG failed to immunoprecipitate the LRP1 promoter. In addition, APP, Fe65 and Tip60 antibodies did not precipitate a control Notch target promoter, HES1 (Figure 7D). Further, the association of Fe65 and Tip60 with LRP1 promoter was greatly reduced in APP-KO mouse brain (Figure 7D). These results indicate that AICD, together with Fe65 and Tip60, bind directly to the LRP1 promoter to suppress its activation.
Figure 7. AICD Binds To and Suppresses LRP1 Promoter Activation.
(A) Schematic diagram of LRP1 promoter-luciferase construct. (B) BHK570 cells were transiently co-transfected with the LRP1 promoter-Luc construct together with control vector, AICD, Fe65, AICD/Fe65, AICD mutant (Y682G), AICD mutant/Fe65, or NICD. LRP1 promoter-driven luciferase activity was significantly decreased by AICD and Fe65 and further by the co-expression of both, but not by an AICD mutant or NICD. (C) APP/APLP2-DKO MEF cells were transiently co-transfected with the LRP1 promoter-Luc construct together with control vector, AICD, Fe65, or both, and the luciferase activity was measured as in (B). (D) ChIP assay showed that antibodies to AICD, Fe65 and Tip60, but not control IgG, immunoprecipitate LRP1 promoter DNA fragment. Notch target HES1 promoter was used as a negative control. The ability of anti-Fe65 and anti-Tip60 to immunoprecipitate LRP1 promoter DNA is greatly reduced in APP-KO mouse brain. (E) Quantitative real-time PCR analysis of LRP1 promoter DNA immunoprecipitated by control IgG or antibodies to AICD, Fe65 and Tip60.
AICD Rescues ApoE and Cholesterol Defects in Cells Lacking APP/APLP2 or PS
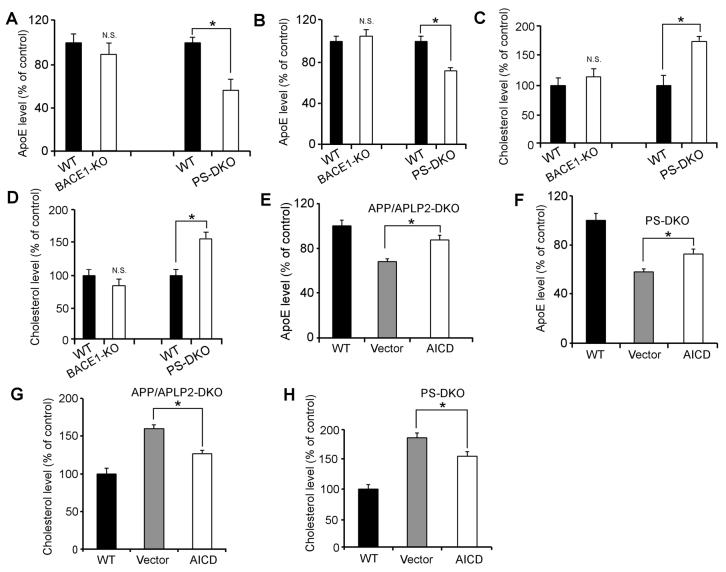
Having demonstrated that LRP1 is a major receptor that regulates apoE and cholesterol metabolism, and that γ-secretase activity is required for APP/APLP2-mediated regulation of LRP1 expression, we were prompted to evaluate potential alterations in apoE and cholesterol levels in PS-DKO MEF cells and mouse brain. As expected, we found that apoE levels were decreased while cholesterol levels were increased in PS-DKO MEF cells (Figures 8A and 8C) and mouse brain (Figures 8B and 8D) when compared to their WT controls. Normal levels of apoE and cholesterol were found in BACE1-KO MEF cells and BACE1-KO mouse brain (Figures 8A-8D) demonstrating that BACE1 is not involved in apoE and cholesterol metabolism. Since AICD rescued LRP1 expression in MEF cells devoid of either APP/APLP2 or PS, we then analyzed whether forced expression of AICD could restore apoE and cholesterol levels in these cells. Forced expression of AICD in APP/APLP2-DKO (Figure 8E) and PS-DKO (Figure 8F) MEF cells significantly increased apoE levels when compared to MEF cells infected with vector alone, albeit not to the levels observed in WT control cells. Likewise, AICD, but not vector alone, partially restored cholesterol levels in APP/APLP2-DKO (Figure 8G) and PS-DKO (Figure 8H) MEF cells. These results confirmed that AICD-mediated control of LRP1 expression is a key regulatory pathway in brain apoE and cholesterol homeostasis.
Figure 8. AICD Rescues ApoE and Cholesterol Defects in Cells Deficient in PS or APP.
(A-D) ApoE (A and B) and cholesterol levels (C and D) were measured in WT, PS-DKO and BACE1-KO MEF cells (A and C) and mouse brains (B and D). ApoE levels were decreased while cholesterol levels were increased in PS-DKO but not BACE1-KO MEF cells and mouse brain when compared to their WT controls. (E-H) APP/APLP2-DKO (E and G) and PS-DKO (F and H) MEF cells were transiently infected with retroviral vector control or AICD cDNA. LRP1 expression levels were then analyzed by Western blot, quantified by densitometry, and compared with those of WT MEF cells. Expression of AICD partially rescued apoE and cholesterol levels in both APP/APLP2-DKO and PSDKO MEF cells.
DISCUSSION
It has been postulated that the amyloidogenic processing of APP to Aβ, particularly of the aggregation-prone Aβ42, plays a central role in the pathogenesis of AD (Hardy and Selkoe, 2002). Accordingly, inhibiting APP processing to Aβ is being actively explored as a therapeutic strategy to treat AD. Here we described a novel mechanism by which the C-terminal fragment of APP, named AICD, modulates brain apoE and cholesterol metabolism by directly regulating the expression and function of the lipoprotein receptor LRP1. Knockout of APP/APLP2 or components of the γ-secretase complex significantly affected the expression of LRP1 as well as apoE and cholesterol levels, and these alterations were partially restored by forced expression of AICD. We also show that AICD, together with the adaptor proteins Fe65 and Tip60, regulates LRP1 promoter function. Our results establish, for the first time, a strong biological relationship between APP processing and apoE/cholesterol metabolism with significant relevance for the pathogenesis of AD.
Several biological functions for APP and its processing products have been described (Zheng and Koo, 2006). The notion that APP is a cell surface receptor has long been speculated but remains unproven due to the lack of a bona fide ligand. The function of APP is further complicated by the presence of two APP-related genes, APLP1 and APLP2 (Zheng and Koo, 2006). Deletion of APLP2 and either APP or APLP1 results in early postnatal lethality (Zheng and Koo, 2006), suggesting redundancy between APLP2 and the other two family members. APP ectodomain has been shown to participate in cell adhesion, neurite outgrowth and synaptogenesis (Zheng and Koo, 2006). APP intracellular domain (AICD), highlighted with an YxNPxY motif for binding of an array of interacting proteins, modulates cell migration, axonal transport and cell signaling (Zheng and Koo, 2006). The most relevant interacting protein is Fe65, which modulates APP processing and AICD nuclear translocation (Cao and Sudhof, 2001). Knockout of Fe65 and its homologues Fe65L1 results in cortical dysplasia and compromised integrity of the pial basement membrane (Guenette et al., 2006). Intriguingly, this phenotype closely resembles that seen in triple mutant mice lacking the APP family members APP, APLP1 and APLP2 (Guenette et al., 2006; Herms et al., 2004), strongly suggesting a common signaling pathway that requires the function of both Fe65 and APP. Mice lacking both APP and APLP2 show defective neuromuscular synapses (Wang et al., 2005), while mice lacking APP alone exhibit increased synapses and associated synaptic function (Priller et al., 2006). In contrast, over-expression of APP in transgenic mice results in deficits of synaptic transmission and learning (Saganich et al., 2006) and dendritic spine abnormalities (Spires et al., 2005). Whether some of these reported phenotypes relate to altered apoE and cholesterol metabolism reported herein requires further investigations.
Although the ε4 allele of the APOE gene was discovered as a strong genetic risk factor for late-onset AD over a decade ago (Corder et al., 1993), the mechanism by which apoE4 contributes to AD pathogenesis is still largely unclear. Furthermore, whether APP and apoE regulate common biological pathways is unknown. This study provides the first evidence that both APP and apoE participate in brain cholesterol metabolism. Cholesterol is an essential component of the cellular membrane and plays pivotal roles in development and maintenance of neuronal plasticity and function (Ledesma and Dotti, 2006). In the adult brain, neuronal cholesterol is supplied primarily by apoE/lipoprotein particles synthesized and secreted by glial cells (Pfrieger, 2003; Puglielli et al., 2003). Uptake of apoE/lipoprotein particles by neurons is mediated by lipoprotein receptors of the LDLR family (Herz and Bock, 2002). Endocytosed cholesterol-containing lipoprotein particles are hydrolyzed in neuronal lysosomes allowing degradation of apoE and intracellular release of free cholesterol which can be stored or incorporated into lipoprotein particles or cellular membranes. A function for LRP1 in brain apoE/cholesterol metabolism has been postulated (Pfrieger, 2003) but direct biological evidence had been lacking until now. Our studies provide the first evidence that deletion of LRP1 in forebrain neurons of adult mice significantly alters brain apoE and cholesterol levels. While apoE levels were increased in LRP1 forebrain knockout mice, cholesterol levels were conversely decreased. Consistent with these findings, our previous work has shown that over-expression of an LRP1 minireceptor in the brain results in a decrease in brain apoE level (Zerbinatti et al., 2006). Together, these results strongly support a role for LRP1 in brain apoE and cholesterol metabolism and establish LRP1 as a neuronal receptor essential for the proper endocytosis and catabolism of apoE. Although the LDLR also functions as a brain apoE receptor (Fryer et al., 2005), our results showed that its expression is not regulated by APP nor γ-secretase.
The role of cholesterol in AD remains controversial (Ledesma and Dotti, 2006; Puglielli et al., 2003; Shobab et al., 2005). Several initial studies suggested a beneficial role of the cholesterol-lowering statins in reducing the risk of AD, and in vitro studies have identified a role for cholesterol in promoting Aβ production. However, several recent studies do not support these conclusions (Shobab et al., 2005). Additionally, mice treated with lovastatin, the most brain penetrant statin currently available, exhibit increased Aβ production and amyloid plaque deposition (Park et al., 2003). Furthermore, neuronal membrane cholesterol loss was recently found to enhance Aβ production (Abad-Rodriguez et al., 2004). Interestingly, a significant reduction of brain cholesterol in AD patients was observed, particularly in areas loaded with amyloid plaques (Ledesma, 2003). These findings suggest that loss of neuronal cholesterol may contribute to synaptic dysfunction and excess Aβ production in AD. Because LRP1 levels are also significantly reduced in AD (Kang et al., 2000), we can speculate that decreased LRP1 levels in AD are directly responsible for cholesterol loss and related synaptic dysfunction. Accordingly, γ-secretase inhibitor treatment, a strategy actively explored for AD therapy, will likely increase the expression and function of LRP1 in cholesterol metabolism, which could in turn support synaptic integrity and function. Because LRP1 also has a role in Aβ clearance by neurons (Zerbinatti and Bu, 2005) and across the blood brain barrier (Deane et al., 2004), restoring LRP1 expression and function in the AD brain could be explored independently as a therapeutic strategy to treat AD. Interestingly, the expression of another member of the LDLR family, SorLA, that regulates APP trafficking and processing to Aβ is also decreased in AD brains (Andersen et al., 2006; Rogaeva et al., 2007) and that inherited variants in the SorLA gene, SORL1, are associated with late-onset AD (Rogaeva et al., 2007). Together, these studies point to multiple pathways by which LDLR family members play roles in the pathogenesis of AD.
Our results reveal a novel role for γ-secretase-dependent APP processing in the regulation of brain cholesterol levels via transcriptional repression of LRP1. Presenilin-dependent cleavage of APP results in the release of AICD, which has been shown to interact with Fe65 and Tip60 and has been suggested to function in nuclear signaling (Baek et al., 2002; Cao and Sudhof, 2001). Subsequent work by Cao and Sudhof (Cao and Sudhof, 2004) demonstrated that nuclear translocation of AICD may be dispensable; raising the possibility that AICD could function by modulating activation of Fe65 rather than functioning as a transcriptional regulator itself. Several studies have reported putative target genes differentially regulated by an AICD-containing complex, including the prostate cancer anti-metastasis gene KAI1 (Baek et al., 2002), APP, GSK3β (Von Rotz et al., 2004), neprilysin (Pardossi-Piquard et al., 2005), regulators of actin dynamics (Muller et al., 2007), and the EGF receptor (Zhang et al., 2007). However, the exact role of AICD in transcriptional regulation of target genes remains controversial (Hebert et al., 2006; Chen and Selkoe, 2007; Pardossi-Piquard et al., 2007). Specifically, work by De Strooper and colleagues (Hebert et al., 2006) has shown that expression of several previously defined AICD targets genes was at best indirectly and weakly affected by APP processing. Further, the role of AICD in regulating neprilysin expression (Pardossi-Piquard et al., 2005; Pardossi-Piquard et al., 2007) was not reproduced by another report (Chen and Selkoe, 2007). Finally, a recent study has shown that secreted APP ectodomain APPsα is sufficient to rescue several anatomical, behavioral, and electrophysiological abnormalities seen in APP-KO mice (Ring et al., 2007). The discrepancy in AICD function could be due to different experimental systems and/or approaches (Herz, 2007). In line with a biological function of AICD, our results provide direct evidence that AICD binds to the LRP1 promoter and regulates its transcriptional function. Although our studies do not exclude the possibility that APP itself or other APP processing products (e.g. soluble APP and Aβ) may also regulate LRP1 expression and function, our findings that LRP1 expression and apoE/cholesterol metabolism are unchanged in BACE1-knockout mice argues against a role for Aβ in regulating LRP1 expression. Supporting an Aβ-independent function of γ-secretase, recent work has demonstrated that a complete loss of presenilin function in the forebrain leads to memory deficits, synaptic dysfunction, and neurodegeneration without generation of amyloid plaques (Saura et al., 2004). These results suggest that at least some of the neurodegenerative pathology seen in AD might result from partial loss of γ-secretase activity or AICD nuclear signaling functions independent of Aβ production (Shen and Kelleher, 2007). It is interesting to note that a recent study demonstrates a role for Aβ in regulating cholesterol biosynthesis and sphingomyelin degradation (Grimm et al., 2005). Because AICD did not completely restore apoE and cholesterol levels in PS-DKO cells, it is possible that other γ-secretase cleavage products may also regulate apoE/cholesterol metabolism via LRP1-dependent and/or independent mechanisms. Regulation of cholesterol synthesis by Aβ, as well as AICD-mediated modulation of LRP1 expression, are likely to be key events in the proper maintenance of brain cholesterol levels.
In summary, this study uncovers a novel biological function of APP in modulating brain apoE and cholesterol homeostasis. We further demonstrate an important role of the APP processing product AICD in modulating the promoter activity of LRP1, an essential lipoprotein receptor for brain apoE and cholesterol metabolism. Our results provide important insights into APP biological function and its potential implications for neuronal dysfunction in AD, and may lead to the design of better therapeutic strategies to treat this devastating disease.
EXPERIMENTAL PROCEDURES
Materials
Human recombinant RAP was expressed in a glutathione S-transferase expression vector and isolated as described previously (Bu et al., 1993). All tissue culture media and serum were from Sigma. Anti-APP C-terminal antibody was purchased from Invitrogen; anti-Fe65 was from Abcam; anti-Tip60 was from Calbiochem; anti-actin from Sigma and anti-NeuN was from Chemicon. In house anti-LRP1 and anti-LDLR antibodies have been described previously (Bu et al., 1995; Li et al., 2005; Zerbinatti et al., 2004). Peroxidase-labeled anti-mouse antibody and ECL system were from GE Healthcare. Carrier-free Na125I was purchased from Perkin Elmer Lifescience. The γ-secretase inhibitors L685,458 and DAPT were from Calbiochem and DFK167 was from Enzyme Systems.
Animals and Tissue Preparation
LRP1 forebrain knockout mice were generated by breeding the LRP1 loxP mice (Rohlmann et al., 1998) with (α-calcium-calmodulin-dependent kinase II-driven Cre recombinase mice [Tsien, 1996 #1326). Littermates of LRP1 forebrain knockout (LRP1flox+/+, Cre+/−) or WT controls (LRP1flox+/+, Cre−/−) at 11 months of age were used for Western blotting, immunofluorescence staining and apoE/cholesterol assays. APP-KO, APP/APLP2-DKO, and WT littermate control mice have been described previously and were used within 24 h after birth due to potential lethality of the APP/APLP2 mice. APP-KO mice were also analyzed at 4 months of age. PS1/2-DKO mice were generated by Cre-lox conditional deletion of the PS1 gene in forebrain of the PS2-KO mice (Feng et al., 2004) and were used at 4 months of age. BACE1-KO mice (Luo et al., 2001) were described in previous work and were used at 2 months of age. Animals were perfused with PBS-heparin (3 units/ml) and brain tissues were dissected and kept frozen at −80°C until further analysis. All animal procedures were approved by the Animal Study Committee at Washington University School of Medicine and in accordance with the regulations of the American Association for the Accreditation of Laboratory Animal Care.
Reverse Transcriptase Real-time PCR
Total RNA was isolated from tissues or cells using the SV Total RNA Isolation System (Promega) and subjected to DNase I digestion to remove contaminating genomic DNA. Total RNA was dissolved in nuclease-free water and stored at −80°C. Reverse transcription was performed using a SuperScript II RNase H-reverse transcriptase (Invitrogen), and the reaction mix was subjected to quantitative real-time PCR to detect levels of the corresponding actin, LRP1, or LDLR. The set of actin primers was used as an internal control for each specific gene amplification. The relative levels of expression were quantified and analyzed by using Bio-Rad iCycler iQ software. The real-time value for each sample was averaged and compared using the CT method, where the amount of target RNA (2−ΔΔCT) was normalized to the endogenous actin reference (ΔCT) and related to the amount of target gene in tissue cells, which was set as the calibrator at 1.0.
ApoE ELISA
The sandwich ELISA for mouse apoE has been described previously (Wahrle et al., 2004). Briefly, 96-well plates were coated overnight with apoE antibody (WU E4), washed with PBS, blocked with 1% milk in PBS, and then washed again. Cells or brain samples were sonicated in 5 M guanidine HCl with 1X Complete protease inhibitor mixture (Roche Applied Science), debris was pelleted by centrifugation at 10,000 X g, and the supernatant was diluted in 0.1% BSA, 0.025% Tween-20 in PBS. Following sample incubation, the plate was washed, and 3 μg/well of biotinylated goat anti-apoE (Calbiochem) was added. After incubation with the secondary antibody, the plate was washed, and poly-horseradish peroxidase streptavidin (Pierce) was added at 1:6000 dilution and incubated. The plate was then washed, developed with tetramethylbenzidine (Sigma), and read at 650 nm with a Biotek 600 plate reader (Bio-Tek Instruments).
Cholesterol Analyses
Cells or brain samples were prepared for cholesterol analysis by sonication in PBS with 1X Complete protease inhibitor mixture. The homogenized whole cell or brain suspension was then subjected to enzymatic analysis for total cholesterol using the Amplex Red Cholesterol Kit (Invitrogen) (Wahrle et al., 2004).
Immunofluorescence Staining
Frozen tissue sections were blocked with 0.1% Tween-20, 5% BSA in PBS for 30 min and stained for 2 h at room temperature with anti-LRP1 antibody. Primary antibody was then visualized using Alexa488-labeled goat anti-mouse secondary antibody (Invitrogen). Neurons were counterstained with anti-NeuN (Calbiochem) and Alexa633 labeled secondary antibody (Invitrogen). Fluorescent images were captured with a confocal microscope (Olympus Fluoview 500).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using a chromatin immunoprecipitation (ChIP) assay kit (Upstate) according to the manufacturer's instructions with minor modifications. Briefly, brain tissue from WT C57BL/6J mice were minced into small pieces with a razor and 1% formaldehyde was added directly to the tissue mixture to cross-link proteins to DNA. Tissue was then lysed in SDS lysis buffer and sonicated to shear DNA to a size range of 200-1000 bp. After centrifugation, the supernatant was diluted 10-fold in ChIP dilution buffer and incubated overnight at 4°C with anti-APP, anti-Fe65, anti-Tip60, or normal rabbit IgG. Protein A-agarose beads were used to immunoprecipitate the antibody/protein/DNA complexes. After washing, the complex was incubated at 65°C for 4 h to reverse the protein/DNA cross-links. The DNA was then purified using PCR Purification kit (Qiagen) and used as template for PCR amplification. LRP1-F (5'-TCGGGTGTCCCTGTTTAC-3') and LRP1-R (5'-GAAAGCGGTCCAAGAGTG-3') primers were used to amplify the LRP1 promoter by RT-PCR. LRP1-F (5'-GGGAGCCTGAAATCCTAGAG-3') and LRP1-R (5'-GGAAAGCGGTCCAAGAGTG-3') primers were used to amplify LRP1 promoter by real-time PCR. Primers for HES1 promoter amplification are: HES1-F, 5'-CGTGTCTCTTCCTCCCATTG-3'; HES1-R, 5'- GATCCAGTGTGATCCGCAGG-3'. PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining.
Luciferase Assay
BHK570 cells or MEF APP/APLP2-DKO cells were transfected with the appropriate cDNAs: empty vector (pGL3-luc), LRP1 promoter-luc, AICD, and/or Fe65. A β-gal reporter cDNA was co-transfected to normalize data for transfection efficiency. Twenty-four hours after transfection, cells were rinsed, gently scraped into PBS (pH 7.4), and pelleted. Cells were then lysed in lysis buffer, and the luciferase activity and β-gal activity were measured by the Luciferase Assay System and β-gal Assay System following the manufacturer's instructions (Promega).
Statistical Analysis
All quantified data represent an average of at least triplicate samples. Error bars represent standard error of the mean. Statistical significance was determined by Student's t-test and P<0.05 was considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jane Knisely and Alan Schwartz for the critical reading of the manuscript and members of the Bu lab for comments and suggestions. We also thank Suzanne Wahrle, David Holtzman, Raphael Kopan, Sheila Stewart and Bart De Strooper for providing valuable reagents. This work was supported by NIH grant R01 AG027924, a grant from the Alzheimer's Association, and a grant from the American Health Assistant Foundation to G.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abad-Rodriguez J, Ledesma MD, Craessaerts K, Perga S, Medina M, Delacourte A, Dingwall C, De Strooper B, Dotti CG. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J. Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G, Geuze HJ, Strous GJ, Schwartz AL. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 1995;14:2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G, Maksymovitch EA, Schwartz AL. Receptor-mediated endocytosis of tissue-type plasminogen activator by low density lipoprotein receptor-related protein on human hepatoma HepG2 cells. J. Biol. Chem. 1993;268:13002–13009. [PubMed] [Google Scholar]
- Cam JA, Zerbinatti CV, Li Y, Bu G. Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the β-amyloid precursor protein. J. Biol. Chem. 2005;280:15464–15470. doi: 10.1074/jbc.M500613200. [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC. Dissection of amyloid-β precursor protein-dependent transcriptional transactivation. J. Biol. Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- Chen AC, Selkoe DJ. Response to: Pardossi-Piquard et al., “Presenilin-dependent transcriptional control of the abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP.” Neuron 46, 541-554. Neuron. 2007;53:479–483. doi: 10.1016/j.neuron.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, Xu F, et al. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Feng R, Wang H, Wang J, Shrom D, Zeng X, Tsien JZ. Forebrain degeneration and ventricle enlargement caused by double knockout of Alzheimer's presenilin-1 and presenilin-2. Proc. Natl. Acad. Sci. USA. 2004;101:8162–8167. doi: 10.1073/pnas.0402733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Demattos RB, McCormick LM, O'Dell M,A, Spinner ML, Bales KR, Paul SM, et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J. Biol. Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- Grimm MOW, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape J-A, et al. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat. Cell Bio. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- Guenette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J. 2006;25:420–431. doi: 10.1038/sj.emboj.7600926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka H, Katoh-Fukui Y, Suzuki K, Kobayashi M, Suzuki R, Motegi Y, Nakahara Y, et al. Altered cholesterol metabolism in human apolipoprotein E4 knock-in mice. Hum. Mol. Genet. 2000;9:353–361. doi: 10.1093/hmg/9.3.353. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu. Rev. Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- Herz J, Chen Y. Reelin, lipoprotein receptors and synaptic plasticity. Nat. Rev. Neurosci. 2006;7:850–859. doi: 10.1038/nrn2009. [DOI] [PubMed] [Google Scholar]
- Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- Herz J. Overview: the long and winding road to understanding Alzheimer's disease. Neuron. 2007;53:477–479. doi: 10.1016/j.neuron.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Molecular and cellular mechanisms of apolipoprotein E4 neurotoxicity and potential therapeutic strategies. Curr. Opin. Drug Discov. Devel. 2006;9:627–641. [PubMed] [Google Scholar]
- Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, et al. Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J. Clin. Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to β-amyloid. J. Biol. Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- Ledesma MD, Abad-Rodriguez J, Galvan C, Biondi E, Navarro P, Delacourte A, Dingwall C, Dotti CG. Raft disorganization leads to reduced plasmin activity in Alzheimer's disease brains. EMBO Rep. 2003;4:1190–1196. doi: 10.1038/sj.embor.7400021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma MD, Dotti CG. Amyloid excess in Alzheimer's disease: What is cholesterol to be blamed for? FEBS Lett. 2006;580:5525–5532. doi: 10.1016/j.febslet.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen J, Lu W, McCormick LM, Wang J, Bu G. Mesd binds to mature LDL-receptor-related protein-6 and antagonizes ligand binding. J. Cell Sci. 2005;118:5305–5314. doi: 10.1242/jcs.02651. [DOI] [PubMed] [Google Scholar]
- Li Y, Marzolo MP, Kerkhof P, Strous GJ, Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal For LDL receptor-related protein (LRP) J. Biol. Chem. 2000;275:17187–17194. doi: 10.1074/jbc.M000490200. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, et al. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat. Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Moestrup SK, Gliemann J, Pallesen G. Distribution of the alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein in human tissues. Cell Tiss. Res. 1992;269:375–382. doi: 10.1007/BF00353892. [DOI] [PubMed] [Google Scholar]
- Muller T, Concannon CG, Ward MW, Walsh CM, Tirniceriu AL, Tribl F, Kogel D, Prehn JHM, Egensperger R. Modulation of gene expression and cytoskeletal dynamics by the amyloid precursor protein intracellular domain (AICD) Mol. Biol. Cell. 2007;18:201–210. doi: 10.1091/mbc.E06-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardossi-Piquard R, Dunys J, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Sevalle J, Pimplikar S, St George-Hyslop P, Checler F. Response to Correspondence: Pardossi-Piquard et al., “Presenilin-dependent transcriptional control of the abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP.” Neuron 46, 541-554. Neuron. 2007;53:483–486. doi: 10.1016/j.neuron.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Ring S, et al. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Park I-H, Hwang EM, Hong HS, Boo JH, Oh SS, Lee J, Jung MW, et al. Lovastatin enhances Abeta production and senile plaque deposition in female Tg2576 mice. Neurobiol. Aging. 2003;24:637–643. doi: 10.1016/s0197-4580(02)00155-0. [DOI] [PubMed] [Google Scholar]
- Pfrieger FW. Cholesterol homeostasis and function in neurons of the central nervous system. CMLS, Cell. Mol. Life Sci. 2003;60:1158–1171. doi: 10.1007/s00018-003-3018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priller C, Bauer T, Mitteregger G, Krebs B, Kretzschmar HA, Herms J. Synapse formation and function is modulated by the amyloid precursor protein. J. Neurosci. 2006;26:7212–7221. doi: 10.1523/JNEUROSCI.1450-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglielli L, Tanzi RE, Kovacs DM. Alzheimer's disease: the cholesterol connection. Nat. Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Hammer RE, Herz J. Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J. Clin. Invest. 1998;101:689–695. doi: 10.1172/JCI1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saganich MJ, Schroeder BE, Galvan V, Bredesen DE, Koo EH, Heinemann SF. Deficits in synaptic transmission and learning in amyloid precursor protein (APP) transgenic mice require C-terminal cleavage of APP. J. Neurosci. 2006;26:13428–13436. doi: 10.1523/JNEUROSCI.4180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Choi S-Y, Beglopoulos V, Malkani S, Zhang D, Rao BSS, Chattarji S, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ., III The presenilin hypothesis of Alzheimer's disease: Evidence for a loss-of-function pathogenic mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobab LA, Hsiung G-YR, Feldman HH. Cholesterol in Alzheimer's disease. Lancet Neurol. 2005;4:841–852. doi: 10.1016/S1474-4422(05)70248-9. [DOI] [PubMed] [Google Scholar]
- Spires TL, Meyer-Luehmann M, Stern EA, McLean PJ, Skoch J, Nguyen PT, Bacskai BJ, Hyman BT. Dendritic spine abnormalities in amyloid precursor protein transgenic mice demonstrated by gene transfer and intravital multiphoton microscopy. J. Neurosci. 2005;25:7278–7287. doi: 10.1523/JNEUROSCI.1879-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J. Cell Sci. 2004;117:4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, Chang P, Zaidi T, Gong Y-D, Zhao N-M, et al. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-like protein 2. J. Neurosci. 2005;25:1219–1225. doi: 10.1523/JNEUROSCI.4660-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbinatti CV, Bu G. LRP and Alzheimer's disease. Rev. Neurosci. 2005;16:123–135. doi: 10.1515/revneuro.2005.16.2.123. [DOI] [PubMed] [Google Scholar]
- Zerbinatti CV, Wahrle SE, Kim H, Cam JA, Bales K, Paul SM, Holtzman DM, Bu G. Apolipoprotein E and low density lipoprotein receptor-related protein facilitate intraneuronal Abeta42 accumulation in amyloid model mice. J. Biol. Chem. 2006;281:36180–36186. doi: 10.1074/jbc.M604436200. [DOI] [PubMed] [Google Scholar]
- Zerbinatti CV, Wozniak DF, Cirrito J, Cam JA, Osaka H, Bales KR, Zhuo M, Paul SM, Holtzman DM, Bu G. Increased soluble amyloid-beta peptide and memory deficits in amyloid model mice overexpressing the low-density lipoprotein receptor-related protein. Proc. Natl. Acad. Sci. USA. 2004;101:1075–1080. doi: 10.1073/pnas.0305803101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H. Presenilin/γ-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc. Natl. Acad. Sci. USA. 2007;104:10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Koo E. The amyloid precursor protein: beyond amyloid. Molecular Neurodegeneration. 2006;1:5. doi: 10.1186/1750-1326-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. Neurovascular mechanisms of Alzheimer's neurodegeneration. Trends Neurosci. 2005;28:202–208. doi: 10.1016/j.tins.2005.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.