Abstract
We describe a bacterial two-hybrid system that allows an easy in vivo screening and selection of functional interactions between two proteins. This genetic test is based on the reconstitution, in an Escherichia coli cya strain, of a signal transduction pathway that takes advantage of the positive control exerted by cAMP. Two putative interacting proteins are genetically fused to two complementary fragments, T25 and T18, that constitute the catalytic domain of Bordetella pertussis adenylate cyclase. Association of the two-hybrid proteins results in functional complementation between T25 and T18 fragments and leads to cAMP synthesis. Cyclic AMP then triggers transcriptional activation of catabolic operons, such as lactose or maltose, that yield a characteristic phenotype. In this genetic test, the involvement of a signaling cascade offers the unique property that association between the hybrid proteins can be spatially separated from the transcriptional activation readout. This permits a versatile design of screening procedures either for ligands that bind to a given “bait,” as in the classical yeast two-hybrid system, or for molecules or mutations that block a given interaction between two proteins of interest.
Most biological processes involve specific protein–protein interactions. General methodologies to identify interacting proteins or to study these interactions have been developed extensively. Among them, the yeast two-hybrid system currently represents the most powerful in vivo approach to screen for polypeptides that could bind to a given target protein. Originally developed by Fields and coworkers (1, 2), it utilizes hybrid genes to detect protein–protein interactions by means of activation of a reporter-gene expression (3, 4). In essence, the two putative protein partners are genetically fused to the DNA-binding domain of a transcription factor and to a transcriptional activation domain, respectively. A productive interaction between the two proteins of interest will bring the transcriptional activation domain in the proximity of the DNA-binding domain and will trigger the transcription of an adjacent reporter gene (usually lacZ or a nutritional marker), giving a screenable phenotype. Very recently, Rossi et al. (5) described a different approach, a mammalian “two-hybrid” system, which uses β-galactosidase complementation to monitor protein–protein interactions in intact eukaryotic cells.
A bacterial equivalent of the two-hybrid system has not yet been reported. Phage display (6, 7) and double-tagging assay (8) represent alternative approaches to screen complex libraries of proteins for direct interaction with a given ligand. However, these techniques do not allow an in vivo selection of the relevant clones.
We describe here a two-hybrid system in Escherichia coli in which the proteins of interest are genetically fused to two complementary fragments of the catalytic domain of Bordetella pertussis adenylate cyclase (9, 10). Interaction between the two proteins results in functional complementation between the two adenylate cyclase fragments leading to cAMP synthesis, which, in turn, can trigger the expression of several resident genes. Using this assay, one can select specific clones expressing a protein that interacts with a given target, by a simple genetic screening.
MATERIALS AND METHODS
Strain and Growth Media.
DHP1 is an adenylate cyclase-deficient (cya) derivative of DH1 (F−, glnV44(AS), recA1, endA1, gyrA96 (Nalr), thi1, hsdR17, spoT1, rfbD1) (25) and was isolated by using phosphomycin as a selection antibiotic (11). Growth media used were the rich Luria–Bertani (LB) medium or the synthetic medium M63 (12) supplemented with 1% carbon source. Antibiotic concentrations were as follows: 100 μg/ml ampicillin and 30 μg/ml chloramphenicol. Screening for the ability to ferment sugars was performed either on MacConkey agar plates containing 1% maltose, or on LB plates containing 40 μg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 0.5 mM isopropyl-β-d-thiogalactopyranoside.
Plasmids.
The plasmid pCm-AHL1 is an expression vector for the full catalytic domain of adenylate cyclase. It was generated by subcloning a 1.5-kb PvuII fragment from pACTΔC1322 (13) containing the first 384 codons of cyaA under the control of the lac UV5 promoter [cAMP/catabolite gene activator protein (CAP)-independent], into pACYC184 linearized by EcoRV-HincII. Plasmid pT25 is a derivative of pACYC184 that encodes the T25 fragment of CyaA (amino acids 1–224) in frame with a multicloning site sequence, under the control of the lac UV5 promoter. Plasmid pT18 is a derivative of pBluescript II KS (Stratagene), compatible with pT25, that encodes the T18 fragment of CyaA (amino acids 225–399) in frame with the multicloning site sequence of pBluescript II KS. The leucine zipper region of the yeast protein GCN4 was amplified by PCR from plasmid pHB16 (14) (a gift from H. Bedouelle, Institut Pasteur) using primers: lzip1 (CTGCAGGTACCTATCCAGCGTATGAAA) and lzip2 (TGAGGGTACCCCACGTTCACCCACCAG). The amplified sequence was cleaved by KpnI and cloned into the KpnI site of pT25 and pT18 to yield plasmids pT25-zip and pT18-zip, which encode, respectively, the T25 and T18 fragments fused in frame with this 35-aa-long leucine zipper. Plasmids pT25-Tyr and pT18-Tyr express, respectively, the T25 and T18 fragments fused in frame with the first 302 aa of the dimeric tyrosyl-tRNA synthetase from Bacillus stearothermophilus (15). The corresponding part of the tyrRS gene was amplified by PCR from plasmid M13EL (15) (a gift from H. Bedouelle) by using primers tyrS1 (AGAGGTACCGGACATGGATTTGCT) and tyrS2 (GCCGGTACCGCCGCTGTCAAATTGGC), cleaved by KpnI and cloned into the KpnI site of pT25 and pT18. The plasmid pT25-prp11 that expresses the T25 fused to the yeast-splicing factor Prp11 (16) was constructed as follows. First, we constructed pT25–2, a derivative of pT25 by inserting the sequence GCCCGGGG between the PstI and BamHI site of pT25, to change the reading frame of the BamHI site. Then, a 0.9-kb BamHI fragment of plasmid pPL253 (a gift from P. Legrain, Institut Pasteur) encompassing the prp11 gene was subcloned into the corresponding site of pT25–2. To construct plasmid pT18-prp21, expressing T18 fused to the yeast splicing factor Prp21 (16), the gene encoding the Prp21 protein was amplified by PCR from plasmid pPL182 (a gift from P. Legrain) by using primers CCCGGTACCGATGGAACCAGAAGATAC and CCCGGTACCAGTTTTTTACTTTTCTTTAACC, cleaved by KpnI and cloned into the KpnI site of pT18.
Analytical Methods.
β-galactosidase assays were performed on toluenized bacterial suspensions, as described in Pardee et al. (17). One unit of activity corresponds to 1 nmol of o-nitrophenyl β-d-galactoside hydrolyzed per min at 28°C.
Bacterial extracts were prepared from cells grown overnight at 30°C in LB medium supplemented with ampicillin and chloramphenicol. Pelleted cells were resuspended in 8 M urea/20 mM Hepes-Na, pH 7.5, and disrupted by sonication. Adenylate cyclase assays were performed on extracts as described previously (9) in the presence of 1 μM of calmodulin. One unit of activity corresponds to 1 nmol of cAMP formed per min at 37°C. Cyclic AMP measurements were done by an ELISA assay. Briefly, a cAMP-biotinylated-BSA conjugate was coated on ELISA plates, and nonspecific protein-binding sites were blocked with BSA. Boiled bacterial cultures were then added, followed by diluted rabbit anti-cAMP antiserum in 50 mM Hepes, pH 7.5/150 mM NaCl/0.1% Tween 20 (HBST buffer) containing 10 mg/ml BSA. After overnight incubation at 4°C, the plates were washed extensively with HBST, then goat anti-rabbit IgG coupled to alkaline phosphatase (AP) was added and incubated for 1 hr at 30°C. After washing, the AP activity was revealed by 5′-para-nitrophenyl phosphate. cAMP concentrations were calculated from a standard curve established with known concentrations of cAMP diluted in LB medium.
RESULTS
Principle.
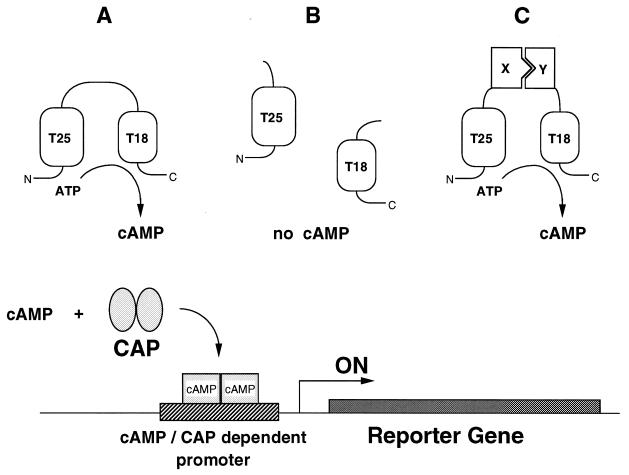
B. pertussis produces a calmodulin-dependent adenylate cyclase toxin encoded by the cyaA gene (18–20). The catalytic domain is located within the first 400 aa of this 1,706-residue-long protein (10, 19). It exhibits a high catalytic activity (kcat = 2,000 s−1) in the presence of calmodulin (CaM), and a low but detectable activity (kcat = 2 s−1) in the absence of this activator (9, 21). Biochemical studies revealed that the catalytic domain can be proteolytically cleaved into two complementary fragments, T25 and T18, which remain associated in the presence of CaM in a fully active ternary complex (9, 10, 22). In the absence of CaM, the mixture of the two fragments did not exhibit detectable activity, suggesting that the two fragments are not able to reassociate to yield basal CaM-independent activity. We reasoned that when expressed in an adenylate cyclase-deficient E. coli strain (E. coli lacks CaM or CaM-related proteins), the T25 and T18 fragments fused to putative interacting proteins would reassociate and lead to cAMP synthesis (Fig. 1).
Figure 1.
Principle of an E. coli two-hybrid system based on functional complementation of CyaA fragments. (Upper) Schematic of the basic principle of in vivo complementation between the two fragments of the catalytic domain of B. pertussis adenylate cyclase. The two boxes represent the T25 and T18 fragments corresponding to amino acids 1–224 and 225–399 of the CyaA protein. In A, the full-length catalytic domain (residues 1–399), when expressed in E. coli, exhibits a basal calmodulin-independent activity that results in cAMP synthesis. In B, the two fragments, T25 and T18, when coexpressed as independent polypeptides, are unable to interact and no cAMP synthesis occurs. In C, the two fragments, fused to two interacting proteins, X and Y, are brought into close proximity, resulting in functional complementation followed by cAMP production. (Lower) Schematic of the readout of the complementation. cAMP, synthesized in an E. coli cya strain by the complementing T25 and T18 pairs, binds to the catabolite gene activator protein, CAP. The cAMP/CAP complex then can recognize specific promoters and switch on the transcription of the corresponding genes. These reporter genes can be either natural E. coli genes, such as lacZ or mal genes, or synthetic ones, such as antibiotic-resistance genes fused to a cAMP/CAP-dependent promoter.
Functional analysis of B. pertussis adenylate cyclase activity can be easily monitored in an E. coli strain deficient in endogenous adenylate cyclase. In E. coli, cAMP bound to the transcriptional activator, CAP, is a pleiotropic regulator of the expression of various genes, including genes involved in the catabolism of carbohydrates such as lactose or maltose (23). Hence, E. coli strains lacking cAMP are unable to ferment lactose or maltose. When the entire catalytic domain of CyaA (amino acids 1–399) is expressed in E. coli cya, under the transcriptional and translational control of lacZ (plasmid pDIA5240), its calmodulin-independent residual activity is sufficient to complement an adenylate cyclase-deficient strain and to restore its ability to ferment lactose or maltose (24). This can be scored either on indicator plates (i.e., LB–X-gal or MacConkey media supplemented with maltose) or on selective media (minimal media supplemented with lactose or maltose as unique carbon source).
Design of a Two-Hybrid System Based on Functional Complementation Between T25 and T18 Fragments of CyaA.
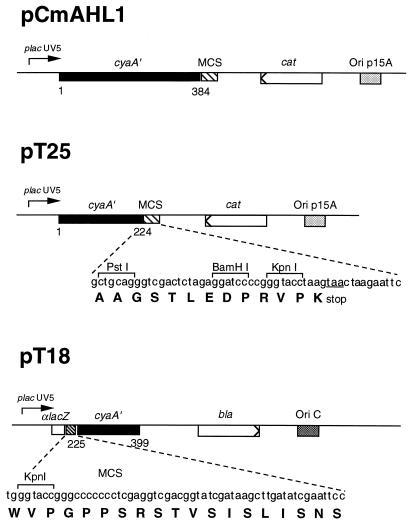
We first constructed two compatible plasmids (derived from pACYC184 and pBluescript II KS) that express either the T25 fragment corresponding to amino acids 1–224 of CyaA or the T18 fragment corresponding to amino acids 225–399. A multicloning site was fused to the C-terminal end of T25 to facilitate construction of fusions with foreign proteins. Similarly, the T18 fragment was fused in frame to αlacZ of pBluescript II KS, downstream of its multicloning site (Fig. 2).
Figure 2.
Schematic representation of plasmids. The open boxes represent the ORFs of β-lactamase (bla) and chloramphenicol acetyl transferase (cat) genes. The solid boxes correspond to the ORF of cyaA′, with codon numbers indicated below. The hatched boxes correspond to the multicloning site sequences (MCS) that are fused at the indicated position of the cya ORF. The origin of replication of the plasmids is indicated by shaded boxes.
The two plasmids, pT25 and pT18, were cotransformed in DHP1, a cya derivative of the E. coli strain DH1 (25), and plated on MacConkey agar supplemented with maltose. As expected, no spontaneous complementation between the two isolated (independently expressed) fragments could be detected in vivo: all the transformants were white (see Table 1). When the DHP1 strain was transformed with a plasmid expressing the full catalytic domain, all colonies were red (Table 1).
Table 1.
Analysis of complementation in DHP1 strain
| Plasmids | Phenotype on MacConkey/maltose | β-Galactosidase, units/mg dry weight bacteria | cAMP, pmol/mg dry weight bacteria | Adenylate cyclase activity*
|
|
|---|---|---|---|---|---|
| +CaM | −CaM | ||||
| None | White | 179 | <10 | <1 | <0.01 |
| pCm-AHL1 | Red/24 hr | 6,650 | 3,400 | 13,000 | 10 |
| pT25 + pT18 | White/72 hr | 130 | <10 | <1 | <0.01 |
| pT25 + pT18-zip | White/72 hr | 183 | <10 | <1 | <0.01 |
| pT25-zip + pT18 | White/72 hr | 178 | <10 | <1 | <0.01 |
| pT25-zip + pT18-zip | Red/30 hr | 4,750 | 1,100 | 10,000 | 4 |
Bacteria were grown in LB at 30°C in the presence of 0.5 mM isopropyl β-d-thiogalactoside plus appropriate antibiotics. The results represent the average values obtained for at least five independent cultures, which differed by less than 10%.
nmol cAMP/min per mg protein; when present in the assays, CaM was at a concentration of 1 μM.
To test whether functional complementation between T25 and T18 could be brought about by fusing them to interacting proteins, we inserted, within the multicloning site of both pT25 and pT18, a DNA sequence that codes for a 35-aa-long leucine zipper derived from protein GCN4, a yeast transcriptional activator (14). When the resulting plasmids, pT25-zip and pT18-zip were cotransformed in DHP1 and plated on MacConkey/maltose media, the resulting colonies became red after 24–30 hr of growth at 30°C (Table 1).
Control experiments were carried out in which pT25-zip was cotransformed with pT18 or pT18-zip was cotransformed with pT25. None of the transformants exhibited complementation, demonstrating that the functional complementation of T25-zip and T18-zip was mediated by the interaction of their leucine zipper motif. The efficiency of complementation could be further quantified by measuring in liquid cultures either cAMP levels or β-galactosidase activities (Table 1).
Adenylate cyclase activities of the different transformants were measured in cell extracts in the presence of CaM that binds tightly to T25 and T18 fragments to form the active adenylate cyclase complex. As shown in Table 1, only the extract from DHP1/pT25-zip/pT18-zip exhibited a significant enzymatic activity. In the absence of CaM, adenylate cyclase activity still could be detected in the extract of DHP1/pT25-zip/pT18-zip, indicating that the noncovalent association of T25-zip and T18-zip, mediated by their leucine zipper moiety, was able to restore the basal enzymatic activity, which was sufficient to sustain in vivo cAMP synthesis (Table 1). The lack of adenylate cyclase activity in the extracts of the three other types of transformants (Table 1) indicates that at least one of the two complementary fragments of adenylate cyclase was missing, most probably as a consequence of its in vivo proteolytic degradation. Therefore, it would appear that the association of T25-zip and T18-zip, through their leucine zipper motif, not only resulted in their functional complementation but also in their stabilization. Stabilization of protein fragments (α and ω peptides) through complementation (26) has also been observed for β-galactosidase (A.U., unpublished results).
Screening for in Vivo Protein–Protein Interactions by Using Functional Complementation of T25 and T18.
We then examined whether the complementation between T25 and T18 could be used to analyze interactions between proteins larger than the 35-residue-long leucine zipper motif. A DNA fragment that encodes the N-terminal part (residues 1–302) of the dimeric tyrosyl tRNA synthetase from Bacillus stearothermophilus (15) was subcloned into the multicloning site of plasmids pT25 and pT18. The resulting plasmids, pT25-TyrRS and pT18-TyrRS, when cotransformed in DHP1, yielded red transformants on MacConkey/maltose. The transformants synthesized cAMP and expressed β-galactosidase (Table 2). Control transformations confirmed that the TyrRS moiety was responsible for the functional complementation between T25-TyrRS and T18-TyrRS (Table 2). Furthermore, as shown in Table 2, no complementation occurred when T25-TyrRS was cotransformed with pT18-zip or vice versa. This demonstrates that the complementation was dictated by the specificity of recognition of the polypeptides fused to the two fragments, T25 and T18. It also demonstrates that functional complementation and stabilization of the chimeric proteins occur only upon specific interactions between the two partners.
Table 2.
Complementation between various chimeric proteins
| Plasmids | Phenotype on MacConkey/maltose | β-Galactosidase, units/mg dry weight bacteria | cAMP, pmol/mg dry weight bacteria |
|---|---|---|---|
| pT25-Tyr + pT18-Tyr | Red/40 hr | 2,800 | 580 |
| pT25-Tyr + pT18 | White/96 hr | 193 | <10 |
| pT25 + pT18-Tyr | White/96 hr | 183 | <10 |
| pT25-Tyr + pT18-zip | White/96 hr | 134 | <10 |
| pT25-zip + pT18-Tyr | White/96 hr | 126 | <10 |
| pT25-prp11 + pT18-prp21 | Red/40 hr | 850 | 65 |
Bacteria were grown in LB at 30°C in the presence of 0.5 mM isopropyl β-d-thiogalactoside plus appropriate antibiotics. The results represent the average values obtained for at least five independent cultures.
We further showed (Table 2) that the bacterial two-hybrid system could detect interaction between the yeast-splicing factors Prp11 and Prp21 (fused to T25 and T18, respectively) that was characterized previously in the yeast two-hybrid assay (16). This demonstrates that this bacterial complementation assay can reveal association between eukaryotic proteins.
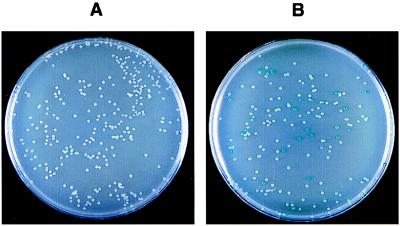
To mimic a screening procedure we mixed plasmids pT18-zip and pT18-TyrRS with about a 5-fold excess of pT18 and cotransformed this mixture in DHP1 with either pT25 or pT25-zip. The transformants were plated on LB–X-gal. All the colonies cotransformed with pT25 were white (Fig. 3). Around 20% of the colonies were blue when the cells were cotransformed with the mixture of pT18 derivatives and pT25-zip. The plasmid DNAs of these clones were further analyzed by restriction mapping. As expected, the blue colonies among the bacteria cotransformed with pT25-zip harbored only pT18-zip. In another series of experiments, pT18-zip was mixed with a 1,000-fold excess of pT18 and this mixture was transformed in DHP1 harboring pT25-zip and plated on MacConkey/maltose. Three red colonies were identified among about 3,000 white ones. Plasmid DNA analysis of the Mal+ clones confirmed the presence of pT18-zip. Transformation of the same mixture of pT18-zip/pT18 into DHP1 harboring pT25 gave no Mal+ clones of 10,000 analyzed (data not shown). These results indicate that the functional complementation between the adenylate cyclase fragments could be used to identify interacting proteins in E. coli.
Figure 3.
Screening of interacting proteins with the bacterial two-hybrid system. DHP1 cells were cotransformed with a mixture of plasmids pTI8, pT18-zip, and pT18-Tyr and either pT25 (A) or pT25-zip (B), plated on LB–X-gal agar plates containing 0.5 mM isopropyl-β-d-thiogalactopyranoside, ampicillin, and chloramphenicol, and incubated for 30 hr at 30°C. Note that the cya+ colonies are larger than the cya ones.
Finally, we examined whether the complementation between T25 and T18 could be used in a selection procedure rather than using the screening described above. DHP1 bacteria cotransformed with complementing plasmids (pT25-zip/pT18-zip or pT25-TyrRS/pT18-TyrRS) were able to grow on minimal medium supplemented with lactose or maltose as unique carbon sources, whereas bacteria cotransformed with noncomplementing plasmids (pT25-zip/pT18-TyrRS or pT25-TyrRS/pT18-zip) did not grow (data not shown). To determine whether this selection could be used to identify interacting proteins among an excess of noninteracting ones, we performed the following “model screening” on selective media. DHP1 bacteria harboring pT25-zip and pT18-zip (expected phenotype: Lac+) were mixed with a 105 excess of DHP1/pT25/pT18 (expected phenotype: Lac−), and then 107 cells from this mixture were plated on minimal medium supplemented with lactose plus antibiotics. After 4–5 days at 30°C, 100–200 Lac+ colonies appeared. Plasmid DNA analysis indicated that 18 of 20 of these colonies tested harbored pT25-zip and pT18-zip. When 107 DHP1/pT25/pT18 cells were plated on minimal medium/lactose, about 10 colonies were detected: these cells appeared to represent spontaneous revertants of DHP1 to a Lac+ phenotype (because of either reversion of cya to cya+ or to cAMP/CAP-independent lac promoter mutations). This “model screening” demonstrates that bacteria expressing specific interacting proteins fused to the adenylate cyclase fragments could be selected among a large number (here a 105-fold excess) of irrelevant clones.
DISCUSSION
We describe here an E. coli two-hybrid system that allows identification of interacting proteins by a simple genetic test (screening and/or selection). In our approach, the in vivo association of two interacting proteins is coupled to the production of a regulatory signaling molecule, cAMP, that in turn triggers the expression of specific reporter genes, giving rise to a selectable phenotype.
We took advantage of the modular structure of the catalytic domain of B. pertussis adenylate cyclase, which is composed of two complementary fragments, T25 and T18, that are both necessary to form an active enzyme, in the presence of CaM (9, 10). As shown here, the two fragments, when expressed in E. coli as separate entities, are unable to recognize each other and cannot reconstitute a functional enzyme. However, when T25 and T18 are fused to peptides or proteins that are able to interact, heterodimerization of these chimeric polypeptides results in a functional complementation between the adenylate cyclase fragments and, therefore, cAMP synthesis. Ultimately, cAMP, upon binding to CAP, is able to activate the transcription of catabolic operons, allowing the bacteria to ferment carbohydrates such as maltose or lactose. We have demonstrated that this bacterial two-hybrid system is able to reveal interactions between small peptides (GCN4 leucine zipper), bacterial (tyrosyl tRNA synthetase), or eukaryotic proteins (yeast Prp11/Prp21 complex).
Several characteristics of this genetic screening and/or selection provide an attractive approach to search for and analyze interacting proteins. Because our two-hybrid system involves the generation of a regulatory molecule, cAMP, the physical association of the two putative interacting proteins, can be spatially separated from the transcriptional events (activation) that are dependent on cAMP synthesis. This means that the protein–protein interaction under study does not need to take place in the vicinity of the transcription machinery as is the case for the yeast two-hybrid system. Hence, it will be possible to analyze protein interactions that occur either in the cytosol (as described here) or at the inner-membrane level. Moreover, because this genetic screen is, in essence, an assay for proximity of the fused T25 and T18 fragments, one could anticipate that this system would be particularly suitable to analyze colocalization of given proteins within multimolecular assemblies.
The readout of the complementation between T25 and T18 fragments is the transcriptional activation of cAMP/CAP-dependent genes. In this work, we took advantage of naturally occurring cAMP/CAP-dependent catabolic genes in E. coli. However, it is easy to design specific reporter cassettes in which any gene of interest is fused to a cAMP/CAP-dependent promoter. We currently are constructing such a system by using an antibiotic-resistance gene. This will facilitate the screening of complex libraries by a simple selection for antibiotic resistance. Alternatively, the reporter gene driven by cAMP/CAP could encode a toxic product. This could be particularly useful to search for chemical compounds or mutations that abolish a given interaction between the two studied proteins. This bacterial system is, therefore, particularly versatile as it offers the possibility of both positive and negative selections.
That this genetic test is carried out in E. coli greatly facilitates the screening as well as the characterization of the interacting proteins. First, it is possible to use the same plasmid constructs to screen a library to identify a putative binding partner to a given “bait,” and then to express the hybrid proteins to characterize their interaction by in vitro binding assays. Second, the high efficiency of transformation that can be achieved in E. coli allows the analysis of libraries of high complexity. This is particularly useful for (i) the screening of peptides from a library made from random DNA sequences that present an affinity for a given bait protein and (ii) the exhaustive analysis of the network of interactions between the proteins of a given organism (27, 28).
In essence, our system exploits one of the fundamental principles of signal transduction, that is, signal amplification. In the system described here we relied only on the basal enzymatic activity of B. pertussis adenylate cyclase, i.e., in the absence of CaM, which nevertheless is sufficient to synthesize enough cAMP to activate the transcription of the lac and mal operons (we estimate, from adenylate cyclase activity measured in bacterial extracts, that about a thousand of T25 and T18 molecules per cell are expressed in the present design). This system could be rendered exquisitely sensitive by using the full catalytic potency of B. pertussis adenylate cyclase, i.e., in the presence of its natural activator. In this case, it is anticipated that reconstitution of only a few hybrid molecules per cell will be sufficient to elicit a detectable signal.
Acknowledgments
We thank H. Bedouelle and P. Legrain for the gift of plasmids, fruitful discussion, and comments on the manuscript. We are grateful to B. Lefévère-Laoide for critical reading of the manuscript and to M. Berlyn for providing strain DH1. Financial support came from the Institut Pasteur and the Centre National de la Recherche Scientifique (URA 1129).
ABBREVIATIONS
- LB
Luria–Bertani
- X-gal
5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- CAP
catabolite gene activator protein
References
- 1.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen J B, Walberg M W, Edwards M C, Elledge S J. Trends Biochem Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 4.Transy C, Legrain P. Mol Biol Rep. 1995;21:119–127. doi: 10.1007/BF00986502. [DOI] [PubMed] [Google Scholar]
- 5.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith G P. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 7.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 8.Germino F J, Wang Z X, Weissman S M. Proc Natl Acad Sci USA. 1993;90:933–937. doi: 10.1073/pnas.90.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladant D. J Biol Chem. 1988;263:2612–2618. [PubMed] [Google Scholar]
- 10.Ladant D, Michelson S, Sarfati R S, Gilles A-M, Predeleanu R, Bârzu O. J Biol Chem. 1989;264:4015–4020. [PubMed] [Google Scholar]
- 11.Alper M D, Ames B N. J Bacteriol. 1978;133:149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 13.Sebo P, Ladant D. Mol Microbiol. 1993;9:999–1009. doi: 10.1111/j.1365-2958.1993.tb01229.x. [DOI] [PubMed] [Google Scholar]
- 14.Blondel A, Bedouelle H. Protein Eng. 1991;4:457–461. doi: 10.1093/protein/4.4.457. [DOI] [PubMed] [Google Scholar]
- 15.Guez-Ivanier V, Bedouelle H. J Mol Biol. 1996;255:110–120. doi: 10.1006/jmbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 16.Legrain P, Chapon C. Science. 1993;262:108–110. doi: 10.1126/science.8211114. [DOI] [PubMed] [Google Scholar]
- 17.Pardee A B, Jacob F, Monod J. J Mol Biol. 1959;1:165–168. [Google Scholar]
- 18.Hewlett E L, Urban M A, Manclark C R, Wolff J. Proc Natl Acad Sci USA. 1976;73:1926–1930. doi: 10.1073/pnas.73.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser P, Ladant D, Sezer O, Pichot F, Ullmann A, Danchin A. Mol Microbiol. 1988;2:19–30. doi: 10.1111/j.1365-2958.1988.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 20.Mock M, Ullmann A. Trends Microbiol. 1993;1:187–192. doi: 10.1016/0966-842x(93)90089-a. [DOI] [PubMed] [Google Scholar]
- 21.Wolff J, Cook G H, Goldhammer A R, Berkowitz S A. Proc Natl Acad Sci USA. 1980;77:3841–3844. doi: 10.1073/pnas.77.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munier H, Gilles A M, Glaser P, Krin E, Danchin A, Sarfati R, Barzu O. Eur J Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 23.Ullmann A, Danchin A. Advances in Cyclic Nucleotide Research. Vol. 15. New York: Raven; 1983. pp. 1–53. [Google Scholar]
- 24.Ladant D, Glaser P, Ullmann A. J Biol Chem. 1992;267:2244–2250. [PubMed] [Google Scholar]
- 25.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 26.Ullmann A, Jacob F, Monod J. J Mol Biol. 1968;32:1–13. doi: 10.1016/0022-2836(68)90140-x. [DOI] [PubMed] [Google Scholar]
- 27.Bartel P L, Roecklein J A, SenGupta D, Fields S. Nature Gen. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 28.Fromont R M, Rain J C, Legrain P. Nature Gen. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]