Abstract
Background
Bifidobacteria are found at varying prevalence in human microbiota and seem to play an important role in the human gastrointestinal tract (GIT). Bifidobacteria are highly adapted to the human GIT which is reflected in the genome sequence of a Bifidobacterim longum isolate. The competitiveness against other bacteria is not fully understood yet but may be related to the production of antimicrobial compounds such as bacteriocins. In a previous study, 34 Bifidobacterium isolates have been isolated from baby faeces among which six showed proteinaceous antilisterial activity against Listeria monocytogenes. In this study, one of these isolates, RBL67, was further identified and characterized.
Results
Bifidobacterium isolate RBL67 was classified and characterized using a polyphasic approach. RBL67 was classified as Bifidobacterium thermophilum based on phenotypic and DNA-DNA hybridization characteristics, although 16S rDNA analyses and partial groEL sequences showed higher homology with B. thermacidophilum subsp. porcinum and B. thermacidophilum subsp. thermacidophilum, respectively. RBL67 was moderately oxygen-tolerant and was able to grow at pH 4 and at a temperature of 47°C.
Conclusion
In order to assign RBL67 to a species, a polyphasic approach was used. This resulted in the classification of RBL67 as a Bifidobacterium thermophilum strain. To our knowledge, this is the first report about B. thermophilum isolated from baby faeces since the B. thermophilum strains were related to ruminants and swine faeces before. B. thermophilum was previously only isolated from animal sources and was therefore suggested to be used as differential species between animal and human contamination. Our findings may disapprove this suggestion and further studies are now conducted to determine whether B. thermophilum is distributed broader in human faeces. Furthermore, the postulated differentiation between human and animal strains by growth above 45°C is no longer valid since B. thermophilum is able to grow at 47°C. In our study, 16S rDNA and partial groEL sequence analysis were not able to clearly assign RBL67 to a species and were contradictory. Our study suggests that partial groEL sequences may not be reliable as a single tool for species differentiation.
Background
Since Tissier discovered the Bifidobacterium spp. in 1899 [1], over 30 species have been isolated and identified [2] and the first genome sequence of a Bifidobacterium is now available [3]. Analyses of amplified partial 16S rDNA sequences assigned to uncultivated bifidobacteria suggest the existence of more new Bifidobacterium species[4,5]. Bifidobacteria are known to be heterofermentative anaerobic, Gram-positive bacteria which belong to the class of Actinobacteria [6] containing genomes with a high G+C content. They mainly colonize the intestines of humans, other mammals and insects [7]. Some bifidobacteria have also been isolated from environmental sources such as sewage [8]. Bifidobacteria have been described as strictly anaerobic bacteria in the sense that they are not able to grow on agar-plates in the presence of air [7]. However, some Bifidobacterium strains were described which were at least partially aerotolerant in the presence of reducing agents in liquid media [9,10]. Genome analyses of Bifidobacterium longum suggests that both growth and survival under oxygen pressure are linked with the presence of a set of oxygen-scavenging NADH oxidases [3]. Strains of a novel species, Bifidobacterium psychraerophilum, were recently reported to be able to grow under air on the surface of solid agar-medium [11]. Based on heat-shock protein HSP60 encoding sequence homologies, this species clusters distinctly from aerotolerant species of the genera Scardovia, Aeriscardovia, Parascardovia and Gardnerella which were related or belonged to the genus Bifidobacterium before the recent reclassification [11,12]. For probiotic use of a particular Bifidobacterium strain, oxygen tolerance is an important characteristic for maintaining cell viability in end products.
The classification of Bifidobacterium species has most often been done in the past mainly by 16S rDNA sequence homology analysis [13] and confirmed by an enzymatic assay of D-fructose-6-phosphate phosphoketolase [14] whose encoding gene xfp is widespread among microorganisms and not unique to Bifidobacterium species [15]. These results were then substantiated by determinations of DNA-DNA relatedness [16] as well as physiological properties such as carbohydrate fermentation profiles in order to discriminate between single species of the genus Bifidobacterium. Recently, phylogenetic trees for bifidobacteria were constructed based on groEL genes encoding heat-shock protein HSP60 [17,18], groES genes encoding chaperon [18] and xfp encoding phosphoketolase [19,20] as alternatives to 16S rDNA-based phylogenetic trees. These trends support the polyphasic approach for species identification as suggested several years ago [21]. The phylogenetic positions of bifidobacteria groEL (synonymly used with HSP60 encoding sequences) seem to generally agree with 16S rDNA-based phylogeny, and in several studies, have been more discriminative than 16S rDNA sequences for species delineation [18]. In a previous study, 34 isolates of Bifidobacterium species from infant faeces have been described, six of which showed bacteriocin-like activity against Listeria monocytogenes which represents a rare property among bifidobacteria [22]. In this work, the taxonomic position of one of these isolates, strain RBL67, whose properties did not match with any of the so far described Bifidobacterium species, was determined using 16S rDNA sequence homology, comparative groEL gene sequence analysis, DNA-DNA genome hybridizations and carbohydrate fermentation patterns. This study aimed to classify RBL67 due to its features which may be used industrially in the future and was not intended to for a complete phylogenetic analysis of Bifidobacterium sp.
Results and discussion
Phylogenetic position of Bifidobacterium sp. RBL67
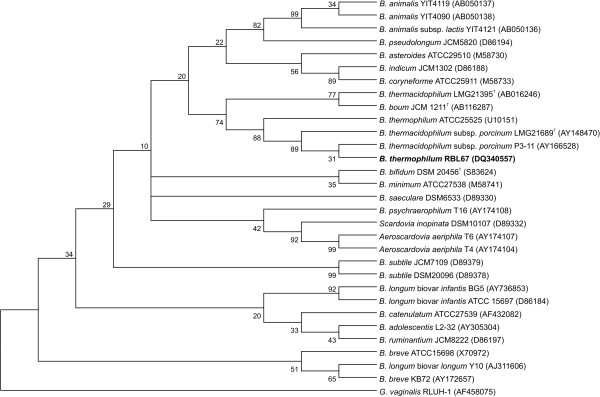
Because 16S rDNA sequences are the most widely used molecules in phylogenetic classification of bacteria, the 16S rDNA fragment of strain RBL67 was amplified by PCR using the lm3/lm26 primer pair (Table 1). The sequence of the resulting fragment (approx. 1.5 kb [Genbank:DQ340557]) showed the highest homologies to 16S rDNA sequences of other bifidobacteria from the GenBank (Table 2) with the highest similarity, 99%, to the sequence of Bifidobacterium thermacidophilum subsp. porcinum LMG21689T [GenBank: AY148470], isolated by Zhu et al. [17], and 94% to that of the Bifidobacterium thermophilum type strain [GenBank: U10151] (Table 2). A phylogenetic tree was constructed from the 16S rDNA sequences using a consensus length of 1392 kb. The tree shows clustering of RBL67 with the Bifidobacterium thermophilum/thermacidophilum/boum branch (Fig. 1), described as the "thermophilic group" by Zhu et al. [17].
Table 1.
Oligonucleotides used in this study
| Primer | Sequence (5'-3') |
| lm3 | CGGGTGCTICCCACTTTCATG [42] |
| lm26 | GATTCTGGCTCAGGATGAACG [42] |
| 520F | CAGGAGTGCCAGCAGCCGCGG [25] |
| 520R | ACCGCGGCTGCTGGC [25] |
| 1100F | CAGGAGCAACGAGCGCAACCC [25] |
| 1100R | AGGGTTGCGCTCGTT [25] |
| H60F | GGNGAYGGNACNACNACNGCNACNG [34] |
| H60R | TCNCCRAANCCNGGNGCYTTNACNGC [34] |
| T7 | TAATACGACTCACTATAGG [Promega] |
| SP6 | ATTTAGGTGACACTATAG [Promega] |
Table 2.
16S rDNA sequence homologies of Bifidobacterium sp. RBL67 with 16S rDNA sequences found in the GenBank
| Bifidobacterium sp. RBL67 [GenBank: DQ340557] compared with* | % Identity {Gaps} |
| Bifidobacterium thermacidophilum subsp. porcinum LMG 21689T complete sequence [GenBank: AY148470] | 99 {2} |
| Bifidobacterium boum JCM1211T 16S rDNA partial sequence [Genbank: D86190] | 97 {2} |
| Bifidobacterium sp. group I-3 16S ribosomal RNA gene, partial sequence [GenBank: AF321295] | 97 {5} |
| B. thermacidophilum subsp. thermacidophilum LMG21395T 16S rDNA complete sequence [GenBank: AB016246] | 96 {12} |
| Bifidobacterium saeculare DSM6533T 16S rDNA partial sequence [GenBank: D89330] | 95 {6} |
| Bifidobacterium subtile JCM 7109T 16S rDNA partial sequence [GenBank: D89329] | 95 {6} |
| Bifidobacterium thermophilum ATCC 25525T 16S rDNA partial sequence [GenBank: U10151] | 94 {8} |
*Accession numbers in brackets []
Figure 1.
Phylogenetic tree based on 16S rDNA sequences. The tree was rooted with Gardnerella vaginalis RLUH-1 and constructed by using the Neighbour-joining method with Jukes-Cantor parameter and a bootstrap value of 1000. The number at each branch point represents percentage bootstrap support. Accession numbers in brackets.
There is insufficient discriminating power to clearly assign a 16S rDNA to a species within the genus Bifidobacterium since similarity values for this gene can range from 93% to 99% between species, as reported in other studies [23-25]. Therefore a polyphasic approach, as suggested by Vandamme et al. [21], was used in this study to identify the unknown strain RBL67 at species level.
Comparative analysis of groEL gene sequences
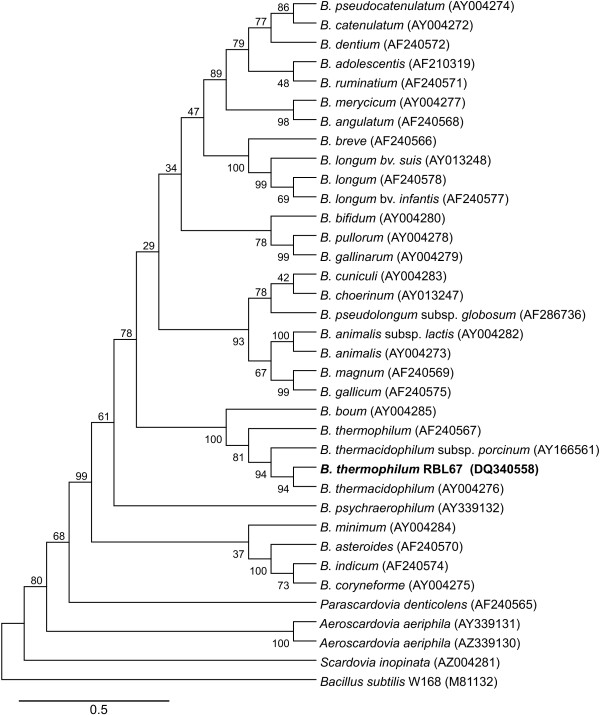
The next step of the classification process was to compare the partial groEL gene sequences (0.59 kb in size) of Bifidobacterium RBL67 [GenBank: DQ340558], B. thermacidophilum subsp. thermacidophilum LMG21395T [GenBank: AY004276] (referred as B. thermacidophilum in Fig. 1 and Fig. 2) and B. thermacidophilum subsp. porcinum LMG21689T [GenBank: AY166561] with those of other bifidobacteria from the GenBank. Based on a consensus length of 0.59 kb, a phylogenetic tree was constructed (Fig. 2) and rooted with Bacillus subtilis W168 [GenBank: M81132]. The tree shows common clustering of strain RBL67 with the Bifidobacterium thermophilum/thermacidophilum/boum branch, which substantiates the 16S rDNA comparisons presented in Fig. 1. According to the 16S rDNA tree, strain RBL67 splits off earlier on the branch and is closer related to B. thermophilum (Fig. 1 [GenBank:U10151 ]) whereas using partial groEL sequences, strain RBL67 is on the same branch as B. thermacidophilum (Fig. 2 [GenBank: AY004276]). The comparison of 16S rDNA sequences and partial groEL sequences showed a discrepancy between the classification of strain RBL67 based on those methods. By comparing the partial groEL gene sequences, Bifidobacterium RBL67 is more closely related to B. thermacidophilum subsp. thermacidophilum LMG21395T (98.25% similarity) than to B. thermacidophilum subsp. porcinum LMG21689T (97.06%, Table 3) whereas based on 16S rDNA sequences strain RBL67 is closer related to B. thermacidophilum subsp. porcinum LMG21689T (99%, Table 2) than to B. thermacidophilum subsp. thermacidophilum LMG21395T (96%). According to the definition of Zhu et al. [17] a similarity of 96.5–100% and 95.5–97% is required for intraspecies and inter-subspecies differentiation by partial groEL sequences, respectively. By applying this definition to partial groEL gene sequencing data (Table 3), Bifidobacterium RBL67 could belong to the species thermacidophilum or it could be a subspecies of B. thermophilum DSM20210T (95.65% similarity), which is contradictory. Clearly sequencing analysis of partial groEL could not provide enough evidence to confirm the phylogenetic classification of Bifidobacterium isolate RBL67.
Figure 2.
Phylogenetic tree based on fragments of the partial groEL gene DNA sequences rooted with Bacillus subtilis W168. The tree was constructed using the neighbour-joining method with Jukes-Cantor parameter and bootstrap values calculated from 1000 trees (represented as percentages at each branch-point). Accession numbers in brackets.
Table 3.
Partial groEL sequence of Bifidobacterium sp. RBL67 compared with closely related Bifidobacterium species
| Bifidobacterium sp. RBL67 groEL [GenBank: DQ340558] gene sequence compared with: | % Similarity |
| B. thermacidophilum subsp. thermacidophilum LMG 21395T [GenBank: AY004276] | 98.25% |
| B. thermacidophilum subsp. porcinum LMG 21689T [GenBank: AY166561] | 97.06% |
| B. thermophilum DSM 20210T [GenBank: AF240567] | 95.65% |
| B. boum DSM 20432T [GenBank: AY004285] | 93.58% |
DNA-DNA hybridization and G+C-content
For the final classification of Bifidobacterium RBL67, DNA-DNA hybridization was performed. The results (Table 4) confirmed that the partial groEL gene sequence was not sufficient to differentiate between species. According to the limit of 70% similarity required for species identification [26], only Bifidobacterium boum DSM20432T could be defined as a different species by DNA-DNA hybridization. B. thermacidophilum subsp. thermacidophilum LMG21395T was on the limit of this definition; its classification as a new species was justified by data from both DNA-DNA hybridization and phylogenetic data reported by Dong et al. [8]. However, our DNA-DNA hybridizations provide evidence that B. thermacidophilum subsp. porcinum LMG21689T belongs to B. thermophilum, showing 82.25% homology to B. thermophilum DSM20210T (Table 4). Bifidobacterium RBL67 showed 86.25% homology with B. thermophilum DSM20210T and could also not be discriminated as a new species. Bifidobacterium RBL67 showed even less similarity to B. thermacidophilum subsp. thermacidophilum LMG21395T and B. thermacidophilum subsp. porcinum LMG21689T than to B. thermophilum DSM20210T (Table 4). Furthermore the homologies between Bifidobacterium RBL67 and both B. thermacidophilum strains were similar (77.9% for LMG21395T and 77.2% for LMG21689T) while the homology between B. thermacidophilum subsp. thermacidophilum LMG21395T and B. thermacidophilum subsp. porcinum LMG21689T was only 71.9%. The result reported by Dong et al. [8] from DNA-DNA hybridization, 58.9% similarity between B. thermacidophilum subsp. thermacidophilum LMG21395T and B. thermophilum DSM20210T, was not confirmed in this study. This might be due to different methods of DNA isolation, DNA-DNA hybridization or interpretation of the data.
Table 4.
Similarity in percent of the DNA-DNA hybridizations of Bifidobacterium RBL67, B. thermacidophilum subsp. porcinum LMG21689T and B. thermophilum DSM2010T with closely related strains
| Bifidobacterium sp. RBL67 | B. thermacidophilum subsp. porcinum LMG21689T | B. thermophilum DSM20210T | |
| Bifidobacterium sp. RBL67 | 100% | n.d. | n.d. |
| B. thermacidophilum subsp. thermacidophilum LMG21395T | 78% | 72% | 71% |
| B. thermacidophilum subsp. porcinum LMG21689T | 77% | 100% | 82% |
| B. thermophilum DSM20210T | 86% | n. d. | 100% |
| B. boum DSM20432T | 48% | n. d. | n. d. |
n. d.: not determined
Based on the data by DNA-DNA hybridization, which is still the strongest method for bacterial species differentiation, we classified strain RBL67 as Bifidobacterium thermophilum.
The G+C content of DNA from Bifidobacterium RBL67 was 59.7 mol-% as determined by DSMZ Germany. This compared well with the G+C determination of B. thermophilum DSM20210T, 60 mol-%, but differed from that of B. thermacidophilum subsp. thermacidophilum LMG21395T and B. thermacidophilum subsp. porcinum LMG21689T, which were 57.7 and 61.5 mol-% G+C, respectively.
Physiological properties of Bifidobacterium sp. RBL67
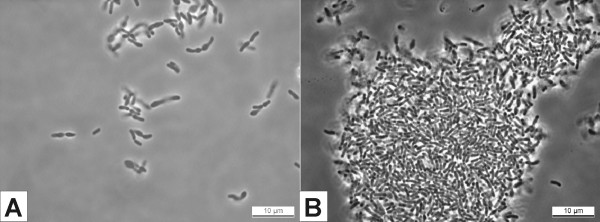
Cells of Bifidobacterium RBL67 grew well on MRS-C agar incubated anaerobically at 37°C overnight and on RB-agar at 40°C in 48 h. They were non-motile, irregularly shaped rods and formed pairs when growing on plates (Fig. 3a). On RB-agar, they formed yellow colonies with a diameter of 1 mm. When growing in liquid culture, agglomeration of cells occurred (Fig. 3b). This agglomeration was pH dependent: the lower the pH, the greater the agglomeration of cells. The formation of agglomerates was inhibited by pH control at pH 7 (data not shown). In contrast to B. thermacidophilum subsp. thermacidophilum LMG21395T, aggregated cell clumps of Bifidobacterium RBL67 were not easily dispersed again, even after extended mixing.
Figure 3.
A: Microscopic picture of Bifidobacterium RBL67 grown on MRS-C agar overnight. White bar indicates 10 μm. B: Microscopic picture of a small agglomerated clump of Bifidobacterium RBL67 cells in MRS-C liquid culture after 24 h growth at 37°C. White bar indicates 10 μm.
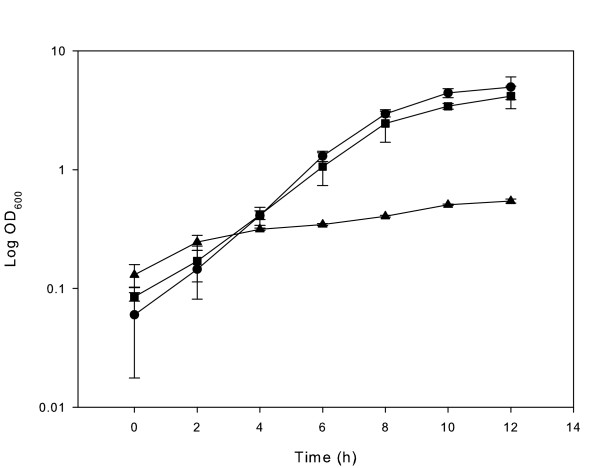
Bifidobacterium RBL67 was moderately oxygen tolerant; therefore it was not necessary to perform dilutions and inoculations anaerobically. However, the strain did not grow on agar plates under aerobic conditions after 7 days. No selected gas atmosphere was necessary to grow the strain in liquid culture. While sparkling the liquid media with oxygen-free nitrogen or carbon dioxide, Bifidobacterium RBL67 ceased to grow. To determine the level of oxygen tolerance, anaerobic media were purged with oxygen as described by Meile et al. [10]. Fig. 4 shows the growth curves for Bifidobacterium sp. RBL67 at different levels of oxygen in reduced medium. At 2.5% oxygen in the bottle growth of this strain was maintained, showing its elevated tolerance to oxygen. Even at 12.5% oxygen, the OD still increased fivefold during incubation for 12 h. B. thermacidophilum subsp. thermacidophilum LMG21395T and B. thermacidophilum subsp. porcinum LMG21689T showed similar growth curves to strain RBL67 under oxidative stress, but seemed to grow slightly better at 12.5% oxygen after 10 h of fermentation (data not shown). Nonetheless, all the strains tested were less oxygen tolerant than B. animalis subsp. lactis, which was shown to tolerate 50 ml of oxygen in comparable reduced medium [10].
Figure 4.
Growth of Bifidobacterium RBL67 under different oxygen concentrations in MRS-C at 37°C. Points are mean of three replicates. ● Growth under anaerobic conditions; ■ Growth with 2.5% oxygen; ▲ Growth with 12.5% oxygen.
Table 5 shows a summary of the fermentation profiles and growth extremes of Bifidobacterium RBL67, RBL68, RBL70, B. thermacidophilum subsp. thermacidophilum LMG21395T, B. thermacidophilum subsp. porcinum LMG21689T, B. animalis subsp. lactis DSM10140T and B. thermophilum DSM20210T. For the determination of the growth limits for pH and temperature a limit of OD600 ≥ 0.4 after 7 days incubation was set. Bifidobacterium RBL67 was found to grow in a very large pH and temperature range (Table 5). Its maximum growth temperature (47°C) was very close to that of B. thermophilum DSM20210T, although the later grew to a higher OD (data not shown). B. thermacidophilum subsp. thermacidophilum LMG21395T was the only strain in this study that grew at 49°C which confirmed the growth characteristics described by Dong et al. [8].
Table 5.
Phenotypic characteristics of the tested Bifidobacterium species: 67:Bifidobacterium sp. RBL67; 68: Bifidobacterium sp. RBL68; 70:Bifidobacterium sp. RBL70; 21689:B. thermacidophilum subsp.porcinum LMG21689T; 21395: B. thermacidophilum subsp.thermacidophilum LMG21395T; 20210: B. thermophilum DSM20210T; 10140: B. animalis subsp. lactis DSM10140T
| 67 | 68 | 70 | 21689 | 21395 | 20210 | 10140 | |
| L-arabinose | - | - | - | - | + | - | + |
| D-ribose | - | - | - | D | - | - | + |
| D-galactose | D | + | D | + | + | + | + |
| D-fructose | + | + | + | + | - | + | D |
| D-mannose | - | - | - | - | - | - | D |
| Methyl-AD-mannopyranoside | D | - | - | - | - | D | - |
| Methyl-AD-glucopyranoside | + | D | - | + | + | + | + |
| Amygdalin | - | - | - | - | D | + | + |
| Arbutin | D | - | - | D | D | + | - |
| Esculin ferric citrate | + | D | - | + | D | + | + |
| Salicin | - | - | - | D | D | + | + |
| D-lactose (bovine) | - | - | - | + | - | w | + |
| D-trehalose | - | - | D | W | - | D | - |
| Inulin | w | D | w | - | - | D | - |
| Gentiobiose | - | - | - | - | - | + | D |
| D-melezitose | - | - | D | - | - | D | - |
| Minimum growth temp. (°C) | ≤25 | ≤25 | n. d. | ≤25 | ≤25 | ≤25 | ≤25 |
| Maximum growth temp. (°C) | 47 | 47 | 47 | 47 | 49 | 47 | 46 |
| Minimum growth pH | ≤4 | ≤4.5 | ≤4 | ≤4.5 | ≤4.5 | ≤4.2 | ≤4.5 |
| DNA G+C content (mol-%) | 59.7 | n. d. | n. d. | 61.5 | 57.7 | 60 | 61.9 |
+, positive reaction; -, negative reaction; w, weak reaction; D, variable reaction → colour could not be assigned to either yellow or blue; n. d.: not determined
As for a lower pH limit, RBL67 and RBL70 were the only strains still growing at pH 4 under the growth conditions of the study. B. thermacidophilum subsp. thermacidophilum LMG21395T did not grow at this pH, which was reported by Dong et al. [8]. The upper pH limit for growth was not determined, but all tested cultures still grew very well at pH 8. The molar ratio of acetate to lactate from glucose for Bifidobacterium RBL67 was determined as 5.91 to 1 in MRS-C medium at 37°C under anaerobic conditions.
Conclusion
In order to classify the Bifidobacterium isolate RBL67 at species level, we used a polyphasic approach since phylogenetic trees based on partial groEL gene (as used by Ventura et al. [18] and Dong et al. [8]) and 16S rDNA sequences generated similar but not identical results for species classification. Berthoud et al. [20] reported similar difficulties when studying the "thermophilic group", which could not be differentiated at the species level by xfp gene sequencing. They proposed the use of 16S rDNA sequencing, as an additional method for species differentiation. However, our DNA-DNA hybridization data (Table 4) revealed a very close relationship between B. thermophilum species and B. thermacidophilum species and questioned the current classification of B. thermacidophilum as a discrete species. Our hybridization data were substantiated by phylogenetic data, molecular characterization and physiological properties and allowed the classification of strain RBL67 into the species Bifidobacterium thermophilum. In contrast with a previous report [8], among our tested strains (Table 4) only B. boum could be clearly classified as a species different from B. thermophilum based on DNA-DNA hybridization. This finding suggests that partial groEL gene sequences may not be reliable as a single tool for Bifidobacterium species differentiation. However, more studies have to be done to confirm this suggestion.
As stated before, we did not intend to perform a complete phylogenetic analysis. However, as more data (16S rDNA sequences, groEL sequences) become available on strains closely related to B. thermophilum RBL67, a complete phylogenetic analysis could be performed which may lead to a new definition of the boum/thermophilum/thermacidophilum branch.
A very important finding of this study is that B. thermophilum could be isolated from human origin (baby faeces). In previous studies, B. thermophilum has been classified as an animal-related species mainly present in ruminant faeces [27,28]. Since it was previously possible to differentiate between animal and human bifidobacteria by species identification, it has been suggested that bifidobacteria should be used to discriminate between animal and human bacterial contamination in foods [27,29]. Because B. thermophilum RBL67 was isolated from baby faeces, it is likely that this species can not be used as animal contamination indicator. On the other hand, Gavini et al. [30] detected B. adolescentis, a species predominantly found in human, in cow dung, swine and rabbit faeces which restricts the number of bifidobacterial species that can be used for human faecal contamination detection.
Another discrimination tool between human and animal bifidobacteria strains was the temperature growth limit of 45°C for strains from human origin [7,31]. B. thermophilum strains are able to grow at 47°C and since they can also be found in human faeces [this study], the use of the temperature is not reliable anymore for the discrimination of the origin of bifidobacteria. However, since only a few species are able to grow above 45°C, the assignment to a Bifidobacterium sp. can be narrowed. Further studies are now undergoing to determine whether B. thermophilum is more widespread within humans.
Methods
Bacterial strains and routine growth conditions
A list of strains used in this study is presented in Table 6. Strains were kept in their respective growth media (see below) supplemented with 30% (v/v) glycerol at -80°C. Prior to use, all bifidobacteria were grown on Raffinose-Bifidobacterium (RB)-agar plates [32], without sodium caseinate, supplemented with 1.5% (v/v) agar (Oxoid). After 5 sub cultivations they were transferred to MRS-C broth consisting of MRS with 0.1% (v/v) Tween 80 (Biolife) and 0.05% (w/v) L-cysteine-hydrochloride (Sigma) or on MRS-C agar (MRS-C broth with 1.5% (w/v) agar). Bifidobacterium bifidum DSM20456T was cultivated only on MRS-C agar as it didn't grow on RB-agar. All strains were incubated anaerobically at 37°C overnight in MRS-C broth and agar, or at 40°C for 48 h on RB-agar.
Table 6.
Strains used in this study
| Species | Strain |
| Bifidobacterium thermacidophilum subsp. thermacidophilum | LMG 21395T |
| Bifidobacterium thermacidophilum subsp. porcinum | LMG 21689T |
| Bifidobacterium adolescentis | DSM 20083T |
| Bifidobacterium bifidum | DSM 20456T |
| Bifidobacterium boum | DSM 20432T |
| Bifidobacterium breve | DSM 20213T |
| Bifidobacterium longum subtype infantis | DSM 20088T |
| Bifidobacterium longum subtype longum | DSM 20219T |
| Bifidobacterium thermophilum | DSM 20210T |
| Bifidobacterium longum subtype suis | DSM 20211 |
| Bifidobacterium animalis subsp. Lactis | DSM 10140T |
| Bifidobacterium | RBL67* |
| Bifidobacterium | RBL68* |
| Bifidobacterium | RBL70* |
| Escherichia coli | XL1-Blue‡ |
BCCM/LMG™: Belgian co-ordinated collections of micro-organisms/Laboratorium voor
Microbiology en microbiele Genetica, Ghent, Belgium
DSM: German Microorganism Collection, Braunschweig, Germany
*Isolated from baby faeces [22]; ‡ referred in Bullock et al. [41]
Growth under oxidative-, heat- and pH-stress conditions
Growth under conditions of oxidative stress was measured after a modified method described by Meile et al. [10]. Volumes of 0, 10 and 50 ml of pure oxygen were added to sterile serum flasks containing 400 ml of MRS-C. The flasks were inoculated with 1% of an overnight culture of either B. longum subtype longum DSM 20219T, Bifidobacterium RBL67, B. thermacidophilum subsp. thermacidophilum LMG 21395T or B. thermacidophilum subsp. porcinum LMG 21689T. The flasks were then incubated in a shaker at 160 rpm and 37°C for 12 h. Samples were taken every 2 h and treated for 3 min in a stomacher to remove clumps prior to measurements of the optical density at 600 nm. Each growth curve was carried out twice.
The temperature and pH extremes of Bifidobacterium RBL67, RBL68 and RBL70, B. thermacidophilum subsp. thermacidophilum LMG 21395T, B. thermacidophilum subsp. porcinum LMG 21689T, B. animalis subsp. lactis DSM 10140T and B. thermophilum DSM 20210T were determined as follows:
25 ml of MRS-C containing 2 mg l-1 resazurin as redox indicator were anaerobically inoculated with 1% of the tested bifidobacteria cultured overnight. The temperature range was evaluated by incubating the strains for 7 days at 12, 25, 46, 47, 48 and 49°C with an initial pH of 7.0. The pH range was determined using pH 4.0, 4.5, 5.0, 5.5 and 8.0 as initial pH for growth. Samples were incubated at 37°C for 7 days. Aliquots were taken daily, treated in a stomacher for 3 min and the OD was measured at 600 nm. Each growth condition was measured twice. If a strain failed to reach OD600 of 0.4 after 7 days, it was declared as not growing under the tested conditions.
Amplification and sequencing of 16S rDNA
Specific amplification of the 16S rDNA of Bifidobacterium RBL67 and other bifidobacteria was done using a slightly modified PCR protocol established by Schürch [33] using the primer pair lm3/lm26 (Table 1). The annealing temperature used was 62°C instead of 60°C. After agarose gel electrophoresis the corresponding band for 16S rDNA at the 1.5 kb position was cut out and purified using the GFX PCR purification kit (Amersham Biosciences). Sequencing of the PCR product was done by Microsynth GmbH using the primers 520F, 520R, 1100F and 1100R (Table 1). Sequence analysis and comparison was done using the GCG software package version 10 as described before [34]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 [35]. The sequences were aligned with ClustalW (version 1.6) and the tree was calculated using the neighbour-joining method with Jukes-Cantor parameter and a bootstrap value of 1000.
Cloning and analysis of groEL sequences
DNA cloning and sequencing of a partial heat shock groEL gene sequence from Bifidobacterium RBL67 were done using a modified method of Jian et al. [34]. The DNA template was extracted using the method of Leenhouts et al. [36]. To amplify part of a gene fragment with PCR, the following reaction mixture (25 μl) was used: 3–30 ng DNA template measured at 260 nm with a Eppendorf-Biophotometer, 2.5 U Taq polymerase (Euroclone), 0.1 mM dNTP's (Amersham Biosciences), 1.5 mM MgCl2, 2 μM each of primer H60R and H60F (Table 1) and 2.5 μl 10 × PCR buffer (Euroclone). The PCR reactions were carried out in a Biometra Tgradient Thermal Cycler using the following protocol: denaturation step 95°C for 5 min followed by 40 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 1 min at a heating rate of 1.5°C min-1. At the end, the temperature was maintained at 72°C for 10 min. A 25-μl aliquot of the reaction mixtures was mixed with 10 μl Orange G loading dye (0.25% Orange G from Fluka in 30% glycerol) and subjected to electrophoresis on agarose (0.8%) gels in 1 × TAE buffer. DNA bands were visualized by 2.5 μg ml-1 ethidium bromide under UV light. Corresponding bands (0.59 kb in size) were then cut out and purified as described above.
Purified PCR fragments were ligated into the pGEM-T easy vector using the Promega PCR cloning kit. Ligation and cloning was performed according to the kit manual. Transformation of E. coli XL-1 blue cells was done using the electroporation method described by Sambrook and Russel [37]. Recombinant plasmids were extracted from transformed cells with the Promega Wizard Plus Midiprep DNA Purification System. Plasmid DNA (100 ng μl-1) carrying partial HSP60 encoding gene sequences from Bifidobacterium RBL67 or B. thermacidophilum subsp. thermacidophilum LMG 21395T was then sequenced by Microsynth GmbH and finally aligned to the corresponding sequences from bifidobacteria and Bacillus subtilis W168 (obtained from Genbank entries) using the CLUSTAL W software (version 1.6). Similarities were calculated and converted into a distance matrix with the Jukes-Cantor parameter and rooted with Bacillus subtilis W168 applying a bootstrap value of 1000 using the software MEGA version 3.1 [35].
DNA-DNA hybridization
Whole genome DNA-DNA hybridizations were carried out externally at DSMZ Germany. Hybridizations were performed with the genomes of Bifidobacterium RBL67, B. thermacidophilum subsp. thermacidophilum LMG21395T, B. thermacidophilum subsp. porcinum LMG21689T, B. thermophilum DSM 20210T and B. boum 20432T. Total DNA was isolated using a French pressure cell according to the method described by Cashion et al. [38], DNA-DNA hybridizations were carried out according to De Ley et al. [39] with modifications of Huss et al. [40] in 2 × SSC and 10% formamide (v/v) at a temperature of 67°C. The analyses were performed with a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostated 6 × 6 multicell changer and a temperature controller with in situ temperature probe.
Carbohydrate fermentation and acid analysis
Carbohydrate fermentation of strain RBL67 was analyzed using API 50 CHL strips (Biomérieux). The tests were carried out in triplicates according to the manufacturer's instructions with a modification in culture preparation. Briefly, 2 ml of an overnight culture in MRS-C broth were centrifuged (14000 g, 5 min, 4°C). The pellet was then washed and resuspended in 1 ml of autoclaved water. This suspension was then mixed with 5 ml CHL50 medium (Biomérieux) and 0.1 ml of this mixture was applied to each tube of the API test. The test strips were then incubated anaerobically for 72 h at 37°C and evaluated after 24, 48 and 72 h.
Molar ratio of acetic and lactic acid was determined using HPLC. 2 ml of an overnight culture (16 h) of either Bifidobacterium RBL67, RBL68 or RBL70, B. thermacidophilum subsp. thermacidophilum LMG 21395T or B. thermacidophilum subsp. porcinum LMG 21689T were centrifuged (14000 g, 5 min, 4°C). The supernatant was 10 × diluted in HPLC-grade water and filtered (0.45 μm) prior to HPLC analysis. This was carried out using an Aminex HPX-87H (300 × 7.8 mm) column (Bio-Rad) in a Merck LaChrom HPLC system. Sulfuric acid (10 mM, Fluka) was used as mobile phase at a flow rate of 0.4 ml min-1. Sugars and acids were detected by a RI detector. Analyses were done in duplicate.
Authors' contributions
UVA did the first identification and characterization experiments, carried out the sequence alignments, prepared the DNA-DNA hybridizations, coordinated the study and drafted the manuscripts. VM did the molecular and phenotypic experiments, statistical analysis and participated in sequence alignment and DNA-DNA hybridization preparation. CL conceived the study and participated in the coordination of the study. LM participated in the coordination of the study and the sequence interpretation and helped in the draft of the manuscript. IF and EEK were responsible for the screening and isolation of bifidobacteria strains from baby faeces. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was supported by a grant from the Swiss National Foundation (Project n°. 3100A0-102256). We thank Dr. G. Dasen for his support on some molecular techniques and Dr. Janice Sych for carefully reading the manuscript.
Contributor Information
Ueli von Ah, Email: ueli.vonah@gmail.com.
Valeria Mozzetti, Email: valeria.mozzetti@ilw.agrl.ethz.ch.
Christophe Lacroix, Email: christophe.lacroix@ilw.agrl.ethz.ch.
Ehab E Kheadr, Email: ehab.kheadr@aln.ulaval.ca.
Ismaïl Fliss, Email: ismail.fliss@aln.ulaval.ca.
Leo Meile, Email: leo.meile@ilw.agrl.ethz.ch.
References
- Tissier MH. La réaction chromophile d'Escherich et Bacterium Coli. C R Seances Soc Biol Fil. 1899;51:943–945. [Google Scholar]
- Klijn A, Mercenier A, Arigoni F. Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev. 2005;29:491–509. doi: 10.1016/j.femsre.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen M-C, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Satokari RM, Vaughan EE, Smidt H, Saarela M, Matto J, de Vos M. Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract. System Appl Microbiol. 2003;26:572–584. doi: 10.1078/072320203770865882. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- Biavati B, Mattarelli P. The family Bifidobacteriaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editor. The prokaryotes: an evolving electronic resource for the microbiological community. 3. New York: Springer; 2005. http://www.springer.com/west/home/life+sci/microbiology?SGWID=4-10037-22-155417496-detailsPage=ppmmedia|aboutThisBook [Google Scholar]
- Dong X, Xin Y, Jian W, Liu X, Ling D. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int J Syst Bacteriol. 2000;50:119–125. doi: 10.1099/00207713-50-1-119. [DOI] [PubMed] [Google Scholar]
- Beerens H, Gavini F, Neut C. Effect of exposure to air on 84 strains of bifidobacteria. Anaerobe. 2000;6:65–67. doi: 10.1006/anae.1999.0317. [DOI] [Google Scholar]
- Meile L, Ludwig W, Rueger U, Gut C, Kaufmann P, Dasen G, Wenger S, Teuber M. Bifidobacterium lactis sp. nov, a moderately oxygen tolerant species isolated from fermented milk. System Appl Microbiol. 1997;20:57–64. [Google Scholar]
- Simpson PJ, Ross RP, Fitzgerald GF, Stanton C. Bifidobacterium psychraerophilum sp. nov. and Aeriscardovia aeriphila gen. nov., sp. nov., isolated from a porcine caecum. Int J Syst Bacteriol. 2004;54:401–406. doi: 10.1099/ijs.0.02667-0. [DOI] [PubMed] [Google Scholar]
- Jian W, Dong X. Transfer of Bifidobacterium inopinatum and Bifidobacterium denticolens to Scardovia inopinata gen. nov., comb. nov., and Parascardovia denticolens gen. nov., comb. nov., respectively. Int J Syst Bacteriol. 2002;52:809–812. doi: 10.1099/00207713-52-3-809. [DOI] [PubMed] [Google Scholar]
- Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol. 1996;46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- Scardovi V, Trovatelli LD. The fructose-6-phosphate shunt as peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann Microbiol Enzim. 1965;15:19–29. [Google Scholar]
- Meile L, Rohr LM, Geissmann TA, Herensperger M, Teuber M. Characterization of the D-xylulose 5-phosphate/D-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J Bacteriol. 2001;183:2929–2936. doi: 10.1128/JB.183.9.2929-2936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Zhu L, Li W, Dong X. Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov. Int J Syst Bacteriol. 2003;53:1619–1623. doi: 10.1099/ijs.0.02617-0. [DOI] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Zink R, Fitzgerald GF, van Sinderen D. Characterization of the gro EL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl Environ Microbiol. 2004;70:6197–6209. doi: 10.1128/AEM.70.10.6197-6209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Chambers JR, Barlow K, Park AS, Wheatcroft R. The gene encoding xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (xfp) is conserved among Bifidobacterium species within a more variable region of the genome and both are useful for strain identification. FEMS Microbiol Lett. 2005;246:251–257. doi: 10.1016/j.femsle.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Berthoud H, Chavagnat F, Haueter M, Casey MG. Comparison of partial gene sequences encoding a phosphoketolase for the identification of bifidobacteria. LMWT. 2005;38:101–105. [Google Scholar]
- Vandamme P, Pot B, Gillis M, Vos Pd, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré R, Kheadr E, Lacroix C, Moroni O, Fliss I. Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes . J Appl Microbiol. 2003;95:1058–1069. doi: 10.1046/j.1365-2672.2003.02085.x. [DOI] [PubMed] [Google Scholar]
- Satokari R. PhD thesis. Tietotie: Wageningen University; 2002. Molecular identification and characterisation of bifidobacteria and lactobacilli in the human gastrointestinal tract. [Google Scholar]
- Vaugien L, Prevots F, Roques C. Bifidobacteria identification based on 16S rRNA and pyruvate kinase partial gene sequence analysis. Anaerobe. 2002;8:341–344. doi: 10.1016/S1075-9964(03)00025-8. [DOI] [PubMed] [Google Scholar]
- Miyake T, Watanabe K, Watanabe T, Oyaizu H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol Immunol. 1998;42:661–667. doi: 10.1111/j.1348-0421.1998.tb02337.x. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- Delcenserie V, Bechoux N, Leonard T, China B, Daube G. Discrimination between Bifidobacterium species from human and animal origin by PCR-restriction fragment length polymorphism. J Food Prot. 2004;67:1284–1288. doi: 10.4315/0362-028x-67.6.1284. [DOI] [PubMed] [Google Scholar]
- Klein G, Pack A, Bonaparte C, Reuter G. Taxonomy and physiology of probiotic lactic acid bacteria. Int J Food Microbiol. 1998;41:103–125. doi: 10.1016/S0168-1605(98)00049-X. [DOI] [PubMed] [Google Scholar]
- Beerens H. Bifidobacteria as indicators of faecal contamination in meat and meat products: detection, determination of origin and comparison with Escherichia coli . Int J Food Microbiol. 1998;40:203–207. doi: 10.1016/S0168-1605(98)00027-0. [DOI] [PubMed] [Google Scholar]
- Gavini F, Delcenserie V, Kopeinig K, Pollinger S, Beerens H, Bonaparte C, Upmann M. Bifidobacterium species isolated from animal feces and from beef and pork meat. J Food Prot. 2006;69:871–877. doi: 10.4315/0362-028x-69.4.871. [DOI] [PubMed] [Google Scholar]
- Gavini F, Pourcher AM, Neut C, Monget D, Romond C, Oger C, Izard D. Phenotypic differentiation of bifidobacteria of human and animal origins. Int J Syst Bacteriol. 1991;41:548–557. doi: 10.1099/00207713-41-4-548. [DOI] [PubMed] [Google Scholar]
- Hartemink R, Kok BJ, Weenk GH, Rombouts FM. Raffinose-Bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J Microbiol Meth. 1996;27:33–43. doi: 10.1016/0167-7012(96)00926-8. [DOI] [Google Scholar]
- Schürch C. PhD thesis. Zurich: Swiss Federal Institute of Technology; 2002. Development of a novel DNA transformation system for bifidobacteria. [Google Scholar]
- Jian W, Zhu L, Dong X. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int J Syst Bacteriol. 2001;51:1633–1638. doi: 10.1099/00207713-51-5-1633. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leenhouts KJ, Kok J, Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: a laboratory manual. 3. Vol. 1. New York: Cold Spring Harbor Laboratory Press;; 2001. [Google Scholar]
- Cashion P, Hodler-Franklin MA, McCully J, Franklin M. A rapid method for base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- Ley JD, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Huss VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. System Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Bullock WO, Fernandez JM, Short JM. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactoside selection. BioTechn. 1987;5:376–379. [Google Scholar]
- Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]