Abstract
Oncogenic mutations of the MLL histone methyltransferase confer an unusual ability to transform non-self-renewing myeloid progenitors into leukemia stem cells (LSCs) by mechanisms that remain poorly defined. Misregulation of Hox genes is likely to be critical for LSC induction and maintenance but alone it does not recapitulate the phenotype and biology of MLL leukemias, which are clinically heterogeneous—presumably reflecting differences in LSC biology and/or frequency. TALE (three-amino-acid loop extension) class homeodomain proteins of the Pbx and Meis families are also misexpressed in this context, and we thus employed knockout, knockdown, and dominant-negative genetic techniques to investigate the requirements and contributions of these factors in MLL oncoprotein-induced acute myeloid leukemia. Our studies show that induction and maintenance of MLL transformation requires Meis1 and is codependent on the redundant contributions of Pbx2 and Pbx3. Meis1 in particular serves a major role in establishing LSC potential, and determines LSC frequency by quantitatively regulating the extent of self-renewal, differentiation arrest, and cycling, as well as the rate of in vivo LSC generation from myeloid progenitors. Thus, TALE proteins are critical downstream effectors within an essential homeoprotein network that serves a rate-limiting regulatory role in MLL leukemogenesis.
[Keywords: Leukemia stem cells, MLL, Meis1, Pbx, TALE homeodomain proteins]
Leukemia stem cells (LSCs) comprise a functionally distinct subpopulation of leukemic cells with the ability to self-renew extensively, and to initiate, sustain, or regenerate disease. In acute myeloid leukemia (AML), LSCs are generally considered to be rare upstream cells that arise out of the normal hematopoietic stem cell (HSC) or primitive progenitor compartments, and are organized in a hierarchy based on quantitative differences in their self-renewal potentials (Passegue and Weisman 2005). This paradigm, however, may not apply to all myeloid leukemias, as suggested by recent studies using a mouse model of AML induced by the MLL-AF9 oncogene (Krivtsov et al. 2006; Somervaille and Cleary 2006), which is typically associated with FAB-M4 or M5 subtypes of human AML (Swansbury 1998). LSCs were found to be very frequent and representative of downstream myeloid lineage cells that ectopically acquired extensive self-renewal and other biologic properties more typical of HSCs. The key regulators and subordinate genetic programs by which MLL converts myeloid progenitors into LSCs are of major interest.
MLL is a histone methyltransferase with features suggestive of a general transcriptional role at most promoters (Guenther et al. 2005); however, Hox genes appear to be particularly dependent on its function (Yu et al. 1995). In leukemias harboring MLL-activating mutations, several Hoxa genes are consistently expressed at high levels, suggesting that MLL oncoproteins inappropriately maintain their expression and prevent their programmed down-regulation that otherwise accompanies terminal myeloid differentiation (Imamura et al. 2002b; Pineault et al. 2002). Since various Hox genes have been implicated in the regulation of normal stem cell self-renewal, their misregulation in MLL leukemias is likely to be important for LSC maintenance. Hoxa9, in particular, has been shown to critically influence MLL oncogenesis (Ayton and Cleary 2003; Kumar et al. 2004; So et al. 2004; Okada et al. 2005; J. Wang et al. 2005). However, Hox gene misregulation alone does not recapitulate the biological and clinical features of MLL leukemia.
Additional candidate factors that may critically regulate LSC potentials in MLL leukemias are TALE (three-amino-acid loop extension) class homeodomain proteins of the Pbx and Meis families. They enhance the relatively nonspecific DNA-binding properties of Hox transcription factors and regulate gene expression as hetero-oligomeric complexes with Hox proteins (Mann 1995). TALE proteins are required for the execution of some Hox-dependent developmental programs and are implicated in leukemogenesis (Azpiazu and Morata 1998; Ryoo et al. 1999; Selleri et al. 2001; Manley et al. 2004; Eklund 2007; Rice and Licht 2007). Notably, mutations of Hoxa9 that prevent interactions with Pbx proteins abrogate its oncogenic properties (Schnabel et al. 2000). Furthermore, coexpression of Meis1 markedly shortens the latency and increases the penetrance of Hoxa9-induced myeloid leukemia, and recapitulates some of the features of MLL leukemia (Kroon et al. 1998; Calvo et al. 2001; Zeisig et al. 2004; G.G. Wang et al. 2005). Since Meis1 is also a transcriptional target of MLL oncoproteins and is consistently highly expressed in MLL leukemias (Lawrence et al. 1999; Imamura et al. 2002b; Milne et al. 2005), simultaneous misregulation of Hoxa9 and Meis1 may be sufficient for MLL oncogenesis (Zeisig et al. 2004). However, MLL leukemias are biologically and clinically diverse, and the specific roles of TALE proteins in regulating LSC properties that underlie these differences have not been defined.
In this report, we demonstrate that TALE homeodomain proteins are essential for the induction and maintenance of MLL leukemogenesis. Meis1, in particular, quantitatively regulates the differentiation arrest, cycling activity, in vivo progression, and self-renewal of MLL leukemia cells, thereby functioning as a critical and rate-limiting determinant of LSC potential.
Results
The latencies of MLL leukemias correlate with Meis1 endogenous expression levels
Twelve different MLL fusion oncoproteins were investigated for their TALE protein-dependent oncogenic properties. These included MLL oncoproteins containing different classes of fusion partners including cytoplasmic proteins (GAS7, AF1P, AF6, and EB1), nuclear proteins of the forkhead family (FKHRL1 and AFX) or AF4 family/complex (LAF4, AF5, ENL, AF9, and AF10), and histone modifiers (CBP), which induce leukemia with widely varying latencies and morphologies in mice (Lavau et al. 1997, 2000; DiMartino et al. 2002; So et al. 2003, 2004; J. Wang et al. 2005; Somervaille and Cleary 2006; our unpublished observations). E2A-HLF served as a control, since it transforms myeloid progenitors through Hox-independent pathways (Ayton and Cleary 2003; So et al. 2004). Retroviral constructs for each of the respective MLL fusion cDNAs expressed proteins that migrated at their predicted molecular weights in Western blot analysis (Supplementary Fig. 1A).
Primary murine myeloid progenitors (c-kit+) transduced by the 12 MLL fusion constructs, but not empty vector, formed colonies in methylcellulose that replated through at least four rounds of culture (data not shown), demonstrating the enhanced self-renewal and impaired differentiation typically induced by MLL oncogenes (Lavau et al. 1997, 2000). Real-time quantitative PCR analysis of cells from fourth-round cultures showed that Hoxa5, Hoxa7, Hoxa9, and Hoxa10 were highly expressed in myeloid cells transduced by MLL oncogenes compared with control (E2A-HLF) cells under our experimental conditions (Supplementary Fig. 1B,C). The relative and absolute levels of these Hox transcripts were fairly uniform, and did not appear to distinguish the different molecular subtypes of MLL-transformed cells.
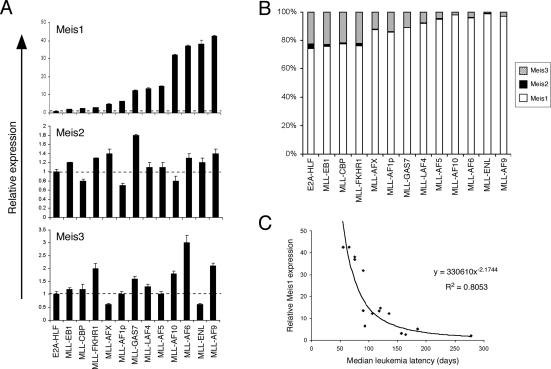
Conversely, absolute Meis1 expression levels varied considerably among the different MLL-transformed cells, from twofold to 40-fold the levels detected in control (E2A-HLF) cells (Fig. 1A). Meis2 and Meis3 were also expressed, but not consistently increased relative to their levels in cells transformed by E2A-HLF, such that Meis1 accounted for 75%–95% of total Meis transcripts in MLL-transformed cells (Fig. 1B). Interestingly, Meis1 transcript levels showed a significant correlation with the latencies for MLL leukemia (Fig. 1C; Supplementary Table 1). Cells transformed by MLL fusion proteins that induce a short latency AML in murine models displayed higher levels of Meis1 expression, whereas those that induce AML with prolonged latencies displayed lower levels of Meis1 expression. Statistical analysis showed a power relationship, indicating that Meis1 transcript levels decreased at a specific rate with respect to increasing time required for leukemia development. Conversely, Hoxa9 expression levels showed no correlation with leukemia latencies (Supplementary Fig. 2). These results suggested that Meis1 likely serves a critical and potentially rate-limiting role in the pathogenesis of MLL leukemia.
Figure 1.
Meis1 expression levels correlate with the latencies of MLL leukemias. (A) The expression levels of Meis1, Meis2, and Meis3 transcripts were determined by real-time PCR analysis of MLL-transformed cells (indicated below) from the fourth round of serial replating (error bars indicate standard deviations of triplicate analyses). Results are expressed relative to levels observed in cells transformed by E2A-HLF (dashed line). (B) Bar graph indicates the total relative levels of Meis transcripts expressed in cells transformed by various oncogenes (indicated at bottom) from the fourth round of plating in methylcellulose cultures. The relative abundance of Meis transcripts was determined using the cycle time (Ct) value method, assuming that all primers were optimized to generate equal PCR efficiencies. (C) The relative expression levels of Meis1 are plotted against median latency times required for leukemia induction by the respective MLL oncogenes. Latency data are derived in part from the current study as well as published studies (Supplementary Table 1). The data display a power trend line best-fit (R2 = 0.8053) indicating that Meis1 expression levels decrease at a specific rate with respect to the time required for leukemia development.
Meis1 is essential for induction and maintenance of MLL-mediated transformation
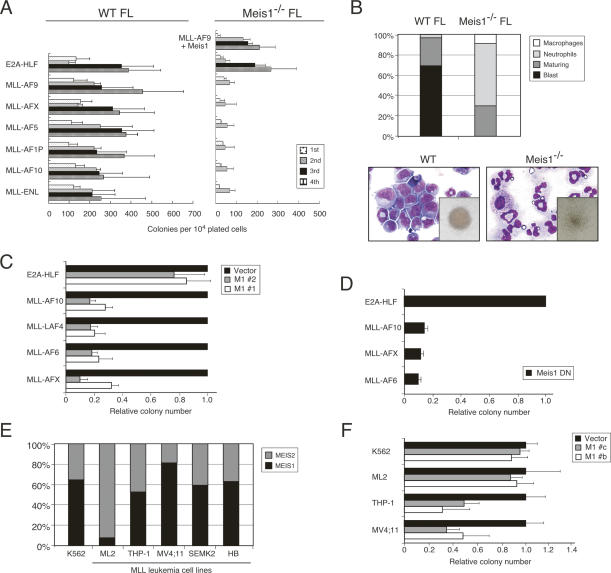
To specifically address the requirement for Meis1 in MLL-mediated oncogenesis, transformation assays were performed using fetal liver (FL) cells isolated from Meis1−/− embryos at embryonic day 13.5 (E13.5), prior to their intrauterine demise at approximately E14 (Hisa et al. 2004). Meis1-deficient FL cells were efficiently transformed by the control E2A-HLF gene; however, none of the MLL oncogenes was capable of inducing sustained replating (Fig. 2A). Rather, the plating capacities of MLL-transduced cells were exhausted after the second round. Meis1−/− second-round colonies were small in size with a diffuse morphology and contained markedly reduced numbers of myeloid blasts when compared with transformed colonies from wild-type FL (Fig. 2B). Conversely, all tested MLL oncogenes induced continued replating of E13.5 FL cells isolated from wild-type mice. Moreover, transformation of Meis1−/− FL cells by MLL-AF9 was rescued by cotransduction of Meis1 (Fig. 2A), demonstrating that the progenitors susceptible to MLL transformation were present in Meis1−/− FLs at E13.5. Thus, in the absence of Meis1, MLL oncogenes are incapable of inducing the enhanced self-renewal and impaired differentiation necessary for myeloid transformation.
Figure 2.
Meis1 is required for initiation and maintenance of MLL-mediated myeloid transformation. (A) FL cells from wild-type (left) or Meis1−/− (right) mice were transduced with fusion oncogenes (indicated on left) and serially plated in methylcellulose cultures every 5 d. Results are shown as colony numbers per 104 plated cells for each round of culture. Data are also shown for Meis1−/− cells cotransduced with MLL-AF9 and Meis1. Error bars indicate standard deviations of three independent experiments. (B) Cytospin preparations of wild-type (left) or Meis1−/− (right) FL cells transduced with MLL-AF9 were analyzed for the proportions of cells at the end of the second round of plating in methylcellulose culture with morphological features of blasts versus differentiation (May Grunwald Giemsa staining). Bar graph indicates the mean number of cells with the indicated morphologic features (n = 4). Insets show colony morphologies at the end of the second round. (C) Myeloid progenitors transformed by various MLL oncogenes (indicated on left) were secondarily transduced with lentiviral vectors encoding shRNAs specific for Meis1 (M1 #1 or M1 #2). Colony numbers are shown relative to cells secondarily transduced with lentiviral vector alone. Error bars indicate standard deviations of at least two independent experiments. (D) Myeloid progenitors transformed by various oncogenes (indicated on left) were secondarily transduced with a dominant-negative Meis1 construct (Meis1 DN). Colony numbers are shown relative to cells transformed by E2A-HLF. Error bars indicate standard deviations of at least two independent experiments. (E) Bar graph indicates the total relative levels of MEIS1 and MEIS2 transcripts expressed in human leukemia cell lines (indicated at bottom). The relative abundance of MEIS transcripts was determined using the Ct value method, assuming that all primers were optimized to generate equal PCR efficiencies. (F) Human leukemia cell lines (indicated on left) were transduced with lentiviral vectors encoding shRNAs specific for MEIS1 (M1 #b or M1 #c). Colony numbers are shown relative to cells transduced with lentiviral vector alone. Error bars indicate standard deviations of at least two independent experiments.
To assess whether Meis1 was required for maintenance of MLL-mediated transformation, wild-type cells stably transformed by MLL oncogenes were secondarily transduced with lentiviral vectors expressing short hairpin RNAs (shRNAs) to specifically silence Meis1 (Supplementary Fig. 3). Expression of two different Meis1 shRNAs resulted in substantial impairment (>70%–80%) of clonogenic capacity compared with MLL-transformed cells secondarily transduced with lentiviral vector alone (Fig. 2C). Similarly, MLL-transformed cells displayed a marked reduction in clonogenic capacity (>80%) when secondarily transduced with a dominantnegative construct that lacked the Meis1 DNA-binding homeodomain, whereas E2A-HLF-transformed cells were unaffected (Fig. 2D). Therefore, Meis1 is essential for both the initiation and maintenance of enhanced self-renewal imposed on myeloid progenitors by MLL oncogenes.
MEIS1 has been reported to be consistently expressed in human MLL leukemias (Imamura et al. 2002b; Ferrando et al. 2003; Quentmeier et al. 2004), suggesting that it may serve a similar role in their maintenance. Quantitative RT–PCR analysis showed that MEIS1 was expressed in leukemia cell lines with MLL chromosomal translocations (Fig. 2E). However, in contrast with murine MLL-transformed cells, MEIS2 was also substantially expressed and ML2 cells were notable for expressing 10-fold more MEIS2 versus MEIS1 (Fig. 2E). Lentiviral transduction of two different MEIS1 shRNAs resulted in substantial impairment (∼40%–60%) of clonogenic capacity for MLL leukemia cell lines compared with control K562 cells with the exception of ML2 cells, which were not significantly compromised (Fig. 2F). Thus, despite the redundancy of MEIS gene expression, our results suggest that it is nevertheless required to maintain human MLL leukemias.
Hyperexpression of Meis1 accelerates the progression of MLL leukemogenesis
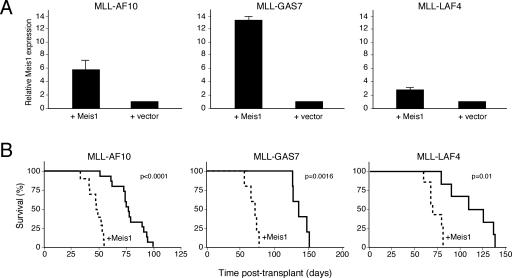
The observed correlation of endogenous Meis1 expression levels with leukemia latency and the requirement of Meis1 for MLL-mediated stem and progenitor cell transformation raised the possibility that Meis1 may directly influence the in vivo generation of LSCs from myeloid progenitors, which requires undefined secondary alterations (Lavau et al. 2000; Somervaille and Cleary 2006). To investigate this possibility, Meis1 was hyperexpressed along with representative MLL oncogenes (MLL-AF10, MLL-GAS7, or MLL-LAF4) to determine whether its enhanced expression may accelerate MLL leukemogenesis. Myeloid progenitors (c-kit+) cotransduced with an MLL oncogene plus Meis1 (MLL/Meis1) displayed Meis1 transcript levels that were several-fold higher than progenitors cotransduced with the respective MLL oncogene plus empty vector (MLL/v) prior to transplantation (Fig. 3A). Following transplantation of equal numbers of cotransduced cells (representing comparable numbers of colony-forming cells [CFCs]) into syngeneic recipient mice, the MLL/Meis1 cohorts developed leukemia with substantially shortened latencies compared with mice transplanted with MLL/v cotransduced cells (Fig. 3B). Leukemias in both cohorts appeared grossly similar pathologically—with effacement of the normal bone marrow architecture, marked splenomegaly, and infiltration of the liver by leukemia cells—and maintained similar levels of Meis1 when compared with the respective primary cotransduced progenitors (data not shown). Therefore, Meis1 serves a rate-limiting role in progression of MLL-associated leukemia.
Figure 3.
Increased Meis1 expression accelerates MLL-mediated leukemogenesis. (A) Meis1 transcript levels in myeloid progenitors (c-kit+) were determined by quantitative real-time PCR 24 h following transduction with retroviruses encoding the MLL oncogenes indicated above the panel, with or without cotransduced Meis1 (indicated below). Data are expressed relative to the level observed in cells transduced with MLL oncogene alone (bars indicate the average of triplicate analyses). (B) Survival curves are shown for cohorts of mice transplanted with cells cotransduced with the indicated MLL oncogene and Meis1 or empty vector. Acute leukemia was confirmed by peripheral blood leukocyte counts, FACS analyses, and/or necropsy.
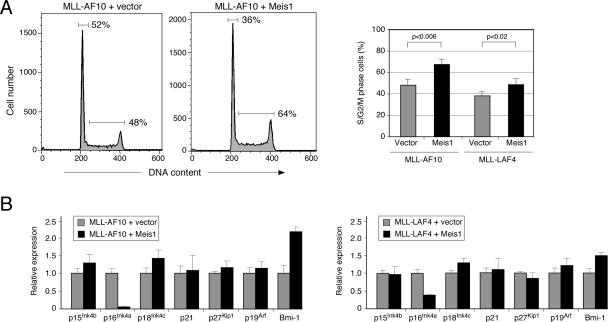
Meis1 modulates the differentiation arrest, self-renewal, and cell cycle activity of MLL leukemia cells
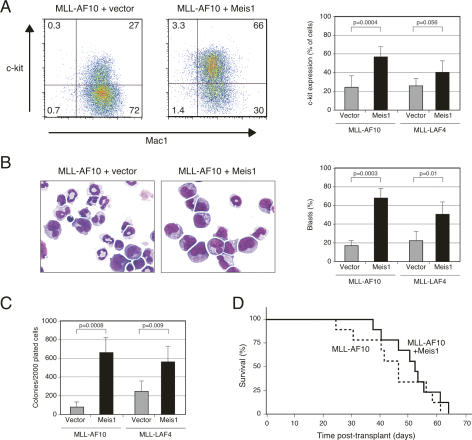
FACS analysis demonstrated that all leukemias were donor-derived (data not shown) and consistently displayed myeloid phenotypes (Mac1+ Gr1+). However, a greater fraction of MLL/Meis1 leukemia cells expressed high-level c-kit and lower Mac1, suggestive of less differentiation (Fig. 4A). Morphologic assessment of splenocytes obtained at necropsy of MLL/Meis1 mice revealed a several-fold higher proportion of cells with cytologic features of blasts and substantially fewer differentiating forms (Fig. 4B) compared with splenocytes from MLL/v mice. Furthermore, in semisolid culture assays, the frequencies of CFCs in the spleens of leukemic mice were up to sevenfold higher for MLL/Meis1 leukemias (Fig. 4C) and colony morphologies (Lavau et al. 1997) were predominantly type I for MLL/Meis1 CFCs, compared with mostly type II/III for MLL/v CFC (72% vs. 21% for MLL-AF10 leukemias) (data not shown). Thus, MLL leukemias where Meis1 was overexpressed exhibited a more pronounced block in differentiation as well as a higher frequency of clonogenic leukemia cells.
Figure 4.
Meis1 modulates myeloid differentiation arrest, as well as CFC and LSC frequencies of MLL leukemias. (A) FACS profiles demonstrate representative c-kit and Mac1 expression on splenocytes from mice with AML induced by transplantation of progenitors transduced with genes indicated at the tops of the panels. Bar graph summarizes data from five mice in each category. (B) Splenocytes from leukemic mice were analyzed for the proportion of cells with morphologic features of blasts versus differentiation following May Grunwald Giemsa staining of cytospin preparations. Bar graph indicates the number of cells with the indicated morphologic features for five mice in each cohort. (C) FACS-sorted Mac1+ leukemic splenocytes were cultured in methylcellulose medium for 6 d to determine CFC frequencies. Bar graph indicates the numbers of clonogenic cells for each of five leukemias in the respective cohorts. (D) Survival curves of sublethally irradiated mice transplanted with single AML colonies (3000 cells) plucked after 7 d in semisolid culture and directly injected into secondary recipients, or expanded in semisolid culture for 5 d prior to injection (2.5 × 105 cells).
Cell cycle analysis of explanted leukemia cells growing in methylcellulose cultures revealed a significant increase in the proportion of S/G2/M-phase cells for MLL/Meis1 versus MLL/v leukemias (Fig. 5A). The higher fraction of cells in cycle correlated with substantially lower transcript levels for the CDK inhibitor p16Ink4a, whereas expression of several other cell cycle regulators including p19Arf, an alternatively spliced Ink4a transcript that codes for a positive regulator of the p53 pathway, did not differ significantly between MLL/v and MLL/Meis1 leukemias (Fig. 5B). However, expression of Bmi-1, a negative regulator of the Ink4a locus (Jacobs et al. 1999), was significantly higher in MLL/Meis1 leukemias (Fig. 5B). The changes in gene expression as well as latency were more pronounced in MLL-AF10 versus MLL-LAF4 leukemias. Therefore, the enabling effects of Meis1 on proliferation and arrested differentiation of MLL leukemia cells correlated with specific perturbations of the Bmi-1/Ink4a axis, which is implicated in maintenance of HSC and LSC self-renewal (Lessard and Sauvageau 2003; Park et al. 2003).
Figure 5.
Altered cell cycle activity induced by Meis1 in MLL-AF10 leukemia. (A) DNA content analysis was determined by FACS analysis of explanted leukemia cells growing in methylcellulose cultures. Bar graph summarizes data for three mice in each cohort. (B) Expression levels of the indicated transcripts were determined by quantitative RT–PCR analysis of splenocytes isolated from leukemic mice. Results are the mean of three leukemias in each category, and are expressed relative to transcript levels in leukemia cells transformed by MLL-AF10.
Meis1 determines MLL LSC frequency
Previous studies of MLL-AF9 AML showed that CFCs present in the spleens of leukemic mice have the biological properties of LSCs (Somervaille and Cleary 2006). To test if the increased CFC frequencies in leukemias induced by coexpressed exogenous Meis1 also correlated with increased numbers of LSCs, individual CFCs (type I or II) were plucked from methylcellulose media and transplanted into syngeneic mice (or expanded in semisolid culture prior to transplant) to assess their potential to initiate AML in secondary recipients, a hallmark feature of LSCs. All transplanted mice succumbed to AML regardless of whether the transplanted CFCs derived from primary leukemias with or without coexpressed exogenous Meis1 (Fig. 4D). These results confirmed that CFCs in each cohort were a reliable indicator of LSCs, and demonstrated sevenfold higher LSC frequencies in MLL/Meis1 leukemias in comparison with MLL/v leukemias. Unlike the case for primary transplant recipients, latencies were similar irrespective of Meis1 status since the cells had achieved full leukemic potential prior to secondary transplant. Nevertheless, the morphologic features and phenotypes of the respective secondary leukemias were similar to those observed in the primary mice—i.e., a less-differentiated phenotype in leukemias coexpressing exogenous Meis1 (data not shown)—demonstrating that Meis1 expression levels stably determine LSC properties in MLL-associated AML.
Meis1 functions as a DNA-binding transcriptional cofactor in MLL transformation
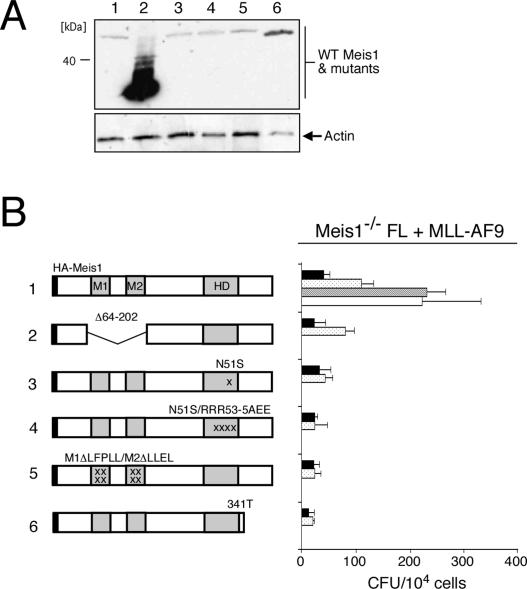
A structure/function analysis was conducted to investigate whether Meis1 may require interactions with Pbx TALE protein partners for MLL-mediated transformation. Meis1 constructs containing either deletion or point mutations (in the M1/M2 domains) that abrogate Pbx interaction (G.G. Wang et al. 2005) were incapable of rescuing MLL transformation of Meis1−/− FL cells (Fig. 6B, constructs #2 and #5), in contrast to wild-type Meis1 (Fig. 6B, construct #1). Meis1 constructs containing homedomain mutations, which have been shown to disrupt Meis1 DNA binding (G.G. Wang et al. 2005), were also unable to complement Meis1 deficiency (Fig. 6A,B, constructs #3 and #4). Furthermore, deletion of the conserved C-terminal tail of Meis1 (Fig. 6A,B, construct #6), which has been implicated in its transcriptional effector properties, abrogated rescue of MLL transformation. Taken together, these analyses suggest that Meis1 associates with Pbx partners in a DNA-binding transcriptional complex to mediate MLL transformation.
Figure 6.
Structure/function analysis of Meis1 requirements in MLL transformation. (A) Western blot analysis of wild-type and mutant Meis1 proteins expressed in retroviral packaging Phoenix cells and detected with an anti-HA antibody. Identities of constructs are indicated at the tops of lanes and correspond to the schematic depictions in B. The enhanced stability of construct #2 lacking Pbx interaction motifs M1/M2 has been reported previously (G.G. Wang et al. 2005). (B) Myeloid transformation assay was performed using Meis1−/− FL cells cotransduced with MLL-AF9 and various Meis1 constructs schematically illustrated on the left. Results from four rounds of plating are shown with each bar representing the mean ± SD of the total number of myeloid colonies per 104 plated cells derived from at least three replicates.
MLL-mediated myeloid transformation is codependent on TALE homeodomain proteins Pbx2 and Pbx3
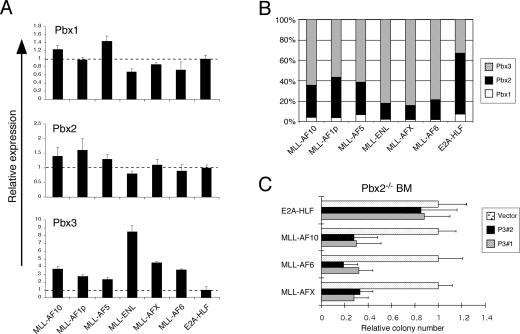
A genetic approach was employed to interrogate the contributions of Pbx proteins in MLL leukemia. Real-time quantitative PCR analysis showed that three Pbx genes (Pbx1, Pbx2, and Pbx3) were expressed in myeloid progenitors immortalized by MLL oncogenes (Fig. 7A; data not shown). Pbx3 was notable for its significantly increased levels (three- to eightfold) compared with control cells transformed by E2A-HLF, consistent with increased Pbx3 levels in human MLL AMLs (Ross et al. 2004). Together, Pbx2 and Pbx3 accounted for >90% of total Pbx transcripts, whereas Pbx1 was minimally expressed (Fig. 7B). To investigate whether specific Pbx functions were required for MLL transformation, serial replating assays were performed using FL cells harvested from Pbx3−/− embryos (E14.5–E18) (Rhee et al. 2004) or bone marrow cells isolated from adult Pbx2−/− mice (Selleri et al. 2004; Capellini et al. 2006). MLL oncoproteins transformed the Pbx-deficient myeloid progenitors as efficiently as wild-type cells (Supplementary Fig. 4), indicating that MLL-mediated transformation was not dependent on the presence of Pbx2 or Pbx3 alone.
Figure 7.
Pbx2 and Pbx3 are required for maintenance of MLL-mediated transformation. (A) The relative expression levels of Pbx1, Pbx2, and Pbx3 transcripts were determined by real-time PCR analysis of MLL-transformed cells (indicated below) from the fourth round of serial replating (error bars indicate standard deviations of triplicate analyses). Results are expressed relative to levels observed in cells transformed by E2A-HLF. (B) Bar graph indicates the total relative levels of Pbx transcripts expressed in cells transformed by various oncogenes (indicated at bottom) from the fourth round of plating in methylcellulose cultures. The relative abundance of Pbx transcripts was determined using the Ct value method, assuming that all primers were optimized to generate equal PCR efficiencies. (C) Pbx2−/− myeloid progenitors transformed by various oncogenes (indicated on left) were secondarily transduced with lentiviral vectors encoding shRNAs specific for Pbx3 (P3 #1 or P3 #2). Colony numbers are shown relative to cells secondarily transduced with lentiviral vector alone. Error bars indicate standard deviations of at least two independent experiments.
To further reduce Pbx levels, Pbx2−/− cells transformed by MLL oncogenes were secondarily transduced with shRNAs targeting Pbx3, resulting in an ∼70%–80% reduction of Pbx3 transcript levels (Supplementary Fig. 3). Following Pbx3 knockdown, the clonogenic potential of Pbx2−/−/Pbx3kd MLL-transformed cells was substantially decreased (>60%) when compared with Pbx2−/− transformed cells secondarily transduced with vector alone (Fig. 7C). This effect was specific to MLL-mediated transformation since Pbx2−/−/Pbx3kd cells transformed by E2A-HLF displayed no significant reduction in clonogenic potential following Pbx3 silencing (Fig. 7C). These results demonstrate that maintenance of MLL-mediated transformation is specifically dependent on Pbx activity, and further support that Meis1 functions in complex with Pbx proteins in myeloid progenitors transformed by MLL oncogenes.
Discussion
MLL leukemias are invariably associated with expression of Hox and TALE homeobox genes, but nevertheless are clinically heterogeneous possibly reflecting differences in LSC biology and/or frequency. Misregulation of Hox genes in MLL leukemias is likely to be critical for LSC maintenance, as suggested by genetic loss-of-function studies (Ayton and Cleary 2003; Kumar et al. 2004; So et al. 2004; Okada et al. 2005; J. Wang et al. 2005) and consistent with their normal roles in HSC self-renewal (Abramovich and Humphries 2005). However, Hox gene misregulation alone does not recapitulate the phenotype and biology of MLL leukemias and is unlikely to support the high LSC frequencies recently demonstrated in a murine model of MLL-AF9 AML that are far in excess of those previously estimated for AML in general (Somervaille and Cleary 2006). Our current studies demonstrate that TALE homeodomain proteins are essential regulators of MLL transformation, and that Meis1 in particular determines LSC frequency and potential by quantitatively regulating the extent of self-renewal, differentiation arrest, and cycling, as well as the rate of in vivo LSC generation from myeloid progenitors.
Meis1 is an essential regulator of LSC biology in MLL leukemia
Genetic analyses revealed a critical dependence on Meis1 for induction and maintenance of myeloid transformation induced by several molecularly and functionally distinct MLL oncogenes. Notably, lack of Meis1 confers a consistent and severe impairment of MLL-mediated transformation in contrast to deficiencies of single Hoxa genes, whose requirements vary for different MLL oncogenes (Ayton and Cleary 2003; Kumar et al. 2004; So et al. 2004; Okada et al. 2005; J. Wang et al. 2005; our unpublished observations). Meis1 is the predominant Meis family gene expressed in murine myeloid progenitors, and its TALE protein product functions as a transcriptional cofactor for multiple Hox proteins, which are redundantly expressed. Thus, Meis1 deficiency likely results in a broad compromise of Hox transcriptional activity, in addition to possible impacts on non-Hox-dependent pathways, and consequently a more severe limitation on MLL transformation than the loss of single Hox genes.
Although Meis1 is consistently expressed and required in murine myeloid cells transformed in vitro by different MLL oncoproteins, its endogenous expression levels varied by >20-fold and correlated with critical features of leukemia biology and LSC frequency. Meis1 dictates the extent of myeloid differentiation arrest induced by MLL oncogenes as well as the fraction of leukemia cells in cycle, and their clonogenic frequency. The similarity of these effects to those observed for Meis1 in Hoxa9-associated leukemogenesis (Kroon et al. 1998; G.G. Wang et al. 2005) provides strong support for previous suggestions that Meis1 and Hoxa9 are major effectors of MLL leukemogenesis (Zeisig et al. 2004). Clonogenic frequency is a surrogate measure of MLL LSCs (Somervaille and Cleary 2006), which are substantially increased (up to sevenfold) in AMLs associated with hyperexpression of exogenous Meis1. This correlates with observations that Meis1 programs expression of HSC-associated genes in Hoxa9-induced leukemia (G.G. Wang et al. 2005). Our studies substantially extend these observations by demonstrating that Meis1 quantitatively regulates functional properties of LSCs in a model of MLL leukemia.
Meis1 is rate-limiting for MLL leukemogenesis
Meis1 appears to serve two distinct but possibly interrelated roles in MLL leukemogenesis. In addition to its requirement to initiate in vitro immortalization, the significant correlation of Meis1 expression levels with latencies required for development of MLL leukemias in our mouse model supports a role for Meis1 in regulating progression of MLL-immortalized cells to LSCs. The genetic and/or epigenetic changes underlying this transition have not yet been defined, although it is characterized in part by acquisition of an enhanced ability to interact with the bone marrow microenvironment (Shah et al. 1998; Somervaille and Cleary 2006). A role in LSC progression is consistent with previous observations that hyperexpression of Meis1 markedly increased the penetrance and reduced the latency for development of Hoxa9-induced AML (Kroon et al. 1998; G.G. Wang et al. 2005). Thus, Meis1 is rate-limiting such that low levels temper or abrogate immortalization of myeloid progenitors in vitro, whereas high levels accelerate their in vivo transition to LSCs.
The critical influence of Meis1 on LSC biology appears to correlate with epigenetic regulation of the RB pathway. Leukemias that developed with shortened latencies following hyperexpression of exogenous Meis1 were associated with increased expression of the Bmi-1 gene, which encodes a polycomb group epigenetic repressor protein. This is consistent with previous observations that BMI-1 levels increased during step-wise transformation and immortalization of human AML (Warner et al. 2005). However, the level of Bmi-1 mRNA increase observed in our studies was relatively modest, presumably because Bmi-1 genetically opposes MLL function through its antagonistic effects on Hox gene expression (Hanson et al. 1999). Bmi-1 is essential for the maintenance of normal adult HSCs as well as LSCs (Lessard and Sauvageau 2003; Park et al. 2003), consistent with the increased LSC frequencies in MLL/Meis1 leukemias. In this capacity, Bmi-1 represses transcription of p16Ink4a, a CDKI at the apex of the RB pathway. Expression of p16Ink4a inversely correlated with Meis1 and Bmi-1 levels as well as the increased fraction of cycling cells in MLL leukemias and the frequency of LSCs, which are highly enriched for cycling cells (T.C.P. Somervaille and M.L. Cleary, unpubl.). It was the only CDKI whose expression was consistently altered, supportive of a specific as opposed to secondary role in regulating LSC potential. Preliminary studies suggest that Bmi-1 is not a direct transcriptional target of Meis1 and is not absolutely essential for MLL-mediated transformation in vitro (P. Wong and M.L. Cleary, unpubl.), consistent with an alternative role in regulating long-term self-renewal that will require assessment by long-term transplantation studies. Taken together, our studies suggest that the contributions of Meis1 in generating MLL LSCs are mediated in part through the RB pathway by epigenetic modulation of Ink4a expression.
Since MLL fusion proteins, in conjunction with their essential cofactor menin, have been shown to associate with the Meis1 promoter (Milne et al. 2005; Yokoyama et al. 2005; Caslini et al. 2007), our observations raise the intriguing possibility that they may vary in their abilities to maintain expression of the Meis1 gene. Consistent with this possibility, MLL-ENL and MLL-FKBP, which differ in their oncogenic potencies, impose different histone modifications at the Meis1 promoter (Milne et al. 2005). Thus, the absolute levels of Meis1 expression may be differentially regulated by the transcriptional effector properties of individual MLL oncoproteins, which in turn dictates the rate of progression of immortalized cells to LSCs that initiate and sustain AML. This contrasts with Hox gene expression levels, which do not show a similar correlation with leukemia latencies or molecular subtypes (Supplementary Fig. 2).
Our studies suggest that TALE proteins may also critically maintain human MLL leukemia, since MEIS1 knockdown impaired the growth of leukemia cell lines. Interestingly, the relative prevalence of different MLL fusion genes in human leukemias is consistent with a potential rate-limiting role for MEIS gene expression comparable with mouse leukemias. Commonly recurring MLL fusion genes such as MLL-AF9, MLL-ENL, MLL-AF6, and MLL-AF10, which were associated with the highest levels of Meis1 expression in our mouse model, comprise the vast majority of spontaneous MLL-associated human AMLs. Conversely, MLL-EB1 and MLL-CBP, which are associated with lower Meis1 expression in our model, have only rarely been observed in human AML (a single case for MLL-EB1) (Fu et al. 2005). Furthermore, MLL-CBP is associated with human MDS (Satake et al. 1997) and induces an MDS-like disorder in mice (J. Wang et al. 2005), which are premalignant conditions characterized by altered self-renewal and differentiation potentials, suggesting that MLL-CBP initiates transformation but does not readily facilitate progression to AML. Thus, MEIS family gene expression levels in subclinical initiated progenitors harboring MLL chromosomal translocations may influence the probability of acquiring necessary secondary mutations for their conversion into LSCs capable of inducing clinical leukemia.
MLL-mediated transformation is dependent on Pbx function
Pbx proteins heterodimerize and bind DNA with Meis proteins, which also regulate Pbx stability and nuclear localization. Consistent with their biochemical interactions, lack of Meis1 partially phenocopies Pbx1 deficiency in hematopoietic development (DiMartino et al. 2001; Hisa et al. 2004). In MLL leukemias, Meis1 contributions are dependent on the integrity of its Pbx dimerization motifs. Unlike the case for Meis1, however, single deficiencies of Pbx2 or Pbx3, the most highly expressed Pbx genes in myeloid progenitors, did not abrogate MLL-mediated transformation. Nevertheless, the observed reduction of clonogenic activity in Pbx2/3 compound-deficient cells indicates that transformation of myeloid progenitors by MLL oncoproteins is dependent on Pbx function. This is a specific requirement and not reflective of a general suppressive effect of Pbx deficiency on myeloid transformation, since no impairment of proliferation or enhanced self-renewal was observed in Pbx2−/−Pbx3KD cells transformed by E2A-HLF, which transforms through a Hox-independent pathway (Ayton and Cleary 2003; So et al. 2004) and would not be expected to require Hox cofactors such as Pbx for oncogenesis.
Dependence on the combined contributions of Pbx2 and Pbx3 may reflect their redundant properties, consistent with previous studies demonstrating that different Pbx proteins exhibit essentially identical cooperative DNA binding with a subset of Hox proteins in vitro (Chang et al. 1995). Although we cannot exclude that Pbx2 and Pbx3 make isoform-specific contributions to MLL oncogenesis, it is more likely that single Pbx deficiencies are functionally compensated by other members of the Pbx protein family, whereas compound deficiencies reduce total Pbx dosage below a critical functional threshold. In support of this, novel compound-deficient Pbx phenotypes have recently been reported for Pbx1 and Pbx2 in limb development (Capellini et al. 2006) despite the fact that Pbx2 deficiency alone results in no phenotype (Selleri et al. 2004). Our data suggest that reduction of total Pbx (Pbx1, Pbx2, Pbx3) expression substantially below half of wild-type levels is limiting for MLL transformation, thus establishing a critical role for Pbx TALE proteins.
In summary, TALE homeodomain proteins are rate-limiting for many of the biological properties that define MLL LSCs. Meis1 in particular regulates LSC frequencies and their origin from immortalized myeloid progenitors. Although deregulated Hox gene expression is consistently induced by MLL fusion oncogenes, Hox protein function may be limited by the availability of TALE protein cofactors, which are more variable in their abundance and correlate with leukemia latency and biologic heterogeneity. The central role of TALE proteins in MLL leukemia maintenance suggests their consideration as potential therapeutic targets, which warrants further investigation.
Materials and methods
Mice
Knockout mice deficient for Pbx2 (Selleri et al. 2004), Pbx3 (Rhee et al. 2004), and Meis1 (Hisa et al. 2004) were maintained on a C57BL/6 genetic background. C57BL/6 mice congenic for CD45 (Ly5.1/Ly5.2) were employed for transplant studies to distinguish donor and recipient cells.
Cell lines
The human leukemia cell lines MV4.11, K562, ML2, THP-1, SEMK2, and HB were obtained from the American Type Culture Collection (ATCC) or generated in our laboratory and were maintained under standard conditions.
Retroviral constructs and hematopoietic progenitor transformation assays
Retroviral constructs encoding MLL-GAS7, MLL-AF1P, MLL-AF6, MLL-ENL, MLL-AF9, MLL-AF10, MLL-FKHRL1, MLL-AFX, MLL-CBP, E2A-HLF, and Meis1 have been described previously (Lavau et al. 1997, 2000; DiMartino et al. 2002; So and Cleary 2002, 2003; So et al. 2003, 2004; Fu et al. 2005; J. Wang et al. 2005; Somervaille and Cleary 2006). Retroviral constructs encoding MLL-LAF4, MLL-AF5, MLL-AF6, and MLL-EB1 were generated by insertion of the respective cDNAs or subtotal fragments with oncogenic potential (encoding LAF4 amino acids 335–1227, AF5 amino acids 727–1163, AF6 amino acids 35–137, and EB1 amino acids 199–269;) (Imamura et al. 2002a; von Bergh et al. 2002; Fu et al. 2005) into the MLL 5′ vector using standard cloning techniques. A dominant-negative Meis1 construct, analogous to known dominant-negative Drosophila and Xenopus Meis isoforms (Dibner et al. 2001; Inbal et al. 2001), was generated by deletion of C-terminal amino acids 280–390, which encode a portion of the homeodomain and downstream residues. Retroviral constructs for Meis1Δ62–202 (Pbx1), M1ΔLFPLL/M2ΔLLEL (Pbx2), HDΔN51S (HD1), HDΔN51S RRR53-5AEE (HD2), and Meis1 341T with a deletion of a conserved C terminus were reported previously (G.G. Wang et al. 2005). Hematopoietic transformation assays were performed essentially as described previously using primary murine myeloid progenitors harvested from bone marrow or FL (So et al. 2004). Cells transduced with retroviral vectors were selected for stable transduction in methylcellulose medium containing the appropriate antibiotic (250 μg/mL hygromycin, 1 μg/mL puromycin, and/or 1mg/mL neomycin).
Lentivirus generation and secondary transduction
The pSicoR lentiviral vector (Ventura et al. 2004) carrying either puromycin resistance or GFP marker genes was used for knockdown studies. shRNAs (Supplementary Table 2) were designed using pSicoOligomaker 1.5 (developed by A. Ventura, Jacks Laboratory, Cambridge, MA) and cloned into HpaI–XhoI-digested pSicoR. Lentiviral stocks were generated essentially as described previously (Ventura et al. 2004). In brief, DNA constructs encoding lentiviral vectors (5 μg), CMV gag-pol-rev 8.74 (4 μg), and VSVG (1 μg) were cotransfected into 293T cells using FuGENE 6 reagent (Roche Diagnostics). Supernatants were collected 36–48 h after transfection, passed through a 0.4-μm filter, and immediately incubated with immortalized cells (2 × 104 to 4 × 104) or human cell lines (1 × 105) for 12 h at 37°C. Cells transduced with pSicoR-GFP vectors were cultured in liquid medium for 48 h and then purified by FACS sorting for GFP expression. Transduced mouse cells (2000 cells) were plated in methylcellulose medium (M3231; Stem Cell Technologies) supplemented with cytokines (20 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, 10 ng/mL GM-CSF) or in MethoCult (H4236; Stem Cell Technologies) for human cells. Cells transduced with pSicoR carrying the puromycin resistance gene were plated 24 h after transduction in 0.9% methylcellulose medium (M3231) supplemented with cytokines and 1 μg/mL puromycin.
Leukemogenicity and long-term in vivo reconstitution assays
Transplantation experiments were performed as described previously (Lavau et al. 2000) with the following minor modifications. For cotransduction experiments, transduced progenitors were incubated in 0.9% methycellulose medium containing cytokines (20 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3) in the presence of puromycin (1 μg/mL) and neomycin (1 mg/mL) for 5 d, and then transplanted (1 × 105 cells) together with a radioprotective dose of total bone marrow cells (2 × 105) into the retro-orbital venous sinus of 6- to 12-wk-old syngeneic C57BL/6 mice that had been lethally irradiated with 9.0 Gy of total body γ irradiation (135Cs). When transplanted mice exhibited signs of ill health (shortness of breath, lethargy, and hunched posture) they were euthanized. Donor and recipient cells were distinguished by FACs analysis of CD45 congenic marker expression. Necropsy tissues were fixed in buffered formalin, sectioned, and stained with hematoxylin and eosin (H&E) for histological analysis. Secondary transplants were performed by retro-orbital injection of leukemia cells (3 × 103 from a single colony or 2.5 × 105 expanded in semisolid medium) into sublethally irradiated (450 cGy) syngeneic C57BL/6 mice.
Flow cytometry analysis
Bone marrow and spleen cells were stained with fluorochrome-conjugated monoclonal antibodies to either c-Kit (2B8 clone), Mac-1 (M1/70 clone), Gr-1 (RB6-8C5 clone), CD19 (1D3 clone), B220 (RA3-6B2 clone), CD45.1 (A20.1.7), or CD45.2 (AL1-4A2). Antibodies were purchased from PharMingen or eBioscience. Procedures employed for cell staining and FACS analysis have been described previously (So et al. 2004). DNA content analysis was performed by PI staining and analyzed by FACS.
Immunoblotting
Transiently transfected Phoenix cells were harvested and lysed in 250 μL of 2× sample buffer. Proteins from ∼20 μL of lysate were fractionated by electrophoresis through 4% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Bio-Rad) using Tris-glycine sodium dodecyl sulfate transfer buffer. After blocking with 5% milk, membranes were incubated with monoclonal antibody N4.4 directed against an MLL N-terminal epitope, and then processed for chemiluminescent detection. HA-tagged Meis1a proteins were detected using the M2 anti-HA monoclonal antibody (Abcam). Anti-actin (mouse monoclonal C4) was obtained from Chemicon International.
Real-time quantitative PCR analysis of gene expression
cDNA was synthesized and subjected to real-time PCR as described previously (Yokoyama et al. 2005). TaqMan probes for the following mouse and human genes were purchased from Applied Biosystems: Meis1 (Mm00487664_m1), Meis2 (Mm00487748_m1), Meis3 (Mm00485209_m1), Pbx1 (Mm00435507_m1), Pbx2 (Mm00479560_m1), Pbx3 (Mm00479413_m1), p19Arf (Mm01257348), Bmi1 (Mm00776122_gH), Cdkn1a (p21) (Mm01303209_m1), Cdkn1b (p27) (Mm00438167_g1), Cdkn2a (Mm00494449_m1), Cdkn2c (Mm00483243_m1), Cdkn4b p15 (Mm00483241_m1), β-Actin (Mm00607939_s1); MEIS1 (Hs00180020_m1), MEIS2 (Hs00542638_m1), MEIS3 (Hs00911770_g1), and ACTB (Hs99999903 _m1). Primers for mouse p16Ink4a were designed previously (Zhang et al. 2003) and purchased from Applied Biosystems. Expression levels of target transcripts relative to that of β-Actin were calculated using a standard curve and relative quantitation methods as described in ABI User Bulletin #2. Relative dosages of Meis, Pbx, and Hox transcripts were calculated using relative cycle time (Ct) value according to the manufacturer’s instructions.
Acknowledgments
We thank P. Ayton for initially suggesting the Meis1 dependence of MLL transformation. We acknowledge M. Ambrus, C. Nicolas, and K. Ochis for technical support, and A. Yokoyama, M. Lin, and J. Sage for technical guidance and assistance. We gratefully acknowledge N. Copeland (Institute of Molecular and Cell Biology, Singapore) for Meis1 knockout mice, and M. Kamps (University of California at San Diego) for Meis1 constructs. These studies were supported by the Children’s Health Initiative of the Packard Foundation, grants from the National Institutes of Health (CA55029 and CA42971), and in part by a Croucher Foundation Research Grant to P.W. T.C.P.S. was supported by a Leukaemia Research Fund (UK) Senior Clinical Fellowship; M.I. was supported in part by the NCI-JFCR Scientist Exchange Program.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1602107
References
- Abramovich C., Humphries R.K., Humphries R.K. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- Ayton P.M., Cleary M.L., Cleary M.L. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes & Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N., Morata G., Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes & Dev. 1998;12:261–273. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo K.R., Knoepfler P.S., Sykes D.B., Pasillas M.P., Kamps M.P., Knoepfler P.S., Sykes D.B., Pasillas M.P., Kamps M.P., Sykes D.B., Pasillas M.P., Kamps M.P., Pasillas M.P., Kamps M.P., Kamps M.P. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: Potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc. Natl. Acad. Sci. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini T.D., Di Giacomo G., Salsi V., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L., Di Giacomo G., Salsi V., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L., Salsi V., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L., Brendolan A., Ferretti E., Srivastava D., Zappavigna V., Selleri L., Ferretti E., Srivastava D., Zappavigna V., Selleri L., Srivastava D., Zappavigna V., Selleri L., Zappavigna V., Selleri L., Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Caslini C., Yang Z., El-Osta M., Milne T.A., Slany R.K., Hess J.L., Yang Z., El-Osta M., Milne T.A., Slany R.K., Hess J.L., El-Osta M., Milne T.A., Slany R.K., Hess J.L., Milne T.A., Slany R.K., Hess J.L., Slany R.K., Hess J.L., Hess J.L. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.P., Shen W.F., Rozenfeld S., Lawrence H.J., Largman C., Cleary M.L., Shen W.F., Rozenfeld S., Lawrence H.J., Largman C., Cleary M.L., Rozenfeld S., Lawrence H.J., Largman C., Cleary M.L., Lawrence H.J., Largman C., Cleary M.L., Largman C., Cleary M.L., Cleary M.L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes & Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Dibner C., Elias S., Frank D., Elias S., Frank D., Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development. 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- DiMartino J.F., Selleri L., Traver D., Firpo M.T., Rhee J., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., Selleri L., Traver D., Firpo M.T., Rhee J., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., Traver D., Firpo M.T., Rhee J., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., Firpo M.T., Rhee J., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., Rhee J., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., Warnke R., O’Gorman S., Weissman I.L., Cleary M.L., O’Gorman S., Weissman I.L., Cleary M.L., Weissman I.L., Cleary M.L., Cleary M.L. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood. 2001;98:618–626. doi: 10.1182/blood.v98.3.618. [DOI] [PubMed] [Google Scholar]
- DiMartino J.F., Ayton P.M., Chen E.H., Naftzger C.C., Young B.D., Cleary M.L., Ayton P.M., Chen E.H., Naftzger C.C., Young B.D., Cleary M.L., Chen E.H., Naftzger C.C., Young B.D., Cleary M.L., Naftzger C.C., Young B.D., Cleary M.L., Young B.D., Cleary M.L., Cleary M.L. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- Eklund E.A. The role of HOX genes in malignant myeloid disease. Curr. Opin. Hematol. 2007;14:85–89. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Armstrong S.A., Neuberg D.S., Sallan S.E., Silverman L.B., Korsmeyer S.J., Look A.T., Armstrong S.A., Neuberg D.S., Sallan S.E., Silverman L.B., Korsmeyer S.J., Look A.T., Neuberg D.S., Sallan S.E., Silverman L.B., Korsmeyer S.J., Look A.T., Sallan S.E., Silverman L.B., Korsmeyer S.J., Look A.T., Silverman L.B., Korsmeyer S.J., Look A.T., Korsmeyer S.J., Look A.T., Look A.T. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- Fu J.F., Hsu H.C., Shih L.Y., Hsu H.C., Shih L.Y., Shih L.Y. MLL is fused to EB1 (MAPRE1), which encodes a microtubule-associated protein, in a patient with acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2005;43:206–210. doi: 10.1002/gcc.20174. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Nakamura T., Croce C.M., Canaani E., Young R.A., Croce C.M., Canaani E., Young R.A., Canaani E., Young R.A., Young R.A. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R.D., Hess J.L., Yu B.D., Ernst P., van Lohuizen M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., Hess J.L., Yu B.D., Ernst P., van Lohuizen M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., Yu B.D., Ernst P., van Lohuizen M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., Ernst P., van Lohuizen M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., van Lohuizen M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., Berns A., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., van der Lugt N.M., Shashikant C.S., Ruddle F.H., Seto M., Shashikant C.S., Ruddle F.H., Seto M., Ruddle F.H., Seto M., Seto M., et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisa T., Spence S.E., Rachel R.A., Fujita M., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Spence S.E., Rachel R.A., Fujita M., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Rachel R.A., Fujita M., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Fujita M., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Nakamura T., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Ward J.M., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Devor-Henneman D.E., Saiki Y., Kutsuna H., Tessarollo L., Saiki Y., Kutsuna H., Tessarollo L., Kutsuna H., Tessarollo L., Tessarollo L., et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T., Morimoto A., Ikushima S., Kakazu N., Hada S., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Morimoto A., Ikushima S., Kakazu N., Hada S., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Ikushima S., Kakazu N., Hada S., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Kakazu N., Hada S., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Hada S., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Tabata Y., Yagi T., Inaba T., Hibi S., Sugimoto T., Yagi T., Inaba T., Hibi S., Sugimoto T., Inaba T., Hibi S., Sugimoto T., Hibi S., Sugimoto T., Sugimoto T., et al. A novel infant acute lymphoblastic leukemia cell line with MLL-AF5q31 fusion transcript. Leukemia. 2002a;16:2302–2308. doi: 10.1038/sj.leu.2402665. [DOI] [PubMed] [Google Scholar]
- Imamura T., Morimoto A., Takanashi M., Hibi S., Sugimoto T., Ishii E., Imashuku S., Morimoto A., Takanashi M., Hibi S., Sugimoto T., Ishii E., Imashuku S., Takanashi M., Hibi S., Sugimoto T., Ishii E., Imashuku S., Hibi S., Sugimoto T., Ishii E., Imashuku S., Sugimoto T., Ishii E., Imashuku S., Ishii E., Imashuku S., Imashuku S. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br. J. Haematol. 2002b;119:119–121. doi: 10.1046/j.1365-2141.2002.03803.x. [DOI] [PubMed] [Google Scholar]
- Inbal A., Halachmi N., Dibner C., Frank D., Salzberg A., Halachmi N., Dibner C., Frank D., Salzberg A., Dibner C., Frank D., Salzberg A., Frank D., Salzberg A., Salzberg A. Genetic evidence for the transcriptional-activating function of Homothorax during adult fly development. Development. 2001;128:3405–3413. doi: 10.1242/dev.128.18.3405. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M., Kieboom K., Marino S., DePinho R.A., van Lohuizen M., Marino S., DePinho R.A., van Lohuizen M., DePinho R.A., van Lohuizen M., van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Wang J., Hahn W.C., Gilliland D.G., Hahn W.C., Gilliland D.G., Gilliland D.G., et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G., Baban S., Buchberg A.M., Sauvageau G., Buchberg A.M., Sauvageau G., Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.R., Hudson W.A., Chen W., Nishiuchi R., Yao Q., Kersey J.H., Hudson W.A., Chen W., Nishiuchi R., Yao Q., Kersey J.H., Chen W., Nishiuchi R., Yao Q., Kersey J.H., Nishiuchi R., Yao Q., Kersey J.H., Yao Q., Kersey J.H., Kersey J.H. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- Lavau C., Szilvassy S.J., Slany R., Cleary M.L., Szilvassy S.J., Slany R., Cleary M.L., Slany R., Cleary M.L., Cleary M.L. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Du C., Thirman M., Zeleznik-Le N., Du C., Thirman M., Zeleznik-Le N., Thirman M., Zeleznik-Le N., Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence H.J., Rozenfeld S., Cruz C., Matsukuma K., Kwong A., Komuves L., Buchberg A.M., Largman C., Rozenfeld S., Cruz C., Matsukuma K., Kwong A., Komuves L., Buchberg A.M., Largman C., Cruz C., Matsukuma K., Kwong A., Komuves L., Buchberg A.M., Largman C., Matsukuma K., Kwong A., Komuves L., Buchberg A.M., Largman C., Kwong A., Komuves L., Buchberg A.M., Largman C., Komuves L., Buchberg A.M., Largman C., Buchberg A.M., Largman C., Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- Lessard J., Sauvageau G., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Manley N.R., Selleri L., Brendolan A., Gordon J., Cleary M.L., Selleri L., Brendolan A., Gordon J., Cleary M.L., Brendolan A., Gordon J., Cleary M.L., Gordon J., Cleary M.L., Cleary M.L. Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 2004;276:301–312. doi: 10.1016/j.ydbio.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Mann R.S. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Milne T.A., Martin M.E., Brock H.W., Slany R.K., Hess J.L., Martin M.E., Brock H.W., Slany R.K., Hess J.L., Brock H.W., Slany R.K., Hess J.L., Slany R.K., Hess J.L., Hess J.L. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y., Li Y., Coffield V.M., Su L., Xu G., Zhang Y., Coffield V.M., Su L., Xu G., Zhang Y., Su L., Xu G., Zhang Y., Xu G., Zhang Y., Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Weissman I.L., Morrison S.J., Clarke M.F., Morrison S.J., Clarke M.F., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Passegue E., Weisman I.L., Weisman I.L. Leukemic stem cells: Where do they come from? Stem Cell Rev. 2005;1:181–188. doi: 10.1385/SCR:1:3:181. [DOI] [PubMed] [Google Scholar]
- Pineault N., Helgason C.D., Lawrence H.J., Humphries R.K., Helgason C.D., Lawrence H.J., Humphries R.K., Lawrence H.J., Humphries R.K., Humphries R.K. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- Quentmeier H., Dirks W.G., Macleod R.A., Reinhardt J., Zaborski M., Drexler H.G., Dirks W.G., Macleod R.A., Reinhardt J., Zaborski M., Drexler H.G., Macleod R.A., Reinhardt J., Zaborski M., Drexler H.G., Reinhardt J., Zaborski M., Drexler H.G., Zaborski M., Drexler H.G., Drexler H.G. Expression of HOX genes in acute leukemia cell lines with and without MLL translocations. Leuk. Lymphoma. 2004;45:567–574. doi: 10.1080/10428190310001609942. [DOI] [PubMed] [Google Scholar]
- Rhee J.W., Arata A., Selleri L., Jacobs Y., Arata S., Onimaru H., Cleary M.L., Arata A., Selleri L., Jacobs Y., Arata S., Onimaru H., Cleary M.L., Selleri L., Jacobs Y., Arata S., Onimaru H., Cleary M.L., Jacobs Y., Arata S., Onimaru H., Cleary M.L., Arata S., Onimaru H., Cleary M.L., Onimaru H., Cleary M.L., Cleary M.L. Pbx3 deficiency results in central hypoventilation. Am. J. Pathol. 2004;165:1343–1350. doi: 10.1016/S0002-9440(10)63392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice K.L., Licht J.D., Licht J.D. HOX deregulation in acute myeloid leukemia. J. Clin. Invest. 2007;117:865–868. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.E., Mahfouz R., Onciu M., Liu H.C., Zhou X., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Mahfouz R., Onciu M., Liu H.C., Zhou X., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Onciu M., Liu H.C., Zhou X., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Liu H.C., Zhou X., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Zhou X., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Song G., Shurtleff S.A., Pounds S., Cheng C., Ma J., Shurtleff S.A., Pounds S., Cheng C., Ma J., Pounds S., Cheng C., Ma J., Cheng C., Ma J., Ma J., et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- Ryoo H.D., Marty T., Casares F., Affolter M., Mann R.S., Marty T., Casares F., Affolter M., Mann R.S., Casares F., Affolter M., Mann R.S., Affolter M., Mann R.S., Mann R.S. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- Satake N., Ishida Y., Otoh Y., Hinohara S., Kobayashi H., Sakashita A., Maseki N., Kaneko Y., Ishida Y., Otoh Y., Hinohara S., Kobayashi H., Sakashita A., Maseki N., Kaneko Y., Otoh Y., Hinohara S., Kobayashi H., Sakashita A., Maseki N., Kaneko Y., Hinohara S., Kobayashi H., Sakashita A., Maseki N., Kaneko Y., Kobayashi H., Sakashita A., Maseki N., Kaneko Y., Sakashita A., Maseki N., Kaneko Y., Maseki N., Kaneko Y., Kaneko Y. Novel MLL-CBP fusion transcript in therapy-related chronic myelomonocytic leukemia with a t(11;16)(q23;p13) chromosome translocation. Genes Chromosomes Cancer. 1997;20:60–63. [PubMed] [Google Scholar]
- Schnabel C.A., Jacobs Y., Cleary M.L., Jacobs Y., Cleary M.L., Cleary M.L. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- Selleri L., Depew M.J., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Depew M.J., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Chanda S.K., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Tsang K.Y., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Cheah K.S., Rubenstein J.L., O’Gorman S., Cleary M.L., Rubenstein J.L., O’Gorman S., Cleary M.L., O’Gorman S., Cleary M.L., Cleary M.L. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Selleri L., DiMartino J., van Deursen J., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., DiMartino J., van Deursen J., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., van Deursen J., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., Sanyal M., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., Boon E., Capellini T., Smith K.S., Rhee J., Popperl H., Capellini T., Smith K.S., Rhee J., Popperl H., Smith K.S., Rhee J., Popperl H., Rhee J., Popperl H., Popperl H., et al. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell. Biol. 2004;24:5324–5331. doi: 10.1128/MCB.24.12.5324-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N., Oseth L., LeBien T.W., Oseth L., LeBien T.W., LeBien T.W. Development of a model for evaluating the interaction between human pre-B acute lymphoblastic leukemic cells and the bone marrow stromal cell microenvironment. Blood. 1998;92:3817–3828. [PubMed] [Google Scholar]
- So C.W., Cleary M.L., Cleary M.L. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell. Biol. 2002;22:6542–6552. doi: 10.1128/MCB.22.18.6542-6552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.W., Cleary M.L., Cleary M.L. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood. 2003;101:633–639. doi: 10.1182/blood-2002-06-1785. [DOI] [PubMed] [Google Scholar]
- So C.W., Lin M., Ayton P.M., Chen E.H., Cleary M.L., Lin M., Ayton P.M., Chen E.H., Cleary M.L., Ayton P.M., Chen E.H., Cleary M.L., Chen E.H., Cleary M.L., Cleary M.L. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- So C.W., Karsunky H., Wong P., Weissman I.L., Cleary M.L., Karsunky H., Wong P., Weissman I.L., Cleary M.L., Wong P., Weissman I.L., Cleary M.L., Weissman I.L., Cleary M.L., Cleary M.L. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- Somervaille T.C., Cleary M.L., Cleary M.L. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Swansbury G.J. The proportion of clonal divisions varies in different hematologic malignancies. Cancer Genet. Cytogenet. 1998;104:139–145. doi: 10.1016/s0165-4608(97)00477-9. [DOI] [PubMed] [Google Scholar]
- Ventura A., Meissner A., Dillon C.P., McManus M., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T., Meissner A., Dillon C.P., McManus M., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T., Dillon C.P., McManus M., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T., McManus M., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T., Van Parijs L., Jaenisch R., Jacks T., Jaenisch R., Jacks T., Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergh A.R., Beverloo H.B., Rombout P., van Wering E.R., van Weel M.H., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., Beverloo H.B., Rombout P., van Wering E.R., van Weel M.H., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., Rombout P., van Wering E.R., van Weel M.H., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., van Wering E.R., van Weel M.H., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., van Weel M.H., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., Beverstock G.C., Kluin P.M., Slater R.M., Schuuring E., Kluin P.M., Slater R.M., Schuuring E., Slater R.M., Schuuring E., Schuuring E. LAF4, an AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2002;35:92–96. doi: 10.1002/gcc.10091. [DOI] [PubMed] [Google Scholar]
- Wang G.G., Pasillas M.P., Kamps M.P., Pasillas M.P., Kamps M.P., Kamps M.P. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Iwasaki H., Krivtsov A., Febbo P.G., Thorner A.R., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Iwasaki H., Krivtsov A., Febbo P.G., Thorner A.R., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Krivtsov A., Febbo P.G., Thorner A.R., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Febbo P.G., Thorner A.R., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Thorner A.R., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Ernst P., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Anastasiadou E., Kutok J.L., Kogan S.C., Zinkel S.S., Kutok J.L., Kogan S.C., Zinkel S.S., Kogan S.C., Zinkel S.S., Zinkel S.S., et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.K., Wang J.C., Takenaka K., Doulatov S., McKenzie J.L., Harrington L., Dick J.E., Wang J.C., Takenaka K., Doulatov S., McKenzie J.L., Harrington L., Dick J.E., Takenaka K., Doulatov S., McKenzie J.L., Harrington L., Dick J.E., Doulatov S., McKenzie J.L., Harrington L., Dick J.E., McKenzie J.L., Harrington L., Dick J.E., Harrington L., Dick J.E., Dick J.E. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia. 2005;19:1794–1805. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Somervaille T.C., Smith K.S., Rozenblatt-Rosen O., Meyerson M., Cleary M.L., Somervaille T.C., Smith K.S., Rozenblatt-Rosen O., Meyerson M., Cleary M.L., Smith K.S., Rozenblatt-Rosen O., Meyerson M., Cleary M.L., Rozenblatt-Rosen O., Meyerson M., Cleary M.L., Meyerson M., Cleary M.L., Cleary M.L. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess J.L., Horning S.E., Brown G.A., Korsmeyer S.J., Hess J.L., Horning S.E., Brown G.A., Korsmeyer S.J., Horning S.E., Brown G.A., Korsmeyer S.J., Brown G.A., Korsmeyer S.J., Korsmeyer S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zeisig B.B., Milne T., Garcia-Cuellar M.P., Schreiner S., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Milne T., Garcia-Cuellar M.P., Schreiner S., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Garcia-Cuellar M.P., Schreiner S., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Schreiner S., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Martin M.E., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Fuchs U., Borkhardt A., Chanda S.K., Walker J., Soden R., Borkhardt A., Chanda S.K., Walker J., Soden R., Chanda S.K., Walker J., Soden R., Walker J., Soden R., Soden R., et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qian X., Redman C., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A., Qian X., Redman C., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A., Redman C., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A., Ramsay E.S., Lowy D.R., Mock B.A., Lowy D.R., Mock B.A., Mock B.A. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene. 2003;22:2285–2295. doi: 10.1038/sj.onc.1206257. [DOI] [PubMed] [Google Scholar]