Abstract
FoxO transcription factors play critical roles in cell cycle control and cellular stress responses, and abrogation of FoxO function promotes focus formation by Myc in vitro. Here we show that stable introduction of a dominant-negative FoxO moiety (dnFoxO) into Eμ-myc transgenic hematopoietic stem cells accelerates lymphoma development in recipient mice by attenuating Myc-induced apoptosis. When expressed in Eμ-myc; p53+/− progenitor cells, dnFoxO alleviates the pressure to inactivate the remaining p53 allele in upcoming lymphomas. Expression of the p53 upstream regulator p19Arf is virtually undetectable in most dnFoxO-positive Myc-driven lymphomas. We find that FoxO proteins bind to a distinct site within the Ink4a/Arf locus and activate Arf expression. Moreover, constitutive Myc signaling induces a marked increase in nuclear FoxO levels and stimulates binding of FoxO proteins to the Arf locus. These data demonstrate that FoxO factors mediate Myc-induced Arf expression and provide direct genetic evidence for their tumor-suppressive capacity.
[Keywords: Arf, FoxO, lymphoma, mouse model, Myc]
The FoxO subclass of forkhead-box transcription factors—consisting of FoxO1 (FKHR), FoxO3a (FKHRL1), FoxO4 (AFX), and FoxO6—regulates numerous cellular functions including proliferation, stress sensitivity, and survival; it has also been implicated in the regulation of organism life span (for review, see Birkenkamp and Coffer 2003; Accili and Arden 2004; Greer and Brunet 2005). The members of this family activate gene expression via interaction with a specific DNA sequence, and known targets include the cell cycle regulating Kip1 (Medema et al. 2000), the proapoptotic Bim (Dijkers et al. 2000), the DNA damage-responsive Gadd45a (Tran et al. 2002), and the oxidative stress-protective manganese superoxide dismutase (Kops et al. 2002) genes. In addition, FoxO proteins can repress several cell cycle promoting genes (e.g., cyclin D1 and cyclin D2) in a manner that might be independent of direct DNA binding (Ramaswamy et al. 2002; Bouchard et al. 2004).
In response to growth factor signaling and to oxidative stress, FoxO proteins are post-translationally modified by phosphorylation, acetylation, and ubiquitination; collectively, these modifications regulate FoxOs’ subcellular localization, transcriptional activity, and stability (Brunet et al. 1999, 2004; Motta et al. 2004; van der Horst et al. 2006). Notably, all FoxO proteins are inhibited by protein kinase B/Akt-mediated phosphorylation that promotes their nuclear export and subsequent proteolytic degradation via ubiquitination by the SCFSkp2 complex (Brunet et al. 1999; Huang et al. 2005). As a consequence, FoxO proteins mediate the induction of p27Kip1 and Bim expression in response to inhibition of the phosphatidylinositol-3-OH (PI3)-kinase/Akt pathway (Medema et al. 2000; Nakamura et al. 2000; Stahl et al. 2002).
Conditional codeletion of the FoxO1, FoxO3, and FoxO4 alleles uncovers a context-dependent cancer-prone phenotype characterized by thymic lymphomas forming in some and hemangiomas developing in most animals after a long latency (Paik et al. 2007), suggesting that FoxO proteins exert their tumor-suppressive capability in the presence of additional oncogenic mutations. In support of this view, we recently identified Akt-mediated phosphorylation of FoxO proteins as the critical PI3-kinase signaling component that substitutes for oncogenic Ras in Myc-induced proliferation and focus formation in vitro (Land et al. 1983; Bouchard et al. 2004). Furthermore, constitutive Akt signaling cooperates with Myc to accelerate B-cell lymphomagenesis (Wendel et al. 2004); however, it remains unclear whether Akt-mediated phosphorylation of FoxO proteins contributes to Eμ-myc transgenic lymphoma formation in this setting.
Proapoptotic Arf/p53 signaling is known as the pivotal Myc-induced tumor-suppressive barrier (Sherr 2006), which, in turn, must be attenuated in full-blown Myc-driven malignancies (Zindy et al. 1998; Eischen et al. 1999; Bertwistle and Sherr 2006). Eμ-myc transgenic mice lacking one p53 allele develop lymphomas that inactivate the remaining wild-type allele (Hsu et al. 1995; Schmitt et al. 1999). Likewise, Eμ-myc; Arf+/− or Eμ-myc; Ink4a/Arf+/− mice produce tumors that lack expression of p19Arf (Eischen et al. 1999; Schmitt et al. 2002a). Primary Arf deletions protect cells from acquiring p53 mutations during lymphoma development (Eischen et al. 1999; Sherr et al. 2005). Similarly, introduction of strictly anti-apoptotic genes such as bcl2 or a dominant-negative form of caspase 9 into Eμ-myc; p53+/− hematopoietic stem cells alleviates the pressure to inactivate p53, thereby underscoring apoptosis as the critical p53-governed tumor suppressor function in Myc-driven lymphomagenesis (Schmitt et al. 2002b).
Previous work has shown that p53 and FoxO3a share target genes and that FoxO3a can activate transcription via p53 sites, suggesting a potential collaboration of FoxO3a and p53 in tumor suppression (Nemoto et al. 2004; You et al. 2006). Although a direct interaction between FoxO3a and p53 proteins has been demonstrated under conditions of overexpression (Nemoto et al. 2004), the observed collaboration would be consistent with an as-yet-unidentified FoxO target acting upstream of p53. We report here that FoxO factors elicit their tumor-suppressive potential as critical inducers of Arf during Myc-driven lymphomagenesis, providing further evidence for a close link between the FoxO and p53 tumor suppressor pathways.
Results
FoxO inactivation promotes Myc-driven lymphomagenesis by disabling p53-mediated apoptosis
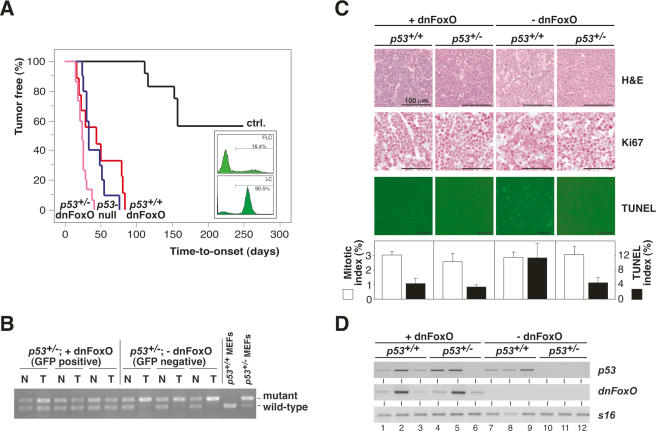
To examine the tumor-suppressive potential of FoxO transcription factors in vivo, we used a dominant-negative FoxO allele (dnFoxO) that comprises the conserved DNA-binding domain of FoxO4. This fragment binds DNA but is incapable of activating FoxO target genes due to lack of the transactivation domain, and has been shown to act as a dominant-negative allele for FoxO subclass members in several experimental systems (Medema et al. 2000; van den Heuvel et al. 2005). FoxO-inactivated Myc-driven B-cell lymphomas were generated via infection of Eμ-myc transgenic fetal liver cells, a source of hematopoietic progenitors, with a retrovirus encoding dnFoxO together with an IRES (internal ribosomal entry side)-linked green fluorescent protein (GFP) and their subsequent propagation in lethally irradiated recipient mice (Fig. 1A, insert). Extensive microarray analysis confirmed that these lymphomas expressed lower mRNAs levels of several FoxO target genes relative to lymphomas not expressing dnFoxO, confirming that the introduced dnFoxO moiety exerts a dominant-negative function in vivo (Supplementary Table 1; Supplementary Fig. 1). Expression of dnFoxO significantly accelerated the development of Eμ-myc lymphomas relative to lymphomas forming in animals reconstituted with empty vector-transduced cells (hereafter referred to as controls; P < 0.0001) (Fig. 1A). To determine whether p53-mediated signaling is still selected against during Myc-driven lymphomagenesis when FoxO function has been ablated, Eμ-myc; p53+/− fetal liver cells transduced with retroviruses encoding dnFoxO-IRES-GFP or GFP-only as a control were propagated in recipient mice as well. In both groups, lymphoma manifestation occurred much faster when compared with controls (both P < 0.0001), but lymphoma latency did not overtly differ between the two p53+/− groups. As expected, mock-transduced Eμ-myc; p53+/− fetal liver cells gave rise to lymphomas that deleted the remaining p53 wild-type allele (considered p53-null; 10 out of 10 cases tested by allele-specific genomic PCR for allelic p53 loss) (see also Schmitt et al. 2002b; three cases are shown in Fig. 1B). In contrast, all GFP-positive dnFoxO-lymphomas retained the remaining p53 allele (15 out of 15 cases tested; P < 0.0001) (three cases are shown in Fig. 1B). Thus, the dnFoxO moiety alleviates the selective pressure to inactivate p53 during Eμ-myc-driven lymphomagenesis.
Figure 1.
The dnFoxO moiety accelerates Myc-driven lymphomagenesis by blocking p53-dependent apoptosis. (A) Lymphoma incidence in recipient mice of Eμ-myc; p53+/+ or Eμ-myc; p53+/− hematopoietic stem cells stably transduced with dnFoxO (p53+/+;dnFoxO; n = 9 [red]; and p53+/−;dnFoxO; n = 15 [purple]) or mock-infected (p53+/+;mock [ctrl.]; n = 12 [black]; and p53+/−;mock [p53-null]; n = 10 [blue]). (Insert) Representative flow cytometric GFP scans of Eμ-myc; p53+/+ fetal liver cells (FLC) infected with the MSCV-dnFoxO-IRES-GFP retrovirus (top) and of lymphoma cells (LC) arising from this population after stem cell transplantation (percentage reflects fraction of GFP-positive cells), indicating positive selection of the dnFoxO moiety (bottom). (B) Allele-specific p53 PCR from genomic DNA extracted from representative p53+/−; dnFoxO (+dnFoxO) and p53+/−; mock (−dnFoxO) lymphoma samples and p53+/+ and p53+/− MEFs as internal PCR control (normal tissue [N], lymphoma tissue from the same animal [T]). (C) Lymph node histopathology of the indicated genotypes sampled at lymphoma diagnosis to visualize histomorphology and mitotic figures by hematoxylin/eosin (H&E), proliferation by Ki67, and spontaneous apoptosis by TUNEL staining in situ, and their respective quantifications. (D) Expression analysis of p53 and dnFoxO transcripts by RT–PCR in individual lymphomas (n = 3 per genotype) with S16 as a control.
No significant differences could be found regarding the proliferative capacity of lymphomas with different genotypes as assessed by Ki67 immunostaining and the frequency of mitotic figures in situ (Fig. 1C). However, spontaneous apoptosis, visualized by hematoxilin/eosin staining and quantified by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling), was detectable at significantly higher levels in control lymphomas when compared with lymphomas lacking intact p53 alleles or expressing dnFoxO (Fig. 1C). Hence, inactivation of FoxO family members accelerates Myc-driven lymphomagenesis primarily by suppressing p53-mediated apoptosis. Given the reduced levels of spontaneous apoptosis in the presence of p53 alleles in dnFoxO-driven lymphomas, we next addressed whether p53 is expressed in these genotypes. Comparable with the demonstration of p53 transcripts in nine out of nine control lymphomas, p53 transcripts remained detectable in seven out of eight Eμ-myc; p53+/− lymphomas with confirmed expression of the dnFoxO moiety (Fig. 1D; Supplementary Fig. 2A). In contrast, p53 expression was lost in four out of four Eμ-myc; p53+/− lymphomas, reflecting the loss of the second p53 allele (P = 0.0101) (three cases are shown in Fig. 1B,D). Sequence analysis of exons 4–8, spanning the DNA-binding domain, of the p53 transcripts present in two p53+/−; dnFoxO lymphomas (tumors 4 and 5) failed to uncover mutations, suggesting that the low levels of spontaneous apoptosis in these tumors are not caused by mutant p53.
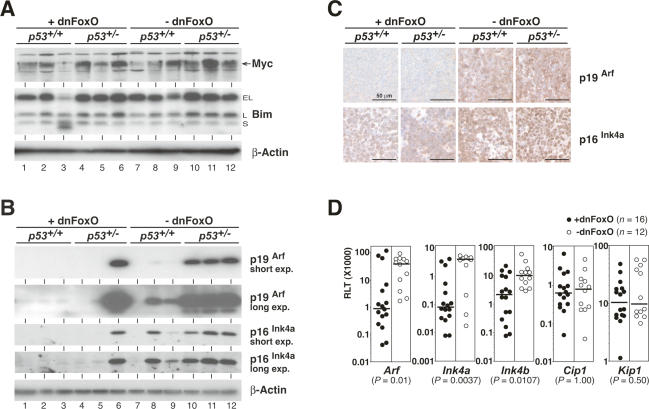
FoxO transcription factors enhance expression of the proapoptotic Bcl2 family member and BH3-only protein Bim in certain cell types (Stahl et al. 2002). Moreover, Bim is an essential mediator of Myc-induced cell death in B-cell lymphomagenesis (Egle et al. 2004), where Bim deletion protects against p53 loss (Hemann et al. 2005). Consequently, repression of Bim in lymphomas expressing dnFoxO might explain the observed phenotypes. However, no overt differences in the expression levels of the differentially spliced isoforms of Bim were observed throughout the genotypes (Fig. 2A; Supplementary Fig. 2B). Moreover, Myc levels were unaffected by the dnFoxO moiety, thereby excluding the possibility that ablation of FoxO function may allow lymphomas to form with lower Myc levels, which, in turn, might fail to activate p53 (Fig. 2A).
Figure 2.
Impact of dnFoxO on critical growth restraints in Eμ-myc transgenic lymphomas arising from a p53+/+ or p53+/− background. (A) Expression levels of Myc (depicted by arrow) and Bim (extra long [EL], long [L], and short [S] variants) and β-actin as a loading control by immunoblot analysis (three individual lymphomas per genotype). Note that p53+/− lymphomas without dnFoxO are in fact p53-null. (B) p19Arf and p16Ink4a protein expression (as in A; shown are both a short exposure [short exp.] and a long exposure [long exp.] of the same blot). (Lane 6) Please note that the only tumor that expresses both dnFoxO and detectable levels of p19Arf has lost p53 expression (cf. Fig. 1D). (C) Expression analysis of p19Arf and p16Ink4a proteins by immunohistochemistry in representative lymph node sections of the indicated genotypes. (D) RQ-PCR analyses of the indicated cell cycle inhibitors plotted as relative level of transcript (RLT) in 28 lymphomas with (closed circles; n = 16) and without (open circles; n = 12) the dnFoxO moiety. For each of these groups, the horizontal line represents the median of the relative expression level.
Increased expression of the translation factor eIF4E, which is released by Akt/mTORC1-mediated phosphorylation of 4EBP1 (for review, see Hay and Sonenberg 2004; Guertin and Sabatini 2007), can substitute for phosphorylation-activated Akt to accelerate Myc-driven lymphoma formation via suppression of apoptosis (Ruggero et al. 2004; Wendel et al. 2004). However, we observed neither reduced levels of 4EBP1 nor increased amounts of eIF4E transcripts in dnFoxO-lymphomas compared with lymphomas not carrying the dnFoxO allele (Supplementary Fig. 2C). Furthermore, we used a panel of antibodies directed against phospho-mTOR, phospho-Akt, and phospho-4EBP1 together with appropriate controls to judge the activity of the Akt/mTOR pathway throughout the lymphoma panel. There was no detectable impact of dnFoxO on phosphorylation or expression levels of any of these proteins, and, in particular, indistinguishable amounts of eIF4E protein were observed (Supplementary Fig. 2D). Thus, in cooperation with Myc, FoxO factors control neither directly (as transcriptional regulators) nor indirectly the activity of the Akt/mTORC1/4EBP/eIF4E pathway. Notably, our findings do not question the previously reported collaborative role of activated Akt or eIF4E in Myc-driven lymphomagenesis (Ruggero et al. 2004; Wendel et al. 2004), but argue against eIF4E as the critical mediator of FoxO function in Myc-driven lymphomagenesis.
DnFoxO-driven Myc-lymphomas display reduced Ink4a/Arf expression levels
Reduced apoptosis despite intact p53 genes has been demonstrated in Eμ-myc lymphomas harboring Ink4a/Arf defects (Eischen et al. 1999; Jacobs et al. 1999; Schmitt et al. 1999). While control lymphomas—except those that spontaneously selected for a biallelic deletion at the Ink4a/Arf locus (tumor 7 in Supplementary Fig. 2E)—and p53-null lymphomas express moderate or high levels of p19Arf and of p16Ink4a, p53-proficient dnFoxO-lymphomas (tumors 1–5) virtually lacked p19Arf expression and displayed markedly reduced protein levels of p16Ink4a (Fig. 2B,C), although no gross deletions at the gene locus were found in these cases (Supplementary Fig. 2E). Even when comparing only those lymphomas that retain expression of wild-type p53 (e.g., tumors 2 or 4 vs. 7–9, as verified by sequence analysis), average p19ARF levels were markedly higher in the absence of dnFoxO, suggesting a dnFoxO-dependent mechanism of Arf repression beyond the established p53/p19Arf negative feedback loop (Kamijo et al. 1998). Quantitative real-time reverse-transcriptase PCR (RQ-PCR) showed that the reduced expression of p19Arf and p16Ink4a was paralleled by lower (but detectable) Arf and Ink4a mRNA levels in lymphomas expressing dnFoxO (Fig. 2D). In addition, these lymphomas expressed moderately reduced levels of Ink4b mRNA, consistent with previous observations that all three genes encoded at the Ink4b/Ink4a/Arf locus are coregulated in several biological settings (Gil and Peters 2006). Further analyses showed that these effects were specific for this locus, since transcript levels of other cell cycle inhibitors, p21Cip1 and p27Kip1, remained unaffected by the dnFoxO moiety in Myc-induced lymphomas (Fig. 2D). Thus, FoxO inactivation correlates with greatly reduced levels of the Ink4a/Arf gene products in the context of constitutive Myc expression.
To extend our observation of dnFoxO’s ability to alleviate selective pressure against p53 alleles in Myc-driven lymphomagenesis to the Arf locus, Eμ-myc; Arf+/− fetal liver cells stably transduced with dnFoxO-IRES-GFP at ∼10% infection efficiency were transplanted into recipient animals, and upcoming lymphomas were examined by flow cytometry to identify the GFP-positive subset of lymphomas that formed with the dnFoxO-IRES-GFP cassette expressed (four out of seven cases). Independent of their dnFoxO status, lymphomas developed rapidly within the expected latency (data not shown; cf. Schmitt et al. 2002a). Importantly, all four dnFoxO-positive Eμ-myc; Arf+/− lymphomas were found to express virtually no p19Arf protein in accordance with low but detectable Arf transcript levels despite a lack of overt genomic defects at the Ink4a/Arf locus (three cases shown in Supplementary Fig. 3A–C). In contrast, all three GFP-negative and, hence, dnFoxO-negative Eμ-myc; Arf+/− lymphomas selected for Ink4a/Arf deletions that rendered them genomically Arf-null (one case shown in Supplementary Fig. 3A–C). These data and the previous observation that deletion of Arf, but not of Ink4a, at this locus cooperates with Myc in lymphoma development via interrupting oncogenic signaling to p53 (Eischen et al. 1999; Krimpenfort et al. 2001) strongly suggest that dnFoxO facilitates Myc-induced lymphomagenesis through reduction of Arf expression.
Myc-lymphomas expressing the dnFoxO moiety functionally recapitulate Arf−/− lymphomas
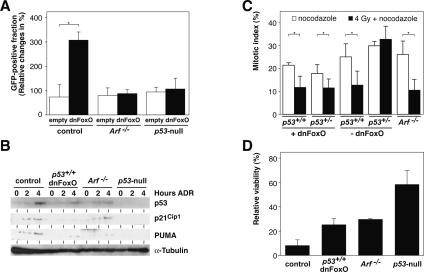
In order to test whether reduction of Arf levels is the crucial mechanism by which dnFoxO accelerates the formation of p53-proficient Myc-driven lymphomas, we infected cultured control, Arf−/−, and p53-null Eμ-myc lymphomas cells with retroviruses encoding either GFP only or the dnFoxO-IRES-GFP moiety. In these experiments, the fraction of GFP-only-infected cells remained unchanged over time. Consistent with the in vivo observations, expression of the dnFoxO moiety provided a selective advantage to control lymphomas, leading to a strong expansion of this subpopulation during the observation period (Fig. 3A). Importantly, no such expansion was observed in Arf−/− or p53-null lymphomas, demonstrating that inactivation of FoxO transcription factors fails to provide a specific growth advantage in Eμ-myc lymphomas that lack functional p19Arf or p53. Furthermore, dnFoxO-driven lymphomas, like p19Arf-deficient lymphomas, displayed a pseudo-diploid DNA content, while the chromosomally instable p53-deficient lymphomas exhibited overt aneuploidy (three out of three cases tested for each genotype) (data not shown; see also Eischen et al. 1999; Schmitt et al. 1999).
Figure 3.
Inactivation of FoxO transcription factors promotes development of Myc-lymphomas that retain an intact DNA damage response. (A) Relative changes of the GFP-positive fraction of control, Arf−/−, and p53-null lymphoma cells 48 h after infection with either a GFP-only (empty) or a dnFoxO-IRES-GFP-encoding (dnFoxO) retrovirus; asterisk denotes a significant P-value of <0.05. (B) Representative immunoblot analysis to detect p53, p21Cip1, and PUMA expression levels (with α-tubulin as a loading control) in lymphoma cells of the indicated genotypes exposed to 0.5 μg/mL DNA-damaging agent ADR for 2 or 4 h or left untreated. (C) Percentage of freshly isolated lymphoma cells of the indicated genotypes trapped in mitosis (i.e., displaying nuclei with condensed, homogeneously Hoechst-stained chromosomes) after 20 h of exposure to 0.1 μg/mL mitotic spindle poison nocodazole alone or applied 2 h after an initial γ-irradiation of 4 Gy (n = 3 each). (D) Viability analysis by trypan blue dye exclusion of the indicated lymphoma cell populations exposed in vitro for 24 h to 0.5 μg/mL ADR relative to untreated cells of the same genotypes (n = 3 each).
p19Arf primarily acts via physical binding and degradation of the p53 E3 ubiquitin ligase Mdm2 (Zhang et al. 1998), thereby controlling p53 protein levels independent of its activation via post-translational modifications in response to DNA damage (Kamijo et al. 1999). Upon exposure to the DNA-damaging anti-cancer agent adriamycin (ADR), dnFoxO-expressing and Arf−/− lymphomas displayed similar levels of p53 activation and induction of the p53 targets Cip1 and PUMA, encoding a proapoptotic BH3-only protein (Fig. 3B; Nakano and Vousden 2001). The induced protein levels were expectedly less pronounced when compared with control lymphoma cells not expressing the dnFoxO moiety due to the much lower basal p53 levels in Arf−/− and dnFoxO-lymphomas. No induction of p21Cip1 or of PUMA could be detected in ADR-treated p53-null lymphoma cells (see also Schmitt et al. 1999). To directly test the functionality of this DNA damage-triggered p53 response, we subjected lymphoma cells to a “mitotic trap” assay, in which p53-proficient cells are expected to arrest in response to γ-irradiation at G1 or G2 checkpoints, while p53-dysfunctional cells can progress into an M-phase block set by the spindle poison nocodazole (Bunz et al. 1998; Schmitt et al. 2002b). Indeed, p53-null lymphoma cells, in contrast to dnFoxO-driven, control, or likewise, Arf−/− lymphoma cells, failed to properly halt in cycle prior to the M-phase block, again demonstrating the integrity and DNA damage responsiveness of the p53 program in dnFoxO-lymphomas comparable with Arf−/− lymphomas (Fig. 3C). Ultimately, we asked whether dnFoxO-lymphomas—irrespective of their markedly reduced spontaneous apoptotic activity—might still be susceptible to drug-induced cell death. Although less sensitive when compared with controls, dnFoxO-lymphoma cells died at a significantly higher rate than p53-null lymphomas following exposure to ADR in a short-term cytotoxicity assay (P < 0.0003), while the extent of cell death was indistinguishable between dnFoxO- and Arf−/− lymphoma cells (Fig. 3D). Therefore, inactivation of FoxO transcription factors by the dnFoxO moiety promotes early-onset Myc-lymphomas that display functional deficits reminiscent of those detectable in lymphomas that formed in the absence of both Arf alleles.
FoxO transcription factors induce Arf expression
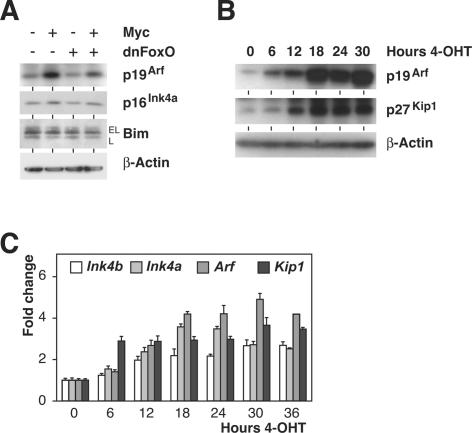
To demonstrate that Myc-induced Arf expression depends on FoxO action in cellular settings outside the lymphoid compartment, we expressed Myc, dnFoxO, or both in primary mouse embryo fibroblasts (MEFs; infected immediately after plating without further passaging) using retroviral gene transfer (Supplementary Fig. 4A). In line with our previous observation that dnFoxO strongly enhances colony formation by Myc in MEFs (Bouchard et al. 2004), coexpression of dnFoxO reduced induction of p19Arf by Myc, while protein levels of p16Ink4a and Bim remained unchanged in response to Myc or dnFoxO action (Fig. 4A). Similar experiments were carried out in primary B-cells and in hematopoietic stem cells; however, p19Arf protein levels were extremely low, and ectopic expression of Myc failed to up-regulate expression of p19Arf in both cell types in short-term assays, thereby precluding a robust analysis of the effects of dnFoxO in these settings (data not shown).
Figure 4.
FoxO transcription factors induce Arf expression. (A) Immunoblot analyses of p19Arf, p16Ink4a, and Bim levels (using β-actin as a loading control) in pools of primary MEFs infected with retroviruses expressing Myc, dnFoxO, or both. Cells were harvested immediately after selection with puromycin (4 d after infection). (B) Expression of p19Arf, p27Kip1, and β-actin as a loading control by immunoblot analysis in FoxO3aA3-ER cells following activation of FoxO3aA3 in response to 4-OHT. (C) RQ-PCR time-course analysis documenting levels (relative to S16 transcripts) of the indicated mRNAs after FoxO3aA3-ER activation by addition of 4-OHT.
To exclude that the effects observed reflect a gain of function beyond FoxO inactivation exerted by the dnFoxO moiety, we used a p19Arf/p53-proficient MEF-derived 3T3 cell line (hereafter referred to as 3T3 cells) (Supplementary Fig. 4B; B. Herkert and M. Eilers, unpubl.) engineered to stably express an inducible FoxO3a estrogen receptor (ER) fusion protein, in which the Akt-phosphorylable sites are mutated to alanine residues (FoxO3aA3-ER) (Supplementary Fig. 4C). The resulting protein can be activated by addition of 4-hydroxy-tamoxifen (4-OHT) independently of PI3-kinase or Akt activity (Tran et al. 2002). Addition of 4-OHT induced expression of Arf in several independent clones of cells expressing FoxO3aA3-ER, but not in control 3T3 cells (Fig. 4B,C; data not shown), demonstrating that activation of FoxO3a is sufficient to induce expression of p19Arf. The kinetics of the induction and the magnitude of the response were similar to those observed for p27Kip1 (Fig. 4B). RQ-PCR analysis revealed that the amounts of p19Arf protein precisely followed the 4-OHT-dependent elevation of Arf mRNA levels (Fig. 4B,C). Consistent with the reported findings in lymphoma cells, activation of FoxO3a also enhanced expression of Ink4a and Ink4b transcripts, albeit to a lower extent (Fig. 4C; cf. Fig. 2D). Importantly, when cycloheximide was added prior to 4-OHT treatment, FoxO3a was still capable of inducing Arf transcription, demonstrating that no intermediate protein has to be synthesized to mediate the effects of FoxO3a on Arf expression (Supplementary Fig. 4D). Hence, mirroring the inhibitory action of dnFoxO on Arf expression, acute activation of FoxO3a produces an immediate and substantial increase of Arf mRNA and p19Arf protein expression.
FoxO transcription factors directly act at the Arf locus
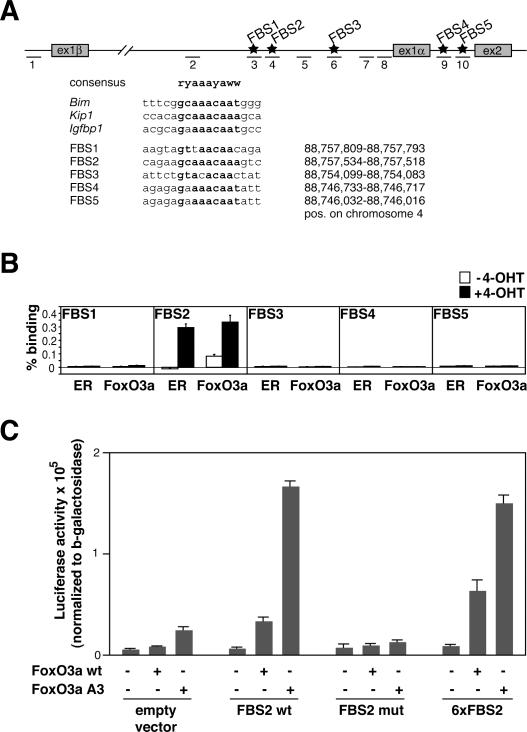
Inspection of the genomic sequence of the murine Ink4a/Arf locus revealed several potential FoxO-binding sites (FBS) localized in the first intron of the Arf gene and in the first intron of the Ink4a gene (Fig. 5A). To determine whether FoxO proteins bind to these sites in vivo, we performed chromatin immunoprecipitations (ChIPs) from 3T3 cells expressing FoxO3aA3-ER before and after addition of 4-OHT (Fig. 5B). Using either anti-ER or anti-FoxO3a antibodies, we detected in vivo binding of FoxO3a to a single site (FBS2; primer pair 4) in the first intron of the Arf gene. In contrast, no binding was detected at any of the other potential binding sites or at several control sites within the Arf locus (illustrated in Fig. 5A), arguing that the binding is highly specific. This binding increased profoundly in response to 4-OHT, further demonstrating that binding at this site is indeed mediated by the FoxO3aA3-ER protein (Fig. 5B). To provide direct evidence that the FBS2 site mediates activation by FoxO proteins, we cloned an oligonucleotide spanning 58 nucleotides (nt) surrounding this element in front of a minimal promoter into a luciferase reporter plasmid. Transient transfection assays revealed that a single copy of this element mediated a robust activation by FoxO3aA3 and a moderate induction by FoxO3a (Fig. 5C). Furthermore, introduction of six point mutations that disrupt the consensus binding site completely abolished induction by either FoxO3a or FoxO3aA3 (Fig. 5C). Finally, upon introduction of an oligonucleotide containing six copies of the FBS2-binding sequence (see Fig. 5A), we observed only a moderately enhanced activation by FoxO3a and no change in activation FoxO3aA3, further documenting that a single copy of the element is fully sufficient to mediate activation by FoxO proteins (Fig. 5C).
Figure 5.
FoxO transcription factors bind to the murine Arf locus and mediate Myc-induced Arf expression. (A, top) Scheme of the mouse Ink4a/Arf locus, showing the position of the putative FBS (FBS1–FBS5; stars) and of the primer pairs specific for the FBS, the control region, and the promoter regions used for RQ-PCR analysis (dashes 1–10). (Bottom) Alignment of FBS1–FBS5 with consensus sequences (in bold) of known FoxO targets and their positions on chromosome 4. (B) ChIP assays documenting in vivo binding of FoxO3aA3-ER proteins to FBS2 within the Ink4a/Arf locus. 3T3 cells expressing the FoxO3aA3-ER chimera were left unstimulated or induced by addition of 500 nM 4-OHT 6 h prior to ChIP with the indicated antibodies, followed by RQ-PCR with primer sets specific for FBS1–FBS5 (primer pairs 3, 4, 6, 9, and 10). Plotted are the percentages of binding based on ΔCt(FoxO3a [or ER, respectively] IP) − ΔCt(CT IP). Please note that the additional control primer pairs shown in A revealed no binding of FoxO3a to these sites (data not shown). (C) Luciferase reporter assays of HeLa cells transfected with CMV-driven expression plasmids encoding wild-type FoxO3a (FoxO3a wt) or FoxO3aA3 together with luciferase reporter plasmids that contain the indicated elements in front of a minimal promoter derived from the SV40 early promoter. “FBS2 wt” contains a single copy of a 58mer oligonucleotide spanning the FBS2 element; “FBS2 mut” carries the same element with six point mutations that disrupt the FoxO-binding sequence (see A, bottom), and “6xFBS2” carries six copies of the actual FoxO-binding sequence as shown in A.
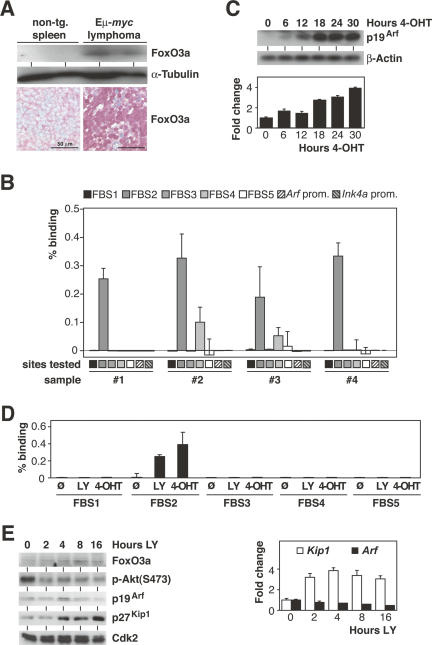
Consistent with a role for FoxO3a protein in mediating induction of Arf during Myc-driven lymphomagenesis, Eμ-myc control lymphomas displayed elevated levels of nuclear FoxO3a relative to nontransgenic lymphoid tissue (Fig. 6A). Furthermore, ChIP analyses from four different lymphomas detected robust binding of endogenous FoxO3a to FBS2 in the Arf locus in each lymphoma (primer pair 4 in Fig. 5A); in addition, low, but significant binding was detected to a second site located further downstream (FBS4; primer pair 9) (Fig. 6B). Similar to the results obtained in cells expressing the FoxO3aA3-ER chimera, no binding of FoxO was detected at either the Arf or Ink4a promoters (primer pairs 1 and 8) in the lymphoma samples.
Figure 6.
Oncogenic Myc signaling activates FoxO transcription factors. (A) FoxO3a protein expression by immunoblot analysis of lysates from immunobead-selected splenic nontransgenic B-lymphocytes and Eμ-myc transgenic control lymphoma cells (n = 2 each) with α-tubulin as a loading control (top), and by immunohistochemistry in representative tissue sections of nontransgenic wild-type spleen and Eμ-myc transgenic control lymphomas (bottom). (B) ChIP assay of four individual Eμ-myc transgenic control lymphomas (#1–#4) demonstrating in vivo binding of endogenous FoxO3a to FBS2 and to a lesser degree to FBS4. Plotted are the percentages of binding based on ΔCt(FoxO3a IP) − ΔCt(CT IP). “Arf prom.” and “Ink4a prom.” refer to primer pairs 1 and 8 that span the murine Arf and Ink4a promoters located at nucleotide positions −431/−376 and −237/−177 relative to the respective start codons. (C) p19Arf protein induction by immunoblot analysis with β-actin as a loading control (top) and Arf mRNA induction (relative to S16 transcripts) by RQ-PCR analysis (bottom) in a time-course experiment conducted in subconfluent Myc-ER 3T3 cells after 4-OHT addition for the indicated hours. (D) ChIP assay of Myc-ER 3T3 cells either left untreated, treated with the PI3-kinase inhibitor LY294002, or induced with 500 nM 4-OHT followed by RQ-PCR with the same primer sets as in B. Plotted are the percentages of binding based on ΔCt(FoxO3a IP) − ΔCt(CT IP). (E) Time-course immunoblot analyses of FoxO3a, phospho-Akt [p-Akt(Ser473)], p19Arf, p27Kip1, and Cdk2 (as a loading control) protein expression levels (left), and RQ-PCR analysis (relative to S16 transcripts) documenting Arf and Kip1 mRNA levels (right) in subconfluent 3T3 cells after LY294002 addition for the indicated hours.
Similar to the Arf induction seen upon forced nuclear expression of FoxO3aA3 (cf. Fig. 4B,C), we found a time-dependent increase of Arf expression levels in 3T3 cells engineered to activate an inducible Myc protein (Myc-ER) after addition of 4-OHT (Fig. 6C). To directly demonstrate that oncogenic Myc signaling renders endogenous FoxO3a capable of activating Arf expression, ChIP analyses were conducted in 3T3 Myc-ER cells. In the absence of 4-OHT, pharmacological inhibition of PI3-kinase by addition of LY294002 enhanced binding of endogenous FoxO3a protein to FBS2 in vivo (Fig. 6D). Importantly, activation of Myc-ER by addition of 4-OHT stimulated binding of endogenous FoxO3a protein to the same site to a similar extent as addition of LY294002. Of note, no binding of endogenous FoxO3A was detected to any other tested site within the Arf locus, confirming the specificity of binding (Fig. 6D). However, inhibition of PI3-kinase without activating Myc was insufficient to induce Arf expression, while other target genes of FoxO factors such as Kip1 and Bim were activated under these experimental conditions (Fig. 6E; data not shown). In conjunction with their ability to induce Arf transcription without further protein synthesis, our data show that FoxO proteins bind to the Ink4a/Arf locus in vivo and strongly suggest that FoxO proteins directly transactivate Arf expression in response to activation of Myc.
Discussion
In this study, we show that disruption of FoxO function dramatically accelerates Myc-driven lymphomagenesis by compromising Arf induction, thereby providing evidence for a tumor-suppressive role of FoxO transcription factors in response to oncogene-initiated tumor development in vivo.
Elucidating the role of FoxO transcription factors as putative tumor suppressors has been difficult due to the redundant functions of the family members. Recently, mouse models lacking expression of FoxO1, FoxO3, and FoxO4 uncovered a predisposition to certain cancers and benign tumors with a long latency, implying the requirement for additional oncogenic lesions to occur (Paik et al. 2007). Consistent with this view, we report here a profound and immediate impact of FoxO inactivation on Myc-driven tumor formation in a mouse model closely recapitulating the pathogenesis of human Burkitt’s lymphoma, which is initiated by a Myc-activating translocation in the vast majority of the cases (Hummel et al. 2006), and in which the Arf/p53 axis represents a particularly important tumor-suppressive growth constraint (Lindstrom et al. 2001). Our approach identifies transcriptional activation of Arf as the functionally critical node linking Myc and FoxO action. Regulation of Arf expression by FoxO proteins was detectable both in a loss-of-function (i.e., expressing the dnFoxO moiety) setting under constitutive Myc expression in the lymphoid compartment in vivo and was confirmed by a gain-of-function (i.e., the inducible FoxO3aA3 activity) approach in fibroblasts in vitro.
Importantly, lymphomas arising in the presence of a dnFoxO moiety retain functional p53 and remain responsive to DNA damage. We suggest, therefore, that FoxO factors act in a specific pathway that mediates activation of p53 via p19Arf in response to oncogenic stress, but have little or no role in regulating p53 function in response to DNA damage. Our data also provide a potential mechanistic link to recent elegant work from several laboratories using conditional alleles of p53, demonstrating that restoration of p53 in established tumors induces tumor regression in a manner that depends on p19Arf, but not on the presence of acute DNA damage (Martins et al. 2006; Ventura et al. 2007). In this view, FoxO proteins serve as cofactors that regulate expression of the Ink4a/Arf locus in response to specific oncogenic signals, but barely affect p53 function per se, similar to, for example, Dmp1, which mediates activation of Arf in response to deregulated MAP kinase activity (Inoue et al. 2000; Sreeramaneni et al. 2005). However, FoxO proteins differ from other known Ink4a/Arf regulators such as Dmp1 in that inhibition of their function does not abrogate cellular senescence in primary MEFs, and has little impact on Arf induction in response to oncogenic Ras (V. Paulus-Hock and M. Eilers, unpubl.). Therefore, our findings imply that FoxO factors activate Arf expression in response to a physiological stimulus that is distinct from deregulated Ras or MAP kinase activity.
Our data show that FoxO proteins have an instructive role in regulating Arf expression during Myc-induced lymphomagenesis. Myc-driven B-cell lymphomas may present with elevated nuclear FoxO3a levels at diagnosis, and mere constitutive expression of nucleus-retained FoxO3aA3 in tissue culture is sufficient to induce Arf expression. How FoxO proteins exactly “sense” activation of Myc to activate Arf remains open at present. Since PI3-kinase/Akt signaling negatively regulates FoxO function, one possibility would be a Myc-induced drop in PI3-kinase activity. Indeed, inhibition of PI3-kinase, like activation of Myc, promotes loading of endogenous FoxO to the Arf locus. However, inhibition of PI3-kinase without activating Myc does not license induction of Arf, despite the fact that other target genes of FoxO factors are activated by PI3-kinase inhibition only. Therefore, additional Myc-induced signals, potentially including Myc-induced oxidative stress (Vafa et al. 2002), must exist that license the relatively low amounts of endogenous FoxO factors to activate expression of Arf, although p19Arf levels that increased upon Myc activation in Myc-ER-MEFs exposed to 4-OHT remain unaffected when cotreated with N-acetylcysteine to scavenge oxygen radicals (Reimann et al. 2007). The identification of FoxO proteins as critical mediators of Myc-induced Arf expression will now allow a systematic analysis of such signals.
Our observations may also have important ramifications for the clinical use of mTORC1 inhibitors such as rapamycin as anti-cancer agents with the intention to target elevated or constitutive PI3-kinase/Akt signaling, a very common finding in various cancer entities (Vivanco and Sawyers 2002). While rapamycin was shown to inhibit cell cycle progression (Fingar et al. 2004), it was recently noted that it may lead to elevated Akt activity (Skeen et al. 2006). Certainly, inhibition of the mTOR pathway will limit tumor cell proliferation in many cancer types and has been shown to reverse chemoresistance to ADR in Akt-driven Eμ-myc transgenic mouse lymphomas in vivo (Wendel et al. 2004), but enhanced inactivation of the tumor-suppressive FoxO transcription factors via elevated Akt activity as a potentially pro-oncogenic side effect of rapamycin warrants further elucidation, particularly since rapamycin is in clinical long-term use as an immunosuppressant in nonmalignant disease settings. In this regard, novel therapeutic strategies combining mTOR with PI3-kinase inhibition (Sun et al. 2005), which may exert their efficacy, at least in part, by blocking FoxO inactivation as well, might be a particularly safe choice of targeting constitutive PI3-kinase/Akt signaling in cancer.
Materials and methods
Mice, fetal liver cell transplantation, and assessment of lymphoma growth
As a source of primary lymphomas or fetal liver cells with defined genetic lesions, Eμ-myc transgenic mice (Adams et al. 1985) were intercrossed to mice harboring targeted deletions at the p53 (Jacks et al. 1992) or Arf (Kamijo et al. 1997) locus. All animals were bred in a pure C57BL/6 strain background. Fetal liver cells were retrovirally transduced in vitro (see below) and subsequently transplanted into lethally irradiated, nontransgenic wild-type mice (Schmitt et al. 2002b). Monitoring of lymph nodes, processing and fixation of tissues, and isolation of lymphoma cells were performed as described (Schmitt et al. 2002a; Braig et al. 2005). Apoptosis-related DNA fragmentation was visualized in situ using a fluorescence-based TUNEL assay (Roche) with DAPI (4′,6-diamidino-2-phenylindole) as a nuclear counterstain for quantification. DNA content was measured after propidium iodide staining in a flow cytometer (Schmitt et al. 1999). Proliferation was assessed as the relative frequency of cells with mitotic nuclear morphology in situ (Schmitt et al. 1999). For quantification, at least 200 cells were counted per section, and at least three independent samples per genotype were evaluated.
Cells and in vitro treatments
Primary lymphoma cells and fetal liver cells were cultured as reported (Schmitt et al. 1999, 2002b). Wild-type primary MEFs were isolated from embryonic day 13.5 (E13.5) embryos and cultured in DMEM medium containing 10% fetal calf serum (FCS; Sigma), 2 mM L-glutamine, 100 U/mL penicillin/streptomycin (GIBCO), and 50 μM β-mercaptoethanol (Roth). Mouse 3T3 fibroblasts were derived from MEFs subjected to a 3T3 protocol (Todaro and Green 1963). For time-course experiments, subconfluent mouse 3T3 FoxO3aA3-ER cells or 3T3 Myc-ER were incubated with 500 nM 4-OHT (Sigma) for the indicated times. Where stated, 50 μg/mL cycloheximide (CHX; Sigma) or a solvent control were added to the cells 10 min prior to the 4-OHT treatment (Bouchard et al. 2001). In short-term cytotoxicity assays, cell viability was analyzed by trypan blue dye exclusion after exposure to the DNA-damaging topoisomerase II inhibitor ADR (alias doxorubicin; Sigma) at various concentrations for 24 h (Schmitt et al. 1999). The “mitotic trap” assay was carried out 20 h after the addition of 0.1 μg/mL nocodazole (Fluka) and, in some experiments, with a 4-Gy γ-irradiation preceding nocodazole by 2 h (Bunz et al. 1998; Schmitt et al. 2002b). At least 200 cells were evaluated for mitotic nuclear morphology per lymphoma preparation, and at least three independent preparations were analyzed per genotype.
Plasmids and retroviral infections
For cloning of an estrogen-inducible allele of human FoxO3aA3 into the retroviral vector pBabe-puro (pBabe-FoxO3aA3-ER-puro), a 2-kb BamHI fragment of the human FoxO3aA3 was isolated from the previously described pBabe-FoxO3aA3-puro (Bouchard et al. 2004) and inserted into BamHI-digested pBabe-Myc-ER-puro. pBabe-HA-dnFoxO-puro has been described before (Medema et al. 2000) and was used to shuttle the dnFoxO fragment into the murine stem cell (retro-)virus MSCV-IRES-GFP (itself serving as the empty control vector), coexpressing GFP via an internal ribosomal entry site (MSCV-dnFoxO-IRES-GFP). Retroviral supernatants were generated by transient transfections of Phoenix cells and were used to transduce MEFs, 3T3 cells, and Eμ-myc transgenic fetal liver cells (Schmitt et al. 2002b). Infected cells were selected with 2 μg/mL puromycin (InvivoGen) and 80 μg/mL hygromycin (Calbiochem). Fetal liver cell populations infected with GFP-coencoding MSCV retroviruses were transplanted without further selection. For enrichment experiments based on GFP-coexpressing vectors, freshly infected lymphoma cells were adjusted to ∼15% GFP-positive cells by admixing uninfected cells of the same lymphoma population, and relative changes of the fractions of GFP-positive cells were reassessed by flow cytometry after 48 h in culture.
RT–PCR, RQ-PCR, and genomic PCR
Two micrograms of total RNA isolated with the peqGOLD TriFast reagent (Peqlab) were transcribed into cDNA using MoMuLV reverse transcriptase and random primers. Standard PCR experiments were performed with RedTaq polymerase and specific primers for mouse p53, dnFoxO-IRES, HA-dnFoxO, human Myc, and s16. RQ-PCR experiments were performed on an Mx3000p thermo cycler (Stratagene), with the Hot Gold Star kit (Eurogentec) and specific primers for mouse Arf, Ink4a, Ink4b, Cip1, Kip1, Bim, eIF4E, 4EBP1, and s16. Data were plotted as ΔCt values (the difference in cycle numbers at which the fluorescence threshold was crossed) relative to a control mRNA (encoding the ribosomal protein s16) or as relative level of transcript (RLT) based on 2(−ΔCt). Primer sequences are available on request. RT–PCR products of p53 exons 4–8 were sequenced as described (Schmitt et al. 1999).
ChIP
Cells expressing FoxO3aA3-ER or Myc-ER were stimulated by addition of 500 nM 4-OHT, 50 μM LY294002 (Calbiochem), or a solvent control. After 6 h, cells were harvested and processed for ChIP. Primary lymphoma cells were expanded, harvested, and subjected to ChIP analyses. ChIP assays were performed as described (Bouchard et al. 2001) using the polyclonal rabbit anti-ER (M-20; sc-542; Santa Cruz Biotechnology), anti-FoxO3a (H-144; sc-11351; Santa Cruz Biotechnology; no cross-reactivity with other FoxOs), and control (CT) IgG (Sigma) antibodies. Immunoprecipitated DNA samples and inputs were PCR-amplified and quantified with primer pairs specific for the potential FBS (FBS1–FBS5; primer pairs 3, 4, 6, 9, and 10), for promoter regions (primer pairs 1 and 8) (Bracken et al. 2007), and for control regions (primer pairs 2, 5, and 7) of the murine Ink4a/Arf locus. For each antibody, the percentage of binding was calculated based on ΔCt(relevant IP) − ΔCt(CT IP), where ΔCt were obtained from the input samples as a reference (Nelson et al. 2006). Primer sequences are available on request.
Microarray analyses
Expression analysis was performed on RNA isolated from individual lymphomas using a 22.5 K mouse cDNA array. Details about the protocols and the array design can be found at http://www.ebi.ac.uk/arrayexpress.
Immunoblotting, immunohistochemistry, and immunofluorescence
For immunoblotting, cells were lysed by three rounds of freezing and thawing in a buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, and a cocktail of protease inhibitors (Sigma). Immunoblotting has been described previously (Steiner et al. 1995). For immunohistochemical analysis (of three to five mice examined per genotype), formalin-fixed and paraffin-embedded sections of lymph nodes and spleens were stained according to published procedures or manufacturers’ recommendations (Schmitt et al. 1999; Braig et al. 2005). For immunofluorescence, cells were fixed in 2% paraformaldehyde/PBS and permeabilized in 0.2% Triton-X/PBS before application of antibodies as described (Reimann et al. 2007). The antibodies used to detect the following antigens were β-actin (A5441; Sigma); p19Arf (R562 and ab80; Abcam); p16Ink4a (sc-1207 and F-12; Santa Cruz Biotechology); p21Cip1 (sc-397; Santa Cruz Biotechnology); p27Kip1 (K25020; Transduction Laboratories); p53 (CM5; Novocastra); PUMA (ab9645; Abcam); Akt (9272), phospho-Ser473-Akt (9271), mTOR (2972), phospho-Ser2481-mTOR (2974), Raptor (4978), 4EBP1 (9452), phospho-Thr70-4EBP1 (9455), and eIF4E (9742) (all: Cell Signaling Technology); Bim (AAP-30; Stressgen; and ab15184; Abcam); Cdk2 (sc-163; Santa Cruz Biotechnology); ER (sc-542; Santa Cruz Biotechnology); FoxO3a (F2178; Sigma); Ki67 (Tec-3; Dako); Myc (9E10; Upstate Biotechnology; and sc-764; Santa Cruz Biotechnology); and α-tubulin (T5168; Sigma).
Statistical evaluation
Tumor onset data reflect the time between transplantation of fetal liver cells and first-time palpability of enlarged lymph nodes in recipient mice. Statistical comparison of Kaplan-Meier curves is based on the log-rank test; the unpaired t-test was applied for comparisons of means and standard deviations, and the Fisher’s exact test was used to statistically compare categorical variables (SD; asterisk denoting a significant P-value of <0.05). All error bars shown denote the SD.
Luciferase assay
Oligonucleotides were annealed and ligated into an XmaI/BglII-digested pGL2-promoter vector (Promega). The FBS2 wt sequence (5′-ccgggggttaatactttctatatcactgacTTTGTTTGCttctgga cttaggagcaaacatacc-3′) contains 58 nt surrounding the 9-base- pair (bp) consensus sequence (FBS2); FBS2 mut (5′-ccgggggttaatac tttctatatcactgacTTGTCGACCttctggacttaggagcaaacatacc-3′) comprises the 58mer sequence with a disrupted FBS; and 6xFBS2 (5′-ccgggT T T G T T T G C T T T G T T T GC TTTGTT T GCT T TGTT TGCTTTGTTTGCTTTGTTTGCc-3′) contains six times the 9-bp sequence that matches the FoxO consensus binding sequence. Transfections of HeLa cells were carried out in triplicate in six-well dishes containing 250,000 cells per well. Cells were transfected by using CaPO4 with 2 μg of the respective reporter (pGL2-promoter-empty, -FBS2 wt, -FBS2 mut, or -6xFBS2) plus 1 μg of pcDNA3.1-empty or pECE-FoxO3a wt or pcDNA3.1-FoxO3aA3. In each transfection, 100 ng of CMV-lacZ were transfected to normalize different transfection efficiencies. The total amount of DNA was kept constant in each transfection by adding equal amounts of expression plasmids. Cells were harvested after 30 h, and luciferase assays were performed as described previously (Bouchard et al. 1999).
Acknowledgments
We are indebted to A. Harris for Eμ-myc transgenic mice, and we thank T. Jacks and C. Sherr for p53−/− and Arf−/− mice; S. Spiekermann and B. Teichmann for excellent technical assistance; Nicole Forster for help and advice; and members of the Eilers and Schmitt laboratories for critical discussions and editorial advice. This work was supported by grants from the Deutsche Krebshilfe and the Deutsche Forschungsgemeinschaft (to C.A.S.), and by the Transregio 17 of the Deutsche Forschungsgemeinschaft (to M.E.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.453107
References
- Accili D., Arden K.C., Arden K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Adams J.M., Harris A.W., Pinkert C.A., Corcoran L.M., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L., Harris A.W., Pinkert C.A., Corcoran L.M., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L., Pinkert C.A., Corcoran L.M., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L., Corcoran L.M., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L., Cory S., Palmiter R.D., Brinster R.L., Palmiter R.D., Brinster R.L., Brinster R.L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bertwistle D., Sherr C.J., Sherr C.J. Regulation of the Arf tumor suppressor in Eμ-Myc transgenic mice: Longitudinal study of Myc-induced lymphomagenesis. Blood. 2006;15:792–794. doi: 10.1182/blood-2006-07-033985. [DOI] [PubMed] [Google Scholar]
- Birkenkamp K.U., Coffer P.J., Coffer P.J. FOXO transcription factors as regulators of immune homeostasis: Molecules to die for? J. Immunol. 2003;171:1623–1629. doi: 10.4049/jimmunol.171.4.1623. [DOI] [PubMed] [Google Scholar]
- Bouchard C., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M., Reed S., Sicinski P., Bartek J., Eilers M., Sicinski P., Bartek J., Eilers M., Bartek J., Eilers M., Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Dohmann K., Menkel A., Eilers M., Luscher B., Menkel A., Eilers M., Luscher B., Eilers M., Luscher B., Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Marquardt J., Bras A., Medema R.H., Eilers M., Marquardt J., Bras A., Medema R.H., Eilers M., Bras A., Medema R.H., Eilers M., Medema R.H., Eilers M., Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., Minucci S., Porse B.T., Marine J.C., Porse B.T., Marine J.C., Marine J.C., et al. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Stein H., Dorken B., Jenuwein T., Schmitt C.A., Dorken B., Jenuwein T., Schmitt C.A., Jenuwein T., Schmitt C.A., Schmitt C.A. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E., Arden K.C., Blenis J., Greenberg M.E., Blenis J., Greenberg M.E., Greenberg M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Ross S.E., Mostoslavsky R., Cohen H.Y., Mostoslavsky R., Cohen H.Y., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Sedivy J.M., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Dijkers P.F., Medema R.H., Lammers J.W., Koenderman L., Coffer P.J., Medema R.H., Lammers J.W., Koenderman L., Coffer P.J., Lammers J.W., Koenderman L., Coffer P.J., Koenderman L., Coffer P.J., Coffer P.J. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- Egle A., Harris A.W., Bouillet P., Cory S., Harris A.W., Bouillet P., Cory S., Bouillet P., Cory S., Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen C.M., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L., Roussel M.F., Sherr C.J., Cleveland J.L., Sherr C.J., Cleveland J.L., Cleveland J.L. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar D.C., Richardson C.J., Tee A.R., Cheatham L., Tsou C., Blenis J., Richardson C.J., Tee A.R., Cheatham L., Tsou C., Blenis J., Tee A.R., Cheatham L., Tsou C., Blenis J., Cheatham L., Tsou C., Blenis J., Tsou C., Blenis J., Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J., Peters G., Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Greer E.L., Brunet A., Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Guertin D.A., Sabatini D.M., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W., Cleveland J.L., Tansey W.P., Lowe S.W., Tansey W.P., Lowe S.W., Lowe S.W. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu B., Marin M.C., el-Naggar A.K., Stephens L.C., Brisbay S., McDonnell T.J., Marin M.C., el-Naggar A.K., Stephens L.C., Brisbay S., McDonnell T.J., el-Naggar A.K., Stephens L.C., Brisbay S., McDonnell T.J., Stephens L.C., Brisbay S., McDonnell T.J., Brisbay S., McDonnell T.J., McDonnell T.J. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene. 1995;11:175–179. [PubMed] [Google Scholar]
- Huang H., Regan K.M., Wang F., Wang D., Smith D.I., van Deursen J.M., Tindall D.J., Regan K.M., Wang F., Wang D., Smith D.I., van Deursen J.M., Tindall D.J., Wang F., Wang D., Smith D.I., van Deursen J.M., Tindall D.J., Wang D., Smith D.I., van Deursen J.M., Tindall D.J., Smith D.I., van Deursen J.M., Tindall D.J., van Deursen J.M., Tindall D.J., Tindall D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M., Bentink S., Berger H., Klapper W., Wessendorf S., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Bentink S., Berger H., Klapper W., Wessendorf S., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Berger H., Klapper W., Wessendorf S., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Klapper W., Wessendorf S., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Wessendorf S., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Barth T.F., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Bernd H.W., Cogliatti S.B., Dierlamm J., Feller A.C., Cogliatti S.B., Dierlamm J., Feller A.C., Dierlamm J., Feller A.C., Feller A.C., et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Inoue K., Wen R., Rehg J.E., Adachi M., Cleveland J.L., Roussel M.F., Sherr C.J., Wen R., Rehg J.E., Adachi M., Cleveland J.L., Roussel M.F., Sherr C.J., Rehg J.E., Adachi M., Cleveland J.L., Roussel M.F., Sherr C.J., Adachi M., Cleveland J.L., Roussel M.F., Sherr C.J., Cleveland J.L., Roussel M.F., Sherr C.J., Roussel M.F., Sherr C.J., Sherr C.J. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes & Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A., Bronson R.T., Goodell M.A., Weinberg R.A., Goodell M.A., Weinberg R.A., Weinberg R.A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Scheijen B., Voncken J.W., Kieboom K., Berns A., van Lohuizen M., Scheijen B., Voncken J.W., Kieboom K., Berns A., van Lohuizen M., Voncken J.W., Kieboom K., Berns A., van Lohuizen M., Kieboom K., Berns A., van Lohuizen M., Berns A., van Lohuizen M., van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes & Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T., Zindy F., Roussel M.F., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J., Zindy F., Roussel M.F., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J., Roussel M.F., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J., Ashmun R.A., Grosveld G., Sherr C.J., Grosveld G., Sherr C.J., Sherr C.J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Weber J.D., Zambetti G., Zindy F., Roussel M.F., Sherr C.J., Weber J.D., Zambetti G., Zindy F., Roussel M.F., Sherr C.J., Zambetti G., Zindy F., Roussel M.F., Sherr C.J., Zindy F., Roussel M.F., Sherr C.J., Roussel M.F., Sherr C.J., Sherr C.J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T., de van Kamp E., Chong M.J., Zindy F., Diehl J.A., Sherr C.J., McKinnon P.J., de van Kamp E., Chong M.J., Zindy F., Diehl J.A., Sherr C.J., McKinnon P.J., Chong M.J., Zindy F., Diehl J.A., Sherr C.J., McKinnon P.J., Zindy F., Diehl J.A., Sherr C.J., McKinnon P.J., Diehl J.A., Sherr C.J., McKinnon P.J., Sherr C.J., McKinnon P.J., McKinnon P.J. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res. 1999;59:2464–2469. [PubMed] [Google Scholar]
- Kops G.J., Dansen T.B., Polderman P.E., Saarloos I., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Dansen T.B., Polderman P.E., Saarloos I., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Polderman P.E., Saarloos I., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Saarloos I., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Huang T.T., Bos J.L., Medema R.H., Burgering B.M., Bos J.L., Medema R.H., Burgering B.M., Medema R.H., Burgering B.M., Burgering B.M. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., Quon K.C., Mooi W.J., Loonstra A., Berns A., Quon K.C., Mooi W.J., Loonstra A., Berns A., Mooi W.J., Loonstra A., Berns A., Loonstra A., Berns A., Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L.F., Weinberg R.A., Parada L.F., Weinberg R.A., Weinberg R.A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lindstrom M.S., Klangby U., Wiman K.G., Klangby U., Wiman K.G., Wiman K.G. p14ARF homozygous deletion or MDM2 overexpression in Burkitt lymphoma lines carrying wild type p53. Oncogene. 2001;20:2171–2177. doi: 10.1038/sj.onc.1204303. [DOI] [PubMed] [Google Scholar]
- Martins C.P., Brown-Swigart L., Evan G.I., Brown-Swigart L., Evan G.I., Evan G.I. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Medema R.H., Kops G.J., Bos J.L., Burgering B.M., Kops G.J., Bos J.L., Burgering B.M., Bos J.L., Burgering B.M., Burgering B.M. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- Motta M.C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Gu W., Bultsma Y., McBurney M., Guarente L., Bultsma Y., McBurney M., Guarente L., McBurney M., Guarente L., Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Ramaswamy S., Vazquez F., Signoretti S., Loda M., Sellers W.R., Ramaswamy S., Vazquez F., Signoretti S., Loda M., Sellers W.R., Vazquez F., Signoretti S., Loda M., Sellers W.R., Signoretti S., Loda M., Sellers W.R., Loda M., Sellers W.R., Sellers W.R. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Vousden K.H., Vousden K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Sova P., Bomsztyk K., Denisenko O., Sova P., Bomsztyk K., Sova P., Bomsztyk K., Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S., Fergusson M.M., Finkel T., Fergusson M.M., Finkel T., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Tothova Z., Horner J.W., Carrasco D.R., Horner J.W., Carrasco D.R., Carrasco D.R., et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Nakamura N., Sansal I., Bergeron L., Sellers W.R., Nakamura N., Sansal I., Bergeron L., Sellers W.R., Sansal I., Bergeron L., Sellers W.R., Bergeron L., Sellers W.R., Sellers W.R. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- Reimann M., Loddenkemper C., Rudolph C., Schildhauer I., Teichmann B., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Loddenkemper C., Rudolph C., Schildhauer I., Teichmann B., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Rudolph C., Schildhauer I., Teichmann B., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Schildhauer I., Teichmann B., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Teichmann B., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Stein H., Schlegelberger B., Dorken B., Schmitt C.A., Schlegelberger B., Dorken B., Schmitt C.A., Dorken B., Schmitt C.A., Schmitt C.A. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood. 2007 doi: 10.1182/blood-2007-02-075614. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P., Londei P., Cordon-Cardo C., Pandolfi P.P., Cordon-Cardo C., Pandolfi P.P., Pandolfi P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W., Wallace-Brodeur R.R., Lowe S.W., Lowe S.W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes & Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman J.S., Yang M., Lee S., Baranov E., Hoffman R.M., Lowe S.W., Fridman J.S., Yang M., Lee S., Baranov E., Hoffman R.M., Lowe S.W., Yang M., Lee S., Baranov E., Hoffman R.M., Lowe S.W., Lee S., Baranov E., Hoffman R.M., Lowe S.W., Baranov E., Hoffman R.M., Lowe S.W., Hoffman R.M., Lowe S.W., Lowe S.W. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002a;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., Yang M., Fridman J.S., Baranov E., Hoffman R.M., Lowe S.W., Yang M., Fridman J.S., Baranov E., Hoffman R.M., Lowe S.W., Fridman J.S., Baranov E., Hoffman R.M., Lowe S.W., Baranov E., Hoffman R.M., Lowe S.W., Hoffman R.M., Lowe S.W., Lowe S.W. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002b;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. Divorcing ARF and p53: An unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sherr C.J., Bertwistle D., den Besten W., Kuo M.L., Sugimoto M., Tago K., Williams R.T., Zindy F., Roussel M.F., Bertwistle D., den Besten W., Kuo M.L., Sugimoto M., Tago K., Williams R.T., Zindy F., Roussel M.F., den Besten W., Kuo M.L., Sugimoto M., Tago K., Williams R.T., Zindy F., Roussel M.F., Kuo M.L., Sugimoto M., Tago K., Williams R.T., Zindy F., Roussel M.F., Sugimoto M., Tago K., Williams R.T., Zindy F., Roussel M.F., Tago K., Williams R.T., Zindy F., Roussel M.F., Williams R.T., Zindy F., Roussel M.F., Zindy F., Roussel M.F., Roussel M.F. p53-dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- Skeen J.E., Bhaskar P.T., Chen C.C., Chen W.S., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Bhaskar P.T., Chen C.C., Chen W.S., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Chen C.C., Chen W.S., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Chen W.S., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Peng X.D., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Nogueira V., Hahn-Windgassen A., Kiyokawa H., Hay N., Hahn-Windgassen A., Kiyokawa H., Hay N., Kiyokawa H., Hay N., Hay N. Akt deficiency impairs normal cell proliferation and suppresses oncogenesis in a p53-independent and mTORC1-dependent manner. Cancer Cell. 2006;10:269–280. doi: 10.1016/j.ccr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Sreeramaneni R., Chaudhry A., McMahon M., Sherr C.J., Inoue K., Chaudhry A., McMahon M., Sherr C.J., Inoue K., McMahon M., Sherr C.J., Inoue K., Sherr C.J., Inoue K., Inoue K. Ras–Raf–Arf signaling critically depends on the Dmp1 transcription factor. Mol. Cell. Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M., Dijkers P.F., Kops G.J., Lens S.M., Coffer P.J., Burgering B.M., Medema R.H., Dijkers P.F., Kops G.J., Lens S.M., Coffer P.J., Burgering B.M., Medema R.H., Kops G.J., Lens S.M., Coffer P.J., Burgering B.M., Medema R.H., Lens S.M., Coffer P.J., Burgering B.M., Medema R.H., Coffer P.J., Burgering B.M., Medema R.H., Burgering B.M., Medema R.H., Medema R.H. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- Steiner P., Philipp A., Lukas J., Godden-Kent D., Pagano M., Mittnacht S., Bartek J., Eilers M., Philipp A., Lukas J., Godden-Kent D., Pagano M., Mittnacht S., Bartek J., Eilers M., Lukas J., Godden-Kent D., Pagano M., Mittnacht S., Bartek J., Eilers M., Godden-Kent D., Pagano M., Mittnacht S., Bartek J., Eilers M., Pagano M., Mittnacht S., Bartek J., Eilers M., Mittnacht S., Bartek J., Eilers M., Bartek J., Eilers M., Eilers M. Identification of a Myc-dependent step during the formation of active G1 cyclin–cdk complexes. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.Y., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Rosenberg L.M., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Wang X., Zhou Z., Yue P., Fu H., Khuri F.R., Zhou Z., Yue P., Fu H., Khuri F.R., Yue P., Fu H., Khuri F.R., Fu H., Khuri F.R., Khuri F.R. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- Todaro G.J., Green H., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H., Brunet A., Grenier J.M., Datta S.R., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E., Brunet A., Grenier J.M., Datta S.R., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E., Grenier J.M., Datta S.R., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E., Datta S.R., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E., DiStefano P.S., Chiang L.W., Greenberg M.E., Chiang L.W., Greenberg M.E., Greenberg M.E. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- Vafa O., Wade M., Kern S., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M., Wade M., Kern S., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M., Kern S., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M., Pandita T.K., Hampton G.M., Wahl G.M., Hampton G.M., Wahl G.M., Wahl G.M. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- van den Heuvel A.P., Schulze A., Burgering B.M., Schulze A., Burgering B.M., Burgering B.M. Direct control of caveolin-1 expression by FOXO transcription factors. Biochem. J. 2005;385:795–802. doi: 10.1042/BJ20041449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., de Vries-Smits A.M., Brenkman A.B., van Triest M.H., van den Broek N., Colland F., Maurice M.M., Burgering B.M., de Vries-Smits A.M., Brenkman A.B., van Triest M.H., van den Broek N., Colland F., Maurice M.M., Burgering B.M., Brenkman A.B., van Triest M.H., van den Broek N., Colland F., Maurice M.M., Burgering B.M., van Triest M.H., van den Broek N., Colland F., Maurice M.M., Burgering B.M., van den Broek N., Colland F., Maurice M.M., Burgering B.M., Colland F., Maurice M.M., Burgering B.M., Maurice M.M., Burgering B.M., Burgering B.M. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T., Newman J., Reczek E.E., Weissleder R., Jacks T., Reczek E.E., Weissleder R., Jacks T., Weissleder R., Jacks T., Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C.L., Sawyers C.L. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wendel H.G., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W., Cordon-Cardo C., Pelletier J., Lowe S.W., Pelletier J., Lowe S.W., Lowe S.W. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T.W., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T.W., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T.W., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T.W., Hacker G., Erlacher M., Villunger A., Mak T.W., Erlacher M., Villunger A., Mak T.W., Villunger A., Mak T.W., Mak T.W. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y., Yarbrough W.G., Xiong Y., Yarbrough W.G., Yarbrough W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF–INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zindy F., Eischen C.M., Randle D.H., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F., Eischen C.M., Randle D.H., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F., Randle D.H., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F., Cleveland J.L., Sherr C.J., Roussel M.F., Sherr C.J., Roussel M.F., Roussel M.F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]