Abstract
The interactions of numerous regulatory small RNAs (sRNAs) with target mRNAs have been characterized, but how sRNAs can regulate multiple, structurally unrelated mRNAs is less understood. Here we show that Salmonella GcvB sRNA directly acts on seven target mRNAs that commonly encode periplasmic substrate-binding proteins of ABC uptake systems for amino acids and peptides. Alignment of GcvB homologs of distantly related bacteria revealed a conserved G/U-rich element that is strictly required for GcvB target recognition. Analysis of target gene fusion regulation in vivo, and in vitro structure probing and translation assays showed that GcvB represses its target mRNAs by binding to extended C/A-rich regions, which may also serve as translational enhancer elements. In some cases (oppA, dppA), GcvB repression can be explained by masking the ribosome-binding site (RBS) to prevent 30S subunit binding. However, GcvB can also effectively repress translation by binding to target mRNAs at upstream sites, outside the RBS. Specifically, GcvB represses gltI mRNA translation at the C/A-rich target site located at positions −57 to −45 relative to the start codon. Taken together, our study suggests highly conserved regions in sRNAs and mRNA regions distant from Shine-Dalgarno sequences as important elements for the identification of sRNA targets.
[Keywords: Small RNA, riboregulator, post-transcriptional control, translation inhibition, ABC transporter, GcvB]
Small noncoding RNAs (sRNAs) that act as regulators of gene expression have been identified in all kingdoms of life. The enterobacterial sRNAs constitute a structurally diverse class of molecules that are typically 50–200 nucleotides (nt) in length and function as stress-response or virulence gene regulators (Wassarman 2002; Gottesman 2004; Romby et al. 2006; Vogel and Papenfort 2006). Recent systematic searches (Vogel and Sharma 2005) showed that Escherichia coli expresses >80 sRNAs; the total number of sRNAs in a typical enterobacterium may well range in the hundreds (Hershberg et al. 2003; Zhang et al. 2004).
The majority of the ∼20 enterobacterial sRNAs of known function are antisense RNAs that repress trans-encoded target mRNAs (Majdalani et al. 2005; Storz et al. 2005). Many sRNAs mask the ribosome-binding sites (RBS) of their targets, thus inhibiting ribosome entry on mRNA (Storz et al. 2004). Specifically, the E. coli MicA, MicC, and OxyS sRNAs were shown by in vitro toeprinting experiments to directly interfere with 30S ribosome binding of target mRNAs (Argaman and Altuvia 2000; Chen et al. 2004; Udekwu et al. 2005). Other well-characterized E. coli sRNA–target pairs that are likely to use the same mechanism include DsrA-hns, MicF-ompF, RyhB-sodB, SgrS-ptsG, and Spot42-galK (for review, see Wagner and Darfeuille 2006). Since the half-life of bacterial mRNA is strongly affected by the association with ribosomes (Deana and Belasco 2005), translation inhibition will promote the decay of the repressed target; e.g., by accelerating RNase E-mediated mRNA turnover (Massé et al. 2003; Morita et al. 2005). Besides RNase E, the bacterial Sm-like protein Hfq has been identified as a key player in this type of translational silencing (Valentin-Hansen et al. 2004). Hfq binds all of the aforementioned sRNAs with high affinity and is most often required for both their intracellular stability and their interaction with target mRNAs (Zhang et al. 2003; Aiba 2007; Urban and Vogel 2007, and references therein). Hfq and some sRNAs form ribonucleoprotein complexes with RNase E to mediate target mRNA destabilization (Morita et al. 2005).
The enterobacterial sRNAs exhibit short and/or imperfect complementarity to their target(s). For example, nine residues of RyhB sRNA interact with sodB mRNA (Geissmann and Touati 2004), and OxyS sRNA targets fhlA mRNA through the formation of two short kissing complexes of 9 and 7 base pairs (bp), respectively (Argaman and Altuvia 2000). Systematic mutational analysis of SgrS–ptsG RNA duplexes revealed that few (six) residues of this interaction are key to mediating SgrS repression of ptsG mRNA (Kawamoto et al. 2006). The limited sequence complementarity has rendered the identification of new sRNA targets difficult, and for the majority of the sRNAs studied to date, a single mRNA remains the only experimentally validated target. However, it was early recognized that some E. coli sRNAs could regulate multiple mRNAs. For example, DsrA was shown to directly act on the hns and rpoS mRNAs (Lease et al. 1998; Majdalani et al. 1998), and constitutive overexpression of OxyS altered the expression of >40 genes (Altuvia et al. 1997). Recent biocomputational and experimental approaches predicted more sRNAs to target multiple mRNAs. For example, pulse expression of several E. coli and Salmonella sRNAs (Massé et al. 2005; Guillier and Gottesman 2006; Johansen et al. 2006; Papenfort et al. 2006; Tjaden et al. 2006) followed by global transcriptome profiling showed the rapid depletion of many mRNAs. The kinetics of target decay observed in these experiments strongly argues that the regulated mRNAs are direct targets.
Multiple mRNA targeting by sRNAs could help bacteria to balance different transcriptional responses at the post-transcriptional level in response to stress or changes in nutrient availability. This additional layer of gene expression control could mediate the coregulation of mRNAs that belong to different transcriptional regulons. However, evidence for direct interactions of sRNAs with multiple mRNAs has been rare in enterobacteria (Lease et al. 1998) and has come primarily from investigations of RNAIII of the Gram-positive pathogen Staphylococcus aureus (Boisset et al. 2007).
In this work, we have studied GcvB sRNA of Salmonella enterica serovar Typhimurium (from here on Salmonella) and found that it directly binds multiple mRNAs that collectively encode periplasmic substrate-binding proteins of amino acid and peptide transporters. The gcvB gene was originally identified in E. coli and is controlled by GcvA/GcvR, the major transcription factors of the glycine cleavage system (Urbanowski et al. 2000). A gcvB deletion caused constitutive synthesis of OppA and DppA, periplasmic substrate-binding proteins of the two major peptide transport systems (Higgins and Hardie 1983; Abouhamad et al. 1991), which are normally repressed in nutrient-rich growth conditions. Gene fusion experiments indicated that GcvB repressed dppA and oppA at the post-transcriptional level, yet the molecular mechanism remained elusive (Urbanowski et al. 2000). Furthermore, the pleiotropic nature of E. coli and Yersinia gcvB mutants (Urbanowski et al. 2000; McArthur et al. 2006), and biocomputational predictions (Tjaden et al. 2006) suggested that GcvB may have additional mRNA targets.
This study provides biochemical and genetic evidence that a conserved G/U-rich region within GcvB mediates translational repression of seven ABC transporter mRNAs. While GcvB represses some of these targets by covering their RBS, this sRNA can also inhibit mRNA translation by binding to regions upstream of the RBS. This latter mode of action may also apply to other regulatory sRNAs.
Results
Characterization of GcvB sRNA in Salmonella
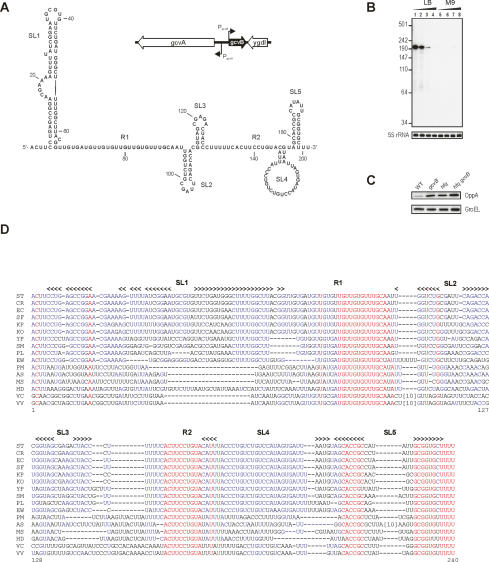
The Salmonella and E. coli gcvB genes are ∼95% identical in the GcvB RNA region (Urbanowski et al. 2000; Argaman et al. 2001). Expression of the ∼200-nt Salmonella GcvB RNA (Fig. 1A) was confirmed on Northern blots of RNA extracted from different growth phases and media. GcvB is expressed in exponentially growing Salmonella in rich medium (L-broth) but not detectable in stationary phase or upon growth in minimal medium (Fig. 1B). This pattern is reminiscent of E. coli gcvB (Argaman et al. 2001) and in keeping with the postulated GcvB repressor function of peptide transporters under nutrient-rich conditions (Urbanowski et al. 2000).
Figure 1.
Salmonella GcvB RNA. (A) Proposed secondary structure as derived from in vitro RNA structure probing (Supplementary Fig. S2), and from alignment of GcvB RNAs of diverse bacteria. Of the five stem–loops (SL), SL5 is predicted to function as a ρ-independent transcription terminator. R1 and R2 denote two single-stranded regions that are strongly conserved in gcvB homologs (see D). The inlet shows the genomic location of the Salmonella gcvB gene in the gcvA–ygdI intergenic region in opposite orientation to these latter two genes. (B) Northern blot (autoradiogram) showing gcvB expression in nutrient-rich (L-broth) or nutrient-limiting (M9) media. Total RNA was prepared from Salmonella (strain JVS-0007) grown to exponential phase (OD600 of 0.4, lanes 1,5), early stationary phase (OD 1, lanes 2,6; OD 2, lanes 3,7), and late stationary phase (lanes 4,8) cells. GcvB was detected with 32P-labeled oligonucleotide JVO-0749 complementary to the GcvB 5′ region. Marker sizes are shown to the left. 5S rRNA probing (panel below) confirmed equal RNA loading. (C) Both GcvB and Hfq act to repress OppA protein synthesis. Protein samples of Salmonella wild-type, hfq or gcvB single deletion, and the hfq gcvB double-deletion strain (JVS-0007, JVS-0255, JVS-0236, and JVS-0617, respectively) grown to exponential phase (OD600 of 0.4) were subjected to Western blot analysis with OppA (top panel) or GroEL antisera (bottom panel; loading control). Quantification of the OppA signals, followed by normalization to GroEL levels, revealed a 6.1-fold, 5.4-fold, or 8.3-fold increase upon gcvB, hfq, or gcvB hfq double deletion, respectively. (D) Alignment of a representative subset of GcvB RNAs identified by computer-based searches (Supplementary Fig. S5). Numbering of residues and the positions of stem–loop structures (SL1 to SL5; indicated by arrowheads) follow the Salmonella GcvB RNA sequence (shown in A). The conserved regions, R1 and R2, common to all known GcvB sequences are indicated. (ST) Salmonella typhimurium; (CR) Citrobacter rodentium; (EC) Escherichia coli K12; (SF) Shigella flexneri; (KP) Klebsiella pneumoniae; (KO) Klebsiella oxytoca; (YP) Yersinia pestis; (SM) Serratia marcescens; (PL) Photorhabdus luminescens; (EW) Erwinia carotovora; (PM) Pasteurella multocida; (AS) Actinobacillus succinogenes; (MS) Mannheimia succiniciproducens; (HD) Haemophilus ducreyi; (VC) Vibrio cholerae; (VV) Vibrio vulnificus.
Several observations suggested that GcvB is an Hfq-dependent sRNA. GcvB coimmunoprecipitates with Hfq in extracts of E. coli (Zhang et al. 2003) and Salmonella (A. Sittka and J. Vogel, in prep.) and is unstable in Δhfq strains of E. coli (Urban and Vogel 2007) and Salmonella (Supplementary Fig. S1). Since repression of the predicted GcvB target, oppA, is abrogated in hfq mutants of both E. coli and Salmonella (Ziolkowska et al. 2006; Sittka et al. 2007), we compared OppA protein levels in Salmonella strains deleted for gcvB, hfq, or both (Fig. 1C). Deletion of gcvB did not impair Salmonella growth in rich broth (data not shown), similar to what was reported for E. coli (Urbanowski et al. 2000). The single gcvB or hfq mutations each elevated OppA levels, whereas the double mutation had no pronounced cumulative effect. This predicts GcvB and Hfq to act in concert, and GcvB to be an Hfq-dependent antisense RNA that regulates trans-encoded mRNA(s).
GcvB targets dppA and oppA mRNAs in vivo and in vitro
To study dppA and oppA mRNA repression by GcvB, we constructed translational fusions to the N terminus of green fluorescent protein, GFP (Urban and Vogel 2007). The Salmonella dppA or oppA 5′ regions spanning the entire 5′ untranslated region (UTR) and 30 bp (dppA) or 50 bp (oppA) of the coding region were cloned into the low-copy gfp fusion vector, pXG10 (Fig. 2A). The cloning strategy used here ensures that the fusions are transcribed from the native +1 site of dppA or oppA without adding additional sequences at the 5′ end. Transcription is driven by a constitutive PLtetO-1 promoter (Lutz and Bujard 1997) to specifically assay post-transcriptional regulation. The Salmonella gcvB gene was placed under control of a constitutive PLlacO-1 promoter on a compatible mid-copy plasmid (Lutz and Bujard 1997), resulting in plasmid pPLgcvB (Fig. 2A).
Figure 2.
Regulation of Salmonella oppA∷gfp and dppA∷gfp fusions by GcvB wild-type and mutant RNAs. (A) Principle approach: GcvB and translational fusions of oppA or dppA to gfp are constitutively expressed from compatible plasmids in an E. coli ΔgcvB recA− strain (Urban and Vogel 2007). The fusions include the entire 5′ UTR (determined by 5′RACE; dppA, 163 bp; oppA, 162 bp) and result in in-frame fusions of the tenth (dppA) or seventeenth (oppA) codon to the N terminus of GFP. (B) Agar plate-based assay of colony fluorescence of E. coli strains carrying control plasmid pXG-1 (left; expresses full-length gfp), the dppA∷gfp (middle; plasmid pJL18-1), or the oppA∷gfp (right; pJL19-1) fusion plasmid, each in combination with control vector pTP11, or plasmids expressing Salmonella wild-type GcvB RNA (pPLgcvB) or four of the mutant alleles (plasmids pPLgcvBΔR1, pPLgcvBΔR2, pPLgcvB5′Δ, pgcvB3′ΔT) shown in C. All strains displayed normal colony morphology (not shown). Reduced colony fluorescence of the dppA∷gfp or oppA∷gfp fusion strains upon sRNA coexpression indicates regulation at the post-transcriptional level. (Left) GcvB has no effect on the expression of gfp alone, confirming that repression is specific to the cloned dppA and oppA regions. (C,D) gcvB mutant alleles and their expression. (C) Horizontal bars below a schematic drawing of GcvB RNA denote the gcvB fragments expressed by mutant alleles; dotted lines denote internal deletions. Alleles gcvBΔR1 and gcvBΔR2 lack residues 66–89 (deletion of the G/U-rich R1 sequence) or 136–144 (deletion of R2), respectively. The truncated gcvB5′Δ and gcvB3′Δ alleles lack residues 1–91 (SL1 and R1) or 135–201 (R2 to SL5), respectively. In allele gcvB3′ΔT, which derives from gcvB3′Δ, SL3 was modified toward a transcription terminator as shown below. (D, lanes 4–9) Northern blots showing expression of gcvB or mutant alleles when borne on a mid-copy plasmid under control of the gcvB promoter in Salmonella. RNA was isolated from Salmonella grown to an OD600 of 1 and, except lane 1 (wild type; JVS-0007), from a ΔgcvB genetic background (JVS-0236). The strains in lanes 3–9 carried control vector, pTP11, or plasmids pgcvB, pgcvB5′Δ, pgcvBΔR1, pgcvBΔR2, pgcvB3′Δ, and pgcvB3′ΔT, respectively. Marker sizes are shown to the right. The top blot was probed with labeled oligo JVO-0749, which is complementary to the GcvB 5′ region (20–4 bp); the bottom blot was probed with JVO-0750 complementary to base pairs 172–150. The asterisk denotes transcriptional read-through to the rrnB terminator located downstream on the plasmids.
The fusion plasmids as well as control plasmid pXG1 expressing full-length GFP were each combined with either pPLgcvB or control vector pTP11 in an E. coli ΔgcvB strain. The specific repression of dppA∷gfp and oppA∷gfp by pPLgcvB was evident from strongly reduced colony fluorescence of these strains on agar plates (Fig. 2B), which established that GcvB regulates dppA and oppA in the 5′ mRNA region.
To map the GcvB interaction sites, we synthesized RNAs of the previously cloned dppA and oppA fragments in vitro and performed structural probing experiments. Gel mobility shift assays showed that GcvB formed singular complexes with either of the two RNAs under standard in vitro conditions (Supplementary Fig. S3). RNA structure probing of these complexes with RNase T1 and lead(II) acetate showed that increasing concentrations of dppA or oppA RNAs resulted in “footprints” on 5′-end-labeled GcvB RNA (Fig. 3A), which were most pronounced with lead(II) probing (Fig. 3A, lanes 6–10). The dppA and oppA RNAs each protect ≥19 residues within the single-stranded, highly G/U-rich region between stem–loops SL1 and SL2 of GcvB (cf. Figs. 3A and 1A).
Figure 3.
Identification of GcvB-binding sites on oppA and dppA mRNAs by in vitro probing. (A) 5′-End-labeled GcvB RNA (∼5 nM) was subjected to RNase T1, lead(II), and RNaseIII cleavage in the absence (lanes 1,6,11) and presence of different concentrations of cold dppA or oppA RNAs (∼50 nM final concentration in lanes 2,4,7,9,12,14; ∼500 nM final concentration in lanes 3,5,8,10,13,15). The synthesized target RNA fragments comprise regions −163/+72 (dppA) and −162/+57 (oppA) relative to the AUG start codon. (Lane C) Untreated GcvB RNA. (Lane T1) RNase T1 ladder of hydrolyzed denatured GcvB RNA. The position of cleaved G residues is given at the left of the gel. (Lane OH) Alkaline ladder. Vertical bars indicate the GcvB region protected by dppA RNA (red) and oppA RNA (blue). Arrows denote specific RNase III cleavage of GcvB in the presence of dppA and oppA RNAs. The approximate positions of stem–loop structures SL1 and SL2 according to the GcvB RNA structure shown in Figure 1A are indicated to the right of the gel. (B,C) 5′-End-labeled dppA RNA treated with lead(II) (B) or RNase III (C) in the absence (lane −) or presence of GcvB wild-type or GcvBΔR1 mutant RNA (lacks residues 66–89) as indicated above the gels. GcvB but not GcvBΔR1 mutant RNA protects an ∼20-nt region (vertical red bar in B) in dppA from lead(II) cleavage, and only GcvB wild type induces strong RNase III cleavage of the dppA RNA (red arrow in C). “G+3” set in bold is the G in the dppA AUG start codon. (D) Predicted RNA duplexes formed by GcvB with the dppA or oppA mRNAs. Vertical arrows denote RNase III cleavage sites. SD and AUG start codon sequences are boxed. The colored residues were protected from lead(II) cleavage upon duplex formation (see A–C; Supplementary Fig. S4).
The reciprocal experiment—i.e., probing of labeled dppA or oppA RNAs in complex with GcvB—identified the GcvB-binding site on the mRNAs. Wild-type GcvB RNA yields a strong footprint on the dppA (Fig. 3B) and oppA (Supplementary Fig. S4) RNAs, which correspond to regions −31 to −14, and −8 to +16, respectively, relative to the AUG start codon (Fig. 3D). In contrast, no footprints were obtained with a mutant RNA, GcvBΔR1, which lacks the G/U-rich region (Fig. 3B; Supplementary Fig. S4).
The duplexes shown in Figure 3D were further supported by cleavage of the GcvB/dppA and GcvB/oppA complexes with the double-strand-specific nuclease, RNase III. While several weak RNase III-dependent bands were observed in GcvB RNA alone (Fig. 3A, lane 11), the enzyme cleaved GcvB strongly and specifically in the G/U-rich region in the presence of either dppA or oppA RNA (Fig. 3A, lanes 12–15). Reciprocally, the dppA and oppA RNAs were specifically cleaved in the proposed GcvB-binding site (Fig. 3C; Supplementary Fig. S4). Collectively, these results indicated the single-stranded, G/U-rich region of GcvB as an important determinant for target recognition.
A conserved G/U-rich region mediates GcvB repression in vivo
The importance of the G/U-rich element was also evident from its strong conservation in GcvB sequences of distantly related species. Computer-based searches predicted gcvB genes in many enterobacteria as well as in Pasteurellaceae and Vibrionaceae (Supplementary Fig. S5). The location of these candidate genes adjacent to and divergent from gcvA and the conservation of promoter and terminator elements argue that these are gcvB homologs. Although the corresponding RNA sequences proved of enormous sequence diversity, we find that much of the G/U-rich linker between stem–loops SL1 and SL2 is highly conserved in all identified gcvB sequences (Fig. 1D). We refer to this linker region as R1 (Conserved Region 1). The single-stranded region between SL3 and SL4 contains another highly conserved sequence (ACUUCCUGUA) found immediately upstream of SL4; this sequence is referred to as R2.
To assess the role of these two regions in target regulation, we constructed mutant alleles, gcvBΔR1, gcvBΔR2, and gcvB5′Δ, the latter being a 5′-truncated gcvB lacking SL1 and R1 (Fig. 2C). Plasmids carrying these alleles expressed distinct GcvB-derived RNAs at levels similar to wild-type gcvB (Fig. 2D). Next, these alleles were cloned under control of the PLlacO-1 promoter to test their ability to regulate the dppA∷gfp and oppA∷gfp fusions. Figure 2B shows that loss of R1 (gcvBΔR1 and gcvB5′Δ) abrogated dppA and oppA fusion repression, whereas loss of R2 (gcvBΔR2) had no effect.
Is the 5′ part of GcvB (including R1) sufficient to confer target repression in vivo? Urbanowski et al. (2000) postulated that SL3 of GcvB, which is followed by a U-stretch in many GcvB species (Fig. 1D), may serve as a ρ-independent transcription terminator, leading to expression of a shorter GcvB RNA. However, truncation of gcvB after residue T134 (allele gcvB3′Δ, expected to yield an ∼134-nt GcvB RNA) (Fig. 2C) did not result in such an RNA (Fig. 2D), whereas modification of SL3 created a functional terminator (allele gcvB3′ΔT) (Fig. 2C) and led to detection of an ∼130-nt RNA (Fig. 2D). Figure 2B shows that the 3′-truncated GcvB RNA of gcvB3′ΔT was fully active in dppA and oppA fusion repression. Taken together, these in vivo experiments further support a key role of the G/U-rich element, R1, of GcvB for dppA and oppA mRNA regulation.
More GcvB targets
The observation that additional periplasmic proteins accumulated in gcvB or hfq mutant strains (Urbanowski et al. 2000; Sittka et al. 2007) prompted us to screen for more GcvB targets. Analysis of periplasmic proteins from a Salmonella gcvB overexpression strain predicted gltI, livJ, argT, and STM4351 as further candidate targets (Fig. 4A). In an independent approach, we used the RNAhybrid algorithm (Rehmsmeier et al. 2004) to search for stable RNA duplexes of the R1 sequence of GcvB with the 5′ regions of all Salmonella genes. This biocomputational search supported gltI, livJ, argT, and STM4351 and further predicted livK as a GcvB target (Fig. 5A). Intriguingly, the predicted GcvB-binding sites on these mRNAs do not match in sequence but are rich in C and A residues, which also holds true for dppA and oppA (Fig. 3D).
Figure 4.
GcvB targets additional periplasmic transporter mRNAs. (A) Salmonella ΔgcvB carrying control plasmid pTP11 (left) or gcvB mid-copy plasmid pTP05 (right) were grown to early stationary phase, and periplasmic protein fractions of these bacteria were resolved on two-dimensional gels. Relevant subsections of the gels are shown. Protein spots indicating repression by GcvB are labeled. (B) Agar plate-based assay of colony fluorescence of E. coli strains carrying either gltI∷gfp (left; plasmid pJL24-1) or livJ∷gfp (right; plasmid pJL20-1). Each fusion plasmid was combined with control plasmid pTP11, or plasmids expressing Salmonella GcvB and mutant RNAs as in Figure 2B. (C) Western blots of target∷GFP fusion proteins prepared from E. coli ΔgcvB recA− carrying the indicated fusion plasmids in combination with control plasmid pTP11, or plasmids expressing wild-type gcvB or the gcvBΔR1 allele. GroEL was probed as the loading control. Fold changes of GFP fusion protein levels (upon normalization to GroEL levels) by gcvB or gcvBΔR1 coexpression relative to the control plasmid were OppA∷GFP, −2.8/−1.2; DppA∷GFP, −3.6/−1.3; GltI∷GFP, −1.8/+2.2; LivJ∷GFP, −5.3/−1.4; LivK∷GFP, −2.1/−1.0; ArgT∷GFP, −1.7/−1.1; STM4351∷GFP, −1.8/−1.3; and GFP alone, −1.1/−1.2.
Figure 5.
Summary of in vitro probing results of GcvB–target complexes. (A) Proposed RNA duplexes formed by GcvB with five periplasmic transporter mRNAs (gltI, livK, livJ, argT, STM4315). Positions in the target sequences are given as distance to the mRNA start codon. Vertical arrows denote in vitro RNase III cleavage of the GcvB–gltI complex, and of GcvB in complex with the four other targets (Supplementary Figs. S6A, S7B). Residues that showed protection in in vitro footprinting experiments (Supplementary Figs. S6, S7A) are set in bold and capitalized. Biocomputational prediction of target sites proposed the formation of longer duplexes around the interaction sites mapped by footprint analysis. (B) Location of GcvB-binding sites on target mRNAs. 5′ UTRs of target genes are drawn to scale. Asterisks indicate promoter positions that were mapped by 5′RACE; “#” indicates promoters according to EcoCyc annotations. SD sequences are shadowed. Positions of the GcvB-binding sites on the mRNAs are marked by a horizontal bar.
The predicted binding sites were subsequently confirmed by in vitro probing experiments as above (summarized in Fig. 5; shown in Supplementary Fig. S6). Although the footprint obtained for gltI mRNA is relatively weak, the predicted interaction is strongly supported by specific RNase III cleavage in the presence of GcvB (Supplementary Fig. S6A). Conversely, R1 is the GcvB region that is most strongly protected upon incubation with the target RNA fragments (Supplementary Fig. S7). When in complex with target RNAs, GcvB was cleaved by RNase III exclusively in the R1 sequence, except with a livK-derived fragment that also promotes cleavage in the GcvB 3′ part (Supplementary Fig. S7). We constructed translational gfp fusions to all of these targets, of which the gltI and livJ fusions showed fluorescence on agar plates (Fig. 4B). Both fusions were strongly repressed by gcvB alleles with an intact R1 sequence (gcvB, gcvBΔR2, and gcvB3′ΔT), whereas the gcvB5′Δ and gcvBΔR1 alleles lacking R1 failed to repress the fusions (Fig. 4B). Western blot analysis of all five target fusions as well as the dppA∷gfp and oppA∷gfp fusions further confirmed that regulation required an intact R1 sequence (Fig. 4C). These results increased the number of GcvB targets to seven mRNAs.
GcvB inhibits translation initiation in vitro
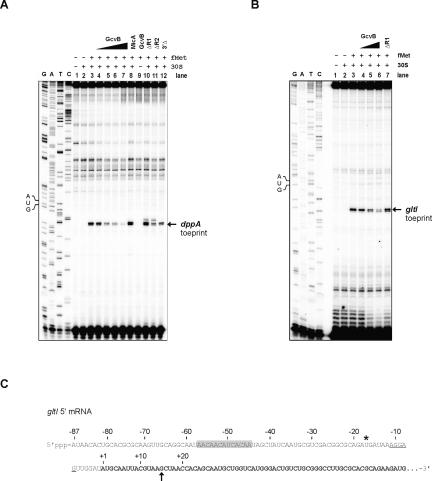
GcvB binds near the Shine-Dalgarno (SD) sequence of dppA and oppA (Fig. 3D) and was thus predicted to prevent ribosome binding to these mRNAs. To test this, we performed 30S ribosome toeprinting assays (Hartz et al. 1988). A dppA mRNA fragment was annealed to an end-labeled primer complementary to the dppA coding region (+58 to +73) and incubated with 30S subunits in the presence or absence of uncharged tRNAfMet, followed by cDNA synthesis. Analysis of the extension products (Fig. 6A) revealed one ribosome-induced, tRNAfMet-dependent termination site at the characteristic +15/+16 positions (start codon A is +1). This “toeprint” signal was decreased when wild-type GcvB RNA was added prior to incubation with 30S/fMet (Fig. 6A, lanes 4–7), suggesting inhibition of 30S binding. Inhibition was also observed with the GcvBΔR2 and GcvB3′Δ mutant RNAs, which have an intact R1 sequence (Fig. 6A, lanes 11,12). In contrast, the GcvBΔR1 mutant RNA (Fig. 6A, lane 10) did not inhibit ternary complex formation.
Figure 6.
GcvB inhibits 30S binding in vitro. (A) Ribosome toeprinting of dppA leader RNA (20 nM) as described in Materials and Methods. “+/−” indicate the presence or absence of 30S subunit (200 nM) and fMet initiator tRNA (1 μM). The dppA AUG start codon position is shown. The arrow indicates the 30S toeprint. Increasing concentrations of GcvB RNA (lanes 4–7: 20, 60, 100, and 200 nM) in the reactions inhibit 30S binding, whereas the nonspecific control RNA, MicA (lane 8, 100 nM), or GcvBΔR1 mutant RNA (lane 10, 100 nM) do not impair binding. (Lanes 11,12) Mutant RNAs GcvBΔR2 and GcvB3′Δ were added at a final concentration of 100 nM. (B) Toeprint on gltI 5′ RNA (20 nM), using 20 nM 30S subunit and 100 nM tRNAfMet. (Lanes 4–6) Increasing GcvB concentrations (20, 100, and 200 nM, respectively) inhibit 30S binding. (Lane 7) GcvBΔR1 mutant RNA (100 nM) does not inhibit the toeprint. (C) mRNA sequence of Salmonella gltI (5′ end). The gltI coding sequence is set in bold. The asterisk denotes the wrongly annotated gltI start codon in the Salmonella LT2 genome sequence. The gltI start codon shown here was confirmed by a specific toeprint signal at position +15, which is indicated by an arrow (cf. B). The SD sequence is underlined, and the C/A-rich GcvB target site is shadowed.
The same assay performed on oppA mRNA confirmed that GcvB inhibited ternary complex formation, and that it required an intact R1 sequence (data not shown). Since GcvB did not inhibit 30S binding to the unrelated ompA mRNA (data not shown), we conclude that it acts as a sequence-specific translation initiation inhibitor by masking the dppA and oppA RBS.
GcvB inhibits gltI translation by binding far upstream of the start codon
GcvB binds to its other five target mRNAs further upstream of SD and the start codon as compared with dppA and oppA (Fig. 5). Notably, GcvB binds to region −57 to −45 of gltI, and −57 to −42 of argT. These interaction sites lie outside the −35 to +19 mRNA region covered by 30S ribosomes (Hüttenhofer and Noller 1994). Toeprinting of 30S on gltI mRNA (Fig. 6B) gave a single toeprint at +15 relative to the gltI AUG in the presence of 30S/fMet (Fig. 6B, lane 3), confirming the gltI start codon shown in Figure 6C (note that the gltI coding region is misannotated in Salmonella and likely in several additional bacteria). Strikingly, GcvB dramatically reduced 30S binding on the gltI mRNA (Fig. 6C, lane 4–6), dependent on an intact R1 sequence (Fig. 6C, lane 7). Thus, GcvB can inhibit mRNA translation by binding upstream of a 30S-binding site.
The C/A-rich target site enhances gltI translation
To further evaluate GcvB regulation at upstream mRNA sites, the C/A-rich target region (−71 to −44) was deleted in gltI∷gfp, yielding fusion gltIΔCA∷gfp. This deletion abolished fusion repression in the presence of pPLgcvB (Fig. 7A), further supporting that GcvB pairs to this region. Intriguingly, loss of this region also resulted in lower GFP fluorescence as compared with the parental gltI∷gfp fusion. Since C/A-rich stretches were reported to enhance mRNA translation (Martin-Farmer and Janssen 1999), we determined whether the GcvB target site acts as an enhancer of gltI translation. In vitro synthesized, full-length gltI∷gfp and gltIΔCA∷gfp fusion mRNAs were translated with reconstituted 70S ribosomes, and GltI∷GFP fusion protein synthesis was monitored over time. Figure 7B shows a linear increase in GltI∷GFP synthesis from both mRNA templates within a 45-min assay. However, the gltI∷gfp mRNA gave approximately threefold higher translation rates compared with that of gltIΔCA∷gfp. Addition of GcvB (10-fold excess) to the reactions strongly inhibited protein synthesis from gltI∷gfp but not gltIΔCA∷gfp mRNA. None of these variations are due to RNA degradation since the gltI∷gfp and GcvB RNAs were stable during the assay (Supplementary Fig. S8). In addition, the inhibitory activity of various GcvB mutants on gltI∷gfp in vitro (Fig. 7C) perfectly correlated with their ability to inhibit the fusion mRNA in vivo (Fig. 4B).
Figure 7.
The C/A-rich upstream element functions both as GcvB target site and as translational enhancer element. (A) Agar plate-based assay of colony fluorescence of E. coli strains carrying either the parental gltI∷gfp (left; plasmid pJL24-1) or the mutant gltIΔCA∷gfp (right; plasmid pJL45-3) fusion. In the latter plasmid, the C/A-rich GcvB target site (−71 to −44) (cf. Fig. 6C) is deleted. The fusion plasmids were combined with control vector pTP11 or GcvB expression plasmid pPLgcvB. (B) In vitro synthesized, full-length mRNAs (40 nM) of the gltI∷gfp and gltIΔCA∷gfp fusions were in vitro translated with reconstituted 70S ribosomes in the presence of 40 nM Hfq as described in Materials and Methods. Syntheses of GltI∷GFP fusion protein levels were determined after incubation for 15 min (lanes 1,4,7,10,13,16), 30 min (lanes 2,5,8,11,14,17), and 45 min (lanes 3,6,9,12,15,18) by Western blot analysis. The reactions in lanes 4–9 and 13–18 contained a 10-fold excess of GcvB or GcvBΔR1 mutant RNAs as indicated above the lanes. Fold regulation upon deletion of the C/A-rich target site or by addition of GcvB according to quantification of GltI∷GFP fusion protein levels is given below the blot. The results of a representative experiment (out of three) are shown. (C) Full-length gltI∷gfp mRNA was in vitro translated as above for 30 min. (Right two lanes) In addition, mutant RNAs GcvBΔR2 and GcvB3′Δ (10-fold excess) were included in the assay. (D) Insertion of the 27-nt C/A-rich element of gltI at position −42 relative to the start codon of an E. coli ompR∷gfp fusion yields mutant ompRCA∷gfp (plasmids pJU-063 and pJL50-11, respectively). (E) In vitro translation assay (reconstituted 70S ribosomes) as in B but with in vitro synthesized ompR∷gfp and ompR∷gfpCA fusion mRNAs (250 nM) and in the presence of 250 nM Hfq. Synthesis of OmpR∷GFP fusion protein was determined on Western blots following incubation for 15 min (lanes 1,4,7,10), 30 min (lanes 2,5,8,11), and 45 min (lanes 3,6,9,12). A 10-fold excess of GcvB RNA over mRNA was added to the reactions where indicated. The results shown are representative of the experiment performed in triplicate.
Creation of an upstream C/A-rich target site permits translational control of an unrelated mRNA
Next, we inserted the C/A-rich GcvB-binding site of gltI at position −42 of an E. coli ompR∷gfp fusion, yielding ompRCA∷gfp (Fig. 7D). The Salmonella gltI and E. coli ompR genes are unrelated in function, and their 5′ mRNA regions have little sequence identity (Supplementary Fig. S9). Insertion of the C/A-rich element resulted in an approximately twofold increase in OmpR∷GFP synthesis in the in vitro translation assay (Fig. 7E; cf. lanes 3 and 9), confirming the stimulatory effect of this element on mRNA translation. When GcvB was present in the reaction, it strongly inhibited translation of the ompRCA∷gfp but not of the parental ompR∷gfp fusion mRNA (Fig. 7E). Intriguingly, GcvB inhibits ompRCA∷gfp translation ∼10-fold; i.e., as strongly as observed with gltI∷gfp. In other words, GcvB effectively represses a structurally unrelated mRNA upon insertion of a C/A-rich target site at an upstream position.
Discussion
ABC transporters constitute a major class of amino acid uptake systems and commonly have at least one periplasmic solute-binding protein to take up substrates upon their diffusion through outer membrane porins (Hosie and Poole 2001; Davidson and Chen 2004). Given the key roles of amino acids in nitrogen and carbon metabolism, regulation of transporter expression and activity is complex. Intriguingly, genes involved in amino acid uptake and metabolism also are the most strongly misregulated class in hfq mutants of E. coli and Salmonella (Guisbert et al. 2007; A. Sittka and J. Vogel, in prep.), suggesting that Hfq-dependent sRNAs are involved in the post-transcriptional control of these pathways. We have here established one such sRNA, GcvB, as a direct regulator of many ABC transporter mRNAs in Salmonella. The conservation of gcvB genes combined with the previous data from the Stauffer laboratory (Urbanowski et al. 2000; McArthur et al. 2006) as well as successful prediction of GcvB–target interactions in related organisms (Supplementary Table S10; Tjaden et al. 2006) strongly argue that the GcvB function in amino acid uptake is conserved in many other bacteria. Interestingly, some enterobacteria encode an additional sRNA, RydC, which regulates the expression of a putative ABC transport system, yejABEF (Antal et al. 2005).
GcvB directly interacts with seven Salmonella mRNAs that belong to various transcriptional regulons but collectively encode periplasmic substrate-binding proteins of ABC transporters (http://www.tcdb.org; http://www.kegg.org). These proteins are known or predicted to bind di- and oligopeptides (DppA, OppA) and polar (GltI, STM4351, ArgT) and branched amino acids (LivK, LivJ). Since GcvB is specifically expressed in rich medium in fast-growing cells (Fig. 1B), its function appears to lie in the general repression of amino acid uptake when nutrients are plentiful. Consistent with this prediction, a Salmonella gcvB deletion strain exhibits elevated protein and mRNA levels of almost all established GcvB targets (Figs. 1C, 4A; Supplementary Fig. S10). The increased steady-state levels of nearly all target mRNAs in ΔgcvB (Supplementary Fig. S10) argue that by repressing mRNA translation, GcvB also promotes the decay of its targets. Western blot quantification of OppA protein levels revealed a sixfold increase in ΔgcvB as compared with wild type (Fig. 1C); the corresponding RT–PCR experiment (Supplementary Fig. S10) suggests a similar increase in oppA mRNA levels. Comparison of the increases in target protein levels (spot intensity on Coomassie-stained 2D gels) (Fig. 4A) and mRNA levels (Supplementary Fig. S10) upon gcvB deletion suggests a similar correlation in mRNA/protein level changes for dppA, gltI, livJ, STM4351, and argT (LivK was not detected on the 2D gels). Whether RNase III, here used as a tool for structure probing in vitro, is the primary nucleolytic activity to degrade the GcvB target mRNA in vivo as shown previously for the IstR-1 sRNA (Vogel et al. 2004) and RNAIII (Huntzinger et al. 2005), remains to be determined. At least dppA, livK, gltI, and oppA each constitutes the first gene of a polycistronic transporter operon. Considering that the downstream cistrons of these polycistronic mRNAs are likely translationally coupled to the direct GcvB targets, the sRNA may, in fact, repress >20 genes. However, since GcvB is intrinsically unstable (half-life 2–3 min) (Vogel et al. 2003; Supplementary Fig. S1), ABC transporter repression will be quickly alleviated upon decreased availability of amino acids.
The sheer number of sRNA genes discovered by systematic searches of bacterial genomes over the last years led to an increasing recognition of the potential impact of these riboregulators on bacterial physiology. It is well established that sRNAs that act to modulate protein activity can control the expression of many genes; e.g., by binding to RNA polymerase (Wassarman and Storz 2000) or to CsrA-like proteins (Babitzke and Romeo 2007). sRNAs can also affect the expression of larger sets of genes by targeting the mRNAs of global transcriptional regulators; e.g., RpoS or FhlA (Lease et al. 1998; Majdalani et al. 1998; Argaman and Altuvia 2000). In contrast, our understanding of how sRNAs could directly control multiple mRNAs by an antisense mechanism has been limited by the low number of validated sRNA–target interactions and hence difficulty to reliably predict new targets. This notwithstanding, the results of several recent studies suggest that multiple targeting may be more common than previously thought, and common denominators are beginning to emerge. Tjaden et al. (2006) suggested that functional relationship of the proteins encoded by target candidates, as it is also seen among the GcvB targets, may add confidence to predictions. Similarly, the OmrA/B and RybB sRNAs were predicted to directly target multiple mRNAs that collectively encode for outer membrane proteins (Guillier and Gottesman 2006; Johansen et al. 2006; Papenfort et al. 2006). By the same token, the iron stress-responding E. coli RyhB sRNA was shown to regulate multiple mRNAs that encode proteins involved in iron metabolism (Massé and Gottesman 2002), and many of the mRNAs demonstrated to be direct targets of S. aureus RNAIII encode bacterial virulence factors (Boisset et al. 2007).
The strong C/A bias of GcvB-binding sites may point to yet another feature of multiple target regulation. CA multimers placed downstream from mRNA start codons were reported to stimulate translation in vivo, and to increase ribosome-binding affinity to mRNAs in vitro (Martin-Farmer and Janssen 1999). While the effect of CA-multimers far upstream of a start codon was not addressed by Martin-Farmer and Janssen (1999), we find that the C/A-rich element has a stimulatory effect on the gltI and ompR fusion mRNAs (Fig. 7). Thus, multiple mRNA targeting by GcvB may have evolved through hijacking a translational enhancer element shared by numerous mRNAs that encode periplasmic transporters. We are thus tempted to speculate that other evolutionarily constrained mRNA regions—e.g., those encoding signal peptides—may also constitute binding sites for sRNAs with multiple targets. However, we emphasize that the effects of GcvB as a translational repressor are clearly much greater than the effects of removing the C/A-rich target site (Fig. 7); thus GcvB does not primarily act by simply blocking any enhancer effect.
In eukaryotes, the recognition that a 5′-terminal seed sequence of the ∼22-nt microRNAs provides the base-pairing specificity to target mRNAs greatly advanced target predictions. In contrast, the target interaction sites of the much longer bacterial sRNAs are not necessarily located at the 5′ end. An alignment of GcvB RNAs (Fig. 1D) revealed two strongly conserved regions, R1 and R2. While R2 may constitute an interaction region for yet another target(s), R1 is a key determinant for base-pairing to seven target mRNAs. Although our structure probing of target mRNAs and GcvB indicates additional, weaker contacts, the C/A-rich motif in the targets and R1 in the sRNA clearly are the anchoring and essential motifs for interaction. Accordingly, giving higher weight to deeply conserved regions in sRNAs is expected to improve the currently available target search algorithms. In support of this, several other regulatory sRNAs—e.g., MicA, MicC, and SgrS—exhibit a higher degree of conservation in their target interaction regions (Chen et al. 2004; Vanderpool and Gottesman 2004; Rasmussen et al. 2005; Udekwu et al. 2005). Unlike these enterobacterial sRNAs, S. aureus RNAIII is not conserved in other species. However, it is still intriguing that a short (∼40-nt) region within the 514-nt RNAIII facilitates base-pairing to multiple mRNAs (Huntzinger et al. 2005; Boisset et al. 2007). Different from GcvB, however, RNAIII typically covers the SD and/or the start codon of a target mRNA (Huntzinger et al. 2005; Boisset et al. 2007).
Competition with ribosome binding explains the inhibitory activity of sRNAs that bind mRNAs within RBS regions (Argaman and Altuvia 2000; Chen et al. 2004; Huntzinger et al. 2005; Udekwu et al. 2005). We show that the same applies to GcvB inhibition of dppA and oppA, whose GcvB-binding sites are very close to the SD (Fig. 3D). However, GcvB binds many of its targets further upstream of the two sequence elements, SD and start codon, that are the key determinants of 30S binding (Fig. 5B). gltI is the mRNA with the most upstream binding site; probing of GcvB/gltI RNA complexes as well as transfer of the GcvB site from gltI to the unrelated ompR mRNA showed that GcvB effectively represses translation by forming a duplex >42 nt upstream of the AUG.
How is translational repression brought about at such upstream sites? Footprinting experiments revealed the maximal ribosome-binding region on mRNAs to range from −39 to +19 relative to the AUG (Hüttenhofer and Noller 1994). Since the GcvB-binding site on gltI lies outside this window, a simple interference model in which GcvB occludes mRNA residues required for base-pairing with 16S ribosomal RNA is unlikely. In eukaryotes, ribosomes generally enter mRNAs at their 5′ ends to subsequently scan for downstream AUG triplets. A requirement for scanning is unknown in prokaryotes, which argues against the possibility that GcvB could act as a roadblock for ribosomes scanning from the 5′ end of gltI mRNA.
Two other bacterial sRNAs, IstR-1 and RyhB, were recently reported to repress translation by binding upstream of the target RBS (Darfeuille et al. 2007; Vecerek et al. 2007), yet the underlying mechanisms do not seem to apply for GcvB either. IstR-1 targets the tisAB mRNA ∼100 nt upstream of the tisB start codon (Vogel et al. 2004). The tisAB mRNA is highly structured, and the tisB SD is entrapped in a stable hairpin. Thus, tisB translation requires ribosomes to bind to an upstream “standby” site, which will be masked upon IstR-1 binding (Darfeuille et al. 2007). Unlike in tisAB, the gltI 5′ UTR is not strongly structured (Supplementary Fig. S6A). Moreover, since GcvB also inhibits the unrelated ompR∷gfpCA mRNA, structure cannot be a primary cause of translational repression. RyhB inhibits fur translation by forming an imperfect duplex with the −96 to −48 region of fur mRNA. Translation of fur is coupled to that of an upstream located reading frame, the direct target of RyhB (Vecerek et al. 2007). We have not found evidence for an upstream reading frame in the gltI mRNA, nor for translation initiation upstream of the determined gltI start codon (Fig. 6B).
The gltI 5′ UTR contains a putative hairpin motif (−40 to −7 region), which could bring the C/A-rich motif in closer proximity to the gltI SD to facilitate repression by GcvB (Supplementary Fig. S11). However, the following observations do not support such a model. Firstly, the SD of ompR∷gfp is not preceded by a hairpin that would bring the GcvB target site and SD in closer proximity; yet transfer of the GcvB site to ompR∷gfp yields the same degree of mRNA repression as observed with the gltI∷gfp construct. Secondly, we disrupted the putative hairpin motif in the gltI 5′ UTR by two point mutations. These mutations did not impair GcvB’s ability to repress the gltI fusion mRNA in vivo (Supplementary Fig. S11).
Consequently, our present results suggest that either the first three GcvB stem–loops or the G/U:C/A-rich helix of the GcvB–target mRNA complex constitute an inhibitory signal for 30S entry on the gltI and argT mRNAs. However, the underlying mechanism may be a more general one, since we have found that Salmonella RybB sRNA also represses several targets by binding upstream of the RBS (F. Mika and J. Vogel, unpubl.). It might therefore be of interest to consider the possibility that target searches are not yet exhausted for those sRNAs that do not show obvious complementarity to RBS sequences.
Materials and methods
Bacterial strains, oligonucleotides, and plasmids
DNA oligonucleotides used for cloning, PCR amplification of T7 templates, and toeprinting assays, and as hybridization probes are provided as Supplemental Material (Supplementary Table S1).
Bacterial strains are listed in Supplementary Table S2. The gcvB deletion strains of Salmonella (JVS-0236) and E. coli (JVS-6081) were constructed using the λ red protocol (Datsenko and Wanner 2000) by replacing residues 17–176 with a kanamycin marker gene, PCR-amplified with primer JVO-0133/-0134 and JVO-0131/-0132, respectively. Mutants were verified by PCR with primers JVO-0135/-0136 or JVO-0137/-0138, respectively. The Salmonella ΔgcvB/Δhfq strain (JVS-0617) was constructed by P22 transduction of Δhfq∷CmR (JVS-0255) into strain JVS-0236. Marker genes were removed with FLP recombinase (Datsenko and Wanner 2000).
Plasmids, details of their construction, and insert sequences are given in Supplementary Tables S3–S6. Control plasmid pTP11 carries a p15A origin and expresses an ∼50-nt nonsense transcript derived from the rrnB terminator. Plasmid pgcvB (pTP05) carries the Salmonella gcvB locus (292 bp upstream of the +1 site to 116 bp downstream from the terminator), while plasmid pPLgcvB (pTP09) expresses the gene from a constitutive promoter. gfp fusions were constructed as described (Urban and Vogel 2007). The dppA, oppA, gltI, livJ, and argT fusions were cloned as 5′RACE cDNA fragments into fusion plasmid pXG-20. The livK and STM4351 fusions were cloned in vector pXG-10. To construct fusion gltIΔCA∷gfp (pJL45-3), base pairs −71 to −44 relative to gltI AUG were deleted from the gltI∷gfp fusion plasmid. The deleted 27-bp C/A-rich element was inserted in the ompR∷gfp fusion (pJU-63) at position −42 according to the ompR AUG to yield ompRCA∷gfp (pJL50-11). The insert sequences of all GFP fusions are given in Supplementary Table S7.
Media and growth conditions
Cells were grown aerobically at 37°C in Lennox broth or M9 minimal medium supplemented with 0.4% glucose. When required, antibiotics were added at 92 μg/μL streptomycin, 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 20 μg/mL chloramphenicol.
RNA and protein detection
RNA preparation and Northern analysis followed previously published protocols (Urban and Vogel 2007). GcvB RNAs were detected with 5′-end-labeled oligos (see figure legends), and 5S rRNA or gfp fusion mRNAs with oligos JVO-0322 or JVO-155, respectively.
OppA protein was detected on a Western blot (protocol in Sittka et al. 2007) using a polyclonal OppA antibody kindly provided by K. Igarashi (Chiba University, Chiba, Japan). GFP fusion and GroEL proteins were detected as described in Urban and Vogel (2007). Periplasmic proteins were prepared from Salmonella cultured to OD 2 as described in Sittka et al. (2007), and analyzed by high-resolution 2D electrophoresis, protein staining, and peptide mass fingerprinting at the MPI-IB protein analysis core facility according to previously published standard protocols (http://info.mpiib-berlin.mpg.de/jungblut).
Colony fluorescence imaging
E. coli carrying gfp fusion plasmids were grown on LB plates overnight. Colonies were photographed with a Fuji LAS-3000 image analyzer with a 510-nm emission filter and excitation at 460 nm.
T7 transcription, purification, and 5′-end labeling of RNA
DNA templates carrying a T7 promoter sequence for in vitro transcription were generated by PCR. Primers and sequences of the T7 transcripts are given in Supplementary Tables S8 and S9. T7 templates of gfp fusion mRNAs were amplified from plasmids using a sense primer that adds a T7 promoter to the +1 site of the 5′ UTR, and antisense oligo pZE-T1, which binds 122 nt downstream from the gfp stop codon. These transcripts end with the rrnB terminator of the fusion plasmids. RNA was in vitro transcribed and quality-checked as described in Sittka et al. (2007). The protocol for 5′-end labeling of RNA is published in Papenfort et al. (2006).
In vitro structure mapping and footprinting
Secondary structure probing and mapping of RNA complexes were conducted on 5′-end-labeled RNA (∼0.1 pmol) in 10-μL reactions. RNA was denatured for 1 min at 95°C and chilled on ice for 5 min, upon which 1 μg of yeast RNA and 10× structure buffer (0.1 M Tris at pH 7, 1 M KCl, 0.1 M MgCl2; Ambion) were added. The concentrations of unlabeled sRNA/mRNA leader added to the reactions are given in the figure legends. Following incubation for 10 min at 37°C, 2 μL of a fresh solution of lead(II) acetate (25 mM; Fluka #15319), 2 μL of RNase T1 (0.01 U/μL; Ambion, #AM2283), or 2 μL of RNase T2 (0.02 L/μL; Invitrogen #18031-013) were added and incubated for 2, 3, or 5 min at 37°C, respectively. RNase III cleavage reactions contained 1 mM DTT and 1.3 U of enzyme (New England Biolabs #M0245S), and were incubated for 6 min at 37°C.
Reactions were stopped with 5 μL of 0.1 M EDTA, precipitated, and dissolved in loading buffer II (95% Formamide; 18 mM EDTA; 0.025% SDS, Xylene Cyanol, and Bromophenol Blue; Ambion), or by direct addition of 12 μL of loading buffer on ice. RNase T1 ladders were obtained by incubating labeled RNA (∼0.2 pmol) in 1× sequencing buffer (Ambion) for 1 min at 95°C. Subsequently, 1 μL of RNase T1 (0.1 U/μL) was added, and incubation was continued for 5 min at 37°C. OH ladders were generated by 5 min of incubation of 0.2 pmol of labeled RNA in alkaline hydrolysis buffer (Ambion) at 95°C. Reactions were stopped with 12 μL of loading buffer. Samples were denatured for 3 min at 95°C prior to separation on 6% polyacrylamide/7 M urea sequencing gels in 1× TBE. Gels were dried and analyzed using a PhosphorImager (FLA-3000 Series; Fuji), and AIDA software.
30S ribosome toeprints
Toeprinting reactions were carried out as described (Hartz et al. 1988; Udekwu et al. 2005) with few modifications. An unlabeled dppA mRNA fragment (0.2 pmol; 236 nt; T7 template amplified with JVO-1034/-1035), and 0.6 pmol of 5′-end-labeled primer JVO-1035 complementary to the dppA coding region was annealed. For inhibition analysis, 0.2, 0.6, 1, and 2 pmol of GcvB RNA or 1 pmol of control RNA (MicA) or GcvB mutant RNAs were added. Nucleic acids were denatured in annealing buffer (10 mM Tris-acetate at pH 7.6, 1 mM DTT, 100 mM potassium acetate) for 1 min at 95°C and chilled for 5 min on ice, upon which Mg2+ acetate and all NTPs were added to final concentrations of 10 mM and 0.5 mM, respectively. All subsequent incubations steps were at 37°C. After 5 min of incubation, 2 pmol of 30S ribosomal subunit (Knud Nierhaus, Max Planck Institute for Molecular Genetics, Berlin, Germany; preactivated for 20 min prior to the assay) were added. Following incubation for 5 min, uncharged tRNAfMet (10 pmol) was added, and incubations continued for 15 min. Reverse transcription was carried by adding SuperScript II (100 U; Invitrogen) for 20 min. cDNA synthesis was terminated with 100 μL of stop buffer (50 mM Tris-HCl at pH 7.5, 0.1% SDS, 10 mM EDTA). Following phenol-chloroform extraction, alkaline hydrolysis of template RNA at 90°C, and ethanol precipitation, cDNA was dissolved in 10 μL of loading buffer II (Ambion). Toeprint analysis on gltI 5′ RNA (161 nt; T7 template amplified with primers JVO-1039/-1040) was performed using 5′-end-labeled primer JVO-1775. See the figure legends for final concentrations of other components.
Sequencing ladders were generated with CycleReader DNA Sequencing Kit (Fermentas #K1711) according to the manufacturer’s protocol on the same DNA template used for T7 transcription and the same 5′-end-labeled primer as in the toeprinting reactions. cDNAs and sequence ladders were separated on a 6% polyacrylamide/7 M urea gel. Autoradiograms of dried gels were obtained as above.
In vitro translation assays
Translation reactions were carried out using PureSystem (Cosmo Bio Co., Ltd, PGM-PURE2048C) according to the manufacturer’s instructions. Ten-microliter (Fig. 7B,C) or 20-μL (Fig. 7E) reactions contained, in addition to 70S ribosomes, mRNA template, Hfq, and—where applicable—GcvB RNAs (see figure legends for final concentrations). Supplementary Tables S8 and S9 list the details of DNA fragments used for T7 transcription. Before addition of PureSystem mix, RNA was denatured for 1 min at 90°C and chilled on ice for 5 min. Hfq was mixed with mRNA (and sRNA) and preincubated for 10 min at 37°C. PureSystem mix was added, and incubation continued at 37°C for the time given in the figure legends. Reactions were stopped with 4 vol of ice-cold acetone and kept on ice for 15 min, and proteins were collected by centrifugation (10,000g, 10 min, 4°C). Proteins were quantified by Western blot analysis with a monoclonal GFP antibody as described in Urban and Vogel (2007).
Prediction of sRNA–target mRNA duplexes
GcvB–target mRNA complexes were predicted with RNAhybrid (Rehmsmeier et al. 2004). 5′ Regions of Salmonella LT2 genes (±50 nt of annotated start codon) were extracted as a multifasta file and used as target sequence input. GcvB or shorter versions thereof (R1 or R2 ±10 nt) were used as miRNA input. For predictions of GcvB/target RNA duplexes, internal and bulge loops were restricted to a size of 1 nt.
Acknowledgments
We thank K. Nierhaus for providing 30S ribosomes; M. Schmid and P. Jungblut for mass spectrometry; K. Igarashi for OppA serum; Marc Rehmsmeier for discussions on target predictions; L. Bui for technical assistance; and E.G. Wagner, K. Nierhaus, and members of our laboratory for critical comments on the manuscript and helpful discussions. C.M.S. was supported by grants from Deutsche Forschungsgemeinschaft, Germany (DFG; VO 875/1-1,3).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.447207
References
- Abouhamad W.N., Manson M., Gibson M.M., Higgins C.F., Manson M., Gibson M.M., Higgins C.F., Gibson M.M., Higgins C.F., Higgins C.F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: Characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 1991;5:1035–1047. doi: 10.1111/j.1365-2958.1991.tb01876.x. [DOI] [PubMed] [Google Scholar]
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G., Weinstein-Fischer D., Zhang A., Postow L., Storz G., Zhang A., Postow L., Storz G., Postow L., Storz G., Storz G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Antal M., Bordeau V., Douchin V., Felden B., Bordeau V., Douchin V., Felden B., Douchin V., Felden B., Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- Argaman L., Altuvia S., Altuvia S. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S., Bejerano G., Wagner E.G., Margalit H., Altuvia S., Wagner E.G., Margalit H., Altuvia S., Margalit H., Altuvia S., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Romeo T., Romeo T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Huntzinger E., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Fechter P., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Bendridi N., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Possedko M., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Chevalier C., Helfer A.C., Benito Y., Jacquier A., Helfer A.C., Benito Y., Jacquier A., Benito Y., Jacquier A., Jacquier A., et al. Staphylococcus aureusRNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes & Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang A., Blyn L.B., Storz G., Zhang A., Blyn L.B., Storz G., Blyn L.B., Storz G., Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille F., Unoson C., Vogel J., Wagner E.G., Unoson C., Vogel J., Wagner E.G., Vogel J., Wagner E.G., Wagner E.G. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.L., Chen J., Chen J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- Deana A., Belasco J.G., Belasco J.G. Lost in translation: The influence of ribosomes on bacterial mRNA decay. Genes & Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Geissmann T.A., Touati D., Touati D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Guillier M., Gottesman S., Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guisbert E., Rhodius V.A., Ahuja N., Witkin E., Gross C.A., Rhodius V.A., Ahuja N., Witkin E., Gross C.A., Ahuja N., Witkin E., Gross C.A., Witkin E., Gross C.A., Gross C.A. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters D.S., Traut R., Gold L., McPheeters D.S., Traut R., Gold L., Traut R., Gold L., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Altuvia S., Margalit H., Altuvia S., Margalit H., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F., Hardie M.M., Hardie M.M. Periplasmic protein associated with the oligopeptide permeases of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1983;155:1434–1438. doi: 10.1128/jb.155.3.1434-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie A.H., Poole P.S., Poole P.S. Bacterial ABC transporters of amino acids. Res. Microbiol. 2001;152:259–270. doi: 10.1016/s0923-2508(01)01197-4. [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C., Lina G., Etienne J., Ehresmann B., Ehresmann C., Etienne J., Ehresmann B., Ehresmann C., Ehresmann B., Ehresmann C., Ehresmann C., et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Noller H.F., Noller H.F. Footprinting mRNA–ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J., Rasmussen A.A., Overgaard M., Valentin-Hansen P., Rasmussen A.A., Overgaard M., Valentin-Hansen P., Overgaard M., Valentin-Hansen P., Valentin-Hansen P. Conserved small non-coding RNAs that belong to the σE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kawamoto H., Koide Y., Morita T., Aiba H., Koide Y., Morita T., Aiba H., Morita T., Aiba H., Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Lease R.A., Cusick M.E., Belfort M., Cusick M.E., Belfort M., Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R., Bujard H., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S., Cunning C., Sledjeski D., Elliott T., Gottesman S., Sledjeski D., Elliott T., Gottesman S., Elliott T., Gottesman S., Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N., Vanderpool C.K., Gottesman S., Vanderpool C.K., Gottesman S., Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Martin-Farmer J., Janssen G.R., Janssen G.R. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 1999;31:1025–1038. doi: 10.1046/j.1365-2958.1999.01228.x. [DOI] [PubMed] [Google Scholar]
- Massé E., Gottesman S., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Escorcia F.E., Gottesman S., Escorcia F.E., Gottesman S., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes & Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Vanderpool C.K., Gottesman S., Vanderpool C.K., Gottesman S., Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur S.D., Pulvermacher S.C., Stauffer G.V., Pulvermacher S.C., Stauffer G.V., Stauffer G.V. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 2006;6:52. doi: 10.1186/1471-2180-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Maki K., Aiba H., Maki K., Aiba H., Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J.C., Vogel J., Pfeiffer V., Mika F., Lucchini S., Hinton J.C., Vogel J., Mika F., Lucchini S., Hinton J.C., Vogel J., Lucchini S., Hinton J.C., Vogel J., Hinton J.C., Vogel J., Vogel J. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.A., Eriksen M., Gilany K., Udesen C., Franch T., Petersen C., Valentin-Hansen P., Eriksen M., Gilany K., Udesen C., Franch T., Petersen C., Valentin-Hansen P., Gilany K., Udesen C., Franch T., Petersen C., Valentin-Hansen P., Udesen C., Franch T., Petersen C., Valentin-Hansen P., Franch T., Petersen C., Valentin-Hansen P., Petersen C., Valentin-Hansen P., Valentin-Hansen P. Regulation of ompA mRNA stability: The role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R., Steffen P., Hochsmann M., Giegerich R., Hochsmann M., Giegerich R., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P., Vandenesch F., Wagner E.G., Vandenesch F., Wagner E.G., Wagner E.G. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Sittka A., Pfeiffer V., Tedin K., Vogel J., Pfeiffer V., Tedin K., Vogel J., Tedin K., Vogel J., Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Opdyke J.A., Zhang A., Opdyke J.A., Zhang A., Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Storz G., Altuvia S., Wassarman K.M., Altuvia S., Wassarman K.M., Wassarman K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Tjaden B., Goodwin S.S., Opdyke J.A., Guillier M., Fu D.X., Gottesman S., Storz G., Goodwin S.S., Opdyke J.A., Guillier M., Fu D.X., Gottesman S., Storz G., Opdyke J.A., Guillier M., Fu D.X., Gottesman S., Storz G., Guillier M., Fu D.X., Gottesman S., Storz G., Fu D.X., Gottesman S., Storz G., Gottesman S., Storz G., Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G., Vogel J., Reimegard J., Holmqvist E., Wagner E.G., Reimegard J., Holmqvist E., Wagner E.G., Holmqvist E., Wagner E.G., Wagner E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes & Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J.H., Vogel J., Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski M.L., Stauffer L.T., Stauffer G.V., Stauffer L.T., Stauffer G.V., Stauffer G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen M., Udesen C., Eriksen M., Udesen C., Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool C.K., Gottesman S., Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vecerek B., Moll I., Blasi U., Moll I., Blasi U., Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Papenfort K., Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Vogel J., Sharma C.S., Sharma C.S. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- Vogel J., Bartels V., Tang T.H., Churakov G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G., Bartels V., Tang T.H., Churakov G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G., Tang T.H., Churakov G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G., Churakov G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G., Hüttenhofer A., Wagner E.G., Wagner E.G. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Argaman L., Wagner E.G., Altuvia S., Argaman L., Wagner E.G., Altuvia S., Wagner E.G., Altuvia S., Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wagner E.G., Darfeuille F., Darfeuille F. Small regulatory RNAs in bacteria. In: Nellen W., Hamann C., Hamann C., editors. Small RNAs: Analysis and regulatory functions. Springer Verlag; Berlin Heidelberg, Germany: 2006. pp. 1–29. [Google Scholar]
- Wassarman K.M. Small RNAs in bacteria: Diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- Wassarman K.M., Storz G., Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S., Rosenow C., Tjaden B.C., Storz G., Gottesman S., Tjaden B.C., Storz G., Gottesman S., Storz G., Gottesman S., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang Z., Ling L., Shi B., Chen R., Zhang Z., Ling L., Shi B., Chen R., Ling L., Shi B., Chen R., Shi B., Chen R., Chen R. Conservation analysis of small RNA genes in Escherichia coli. Bioinformatics. 2004;20:599–603. doi: 10.1093/bioinformatics/btg457. [DOI] [PubMed] [Google Scholar]
- Ziolkowska K., Derreumaux P., Folichon M., Pellegrini O., Regnier P., Boni I.V., Hajnsdorf E., Derreumaux P., Folichon M., Pellegrini O., Regnier P., Boni I.V., Hajnsdorf E., Folichon M., Pellegrini O., Regnier P., Boni I.V., Hajnsdorf E., Pellegrini O., Regnier P., Boni I.V., Hajnsdorf E., Regnier P., Boni I.V., Hajnsdorf E., Boni I.V., Hajnsdorf E., Hajnsdorf E. Hfq variant with altered RNA binding functions. Nucleic Acids Res. 2006;34:709–720. doi: 10.1093/nar/gkj464. [DOI] [PMC free article] [PubMed] [Google Scholar]