Abstract
The chicken genome draft sequence has provided a valuable resource for studies of an important agricultural and experimental model species and an important data set for comparative analysis. However, some of the most gene-rich segments are missing from chicken genome draft assemblies, limiting the analysis of a substantial number of genes and preventing a closer look at regions that are especially prone to syntenic rearrangements. To facilitate the functional and evolutionary analysis of one especially gene-rich, rearrangement-prone genomic region, we analyzed sequence from BAC clones spanning chicken microchromosome GGA28; as a complement we also analyzed a gene-sparse, stable region from GGA11. In these two regions we documented the conservation and lineage-specific gain and loss of protein-coding genes and precisely mapped the locations of 31 major human-chicken syntenic breakpoints. Altogether, we identified 72 lineage-specific genes, many of which are found at or near syntenic breaks, implicating evolutionary breakpoint regions as major sites of genetic innovation and change. Twenty-two of the 31 breakpoint regions have been reused repeatedly as rearrangement breakpoints in vertebrate evolution. Compared with stable GC-matched regions, GGA28 is highly enriched in CpG islands, as are break-prone intervals identified elsewhere in the chicken genome; evolutionary breakpoints are further enriched in GC content and CpG islands, highlighting a potential role for these features in genome instability. These data support the hypothesis that chromosome rearrangements have not occurred randomly over the course of vertebrate evolution but are focused preferentially within “fragile” regions with unusual DNA sequence characteristics.
The draft sequence of the genome of the red jungle fowl, Gallus gallus, provided a first look into the biology of a species that is both a significant agricultural animal and an important developmental model. Chicken sequence alignments also provide a useful tool for human genome annotation, more highly enriched than human–fish comparisons in genes and regulatory elements in rapidly diverging regions, while still providing a stringent filter for detection of evolutionarily conserved DNA regions in slowly evolving genomic intervals (International Chicken Genome Sequencing Consortium 2004). Many striking features of genome architecture and organization are also shared between mammals and birds (Ovcharenko et al. 2005). In particular, relative gene density and other properties, including GC content and recombination rates, are very similar in most homologous segments of the human and chicken genomes (International Chicken Genome Sequencing Consortium 2004). Intriguingly, the majority of human and chicken “gene deserts” have been preserved as intact segments, whereas gene-rich chromosomal regions have undergone repeated rearrangements over evolutionary time (Ovcharenko et al. 2005). Gene-rich segments are also enriched in evolutionary rearrangements that distinguish chromosome structure in different mammals, including many breakpoint sites that have been repeatedly involved in independent genomic rearrangement events. The genomic clustering and repeated reuse of breakpoint sites have been used to argue in favor of a “fragile breakage” model of chromosome evolution over models presuming random distribution of chromosome breaks (Pevzner and Tesler 2003; Bourque et al. 2004; Murphy et al. 2004, 2005). Presumably, the fragility of these putative rearrangement hotspots is related, at least indirectly, to underlying features of DNA structure.
Evidence for the fragile breakage model has been taken mostly from comparisons between mammalian lineages, examining large intervals surrounding reused breaks. To understand the molecular basis of the relative fragility of these genomic regions, it would be useful to examine the rearrangement-prone regions across a deeper evolutionary spectrum and to pinpoint their locations more precisely using solidly assembled mammalian and nonmammalian genomes as anchor points. The chicken genome sequence provides an excellent candidate for this comparison. However, chicken whole genome shotgun (wgs) assembly was particularly challenging in the most gene-rich segments, including chicken DNA segments related to most of humanchromosome 19 (HSA19). HSA19-related sequences are absent or heavily underrepresented in the chicken wgs draft, with 21% of the genes estimated to be missing from the chicken assembly corresponding to HSA19 orthologs (International Chicken Genome Sequencing Consortium 2004).
To provide a solid anchor for genome evolution studies and improved resources for annotation and genetic analysis, we analyzed sequence from deeply overlapping BAC clones spanning the length of a chicken microchromosome, GGA28. GGA28 is evolutionarily related to HSA19p, one of the human genome’s most gene-rich territories (Grimwood et al. 2004), and this region has undergone multiple intrachromosomal rearrangement events during its evolution. As a counterpoint, we also sequenced BACs spanning a segment of GGA11 with homology to a gene-sparse HSA19q region including the flanking syntenic breaks. In both regions, we identified both conserved and novel human and chicken genes, documenting clear instances of gene gain and loss in both the avian and mammalian lineages. We also mapped 31 major human–chicken syntenic breakpoints, including 22 regions that have been used repeatedly in chromosome breakage events throughout vertebrate evolution.
Results
Generating BAC-based chicken sequence
Contiguous BAC maps spanning GGA28- and HSA19q-related regions of GGA11
We isolated overlapping BAC clones from libraries derived from DNA of red jungle fowl (JF), G. gallus gallus, and the White Leghorn (WL) domestic strain, G. gallus domesticus, using probes designed from conserved HSA19 genes (see Methods). We focused on generating complete JF BAC contigs but also generated WL contigs for most regions. The BACs were selected to provide maximum coverage and sequenced to >10× depth. Automated JF sequence assembly was curated manually and checked against parallel sets of assembled WL BACs, providing an independent check of sequence accuracy and completeness. Accession numbers for sequence of individual clones contributing to the final assemblies are provided in Supplemental Table S1.
In the resulting assembly, GGA28 is represented by two nonredundant scaffolds, Jf_g1 and Jf_g2, together spanning 4.7 Mb (1.1 and 3.6 Mb, respectively). The GGA28 sequence is homologous to 10.6 Mb of HSA19p, with a short HSA16p-related region located at the telomeric end (Fig. 1). Another 3.7-Mb scaffold, Jf_g3, spans the entire GGA11/HSA19 homology region and flanking regions related to 5.9 Mb of HSA19q and 0.8 Mb of HSA16q, respectively. All three scaffolds are presented with other data derived from this study on a University of California, Santa Cruz (UCSC)-style browser interface and available for download at our website (http://genome.llnl.gov/chicken.html). The order and spacing of anchoring genetic markers for GGA28 correlates well with physical location in the BAC-based contigs (Supplemental Table S2), providing additional support for the assemblies.
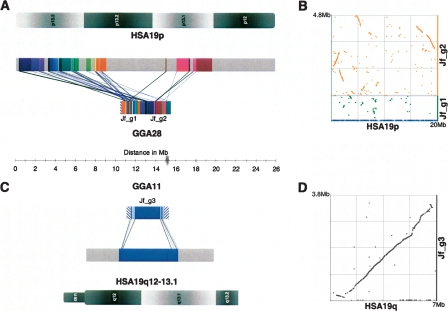
Figure 1.
Two regions of human chromosome 19 (HSA19) homologous to chicken chromosomes 28 and 11 (GGA28 and GGA11). (A) Gene-rich HSA19p13.3-p13.2 and p13.1 is evolutionarily related to GGA28. Each of 30 major human-chicken homology segments is uniquely color-coded to demonstrate intrachromosomal rearrangements between these two lineages. Homology segment starts connected with black lines are oriented in the forward direction, segment starts connected with blue lines are oriented in the reverse direction. HSA19 segments shaded gray are not homologous to GGA28 and were not sequenced; most regions are also not found in wgs assemblies. Internal rearrangements and inversions are not depicted. The GGA28 telomere contains a short region of HSA16p11-related material (hatched). (B) Dot plot of GGA28 vs. HSA19p. (C) Gene-sparse HSA19q12–13.1, including a 2.5-Mb gene desert in q12, is recapitulated on GGA11 in a single homology segment with only localized inversions (light blue) at the ends of the segment. Only the portion of GGA11 sequenced here is depicted; the region related to HSA19q is flanked by HSA16q12 and q22-related material (hatched). (D) Dot plot of GGA11 vs. HSA19q.
Comparing BAC-based and wgs sequence
To link our assemblies to public data sets, we aligned them with the galGal3 chicken wgs sequence (International Chicken Genome Sequencing Consortium 2004; updated assembly released May 2006); BAC-based and wgs assemblies are concordantly arranged (Supplemental Fig. S1; Supplemental Table S3). However, 560 kb of GGA28 sequence assembled securely within Jf_g1 or Jf_g2 is not integrated into the galGal3 assembly. This includes 94 kb assigned to chr28_random, 63 kb assigned to chrUn_ random in multiple fragments, and 403 kb that is not represented in the wgs draft. The galGal3 sequence, on the other hand, includes 56 kb not found in our assemblies and extends BAC contig ends (Supplemental Table S3; Supplemental Fig. S1A). We incorporated loci from these wgs contigs into our GGA28 gene catalog and include them in analyses reported below. GGA11 sequences in the galGal3 wgs and BAC-based assemblies are essentially colinear with the exception of short regions of duplicated sequence in the wgs assembly that are not found in Jf_g3 (Supplemental Fig. S1B).
Conservation, gain, and loss of human and chicken genes
We aligned Jf_g1, Jf_g2, and Jf_g3 contigs with related human regions to identify human–chicken (H/C) evolutionarily conserved regions (ECRs) and used BLAST to map human protein homologies to the chicken sequence. We then generated GeneWise (Birney et al. 2004) gene models around chicken ECR and protein-based anchors and manually curated the models (see Methods). We correlated our annotations with Ensembl chicken gene models and, where appropriate, incorporated those models to provide better gene coverage (Supplemental Tables S1, S4). The resulting chicken gene models, predicted protein sequences, and supporting evidence are displayed within the GGA11 and GGA28 assemblies on our website (http://genome.llnl.gov/chicken.html). Altogether we identified 285 genes in the 4.7-Mb GGA28 sequence and 64 genes in 3.7 Mb of GGA11 DNA (Table 1). These include 72 gene loci newly integrated into GGA28 from regions that were missing or unassigned in the chicken draft assembly. We also identified or improved another 72 GGA28 models and 17 GGA11 models for which sequence was at least partially represented in the wgs draft (Supplemental Table S4).
Table 1.
Gene loci in GGA28, GGA11, and homologous human regions
aSee Supplemental Methods and Supplemental Table S1 for definitions of orthologs, paralogs, and novel genes, and for criteria used to determine rearrangement sites inside major synteny blocks. One GGA11 locus homologous to both HSA19 and HSA16 is tallied for both chromosomes but only once in totals.
bTotal gene counts for HSA19 and HSA16 homology regions equivalent to GGA28 and GGA11 include all known protein-coding RefSeq genes from those regions, plus any hypothetical loci validated by the presence of a chicken ortholog; see Supplemental Table S5 for details.
cExcludes 4 human loci “missing” from chicken that fall near scaffold gaps in partially ordered and oriented sequence.
Of 285 annotated GGA28 genes, 257 were classified as HSA19 orthologs (Table 1). These include chicken orthologs for 250 HSA19 RefSeq loci (Pruitt et al. 2005; http://www.ncbi.nlm.nih.gov/RefSeq) and 7 chicken loci that correspond to, and provide additional validation for, HSA19 ab initio models (Table 1; Supplemental Tables S5, S6). Orthologs for 53 RefSeq loci and 5 hypothetical loci from related HSA19 or HSA16 regions were detected in the GGA11 sequence. However, 39 HSA19 and HSA16 loci were not detected in corresponding GGA28 and GGA11 regions (Table 1; Supplemental Table S7). Reciprocal best-match chicken orthologs for two HSA19 genes, RPL18A and TSSK6, were found in non-HSA19 syntenic contexts but clear orthologs for the remaining genes were not detected in the chicken genome. Many human genes not represented in the chicken sequence are members of gene families that have expanded in mammals (e.g., zinc finger and olfactory receptor genes) and 8 of these genes are not conserved, or not conserved as 1:1 orthologs, even in mouse.
Reciprocally, 33 GGA28 and GGA11 loci correspond to putative chicken genes for which no obvious HSA19 or HSA16 counterpart could be ascertained. Two of these chicken genes, ZAP70 and SS18L2, represent best reciprocal match orthologs for RefSeq genes located in other human genomic contexts. Another 26 chicken loci correspond to lineage-specific paralogs; for 18 of these, we found evidence of orthologous loci in pufferfish, frog, and/or opossum genomes in GGA28 or GGA11 syntenic context, indicating ancient genes lost in the ancestors of present-day placental mammals. Five gene models demonstrate no homology with known human proteins (Supplemental Methods, Supplemental Tables S1, S7).
Syntenic conservation and location of evolutionary breakpoint sites
We compared GGA28 and GGA11 genes and noncoding ECRs to those in related human regions to define homologous synteny blocks (HSBs) and the locations of evolutionary breakpoint regions (EBRs). With the exception of localized rearrangements at the boundaries between HSA19- and HSA16-related DNA (Fig. 1C,D), the order and orientation of both genes and noncoding ECRs in the GGA11/HSA19q homology region is rigidly preserved. In striking contrast, GGA28 and human sequence alignments revealed 31 “major” HSBs (defined as contiguous blocks of conserved genes interrupted only by localized microrearrangements). Thirty GGA28 HSBs carry homology with HSA19p; a single HSB with homology to HSA16p was also detected (Fig. 1A,B).
Of note, more than half of the informative 29 EBRs that define the 31 H/C HSBs in GGA28 are flanked by lineage-specific genes: 13 of the 27 GGA28 loci and 14 of the 36 HSA19p loci that do not detect orthologs in the same syntenic context are positioned at or near 17 H/C syntenic breakpoints (Supplemental Tables S1, S7). Intrigued by this association, we searched regions inside the major homology blocks that surround HSA19 genes that are missing in the chicken sequence, or chicken genes missing from HSA19, for evidence of association with evolutionarily unstable sites. At least 12 of 30 internal sites with local synteny changes in GGA28 showed evidence of gene transposition, inversions, ortholog insertions or deletions, and/or breakpoint reuse in other species (Supplemental Table S8; Supplemental Methods; see below). In total, at least 41 evolutionary rearrangement sites define the differences in gene order and orientation between GGA28 and homologous regions of the human genome (Supplemental Tables S1, S8).
Human–mouse synteny breaks provide evidence for breakpoint reuse
To trace the evolutionary histories of the H/C syntenic rearrangements we analyzed related synteny groups in fish (Fugu), amphibian (frog), nonplacental mammals (opossum), and two additional mammalian genomes (dog and mouse). We focused first on regions that have undergone rearrangements in more recent evolutionary time by examining human–mouse (H/M) EBRs. Most HSA19p H/M rearrangements correspond to interchromosomal translocation events that occurred specifically in the rodent lineage (Dehal et al. 2001; Kim et al. 2001). GGA28 sequence is informative for 9 HSBs flanking one or both sides of 6 H/M major synteny breaks (Supplemental Fig. S2; Supplemental Table S9). All but one of the H/M sites also correspond to H/C synteny breakpoints, and mouse/chicken (M/C) synteny is broken at all of these same sites. However, homology segments that flank the shared H/M, M/C, and H/C EBRs are different in chicken, human, and mouse. The one H/M rearrangement for which H/C synteny nominally appears unbroken (MGC33407-MUC16) is nonetheless structured very differently in human and chicken and is highly prone to rearrangement (discussed below). Furthermore, 8 of 15 informative segment ends flanking 9 H/M inversion breakpoints inside major homology segments bear similar evidence of reuse in chicken, including a site where chicken gene order mimics that of the mouse but the presence of variable, species-specific loci associated with the EBR suggest independent rearrangement events. These data indicate that H/C and H/M rearrangements occurred independently at these sites, the first occurring in an ancestral genome after the divergence of mammals and birds and the second in the rodent lineage.
HSA19- and GGA28-related chromosome rearrangements in six species
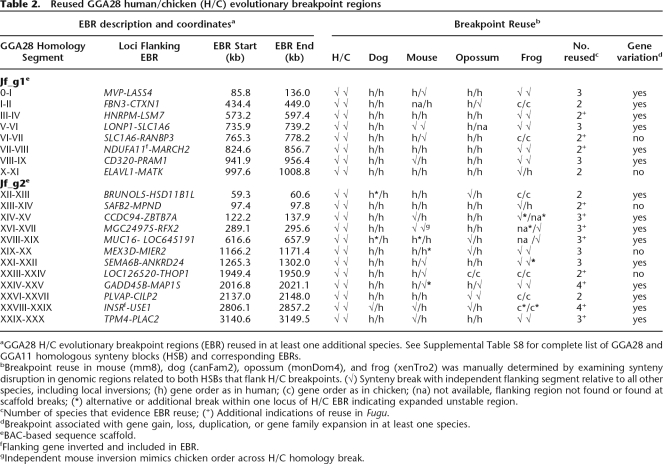
Using HSA19 sequence and BAC-based chicken assemblies as anchor points, we examined gene order and synteny breaks in GGA28-related regions of mouse, dog, opossum, and frog draft assemblies (Supplemental Fig. S2; Supplemental Table S8). Fugu was also examined where genes of interest were found within high-quality sequence scaffolds (Supplemental Table S10). Twenty of 29 informative GGA28 EBRs show evidence of reuse in at least one other species, as do 7 sites internal to the major H/C synteny blocks (Table 2; Supplemental Table S8). Since each EBR is flanked by two genomic regions with independent fates, every break was tallied in both directions.
Table 2.
Reused GGA28 human/chicken (H/C) evolutionary breakpoint regions
aGGA28 H/C evolutionary breakpoint regions (EBR) reused in at least one additional species. See Supplemental Table S8 for complete list of GGA28 and GGA11 homologous synteny blocks (HSB) and corresponding EBRs.
bBreakpoint reuse in mouse (mm8), dog (canFam2), opossum (monDom4), and frog (xenTro2) was manually determined by examining synteny disruption in genomic regions related to both HSBs that flank H/C breakpoints. (√) Synteny break with independent flanking segment relative to all other species, including local inversions; (h) gene order as in human; (c) gene order as in chicken; (na) not available, flanking region not found or found at scaffold breaks; (*) alternative or additional break within one locus of H/C EBR indicating expanded unstable region.
cNumber of species that evidence EBR reuse; (+) Additional indications of reuse in Fugu.
dBreakpoint associated with gene gain, loss, duplication, or gene family expansion in at least one species.
eBAC-based sequence scaffold.
fFlanking gene inverted and included in EBR.
gIndependent mouse inversion mimics chicken order across H/C homology break.
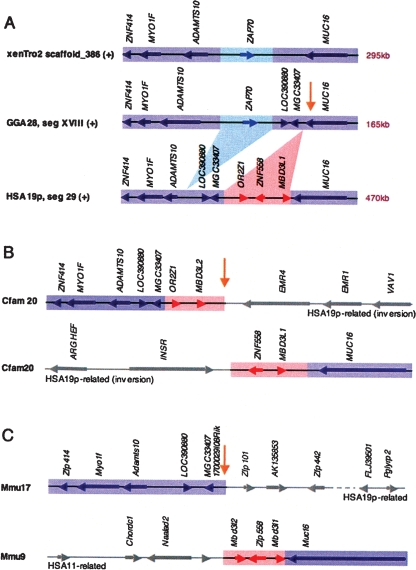
As an example, the presence of ZAP70 between HSA19p-related loci, ADAMTS10 and MUC16, in chicken and frog, and the absence of related sequences in HSA19 but presence of a unique ZAP70 ortholog in HSA2, suggests an ancient rearrangement event at that site (Fig. 2). However, the apparent transposition of ZAP70 is only one of a series of gene gain, gene loss, and rearrangement events that have occurred in the larger ADAMTS10-MUC16 region over evolutionary time. One endpoint of the single inversion that distinguishes GGA28-related human and dog regions is located here, and sequences surrounding both endpoints of this human/dog (H/D) inversion have been used in rodent-specific translocations (Figs. 2, 3). In human, distal and proximal ends of the inverted interval contain paralogous genes, ZNF557 and ZNF558, respectively; ZNF557 is found only in primate genomes (Huntley et al. 2006) and displays 90.7% nucleotide sequence identity with ZNF558 gene, indicating a recent primate duplication. ZNF557 lies adjacent to a series of tandem MBD3L2 duplicates that are >97% identical over the locus lengths, suggesting even more recent primate duplications at this site (Fig. 3).
Figure 2.
Evolutionary rearrangement hotspot inside human/chicken (H/C) homology segment related to other rearrangement hotspots and H/C breakpoint reuse sites. (A) Human, chicken, and frog genomic segments between ZNF414 and MUC16 share overall gene order and orientation (purple). Respective genomic sizes are indicated and locus boundaries are accurately scaled. Gene order is interrupted at two closely spaced internal sites. First, ZAP70 (light blue) is found adjacent to ADAMTS10 in the amphibian, avian, and marsupial lineages (opossum not shown), but is in an alternate HSA2-related context in placental mammals (not shown). Second, synteny between MGC33407 and MUC16 is locally disrupted by lineage-specific gene expansions of OR2Z1, ZNF558, and MBD3L1 in human (red); it is also associated with a known duplication site (Bailey et al. 2001). (B,C). The mammalian expansion site between MGC33407 and MUC16 is reused in all other mammalian species examined. Two chromosomal locations are presented for dog (B) and mouse (C) depicting the alternate synteny contexts for each of the two homology segments flanking the reused breakpoint (yellow arrow). The human and dog rearrangement sharing the evolutionary breakpoint region near MUC16 is part of an inversion related in turn to another heavily reused evolutionary breakpoint region at INSR (see Fig. 3). The expansion associated with the opposite end of the dog inversion contains OR2Z1 and a MBD3L1 duplicate, followed by mucin-domain containing, mammalian-specific duplicates, EMR1 and EMR4. In human, the species-specific expansion at this site includes a second zinc finger ZNF557 and an expansion of MBD3L-like gene loci (Fig. 3). In mouse, the respective genomic regions flanking the breakpoint are again differentially split and rejoined, and one flank contains a different zinc finger expansion. In related H/M rearrangements mouse Emr1 and Emr4 orthologs are also split, one flanking each end of H/M segment II; one end is associated with yet another H/C reuse break while the other flank, H/M segment III, becomes a centromere proximal to mouse Insr (Supplemental Fig. S2, Supplemental Table S9).
Figure 3.
Chromosome rearrangement map for 2 MB of rearrangement prone GGA28-related regions of HSA19p. Using the finished HSA19 sequence (hg18) and the high-quality, BAC-based draft sequence for GGA28 as a base, homology breaks and synteny changes relative to HSA19p are detailed for five vertebrate species. Genes and gene order for mouse, dog, opossum, and frog were manually inspected in related regions using wgs draft assemblies mm8, canFam2, monDom4, and xenTro2 (UCSC Genome Browser; http://genome.ucsc.edu/). Relative order, orientation, and spacing of HSA19 gene loci are represented with a series of arrows; blue arrows indicate a chicken ortholog, gray arrows indicate HSA19 loci that were not found in any GGA28 assembly. The relative location of chicken-specific loci are noted with carats (^). (P) Paralog; (O) non-HSA19 ortholog; (D) duplication. Homology breaks for chicken (orange), frog (green), opossum (purple), dog (brown), and mouse (pink) are indicated by paired vertical lines; each line delineates the end of a genomic region that was found to be flanked by a different homology segment in the other species, two per breakpoint. Where two species share an alternate syntenic configuration relative to human, a single color is used. Thus, breakpoints marked with different colors indicate flanking segments that have been differentially joined, i.e., “reused” in different rearrangement events. Reused breakpoints are marked with a yellow arrow (light outline, 2 species; heavy outline, ≥3 species). Inversion breaks within homology segments are dotted and paired with arrows to track related breakpoints. Scaffold breaks in draft wgs assemblies are presented in gray; at these sites it could not be definitively determined whether the flanking segment is or is not changed relative to human. Differentially expanded gene family regions are marked with color-coded hatched lines. Missing genes and regions are marked with gray cross-hatched lines. Human/chicken (H/C) and human/mouse (H/M) HSBs are presented beneath. See Supplemental Figure S2 for a comprehensive map of all GGA28-related regions of HSA19p and full pictorial legend of annotation marks.
Regions surrounding both ends of the H/D inversion have served as breakpoints for independent rearrangements in every vertebrate genome examined (Figs. 2B, 3). Although the breakpoints are clustered, rearrangement sites in different lineages are not identical and in each case, breakpoint-flanking sequences are fused to different chromosomal sites (Supplemental Table S8). Together these observations indicate that the larger region is prone to rearrangement, and that the rearrangements correspond to lineage-specific events. As evidenced by the recent primate-specific duplications, sequence changes and gene evolution appear to be continuing at these reused breakpoint sites.
Gene duplications and inversions at GGA11 homology breaks
Although the GGA11/HSA19q homology segment is highly conserved, localized inversions and lineage-specific genes and gene duplications flank the boundaries of this HSB, and the breakpoints share features with GGA28 EBRs. For example, orthologs of neighboring GGA11 genes, SHCBP1 and UQCRFS1, are found near the centromeres of HSA16q and HSA19q, respectively. A diverged copy of human SHCBP1, defined by locus BC068609, lies adjacent to human UQCRFS1 in inverted orientation relative to the related chicken gene (Fig. 4A). The arrangement of these paralogous loci suggests that gene duplication and possibly an inversion event occurred prior to, or concomitant with, the syntenic rearrangement at this site. Although SHCBP1 and UQCRFS1 are found together in similar contexts in chicken and opossum, surrounding regions have undergone repeated rearrangements resulting in very different configurations in placental mammalian lineages (Fig. 4B).
Figure 4.
Evolutionary rearrangements distinguishing GGA11 and related regions in human and mouse. (A) Comparative organization of genes in GGA11 and homologous human regions. Complex series of gene duplication and inversion events mark the boundaries of the conserved HSA19q/GGA11 homology segment. Block arrows show orientation of individual genes: HSA16-related (light and dark blue); HSA19-related (light and dark red); unique chicken genes (green); HSA3 ortholog (yellow). Solid bars indicate long syntenic blocks with only anchor loci named. Duplicated genes associated with the evolutionary breakpoints between HSA16- and HSA19-related regions are hatched, e.g., a diverged HSA19 locus related to HSA16 locus SHCBP1 (light blue, hatched), and CES family members (dark blue, hatched). Gray-shaded areas link homologous regions found in inverted order between species; dashed lines denote regions of variable length between segments. Maps not drawn to scale. (B) Breakpoint reuse highlighted by alternate arrangements in the mouse genome. While many HSA19- and HSA16-related genes are similarly ordered in chicken, human, and mouse genomes (Mmu7, Mmu8, respectively), Shcbp1 and Uqcrfs1 have been inserted into alternate contexts in the rodent genome. HSA16q/GGA11 locus Shcbp1 has fused to another site of repeated breakpoint reuse, HSA19p/GGA28 ortholog Lass4 (lilac, Mmu8); in chicken LASS4 is flanked instead by the HSA16p11 locus MVP at the telomere of GGA28 (Table 2, Supplemental Fig. S2). Uqcrfs1 (dark red, Mmu13) is found between HSA6-related H/M HSBs (light and dark brown). The rodent CES gene family (brackets) has expanded dramatically to include at least 15 annotated loci.
On the distal end of the H/C homology segment, a block of three HSA19 genes (WTIP-PDCD2L) is inverted relative to GGA11 counterparts; orthologs of adjacent chicken genes are also found in inverted order in HSA16 (Fig. 4). Between the two inverted segments in chicken lies a family of four carboxylesterase (CES) genes (Supplemental Table S1). Protein alignments clearly show that these four chicken genes, and seven HSA16 CES loci, are ancestrally related (Supplemental Fig. S3). The physical arrangement of evolutionarily related proteins in this family indicates that some ancestors of the human CES paralogs were generated by duplication before the cluster was split by further rearrangement events (Fig. 4A). Expanded CES gene clusters in the same syntenic locations in mouse (Fig. 4B) indicate that segmental duplications have continued within these clusters in mammalian lineages.
Sequence characteristics of rearrangement-prone genomic intervals
To identify properties potentially associated with evolutionary rearrangements, we examined rearrangement-prone regions and specific EBR sites for a series of sequence characteristics, including GC content and density of CpG islands, single-nucleotide polymorphisms (SNPs), dispersed repeats (LTRs and LINEs), simle sequence repeats (SSRs), and duplications. Data were gathered in 10-kb windows for each region, appropriate control groups were selected to account for GC bias, and pairwise comparisons were performed using Welch’s two-sample t-tests as follows.
Analysis of rearrangement-prone versus stable genomic regions
Like most chicken microchromosomes, GGA28 is exceptionally GC rich (Table 3; Supplemental Fig. S4), and since many other sequence features are linked to GC content (International Chicken Genome Sequencing Consortium 2004) we selected a “high-GC control” (HGC) group from the wgs assembly for GGA28 comparisons. The HGC group comprises 15 relatively stable chicken genomic regions with similar GC content and length to GGA28 contigs, but with few evolutionary rearrangements in mammalian comparisons (Supplemental Table S11). To provide a genome-wide context, we also identified 14 regions from elsewhere in the chicken genome with high densities of evolutionary rearrangements including multiple reused breaks (genome-wide rearranged, or GWR regions). We compared the GWR regions to 16 relatively stable 1-Mb chicken genomic regions with GC content within ±2% of the GWR average (genome-wide control, or GWC regions). Finally, we identified a “super-stable control” (SSC) group, consisting of 30 large regions with no or very few evolutionary rearrangements in comparisons with mammals and even with frog; as a whole, this group exhibits GC content slightly lower than genome average (Table 3).
Table 3.
Sequence features of stable and rearranged chicken genomic regions
aShaded boxes contain P values calculated from Welch’s two-sample t-test. Text in italics indicates negative trends.
bGC, CpG, LINE, and SSR densities for GGA28 were calculated with BAC-based assemblies; duplication rate was calculated using wgs assembly galGal3. All other feature values were calculated using galGal3. See Supplemental Tables S11 and S12 for control region and reused EBR details.
cOnly refGene was used to calculate gene density. (NA) Not applicable, see Methods.
dNumber of nucleotide matches in a self-chain alignment calculated in 10-kb windows.
eGGA28 and GWR regions are presented in both tables for ease of comparison.
Comparisons between these GC-matched groups highlighted CpG islands as a feature most strongly associated with the break-prone regions (Table 3). GGA28 CpG island density is significantly higher than in the evolutionarily stable HGC regions, even though the regions share similar overall GC content. Likewise, compared with GWC regions, CpG-island density is significantly higher in the highly rearranged GWR intervals (Table 3). Since high gene density has been linked to rearranged regions in previous studies (Murphy et al. 2005; Ovcharenko et al. 2005), we also examined RefSeq gene density in GGA28, GWR, and their control groups. Not surprisingly, GC-rich GGA28 and HGC control regions both have much higher gene densities than the chicken genome-wide average and SSC group; no difference in gene density was observed between the two types of GC-rich regions. Likewise, GWR and GWC regions are elevated but do not differ in gene content. These data suggest that CpG island density, rather than gene density per se, is significantly associated with rearranged sites.
Compared with GC-matched stable controls, rearranged GGA28 and GWR regions also contain significantly higher numbers of LINEs. However, LINE density in rearranged regions is close to genome-wide GC-matched average; instead, the difference can be attributed to the fact that all groups of “stable control” regions we selected are significantly depleted in LINEs (Supplemental Fig. S5). A similar but less dramatic trend was observed for duplications (Table 3). No significant difference was observed between SSR, SNP, or LTR density in the chicken rearranged regions and corresponding controls (not shown).
Analysis of reused breakpoint sites
Focusing more specifically on breakpoint sites, we first examined sequence content in GGA28 reused EBRs. Compared with GGA28 as a whole, the reused EBRs have higher average local GC content, CpG island density, and LINE density but no indication of increased SSR or duplication frequency (Table 3). However, since the number of GGA28 EBRs is small (n = 20), these differences were not all statistically significant. To provide a genome-wide perspective and a more solid statistical basis, we also analyzed features of 101 additional reused EBRs identified from the GWR regions (Supplemental Table S12). In this larger group, GC and CpG-island density again stood out for their significant enrichment, even compared with the GC-rich genomic surroundings (Table 3). Indeed, many reused EBRs are contained within or closely flanked by chains of closely packed CpG islands (Supplemental Table S12).
Neither SSR nor duplication densities were detected to be significantly elevated relative to surrounding GWR intervals. LINE density is slightly increased in EBRs relative to GWR regions (Table 3) and in both GGA28 (P = 0.03) and genome-wide EBRs (P = 0.024) when compared with GC-matched genome average (Supplemental Fig. S5). In addition, all three features are highly enriched in some EBRs (Supplemental Table S12). Since many of the reused EBRs we analyzed are located on chicken macrochromosomes, these data confirm genome-wide rather than GGA28- or microchromosome-specific trends.
Discussion
The GGA28 and GGA11 sequence assemblies described here, together with the associated annotation of genes, polymorphic markers, orthology relationships, and other features, provide a new resource for the avian genetics and genomics research communities. In particular, GGA28 variation has been associated with important agricultural traits including viral susceptibility (Elleder et al. 2004), fat distribution (Ikeobi et al. 2002), growth characteristics (Deeb and Lamont 2003), and pulmonary hypertension syndrome (Rabie et al. 2005); the assignment of genes and SNP markers to solidly anchored physical locations in our chicken sequence should facilitate molecular and genetic analysis of these and other avian phenotypes.
These assemblies also provide a solid nonmammalian anchor point for analysis of a gene-rich genomic region with an especially dynamic evolutionary history. More than two thirds of the GGA28 H/C EBRs display clear evidence of evolutionary reuse, a frequency that is significantly higher than the 20% breakpoint reuse rate estimated in mammalian genome-wide comparisons (Murphy et al. 2005). The difference may in part reflect our focus on the extremely break-prone GGA28, but the inclusion of nonmammalian vertebrate species in these comparisons also raised reuse counts significantly. Taken together, these data provide strong support for the fragile breakage model of chromosome evolution (Pevzner and Tesler 2003), confirming that selected genomic regions with unusual features have been reused repeatedly over the course of vertebrate evolution.
We found enrichment for several sequences features in GGA28 and other rearrangement-prone regions relative to stable intervals in the chicken genome. Specifically, LINE elements and duplications, both known to play roles in genome instability (Deininger and Batzer 2002; Kazazian and Goodier 2002; Lupski and Stankiewicz 2005) are significantly enriched in rearranged regions compared with GC-matched stable controls. However, these differences are due to the relative depletion of duplications and LINEs in the stable genomic regions rather than special enrichment at evolutionary breakpoints or in surrounding DNA. This observation stands in contrast with results from mammalian comparisons, which detected significant overall enrichment of duplicated sequences in regions surrounding reused EBRs (Murphy et al. 2005). However, as illustrated by the CES gene family and other examples highlighted here, when EBR-linked duplications were found, they were typically more pronounced in mammalian regions compared with homologous chicken DNA. The more general expansion of segmental duplications in mammals relative to chickens (International Chicken Genome Sequencing Consortium 2004) might therefore be linked to a more striking enrichment of duplications at mammalian EBRs.
On the other hand, the common trend of depletion of LINEs and duplications in evolutionarily stable regions is notable and remarkable considering the wide range of properties that characterize the three different stable groups we selected for study. As far as we can discern, the HGC, GWC, and SSC regions we selected share nothing else in common aside from low levels of evolutionary breaks. The HGC and GWC regions contain high gene densities; regions of this type may be particularly sensitive to LINE insertions and duplications since these changes can alter copy number or disrupt structure and expression of essential genes. However, gene-rich rearranged regions are not LINE-poor and the “super stable” regions, which are significantly depleted in both duplications and LINEs, have lower-than-average gene densities. These data suggest a biological difference in the stable regions that transcends gene density and other obvious features and that renders these genomic intervals especially intolerant to insertions and duplications, as well as rearrangements of other types.
Chicken genomic intervals hosting clusters of repeated rearrangement are indeed particularly gene rich, consistent with previous reports (Murphy et al. 2005; Ovcharenko et al. 2005). However, data presented here point to a correlation between rearrangements and high localized GC-content and CpG island density rather than gene density per se. CpG islands are enriched in the larger break-prone regions compared with GC-matched controls, and both GC content and CpG content are further enhanced at reused EBR sites. These features are the only ones we tested that are consistently and specifically concentrated at reused breakpoints and surrounding break-prone regions, pointing to a special role in fragility.
Although CpG-rich sequences have been linked to high substitution rates and recombination frequencies (Webster et al. 2006), a simple molecular explanation for this correlation is not obvious. However, CG repeats, found at high frequency in CpG islands, can form non-B DNA secondary structures that are associated with elevated rates of rearrangements in mammalian cells (Wang et al. 2006). Through their propensity to accumulate double-strand breaks, the hairpin loops, Z-DNA, and other structures generated in CG-rich regions could potentially play key roles in the fragility of reused EBRs. Active transcription increases the mutability of CG repeats (Wang et al. 2006), and CpG islands are typically associated with the promoters of highly and widely expressed genes (for review, see Antequera 2003). As a result, a large fraction of islands reside in domains of open chromatin (Roh et al. 2005; Heintzman et al. 2007). Notably in this regard, HSA19p corresponds to one of the most highly transcribed and hyper-acetylated human chromosomal domains (Caron et al. 2001; Roh et al. 2005), and it is tempting to speculate that high levels of transcriptional activity render these CG-rich regions especially susceptible to DNA rearrangement events.
That regions packed with active genes should be excessively prone to rearrangement seems counterintuitive; if such breaks were randomly distributed they would frequently disrupt functional genes. However, with rearrangements biased near CpG islands, many of which extend upstream of genes for considerable distances, the chances of coding-sequence disruption might be somewhat diminished. On the other hand, given the association between CpG islands and promoters as well as other types of regulatory sequences (for review, see Klose and Bird 2006), breaks in these regions could alter expression patterns and functions for flanking genes. In any case, rearrangements focused repeatedly in the genome’s most gene-dense regions can be expected to serve as significant engines of functional change.
Indeed, data presented here clearly implicate EBRs as major sites of genetic innovation, revealing significant levels of gene duplication, transposition, and loss. The “lost” genes, present in chickens and other vertebrates but missing from mammalian genomes, may have been disrupted by chromosome breakage or rendered nonfunctional by nearby rearrangement events. Alternatively, these genes may have simply been excised concomitant to, or independently of, larger rearrangement events at these unstable sites. Whatever the mechanism, duplication, loss, or disruption of specific genes has also been reported at other mammalian EBRs (Lund et al. 2000; Fitzgerald and Bateman 2004; Fortna et al. 2004), and data presented here suggest the association is both common and global.
No simple combination of factors can yet account conclusively for the genetic instability of all reused breakpoint sites. However these data point to common features, the mechanistic relevance of which should be highlighted more clearly as additional genomes are added to evolutionary comparisons. Since human reused EBRs overlap considerably with fragile sites and rearrangement breakpoints associated with human genetic disease (Ruiz-Herrera et al. 2006), a deeper understanding of the mechanisms involved promises broad scientific and practical benefits.
Methods
BAC mapping
Overgo probes were designed from chicken cDNA sequences BLAST-identified from protein-translated HSA19 genes verified as best-in-genome reciprocal matches. To screen three libraries, one from G. gallus domesticus (5.5× Wageningen chicken BAC library, Crooijmans et al. 2000) and two from G. gallus gallus (4.8× Texas A&M chicken BAC library, segment 1, Ren et al. 2003; 11× CHORI-261, BACPAC Resources, http://bacpac.chori.org/chicken261.htm), 32P-labeled DNA probes were hybridized in pools of 20 to high-density filters, then rescreened individually as previously described (Ashworth et al. 1995; Ross et al. 1999; Kim et al. 2001). BACs were restriction fingerprinted and manually assembled into maps (Kim et al. 2001; Grimwood et al. 2004, Supplemental Methods). Finally, isolated islands of gene-poor GGA11 were joined, and the telomeric end of GGA28 was extended, after identifying spanning BAC clones from the Washington University fpc fingerprints (Wallis et al. 2004).
Sequencing and assembly
BACs selected to create an efficient tiling path were isolated, fragmented, subcloned, and sequenced in both directions as previously described (Dehal et al. 2001, Detter et al. 2002). Detailed protocols are available on-line at http://www.jgi.doe.gov. Sequences were base-called and assigned a quality score using Phred, screened for BAC, pUC18 vector, and E. coli contaminants, assembled with Phrap, and viewed with Consed (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). Using a modified Finisher program, paired end sequences were used to order and orient contigs. All ordered sequences are available at NCBI (http://www.ncbi.nlm.nih.gov). Accession numbers are detailed in Supplemental Table S1.
To create assembled scaffolds, JF BAC clones were separated into component sequence contigs, and overlapping contigs were consolidated into 355 supercontigs (N50 length of 13 kb, longest supercontig was 246 kb). The algorithm then iteratively investigated all pairwise inter- and intra-BAC relationships, using sequence contig order within individual BACs and clone overlaps between BACs to assemble the supercontigs into three nonredundant scaffolds. The scaffolds were then inspected manually and refined as follows. Individual clones were tested against the assembly to verify that they were completely and nonredundantly represented; stretches of redundant sequence were eliminated. Parallel WL BAC sequence assemblies were compared with JF scaffolds to verify gene order and tiling path fidelity. WL sequence was inserted in three regions to fill gaps that were not covered by JF BACs despite exhaustive library screening; it extends the Jf_g1 scaffold (37.4 kb) and crosses two gaps in Jf_g2 (130.3 and 3.9 kb, respectively, Supplemental Table S3). These inserted segments were identified from flanking sequence overlaps and “patched in” as intact segments from solidly assembled WL BAC sequence contigs.
Gene models
Sequence matches to predicted proteins from the original curation of finished HSA19 sequence (Grimwood et al. 2004) and UCSC known human and mouse genes (hg16 and mm4 assemblies) were identified in the chicken sequence using tblastn. BLAST similarity regions were extracted from the chicken sequence, along with the flanking intergenic regions spanning the segments between closest neighbors, and GeneWise (Birney et al. 2004) was executed to build gene models using the matching protein sequence(s) as a guide. Predicted protein-coding loci were manually inspected to verify gene identities and automated predictions were manually curated. Novel chicken genes with no homology with known proteins were also added manually based on chicken EST and mRNA matches. For chicken-specific loci, we required either deep evidence of gene expression (3 or more spliced cDNAs) or 2 or more of the following criteria: gene models, solid reading frame and conserved protein domains, evidence of gene expression, and/or evidence of evolutionary conservation in another species. Non-HSA19 orthologs were identified as best reciprocal matches with human counterparts, with no related sequences in the syntenic chicken region, and no contradictory evidence in cross-species comparisons. More detailed annotation considerations are described in Supplemental Methods.
Homologous synteny blocks and evolutionary break regions
Contiguous blocks of gene neighborhoods common to human and chicken and disrupted only by localized microrearrangements were identified as “major” homologous synteny blocks (HSBs); genomic regions between H/C homology blocks, some containing chicken-specific loci, were designated the “core” evolutionary break regions (EBRs). Inverted or duplicated H/C orthologs flanking breakpoints were included in both HSB and EBR. Localized synteny changes that did not disrupt the overall gene neighborhood were considered “internal” changes. We applied conservative criteria to designate an internal site as rearranged (boxed in Supplemental Tables S1, S7); it had to evidence an inversion, ortholog transposition between species or within the homology segment, or reuse in another species (see Supplemental Methods for details). Reused breakpoints were identified by examining wgs regions (UCSC browser) related to H/C HSBs and EBRs for independent flanking segments in mouse, dog, opossum, and frog (see Table 2; Supplemental Table S8 for details).
Assembly comparisons
Clone-based assemblies were aligned with galGal3 wgs assemblies using Advanced PIP Maker [R] (Schwartz et al. 2000). Both strands were searched for alignments and single coverage (or “all alignments” for GGA11 dot plots), and results were retrieved from the server as concise coordinate text files and dot plots (Penn State University Center for Comparative Genomics and Bioinformatics). Aligned segments ≤1.5 kb in size or <90% identity were filtered out, then segments <50 kb apart in either assembly were merged to form longer “supersegment” alignments. Supersegments <10 kb in length or that had an average identity of <90% over the alignment were discarded. Regions of the clone-based assembly that aligned to wgs sequence on chromosomes other than GGA28 were reported as misassembled and regions that did not align to the wgs sequence on any chromosomes in super-segments were reported as missing.
Sequence analysis of breakpoint regions
GC content of all chicken (galGal3) chromosomes was calculated using 10-kb nonoverlapping windows. Candidate control regions were selected automatically based on GC content and/or evolutionary stability of in chicken (galGal3) chromosomes, then manually screened to select intervals that best fit control group criteria. Candidate HGC regions were identified where average GC content of continuous windows exceeded 51% within a domain of ≥1 Mb; intervals were then screened for minimal synteny disruption in mammals (≤1 EBR). Potential SSC regions were identified using human and mouse net alignments (UCSC browser) with >1 Mb in a continuous level 1 net, less than 10% in a level 2 net relative to human, total alignment gaps less than 90% in human, and no synteny breaks in mouse. Intervals determined to be stable in mammals as well as frog were included in the final set. To identify GWR candidates, the wgs was scanned for 1-Mb intervals with more than 10 net alignment breaks in human and/or mouse level 1 alignments with chicken; regions with ≥6 confirmed EBRs per Mb comprised the final set. For GWC, wgs regions with GC content ±1% of GWR were screened to select 1-Mb intervals with ≤1 EBR in mammals and a final GC content ±2% of GWR. Reused EBRs were identified in GWR intervals as described above for GGA28.
All alignments were done using Advanced PipMaker from Penn State University Center for Comparative Genomics and Bioinformatics (Schwartz et al. 2000; http://pipmaker.bx.psu.edu/cgi-bin/pipmaker?advanced). Sequence features (GC, CpG, LINE, SSR, SNP, LTR) for galGal3 regions were downloaded from the UCSC genome Browser. For GGA28, CpG islands were identified in BAC-based contigs using standard methods (UCSC Browser, http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=cpgIslandExt); repeats were identified using RepeatMasker (http://www.repeatmasker.org/). All feature densities were calculated using 10-kb nonoverlapping windows with sequence gap regions masked. Duplications for all regions, including GGA28, were calculated from galGal3 sequence based on number of nucleotide matches in self-chain alignments using 10-kb windows. RefSeq genes aligned to BAC-based assemblies were counted for GGA28, all other gene densities were calculated directly from galGal3. RefSeq gene density was calculated for each control region as a whole, precluding statistical comparisons. All P values were calculated using Welch’s two-sample t-test. Statistical analyses were performed using the R package version 2.3.1 (http://www.r-project.org/).
Acknowledgments
We thank Elbert Branscomb, Joomyeong Kim, and Alice Yamada for critical reviews of the manuscript, Gawain Lavers for website development, and the following individuals for their contributions to mapping, sequencing, and bioinformatics: Matthew Groza, Elizabeth Fields, Mark Wagner, Tijana Glavina, Heather Kimball, and Andrea Aerts. This work was performed under the auspices of the U. S. Department of Energy (DOE) by the University of California, Lawrence Livermore National Laboratory (LLNL) under Contract No. W-7405-Eng-48.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6775107
References
- Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth L.K., Batzer M.A., Brandriff B., Branscomb E., de Jong P., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Batzer M.A., Brandriff B., Branscomb E., de Jong P., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Brandriff B., Branscomb E., de Jong P., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Branscomb E., de Jong P., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., de Jong P., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Garcia E., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Garnes J.A., Gordon L.A., Lamerdin J.E., Lennon G., Gordon L.A., Lamerdin J.E., Lennon G., Lamerdin J.E., Lennon G., Lennon G., et al. An integrated metric physical map of human chromosome 19. Nat. Genet. 1995;11:422–427. doi: 10.1038/ng1295-422. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Massa H.F., Trask B.J., Eichler E.E., Trask B.J., Eichler E.E., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., Pevzner P.A., Tesler G., Pevzner P.A., Tesler G., Tesler G. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 2004;14:507–516. doi: 10.1101/gr.1975204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Asperen R., Boon K., Voute P.A., Boon K., Voute P.A., Voute P.A., et al. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Crooijmans R.P., Vrebalov J., Dijkhof R.J., van der Poel J.J., Groenen M.A., Vrebalov J., Dijkhof R.J., van der Poel J.J., Groenen M.A., Dijkhof R.J., van der Poel J.J., Groenen M.A., van der Poel J.J., Groenen M.A., Groenen M.A. Two-dimensional screening of the Wageningen chicken BAC library. Mamm. Genome. 2000;11:360–363. doi: 10.1007/s003350010068. [DOI] [PubMed] [Google Scholar]
- Deeb N., Lamont S.J., Lamont S.J. Use of a novel outbred by inbred F1 cross to detect genetic markers for growth. Anim. Genet. 2003;34:205–212. doi: 10.1046/j.1365-2052.2003.00998.x. [DOI] [PubMed] [Google Scholar]
- Dehal P., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Land M., Terry A., Ecale Zhou C.L., Rash S., Terry A., Ecale Zhou C.L., Rash S., Ecale Zhou C.L., Rash S., Rash S., et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Detter J.C., Jett J.M., Lucas S.M., Dalin E., Arellano A.R., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Jett J.M., Lucas S.M., Dalin E., Arellano A.R., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Lucas S.M., Dalin E., Arellano A.R., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Dalin E., Arellano A.R., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Arellano A.R., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Wang M., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Nelson J.R., Chapman J., Lou Y., Rokhsar D., Chapman J., Lou Y., Rokhsar D., Lou Y., Rokhsar D., Rokhsar D., et al. Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics. 2002;80:691–698. doi: 10.1006/geno.2002.7020. [DOI] [PubMed] [Google Scholar]
- Elleder D., Plachy J., Hejnar J., Geryk J., Svoboda J., Plachy J., Hejnar J., Geryk J., Svoboda J., Hejnar J., Geryk J., Svoboda J., Geryk J., Svoboda J., Svoboda J. Close linkage of genes encoding receptors for subgroups A and C of avian sarcoma/leucosis virus on chicken chromosome 28. Anim. Genet. 2004;35:176–181. doi: 10.1111/j.1365-2052.2004.01118.x. [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J., Bateman J.F., Bateman J.F. Why mice have lost genes for COL21A1, STK17A, GPR145 and AHRI: Evidence for gene deletion at evolutionary breakpoints in the rodent lineage. Trends Genet. 2004;20:408–412. doi: 10.1016/j.tig.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Hink R., Burgers S., Hernandez-Boussard T., Burgers S., Hernandez-Boussard T., Hernandez-Boussard T., et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grimwood J., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Goodstein D., Couronne O., Tran-Gyamfi M., Couronne O., Tran-Gyamfi M., Tran-Gyamfi M., et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Van Calcar S., Qu C., Ching K.A., Qu C., Ching K.A., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Huntley S., Baggott D.M., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Baggott D.M., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Kim J., Gordon L., Branscomb E., Stubbs L., Gordon L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeobi C.O., Woolliams J.A., Morrice D.R., Law A., Windsor D., Burt D.W., Hocking P.M., Woolliams J.A., Morrice D.R., Law A., Windsor D., Burt D.W., Hocking P.M., Morrice D.R., Law A., Windsor D., Burt D.W., Hocking P.M., Law A., Windsor D., Burt D.W., Hocking P.M., Windsor D., Burt D.W., Hocking P.M., Burt D.W., Hocking P.M., Hocking P.M. Quantitative trait loci affecting fatness in the chicken. Anim. Genet. 2002;33:428–435. doi: 10.1046/j.1365-2052.2002.00911.x. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICSGC) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Goodier J.L., Goodier J.L. LINE drive, retrotransposition, and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- Kim J., Gordon L., Dehal P., Badri H., Christensen M., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Gordon L., Dehal P., Badri H., Christensen M., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Dehal P., Badri H., Christensen M., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Badri H., Christensen M., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Christensen M., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Groza M., Ha C., Hammond S., Vargas M., Wehri E., Ha C., Hammond S., Vargas M., Wehri E., Hammond S., Vargas M., Wehri E., Vargas M., Wehri E., Wehri E., et al. Homology-driven assembly of a sequence-ready mouse BAC contig map spanning regions related to the 46-Mb gene-rich euchromatic segments of human chromosome 19. Genomics. 2001;74:129–141. doi: 10.1006/geno.2001.6521. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lund J., Chen F., Hua A., Roe B., Budarf M., Emanuel B.S., Reeves R.H., Chen F., Hua A., Roe B., Budarf M., Emanuel B.S., Reeves R.H., Hua A., Roe B., Budarf M., Emanuel B.S., Reeves R.H., Roe B., Budarf M., Emanuel B.S., Reeves R.H., Budarf M., Emanuel B.S., Reeves R.H., Emanuel B.S., Reeves R.H., Reeves R.H. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velocardiofacial syndrome region on chromosome 22q11.2. Genomics. 2000;63:374–383. doi: 10.1006/geno.1999.6044. [DOI] [PubMed] [Google Scholar]
- Lupski J.R., Stankiewicz P., Stankiewicz P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Pevzner P.A., O’Brien S.J., Pevzner P.A., O’Brien S.J., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Hardison R.C., Miller W., Stubbs L., Miller W., Stubbs L., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., Tesler G., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie T.S., Crooijmans R.P., Bovenhuis H., Vereijken A.L., Veenendaal T., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., Crooijmans R.P., Bovenhuis H., Vereijken A.L., Veenendaal T., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., Bovenhuis H., Vereijken A.L., Veenendaal T., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., Vereijken A.L., Veenendaal T., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., Veenendaal T., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., van der Poel J.J., Van Arendonk J.A., Pakdel A., Groenen M.A., Van Arendonk J.A., Pakdel A., Groenen M.A., Pakdel A., Groenen M.A., Groenen M.A. Genetic mapping of quantitative trait loci affecting susceptibility in chicken to develop pulmonary hypertension syndrome. Anim. Genet. 2005;36:468–476. doi: 10.1111/j.1365-2052.2005.01346.x. [DOI] [PubMed] [Google Scholar]
- Ren C., Lee M.K., Yan B., Ding K., Cox B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Lee M.K., Yan B., Ding K., Cox B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Yan B., Ding K., Cox B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Ding K., Cox B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Cox B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Romanov M.N., Price J.A., Dodgson J.B., Zhang H.B., Price J.A., Dodgson J.B., Zhang H.B., Dodgson J.B., Zhang H.B., Zhang H.B. A BAC-based physical map of the chicken genome. Genome Res. 2003;13:2754–2758. doi: 10.1101/gr.1499303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K., Cuddapah S., Zhao K., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.T., LaBrie S., McPherson J., Stanton V.P., LaBrie S., McPherson J., Stanton V.P., McPherson J., Stanton V.P., Stanton V.P. Screening large-insert libraries by hybridization. In: Dracopoli N.C., et al., editors. Current Protocols in Human Genetics. John Wiley and Sons; New York: 1999. pp. 5.6.1–5.6.52. [Google Scholar]
- Ruiz-Herrera A., Castrasana J., Robinson T.J., Castrasana J., Robinson T.J., Robinson T.J. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W., Bouck J., Gibbs R., Hardison R., Miller W., Gibbs R., Hardison R., Miller W., Hardison R., Miller W., Miller W. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.W., Aerts J., Groenen M.A., Crooijmans R.P., Layman D., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Aerts J., Groenen M.A., Crooijmans R.P., Layman D., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Groenen M.A., Crooijmans R.P., Layman D., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Crooijmans R.P., Layman D., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Layman D., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Graves T.A., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Scheer D.E., Kremitzki C., Fedele M.J., Mudd N.K., Kremitzki C., Fedele M.J., Mudd N.K., Fedele M.J., Mudd N.K., Mudd N.K., et al. A physical map of the chicken genome. Nature. 2004;432:761–764. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- Wang G., Christensen L.A., Vasquez K.M., Christensen L.A., Vasquez K.M., Vasquez K.M. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl. Acad. Sci. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.T., Axelsson E., Ellegren H., Axelsson E., Ellegren H., Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol. Biol. Evol. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]