Abstract
We describe a gene network for the Arabidopsis thaliana transcriptome based on a modified graphical Gaussian model (GGM). Through partial correlation (pcor), GGM infers coregulation patterns between gene pairs conditional on the behavior of other genes. Regularized GGM calculated pcor between gene pairs among ∼2000 input genes at a time. Regularized GGM coupled with iterative random samplings of genes was expanded into a network that covered the Arabidopsis genome (22,266 genes). This resulted in a network of 18,625 interactions (edges) among 6760 genes (nodes) with high confidence and connections representing ∼0.01% of all possible edges. When queried for selected genes, locally coherent subnetworks mainly related to metabolic functions, and stress responses emerged. Examples of networks for biochemical pathways, cell wall metabolism, and cold responses are presented. GGM displayed known coregulation pathways as subnetworks and added novel components to known edges. Finally, the network reconciled individual subnetworks in a topology joined at the whole-genome level and provided a general framework that can instruct future studies on plant metabolism and stress responses. The network model is included.
Remarkable conceptual and technical advances in genomics have generated exceptionally large data sets. Global analyses of these collections of data may now be used to construct biological networks that systematically categorize all molecules and describe their functions and interactions (Barabasi and Oltvai 2004). Networks are emerging that, oriented to highlight different levels of complexity and placing emphasis on distinct regulatory, developmental, or metabolic “pathways,” can now integrate biological functions of cells, organs, and organisms (Brazhnik et al. 2002).
Most advanced are gene networks analyzing large-scale microarray hybridizations that monitor transcriptome dynamics (de la Fuente et al. 2002; Yugi et al. 2005). Emerging also are networks extracted from protein–protein interactions or protein complexes (Ito et al. 2001; Gavin et al. 2002), regulatory networks based on ChIP-chip data, which describe the interactions between transcription factors and their targets (Lee et al. 2002; Buck and Lieb 2004), or metabolic networks elucidating effects of the dynamics of metabolites (Baxter et al. 2007; Martins et al. 2007). Synthetic lethal networks extract genetic interactions critical for an organism’s fitness (Tong et al. 2004; Pan et al. 2006).
In contrast to single-cell organisms, network reconstruction of higher organisms has been restricted mainly due to limitations in data availability. Nevertheless, in a complex system such as the plant model Arabidopsis thaliana, expression profiles extracted from microarray data sets offer information on physiological status, in particular, because data from time series and from developmental, genetic intervention, or manipulative treatments are available (Schmid et al. 2005; Kilian et al. 2007).
The assembly of a gene network depends on the mathematical models applied, which, ideally, should describe inferred causal relationships that govern the expression patterns and dynamics of a set of genes. In reality, networks are assembled according to coincidence or coregulation of genes and the magnitude of regulation or statistical significance of the coincidence (Brazhnik et al. 2002). Currently, the most widely used computational method involves calculating standard Pearson correlation coefficients (r) between pairs of genes. A pair of genes with r larger than a preselected threshold is considered to reveal functional interaction, influence, or dependence. Networks based on these interactions are termed relevance network. However, such networks may lead to ambiguous results, especially when the network is heavily connected (Brazhnik et al. 2002). An alternative method, the graphical Gaussian model (GGM), uses partial correlations as the source for a robust assessment of a direct interaction between any gene pair (Whittaker 1990; Toh and Horimoto 2002). Different from Pearson correlation that records correlation between gene pairs without regard to other genes, partial correlation between two genes measures the degree of correlation remaining after removing the effects of other genes. Recent studies have demonstrated that GGM is a useful tool to infer conditional dependency structure and to reconstruct network-like associations among genes (Kishino and Waddell 2000; Toh and Horimoto 2002; Magwene and Kim 2004; Schäfer and Strimmer 2005b; Wille and Buhlmann 2006).
Irrespective of the potential intrinsic to GGM, its application for building network inferences had before been restricted to a small number of genes (Kishino and Waddell 2000; Toh and Horimoto 2002) due to the generally small number of samples (n) available from microarray experiments. This number is typically much smaller than the number of genes (P). Classical GGM theory cannot accommodate settings for P >> n (Schäfer and Strimmer 2005a; Wille and Buhlmann 2006). Recently, GGM with a limited-order partial correlation function, which estimates correlations conditional on one or two, but not all other genes, has been developed to infer gene networks from Arabidopsis and yeast transcript profiles (Magwene and Kim 2004; Wille et al. 2004). Another way to tackle the small sampling problem is to infer GGM with regularization and moderation (Schäfer and Strimmer 2005b). This shrinkage approach to graphical Gaussian modeling, implemented in “GeneNet” in R, is such an approach that is applicable to data sets with P slightly larger than n (Schäfer and Strimmer 2005c; Schäfer et al. 2006).
We have used this regularized GGM to build a gene network for A. thaliana, based on data from more than 2000 Affymetrix ATH1 microarray experiments deposited in the NASC database (Craigon et al. 2004). A pilot study evaluated the method for 2000 genes for which biologically meaningful interactions had been established in single-gene studies. Then, as an exploratory experiment, by using an iterative random sampling strategy, the model was expanded to cover >22,000 Arabidopsis genes, resulting in a network that included 6760 nodes (genes) connected by 18,625 significant edges (interactions).
Results and Discussion
Pilot experiment with 2000 genes
The data for construction of the model (Schäfer and Strimmer 2005c) represented 2466 Affymetrix ATH1 microarray slides deposited at NASC by August 2006. After excluding 421 potentially outlying experiments according to Persson et al. (2005), 2045 chips remained for network construction. The selected conditions reported transcript changes in plants challenged by a spectrum of abiotic and biotic stresses and chemical treatments. In addition, transcript profiles from different tissues or developmental stages were included (Supplemental Table S1).
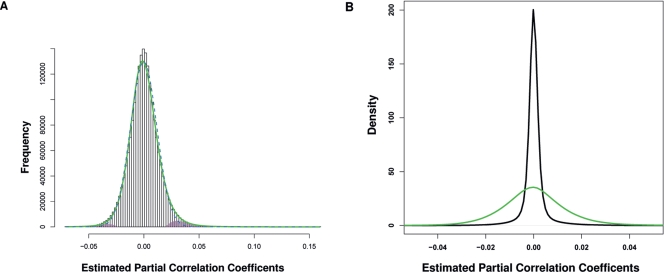
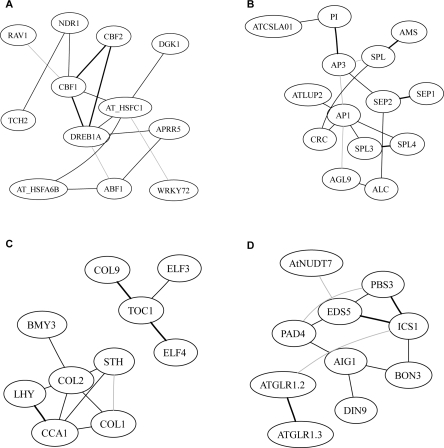
A proof-of-concept experiment started with a collection of ∼5000 named, and to some degree, analyzed genes in Arabidopsis. This collection was filtered by a selection of genes with high regulation by biotic and abiotic stresses and tissue expression characteristics, which reduced the number to ∼2000 genes. Partial correlation (pcor) was estimated for every gene pair among these genes using the “GeneNet” package (Schäfer et al. 2006). Figure 1A shows the histogram for the distribution of the estimated pcor values. According to previous observations, connections within biomolecular networks are typically sparse (Jeong et al. 2001; Yeung et al. 2002). It has been assumed that most estimated pcor identified gene pairs lacking interactions and showed values close to 0 (Schäfer and Strimmer 2005c). These pcor from noninteracting gene pairs provided a basis to infer a null distribution, resulting in an excellent fit with a formula describing the distribution of the sample normal correlation coefficients (Hotelling 1953; Schäfer and Strimmer 2005c). The null model was then used to calculate the P-value for every pcor and determine the probability that it satisfied the null distribution. We focused on 1024 gene pairs with |pcor| ≥0.10, whose P-values were <2.2 × 10−19. Then, gene pairs with Pearson correlation values (r) ranging from –0.25 to 0.35 were eliminated, reasoning that any r close to zero indicated independence. This filter is asymmetric because there were far more significant positive than negative pcors. The asymmetry was further supported by the permutation experiment for the expanded network (see below). This resulted in a network with 820 nodes and 828 edges. An inspection of this network revealed subnetworks of biological significance (Fig. 2). The figure shows networks of genes predicted to interact with CBF1 (cold stress response) (Fowler et al. 2005; Agarwal et al. 2006), AP3 (flower development) (Krizek and Fletcher 2005), CCA1 and TOC1 (circadian rhythmicity) (Ledger et al. 2001; Salome and McClung 2004; Kikis et al. 2005), and phytoalexin-deficient 4, PAD4 (salicylic acid metabolism and pathogen response) (Glazebrook et al. 2003). The predicted networks consistently included experimentally verified genes, demonstrating the ability of this GGM to reveal significant, potentially important gene interactions.
Figure 1.
The distribution of estimated partial correlations. (A) Distribution of pcors for 2000 named and at least partially studied genes. (Bars) Histogram; (green line) distribution of estimated pcor after Fisher’s normalizing z-transformation; (dashed blue line) fitted null distribution; (pink) alternative distribution, inferred by the locfdr algorithm (Efron 2004, 2007). (B) Comparison of the pcor distribution in the pilot experiment (green line) and the expanded calculation (dark line). Shown is the area between –0.05 and 0.05. Densities were calculated using the kernel density estimator embedded in R, with the bandwidth set to 0.001.
Figure 2.
Subnetworks in a pilot experiment with functionally characterized genes. The subnetworks were derived from seeded nodes, and including all other nodes within two connections from seeded nodes (four for PAD4). The seed nodes in each subnetwork are (A) CBF1; (B) AP3; (C) CCA1 and TOC1; (D) PAD4. Within each subnetwork, a link between two nodes indicates direct interactions. Black indicates edges with the highest 20% pcor values, gray lines indicate edges at the lower 20% pcor, and dashed lines indicate negative interactions.
A network for 22,200 Arabidopsis genes
This result provided motivation to expand the network by including ∼22,200 genes of the Arabidopsis transcriptome, represented by 22,266 Affymetrix ATH1 probes with the discrepancy in numbers, due to the fact that some genes were represented by more than one probe set. GGM does not allow for computing the pcor of all input genes simultaneously, because the maximum number of genes that may be analyzed at one time depends on sampling numbers. An iterative process with 2000 iterations was adopted. In each iteration, 2000 genes were randomly selected and used as input for pcor estimation. On average, every gene pair was sampled 16.2 times, and the pcor with the lowest absolute value, representing the one with the largest amount of effects from other genes removed, was chosen as an estimation of the final pcor in the expanded network. Compared with the pilot experiment, these pcors were more narrowly concentrated around zero (Fig. 1B). A null distribution model was then built to estimate the P-values for the edges derived from these final pcors (Supplemental Fig. 1). When setting the cutoff values for pcor at less than –0.10 or larger than 0.10, the corresponding P-value, according to this null model, was lower than 1.92 × 10−190. With this pcor cutoff and after applying the Pearson correlation filter (−0.25–0.35; see Methods), which removed 12.4% of the accepted pcor values, a network for 6760 genes and 18,625 edges was recovered, which retained ∼0.01% of all possible interactions as significant edges. Our selection of the pcor cutoff value at |0.10| represents high stringency. In comparison, a yeast gene network recovered 70,201 interactions between 5205 genes, a human coexpression network identified 220,649 links among 8805 genes, while another yeast network based on first order partial correlation revealed 11,416 connections between 4686 genes (Lee et al. 2004; Magwene and Kim 2004; Yu et al. 2006).
Additionally, two random permutation experiments were conducted to evaluate potential false discovery rates. First, all 22,266 genes were permutated, followed by the analysis described before. After 1000 iterations, all final pcors were in the range of from –0.0002 to +0.0004 and deemed insignificant. Second, 1000 genes were randomly chosen, permutated, combined with the remaining 21,266 genes, and subjected to the analysis with 2000 iterations, resulting in an overall pcor distribution similar to that in Figure 1B. Among the 21,765,500 gene pairs with one or two genes permutated, 4875 pairs showed |pcor| ≥0.05, and 132 pairs had |pcor| ≥0.10. However, the corresponding Pearson correlation for 4873 of these gene pairs ranged from –0.10 to +0.30, and only two pcor values, or 9.18E-08 of the permutated gene pairs, survived the Pearson correlation filter, both with |pcor| >0.10. The result indicated that few (∼23) pcors were attributable to false discovery in analyses without permutations, which are disregarded as we discuss the properties of the expanded network. It should be noted that the permutation was carried out without replications due to the length of computing time; more permutations are required to reach conclusions with highest certainty.
Overall network properties
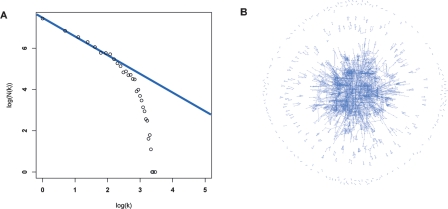
The resulting network was not completely scale free, but exhibited scale-free behavior over a wide range. The average network connectivity for a node was 5.5. Figure 3A shows the connectivity frequency distribution, with k symbolizing connectivity and N(k) the number of nodes with connectivity k. For a typical scale-free network, N(k) observes power-law distribution, and in the plot of log(N(k)) relative to log(k) the dots should fit a straight line (Barabasi and Albert 1999).
Figure 3.
Connectivity and network structure. (A) The connectivity distribution for the expanded network (k) Connectivity; (N(k)) number of nodes with connectivity k. The line indicates the distribution expected for a network following the power law. (B) An overview of the network (with 5000 edges included), as generated by the tYNA platform.
The network seems to follow a truncated power-law distribution (Amaral et al. 2000), with a power-law regime at 1 ≤ k ≤ 11, where the network exhibits certain scale-free behavior, followed by a sharp drop off. Biological networks with similar connectivity distribution have been reported before (Jeong et al. 2001; Giot et al. 2003). A recent analysis indicated that most biological networks were not totally scale free, but rather might better be described as following a truncated power law, while certain scale-free features such as small world and centrality properties hold true (Khanin and Wit 2006). An evident qualitative feature of our network, characteristic of scale-free network models, was the presence of few nodes with many connections, which appeared to constitute major hubs and many nodes with very few connections.
The final overall network (Fig. 3B) was densely organized. When querying the network with selected genes, a number of coherent subnetworks emerged to which biological significance could be attached (Figs. 3, 4, 5, 6, below; Supplemental Fig. 2). We have chosen subnetworks for which biological proof and significance already exists. The resulting network modules, in their majority, defined and organized functions in metabolism or stress responses (Table 1). Subsequently, we included additional edges that then described the Arabidopsis transcriptional response to cold treatment. In these examples, the potential usefulness of the GGM gene network tool may be seen in establishing connections within a subnetwork of known functions with genes not previously associated with a network module or pathway. Often, these novel nodes were functionally unknown, never having been studied before.
Figure 4.
Subnetworks extracted from the expanded network. The examples are centered on (A) APR1; (B) SQD1; (C) DIN4; (D) TRP1; (E) NIA2; and (F) SBE2.1. Symbols as in Figure 2.
Figure 5.
Subnetworks for cell wall biosynthesis. (A) Genes centered on CESA6 (note: this subnetwork is drawn from edges with pcor >0.08, i.e., lower stringency). (B) Genes centered on CESA8. Symbols as in Figure 2.
Figure 6.
Subnetworks for cold stress responses. (A) Overview of the subnetwork centered on CBF1, CBF2, RAV1, and DREB1A. (B–F) Enlargement of different parts of the subnetwork shown in A. Symbols as in Figure 2.
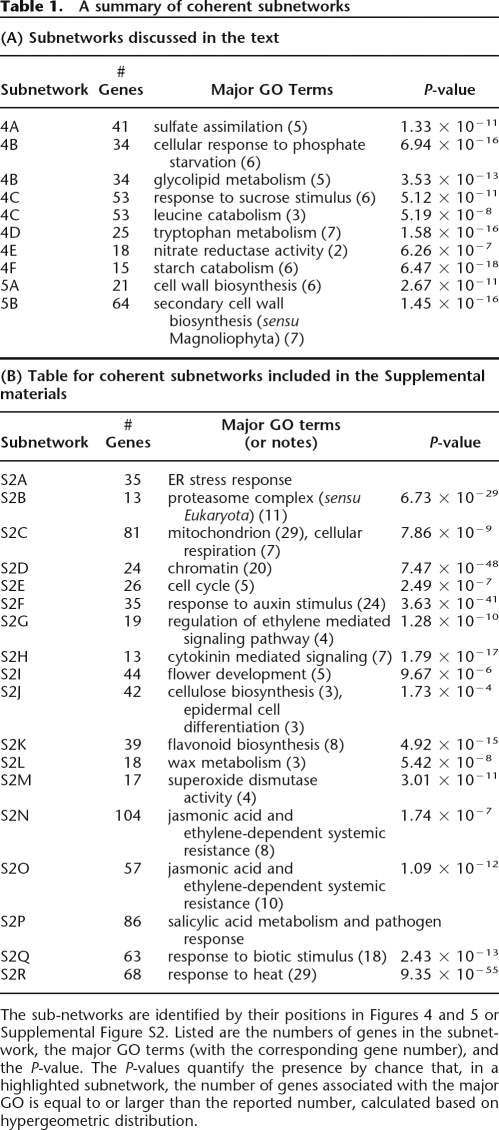
Table 1.
A summary of coherent subnetworks
The sub-networks are identified by their positions in Figures 4 and 5 or Supplemental Figure S2. Listed are the numbers of genes in the subnetwork, the major GO terms (with the corresponding gene number), and the P-value. The P-values quantify the presence by chance that, in a highlighted subnetwork, the number of genes associated with the major GO is equal to or larger than the reported number, calculated based on hypergeometric distribution.
Modules in metabolism reveal coherent network subgraphs
By use of the kCores method in Carey and Long’s RBGL package (version 1.10.0) in Bioconductor, we identified coherent subgraphs (Gentleman et al. 2004). Easily identifiable among these subgraphs were networks assignable to defined metabolic processes. Figure 4 exemplified this for six cases, which were summarized in Table 1A. Table 1B summarizes 18 additional subnetworks (Supplemental Fig. 2), listing enriched GO-terms associated with the genes identified.
Genes centered on APR1, one of three 5′-adenylylsulfate reductase genes in Arabidopsis, identified a coherence group associated with sulfur metabolism (Fig. 4A). Strongly associated with APR1 were APR2 and APR3, two homologs of APR1 in the Arabidopsis genome. Associated genes in this network were ATSERAT2;1, AKN2, APK, AT1G18590, AT1G74090, ATGSTF11, SULTR4;1, and AT1G74100, all of which encode proteins related to sulfur metabolism. Two genes, SUR1 and ASA1, are genes associated with auxin and tryptophan biosynthetic pathways, confirming other reports (Nikiforova et al. 2003, 2005; Dan et al. 2007).
In Figure 4B, genes involved in phosphate starvation reactions are linked. AT3G05630 encodes phospholipase DZ2, which hydrolyzes phospholipids in plasma membranes, thus releasing inorganic phosphate upon phosphate starvation (Cruz-Ramirez et al. 2006). NPC4 is a starvation-induced phosphate lipase C (Nakamura et al. 2005). MGD2, MGDC, SQD2, and SQD1 all participate in converting phospholipids to nonphospholipids, sulfolipid, and galactolipid, releasing phosphate (Yu et al. 2002; Benning and Ohta 2005). SRG3, a glycerophosphoryl diester phospho-diesterase family protein may be involved in a similar process. Other related genes are AT3G17790, specifying a type 5 acid phosphatase, AT2G27190, a purple acid phosphatase (PAP12), PHT5, an inorganic phosphate transporter, and two SPX domain containing proteins (AT2G26660, AT5G20150). Transcript profiling had shown that many of these genes are induced by phosphate starvation (Misson et al. 2005).
Figure 4C highlights genes that participate in branch-chained amino acid degradation. MCCA and MCCB form a complex involved in leucine degradation in mitochondria (Gavin et al. 2002). DIN4, BCE2, AT1G10070, AT1G21400, and BCDH BETA1 encode subunits of branched chain alpha-keto acid dehydrogenase. The expression of several genes in the group, e.g., DIN9, ASN1, DIN2, and AT2G43400, were shown as regulated by senescence and repressed by sugars (Fujiki et al. 2001). Thus, genes in this module could be involved in the regulation of cellular energy levels.
Figure 4D includes genes associated with TRP1. TRP1, TSA1, ASA1, CYP79B2, AT1G25155, PAD3, TSB1, and DHS1 are involved in tryptophan biosynthesis. GLIP1 is an important component of pathogen responses (Oh et al. 2005). In addition, we note that this subgraph is itself strongly connected to the subgraph in Figure 4A, including many genes related to sulfur metabolism.
Shown in Figure 4E and F are subgraphs for nitrogen and starch metabolism, respectively. NIA1 and NIA2 encode nitrate reductases involved in the first step of nitrate assimilation with NIR1 (encoding nitrite reductase) participating in the second step. NIR1 is connected to AT5G13110, AT1G24280, and AT4G05390, genes whose products participate in NADP metabolism. ASN2 encodes an asparagine synthetase converting ammonium into nitrogen-containing compounds. The subnetwork around starch catabolism included 15 genes, seven of which are known to belong to this pathway: SBE2.1, SEX1, AT5G64860, AT4G09020, DPE2, AT3G52180, and AT5G26570. Among them, AT3G52180 (DSP4) has been identified as encoding a protein phosphatase that binds to starch and regulates its accumulation (Sokolov et al. 2006). COR414-TM1 is a known cold-induced gene of unknown function (Breton et al. 2003), whose connection with SEX1 possibly indicates the necessity of increasing the cellular osmotic potential for acquisition of cold tolerance.
The selected seed genes for metabolic functions revealed a structure of the model (Fig. 4) that could be reconciled with established functions in plant metabolism. During phosphate starvation, biochemical studies have established degradation of phospho-, sulfo- and galactolipids (Cruz-Ramirez et al. 2006), requiring the lipases, phosphatases, sulfolipid synthases, or galactolipid synthases that populate this subgraph. Similarly, the subnetworks on sulfur and nitrate metabolism, starch catabolism, and tryptophan biosynthesis are supported by biochemical evidence. The lysine catabolism subgraph included genes with relationships to sulfur and phosphate metabolism and connections to tryptophan biosynthesis and mitochondrial functions (Nikiforova et al. 2003, 2005; Glawischnig et al. 2004; Dan et al. 2007).
Subnetworks describing cell wall biosynthesis and related processes
As another example, we analyzed placement of cellulose synthase genes, CESAn, in the network. Two major subnetworks were identified that covered eight CESA genes. Figure 5, A and B, show that the CESA genes separated into two groups: CESA1, CESA2, CESA3, CESA5, and CESA6 are group I CESAs responsible for primary cell wall synthesis, while CESA4, CESA7 (IRX3), and CESA8 are group II CESAs in charge of secondary cell wall synthesis (Somerville 2006). The Figure 5A subnetwork is drawn from edges with pcor larger than 0.08 instead of 0.10 to slightly increase the population. The analysis agreed with previous studies in which the cell wall-related gene network in Arabidopsis had been analyzed (Brown et al. 2005; Persson et al. 2005).
Of particular interest here were genes related to secondary cell wall synthesis. Covered in Figure 5B were not only group II CESA genes, but also other genes that have been demonstrated to be important for secondary cell wall synthesis, such as SND1 (AT1G32770), a NAM transcription factor (Zhong et al. 2006), IRX6 (AT5G15630), a member of the COBRA family of proteins, and UXS3, encoding an enzyme that produces UDP-xylose for cell wall biosynthesis. Also included were multiple metabolism-related genes, encoding laccases, glycoside hydrolase, and glycogenin glucosyltransferase. Further included were several genes related to vesicle trafficking and microtubule functions, such as ARAC2, RIC2, TUB8, AT1G73640 (G-protein), and AT4G38320 (microtubule associated). According to Genevestigator (Zimmermann et al. 2004), these genes are in their majority induced by salinity, osmotic, and oxidative stresses, in agreement with the need to strengthen secondary cell walls under such challenges and with physiological observations.
In addition to group I and II CESAs, CESA10, one of the cellulose synthases involved in the biosynthesis of primary cell walls (Beeckman et al. 2002), appeared in a subnetwork with relationships to epidermal cell development, including trichomes, root hairs, and seed coats (Supplemental Fig. S2J). Also clustered in separate, but closely related subnetworks were genes related to lignin and wax biosynthesis (Supplemental Fig. S2K,L).
Gene modules related to cell wall synthesis showed substantial overlap with networks based on Pearson correlation coefficients, with the exception that GGM provided more complex structure in as far as additional nodes were inserted. Also, highest correlation with genes reported by Pearson correlation were found only when the subgraphs were extended by several edges. For example, the cellulose synthases CESA4, CESA7 (IRX3), and CESA8, and additional genes in the synthesis of secondary cell walls (Fig. 5B) were arranged similar to structures reported by others (Brown et al. 2005; Persson et al. 2005). The study by Persson et al. (2005) identified AT4G28500, a NAM transcription factor correlated with three group-II CESA genes. The GGM network assigned connections between the CESA genes and this NAM transcription factor mediated through RIC2 (AT1G27380), a protein with rho-binding capacity, and correctly placed SND1 (AT1G32770), another NAM protein recently identified as a regulator of secondary cell wall synthesis (Zhong et al. 2006).
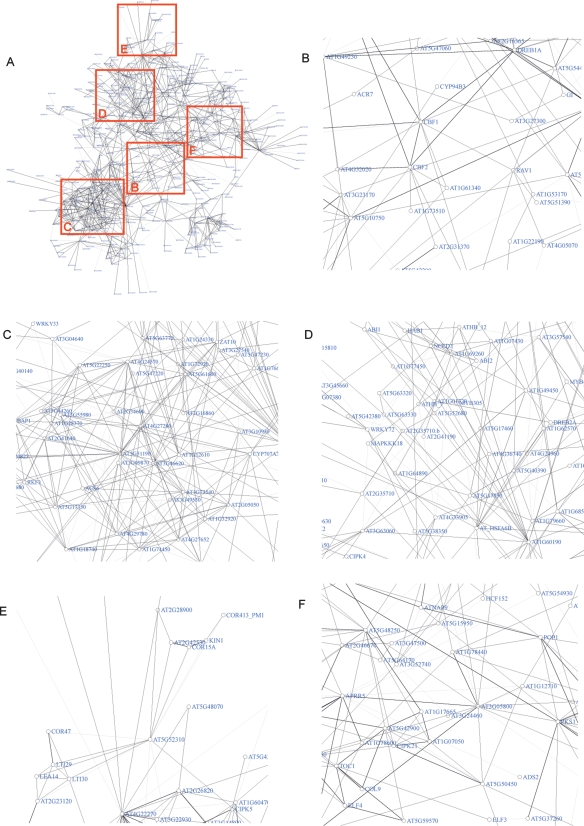
Arabidopsis responses to cold stress
To visualize a network for genes induced by cold stress, we extracted a subnetwork centered on CBF1, CBF2, DREB1A, and RAV1 (Fig. 6A), all known as cold stress-induced transcription factors in this pathway according to Genevestigator (Zimmermann et al. 2004). The network structures distinguished CBFs with similar expression patterns that appear to contribute to fine control of cold stress responses. In addition, the cold response pathways appeared to diverge into at least four different directions (Fig. 6C–F). Several genes that may connect these different parts were identified in the network. The U-box protein encoded by AT1G60190, for example, emerged as one of these connectors (Fig. 6D).
The center of the subnetwork was dominated by DREB-type transcription factors (Fig. 6B). The three CBFs (CBF1, CBF2, and DREB1A) were identified by strong interactions, indicating mutual functional redundancy (Gilmour et al. 2004; Maruyama et al. 2004; Agarwal et al. 2006). Connected to these central, well-studied transcription factors were other DREBs, such as RAV2 (AT1G68840) and AT1G25560. The concentration of transcription factors seems to reflect a complex coregulatory network.
Genes strongly induced by cold stress, and as well by a variety of other stress treatments (Fig. 6C), might be viewed as common or ubiquitous stress-response genes (Glazebrook et al. 2003). Several genes related to calcium (AT4G27280, AT3G25600, PBP1, and TCH2) and ethylene signaling (ACS6), and several zinc finger functions (STZ, C2H2, AT3G46620, AT3G55980, and AT1G20823) appeared in this subnetwork. Many of these genes have been found to be induced by a calcium burst in Arabidopsis. Their expression is also strongly responsive to ROS-based signals (Kaplan et al. 2006; Mittler et al. 2006).
Figure 6D included genes rapidly induced predominantly by cold stress and, somewhat less pronounced, by salinity, osmotic stress, and ABA. Included were multiple PP2Cs (ABI1, ABI2, HAB1, AT1G07430), two homeobox genes (ATHB7 and ATHB12), and NCED3, whose functions in ABA metabolism and response have amply been demonstrated. Interestingly, these genes were connected to AFP (AT1G69260), a negative regulator in ABA signaling, promoting ABI5 protein degradation (Lopez-Molina et al. 2003). Also connected was AT1G60190, encoding an U-box-containing protein that may have similar functions in terminating ABA signaling. AT1G60190 is extremely highly up-regulated by various stresses (Genevestigator) (Zimmermann et al. 2004), similar to the demonstration of protein degradation catalyzed by an E3-ligase (AT5G13530/AT5G13540) as a component of ABA signaling (Stone et al. 2006). A similar collection of known cold response and abiotic stress markers (Fig. 6E) included the functionally unknown COR47, LTI29, LTI30, COR15A, KIN1, and AT2G42530 (COR15B), which were indirectly connected to the key cold response transcription factors.
Other cold stress-induced functions included genes related to the regulation of circadian rhythm (Fig. 6F), TOC1, APRR5, ELF3, ELF4, COL9, and GI (Fig. 6A for GI). The subgraph identified other CONSTANS-like zinc finger proteins, AT1G07050, AT1G78600, and AT5G48250. Their placement into a separate subcluster might indicate regulation different from that of other cold stress-regulated genes, possibly connected to a diurnal cycle. Indeed, cold treatments have been shown to alter the expression of genes involved in the circadian rhythm (Kreps and Simon 1997) and clock and cold regulation has been reported, e.g., for CBF1, CBF2, and DREB1A (Fowler et al. 2005).
Comparison of GGM with a relevance network
Relevance networks based on standard Pearson correlation establish relationships different from GGM, without reference to other genes (Schäfer and Strimmer 2005c). Two genes may demonstrate the difference. ST3 and ST4 list the top 30 genes with the highest Pearson correlation in relationship to genes SQD2, a sulfolipid synthase (Fig. 4B), and AT1G26880, encoding the ribosomal protein RPL34 that does not appear in the GGM network. Notably, gene AT1G26880 showed Pearson correlation coefficients higher than 0.86 with 26 other ribosomal proteins, while the highest Pearson correlation coefficient of SQD2 with other genes was 0.69. While SQD2 would be excluded from a stringent relevance network, the GGM placed SQD2 with genes related to phosphate metabolism.
The complete Arabidopsis data set that had generated the GGM network was then used to construct a relevance network (Supplemental Data File S2). This analysis recovered 134,594 gene-pair interactions among 5745 genes with Pearson correlation coefficients larger or equal to 0.80. We excluded 12 negative interactions, lower than −0.80, in this analysis. Figure 7 shows the intersection between the two models. Among the ∼18,000 interactions in the GGM network, 4279 (22.9%) exhibited Pearson correlation coefficients larger than or equal to 0.8, while the majority of the interactions (96.8%) reported in the relevance network did not appear in the GGM network. One possibility is that a large number of the interactions revealed by relevance networks disappear when only those are considered that show the most robust correlation after excluding all other genes. The stringency achieved using higher order partial correlation appeared to generate networks with high biological support.
Figure 7.
Venn diagram outlining overlap and differences between the GGM and relevance network approaches. Numbers indicate the number of interactions recovered by the two methods.
The relevance network showed node distribution more similar to power-law (Supplemental Fig. S3), but many highly connected nodes in this relevance network were connected internally. Supplemental Figure S4A shows a subnetwork for the 100 most connected nodes, with 1939 interactions. Among these interactions, 1936 were assigned pcors lower than 0.10 and deemed insignificant in GGM, because the corresponding gene pairs shared expression patterns with many other genes, which then explained the low number of highly connected nodes in the GGM network. Additionally, GGM required high similarity in expression pattern for a gene to become connected with a highly populated node. As observed, this constraint in highly connected nodes generated the truncated power-law distribution for the whole network (Amaral et al. 2000). However, GGM continued to identify potential hubs, as shown by the 100 most highly connected nodes. The relevance network sorted these nodes into three potential hubs, while GGM arrived at a much higher number (Supplemental Fig. S4).
The model used is based on a shrinkage approach (Schäfer and Strimmer 2005c) that expanded classical GGM and performed well for the data set with P slightly larger than n, but was still limited, in that large transcriptomes could not be analyzed. By using iterations coupled with random sampling, our procedure allowed for expanding coverage to the genome level for Arabidopsis. The permutation experiments further indicated a low false discovery rate in this expanded network, whose biological significance was supported by case studies. We note, however, that the final pcor closely approached the 1998th-order partial correlation rather than a full partial correlation, because, in each iteration, only effects of 1998 other genes were removed for every gene pair. We present this GGM as an exploratory tool and heuristic model, whose significance is supported by the case studies outlined.
GGM-based gene network structures at the genome level for Arabidopsis have not been presented before, but networks for selected pathways have been constructed (Wille et al. 2004; Nikiforova et al. 2005; Li and Gui 2006; Gutierrez et al. 2007). The models presented here, when queried for nodes in these pathways, revealed significant overlap (data not shown). Recently, coexpression patterns based on Pearson correlation coefficients to infer gene function have been a highly active field in Arabidopsis research, with approaches expanding into two directions. For one, focus on coexpression of genes in selected functions, such as glucosinolate biosynthesis, primary carbon and nitrogen, or secondary metabolism, showed considerable overlap with this GGM network (Williams and Bowles 2004; Gachon et al. 2005; Wei et al. 2006; Hirai et al. 2007). Typically, these studies relied heavily on prior knowledge, such as biochemically established pathway structures, which is not a requirement for the GGM presented here. A second approach established databases that may be queried with individual genes to extract information about coexpressed genes (Zimmermann et al. 2004; Aoki et al. 2007; Obayashi et al. 2007). For one example, the database ATTED-II (Obayashi et al. 2007) lists highly coexpressed genes for every gene. Querying our GGM to the extent of one edge from the seed gene will only reveal a few of the connections identified by ATTED-II, while additional connections appear when the query is extended to include additional edges. Interestingly, these models, based on Pearson correlations alone, have not presented a network for the entire genome, possibly because such a structure would be dominated by genes related to a few dominant functional categories, such as ribosome structure, photosynthesis and carbon fixation, or flowering, while networks of metabolism would be hidden within the immensity of interactions.
The examples (Figs. 4, 5, 6; Supplemental Fig. S2) showed GGM revealing subnetworks that were strongly associated with established biological knowledge, while they invariably incorporated genes with unknown functions. Many modules identified functions that play important roles in the response to various stress conditions and in biochemical pathways. Although the procedures leading to this model generated a gene network that is substantially different from other types of networks, we suggest that in combination with these other models, GGM, which is accessible through the script that is included, could provide hypotheses for future studies.
Methods
Microarray data
All microarray data derived from Affymetrix ATH1 slides. The “Super Bulk Gene Download,” a file with all genes and experiments, was downloaded from NASCarrays (http://affymetrix.arabidopsis.info/narrays/help/usefulfiles.html). By August 2006, the file contained data from 2466 slides recorded as raw intensities. The corresponding experiments are summarized in Table 1. Six slides with missing data were removed and the remaining 2460 slides were subjected to quantile normalization.
A method based on “deleted residuals” was used to screen for potential outlier chips (Persson et al. 2005). Briefly, studentized deleted residuals d* are calculated for each probe set in every chip. The d* from the same chip were expected to observe a t distribution. Problematic chips were featured with significant deviation from t distribution of d*, which should be excluded. The Kolmogorov-Smirnov (K-S) goodness-of-fit test was used to calculate the K-S D-value to decide whether the d* from a chip fit the t distribution. With the K-S D value set at 0.10, we identified 415 chips, around 17% of all chips, as potential outliers.
The raw intensity data (after quantile normalization) from the remaining 2045 chips were rounded to integers (for values ≥10) or to the first digit after the decimal (for values <10), and used for analysis. Of 22,810 Affymetrix ATH1 probe sets, 22,266 were annotated as actual Arabidopsis genes. Data from these 22,266 probe sets were used for the GGM network construction, including both the pilot experiment and the expanded network. We treated each probe set as an individual gene. For probe sets matching more than one gene, we used the name of one of the matched genes. Supplemental Table 2 lists probe sets, corresponding gene names, and annotations.
The pilot experiment
The shrinkage approach (Schäfer and Strimmer 2005c) was used to estimate partial correlation coefficients (pcor) of gene pairs among 2000 chosen genes. The highest 0.01% and lowest 0.01% of pcors were excluded when building the null model. All calculations were conducted via the software package “GeneNet”, version 1.0.1 (Schäfer et al. 2006). Genes used in pilot experiments are listed in Supplemental Table 3.
The GGM network for the entire Arabidopsis genome
In total, 2000 iterations with random sampling were used to expand the network to cover the whole genome. Iteratively, 2000 genes were randomly selected and the “ggm.estmate.pcor” in GeneNet package 1.0.1 was used to estimate the pcor between gene pairs. Pcors from all iterations were recorded. With an average of 3 min, 10 sec per iteration on a PC (Intel Core2 E6420 processor), one round of 2000 iterations consumes ∼4 d. For each gene pair the pcor with the lowest absolute value was chosen as the final value. Supplemental Table 4 lists the significant interactions with absolute values of estimated pcors larger or equal to 0.10 used to construct the network.
Permutation experiment
The raw intensity data set (after quantile normalization) with 22,266 genes from 2045 chips was used. For permutations of a gene, the intensity values for that gene in all 2045 chips were collected, and then randomly and nonrepeatedly assigned as the intensity values for that gene among the 2045 chips. In one experiment, the entire 22,266 genes were permutated, while in a second, 1000 randomly selected genes were permutated. The permutated data set were then subjected to the analysis procedure described before.
Network layout and visualization
Three methods were used. For the complete network (Fig. 3B), layout and visualizations were carried out using the tYNA platform (Yip et al. 2006) with the aiSee graph visualization software (http://www.aisee.com). Subnetworks were extracted by specifying seed node and number of connecting steps by which the subnetwork was expected to expand from the seed node. The extracted subnetworks were saved as dot files, which were visualized with the fdp program (Figs. 2, 4, 5; Supplemental Fig. S2) or the neato program (Fig. 6), both included in the software package Graphviz 2.8 (Gansner and North 2000). When using neato, the algorithm “Stress Majorization,” designed for large size, was used (Gansner et al. 2004). An R script for network query and visualization is included (Supplemental data file S1).
Acknowledgments
We thank the members of NASCArrays and the laboratories providing data for contributing to the database. Advice by Dr. K. Strimmer is gratefully acknowledged. The work was supported by grants from the National Science Foundation Plant Genome Project (DBI-0223905) and University of Illinois at Urbana-Champaign institutional grants. S.M. conceived the experimental approach and performed calculations. S.M., Q.G., and H.J.B. analyzed intermediary approaches to the problem and wrote the article.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6911207
References
- Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K., Agarwal P., Reddy M.K., Sopory S.K., Reddy M.K., Sopory S.K., Sopory S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Amaral L.A., Scala A., Barthelemy M., Stanley H.E., Scala A., Barthelemy M., Stanley H.E., Barthelemy M., Stanley H.E., Stanley H.E. Classes of small-world networks. Proc. Natl. Acad. Sci. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Shibata D., Ogata Y., Shibata D., Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Albert R., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Oltvai Z.N., Oltvai Z.N. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Baxter C.J., Redestig H., Schauer N., Repsilber D., Patil K.R., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Redestig H., Schauer N., Repsilber D., Patil K.R., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Schauer N., Repsilber D., Patil K.R., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Repsilber D., Patil K.R., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Patil K.R., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Nielsen J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Selbig J., Liu J., Fernie A.R., Sweetlove L.J., Liu J., Fernie A.R., Sweetlove L.J., Fernie A.R., Sweetlove L.J., Sweetlove L.J. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 2007;143:312–325. doi: 10.1104/pp.106.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T., Przemeck G.K., Stamatiou G., Lau R., Terryn N., De Rycke R., Inze D., Berleth T., Przemeck G.K., Stamatiou G., Lau R., Terryn N., De Rycke R., Inze D., Berleth T., Stamatiou G., Lau R., Terryn N., De Rycke R., Inze D., Berleth T., Lau R., Terryn N., De Rycke R., Inze D., Berleth T., Terryn N., De Rycke R., Inze D., Berleth T., De Rycke R., Inze D., Berleth T., Inze D., Berleth T., Berleth T. Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 2002;130:1883–1893. doi: 10.1104/pp.102.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C., Ohta H., Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 2005;280:2397–2400. doi: 10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- Brazhnik P., de la Fuente A., Mendes P., de la Fuente A., Mendes P., Mendes P. Gene networks: How to put the function in genomics. Trends Biotechnol. 2002;20:467–472. doi: 10.1016/s0167-7799(02)02053-x. [DOI] [PubMed] [Google Scholar]
- Breton G., Danyluk J., Charron J.B., Sarhan F., Danyluk J., Charron J.B., Sarhan F., Charron J.B., Sarhan F., Sarhan F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 2003;132:64–74. doi: 10.1104/pp.102.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Zeef L.A., Ellis J., Goodacre R., Turner S.R., Zeef L.A., Ellis J., Goodacre R., Turner S.R., Ellis J., Goodacre R., Turner S.R., Goodacre R., Turner S.R., Turner S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.J., Lieb J.D., Lieb J.D. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S., James N., Okyere J., Higgins J., Jotham J., May S., Okyere J., Higgins J., Jotham J., May S., Higgins J., Jotham J., May S., Jotham J., May S., May S. NASCArrays: A repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramirez A., Oropeza-Aburto A., Razo-Hernandez F., Ramirez-Chavez E., Herrera-Estrella L., Oropeza-Aburto A., Razo-Hernandez F., Ramirez-Chavez E., Herrera-Estrella L., Razo-Hernandez F., Ramirez-Chavez E., Herrera-Estrella L., Ramirez-Chavez E., Herrera-Estrella L., Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. 2006;103:6765–6770. doi: 10.1073/pnas.0600863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H., Yang G., Zheng Z.L., Yang G., Zheng Z.L., Zheng Z.L. A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 2007;63:221–235. doi: 10.1007/s11103-006-9084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A., Brazhnik P., Mendes P., Brazhnik P., Mendes P., Mendes P. Linking the genes: Inferring quantitative gene networks from microarray data. Trends Genet. 2002;18:395–398. doi: 10.1016/s0168-9525(02)02692-6. [DOI] [PubMed] [Google Scholar]
- Efron B. Large-scale simultaneous hypothesis testing: The choice of a null hypothesis. J. Am. Stat. Assoc. 2004;99:96–104. [Google Scholar]
- Efron B. Correlation and large-scale simultaneous significance testing. J. Am. Stat. Assoc. 2007;102:93–103. [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F., Cook D., Thomashow M.F., Thomashow M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshikawa Y., Sato T., Inada N., Ito M., Nishida I., Watanabe A., Yoshikawa Y., Sato T., Inada N., Ito M., Nishida I., Watanabe A., Sato T., Inada N., Ito M., Nishida I., Watanabe A., Inada N., Ito M., Nishida I., Watanabe A., Ito M., Nishida I., Watanabe A., Nishida I., Watanabe A., Watanabe A. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 2001;111:345–352. doi: 10.1034/j.1399-3054.2001.1110312.x. [DOI] [PubMed] [Google Scholar]
- Gachon C.M., Langlois-Meurinne M., Henry Y., Saindrenan P., Langlois-Meurinne M., Henry Y., Saindrenan P., Henry Y., Saindrenan P., Saindrenan P. Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: Functional and evolutionary implications. Plant Mol. Biol. 2005;58:229–245. doi: 10.1007/s11103-005-5346-5. [DOI] [PubMed] [Google Scholar]
- Gansner E.R., North S.C., North S.C. An open graph visualization system and its applications to software engineering. Software-Practice & Experience. 2000;30:1203–1233. [Google Scholar]
- Gansner E.R., Koren Y., North S., Koren Y., North S., North S. Graph drawing by stress majorization. Graph Drawing. 2004;3383:239–250. [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Rick J.M., Michon A.M., Cruciat C.M., Michon A.M., Cruciat C.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Ellis B., Gautier L., Ge Y., Gentry J., Gautier L., Ge Y., Gentry J., Ge Y., Gentry J., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Fowler S.G., Thomashow M.F., Fowler S.G., Thomashow M.F., Thomashow M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Ooi C.E., Godwin B., Vitols E., Godwin B., Vitols E., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A., Hansen B.G., Olsen C.E., Halkier B.A., Olsen C.E., Halkier B.A., Halkier B.A. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Chen W., Estes B., Chang H.S., Nawrath C., Metraux J.P., Zhu T., Katagiri F., Chen W., Estes B., Chang H.S., Nawrath C., Metraux J.P., Zhu T., Katagiri F., Estes B., Chang H.S., Nawrath C., Metraux J.P., Zhu T., Katagiri F., Chang H.S., Nawrath C., Metraux J.P., Zhu T., Katagiri F., Nawrath C., Metraux J.P., Zhu T., Katagiri F., Metraux J.P., Zhu T., Katagiri F., Zhu T., Katagiri F., Katagiri F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez R.A., Lejay L.V., Dean A., Chiaromonte F., Shasha D.E., Coruzzi G.M., Lejay L.V., Dean A., Chiaromonte F., Shasha D.E., Coruzzi G.M., Dean A., Chiaromonte F., Shasha D.E., Coruzzi G.M., Chiaromonte F., Shasha D.E., Coruzzi G.M., Shasha D.E., Coruzzi G.M., Coruzzi G.M. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M.Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Araki R., Sakurai N., Suzuki H., Aoki K., Sakurai N., Suzuki H., Aoki K., Suzuki H., Aoki K., Aoki K., et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. New light on the correlation coefficient and its transforms. J. Royal Stat. Soc. Series B-Stat. Meth. 1953;15:193–232. [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N., Mason S.P., Barabasi A.L., Oltvai Z.N., Barabasi A.L., Oltvai Z.N., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H., Galon Y., Knight M.R., Fluhr R., Fromm H., Knight M.R., Fluhr R., Fromm H., Fluhr R., Fromm H., Fromm H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell. 2006;18:2733–2748. doi: 10.1105/tpc.106.042713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanin R., Wit E., Wit E. How scale-free are biological networks. J. Comput. Biol. 2006;13:810–818. doi: 10.1089/cmb.2006.13.810. [DOI] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H., Khanna R., Quail P.H., Quail P.H. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K., Bornberg-Bauer E., Kudla J., Harter K., Kudla J., Harter K., Harter K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kishino H., Waddell P.J., Waddell P.J. Correspondence analysis of genes and tissue types and finding genetic links from microarray data. Genome Inform. Ser. Workshop Genome Inform. 2000;11:83–95. [PubMed] [Google Scholar]
- Kreps J.A., Simon A.E., Simon A.E. Environmental and genetic effects on circadian clock-regulated gene expression in Arabidopsis. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Fletcher J.C., Fletcher J.C. Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Ledger S., Strayer C., Ashton F., Kay S.A., Putterill J., Strayer C., Ashton F., Kay S.A., Putterill J., Ashton F., Kay S.A., Putterill J., Kay S.A., Putterill J., Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Harbison C.T., Thompson C.M., Simon I., Thompson C.M., Simon I., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P., Hsu A.K., Sajdak J., Qin J., Pavlidis P., Sajdak J., Qin J., Pavlidis P., Qin J., Pavlidis P., Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gui J., Gui J. Gradient directed regularization for sparse Gaussian concentration graphs, with applications to inference of genetic networks. Biostatistics. 2006;7:302–317. doi: 10.1093/biostatistics/kxj008. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H., Mongrand S., Kinoshita N., Chua N.H., Kinoshita N., Chua N.H., Chua N.H. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes & Dev. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene P.M., Kim J., Kim J. Estimating genomic coexpression networks using first-order conditional independence. Genome Biol. 2004;5:R100. doi: 10.1186/gb-2004-5-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins A.M., Sha W., Evans C., Martino-Catt S., Mendes P., Shulaev V., Sha W., Evans C., Martino-Catt S., Mendes P., Shulaev V., Evans C., Martino-Catt S., Mendes P., Shulaev V., Martino-Catt S., Mendes P., Shulaev V., Mendes P., Shulaev V., Shulaev V. Comparison of sampling techniques for parallel analysis of transcript and metabolite levels in Saccharomyces cerevisiae. Yeast. 2007;24:181–188. doi: 10.1002/yea.1442. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K., Shinozaki K., Yamaguchi-Shinozaki K., Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- Misson J., Raghothama K.G., Jain A., Jouhet J., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Raghothama K.G., Jain A., Jouhet J., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Jain A., Jouhet J., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Jouhet J., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Block M.A., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Bligny R., Ortet P., Creff A., Somerville S., Rolland N., Ortet P., Creff A., Somerville S., Rolland N., Creff A., Somerville S., Rolland N., Somerville S., Rolland N., Rolland N., et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K., Lee H., Stevenson B., Zhu J.K., Stevenson B., Zhu J.K., Zhu J.K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Awai K., Masuda T., Yoshioka Y., Takamiya K., Ohta H., Awai K., Masuda T., Yoshioka Y., Takamiya K., Ohta H., Masuda T., Yoshioka Y., Takamiya K., Ohta H., Yoshioka Y., Takamiya K., Ohta H., Takamiya K., Ohta H., Ohta H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 2005;280:7469–7476. doi: 10.1074/jbc.M408799200. [DOI] [PubMed] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R., Kempa S., Adamik M., Hesse H., Hoefgen R., Adamik M., Hesse H., Hoefgen R., Hesse H., Hoefgen R., Hoefgen R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- Nikiforova V.J., Daub C.O., Hesse H., Willmitzer L., Hoefgen R., Daub C.O., Hesse H., Willmitzer L., Hoefgen R., Hesse H., Willmitzer L., Hoefgen R., Willmitzer L., Hoefgen R., Hoefgen R. Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J. Exp. Bot. 2005;56:1887–1896. doi: 10.1093/jxb/eri179. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H., Saeki M., Shibata D., Saito K., Ohta H., Shibata D., Saito K., Ohta H., Saito K., Ohta H., Ohta H. ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gki783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh I.S., Park A.R., Bae M.S., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Park A.R., Bae M.S., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Bae M.S., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K., Kang N.Y., Lee S., Cheong H., Park O.K., Lee S., Cheong H., Park O.K., Cheong H., Park O.K., Park O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. 2005;17:2832–2847. doi: 10.1105/tpc.105.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Wang X., Bader J.S., Boeke J.D., Bader J.S., Boeke J.D., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G.P., Somerville C.R., Wei H., Milne J., Page G.P., Somerville C.R., Milne J., Page G.P., Somerville C.R., Page G.P., Somerville C.R., Somerville C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome P.A., McClung C.R., McClung C.R. The Arabidopsis thaliana clock. J. Biol. Rhythms. 2004;19:425–435. doi: 10.1177/0748730404268112. [DOI] [PubMed] [Google Scholar]
- Schäfer J., Strimmer K., Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005a;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- Schäfer J., Strimmer K., Strimmer K. Learning large-scale graphical Gaussian models from genomic data. In: Mendes J., et al., editors. Proceedings of “Science of Complex Networks: From Biology to the Internet and WWW” (CNET 2004) The American Institute of Physics; Aveiro, Portugal: 2005b. [Google Scholar]
- Schäfer J., Strimmer K., Strimmer K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 2005c;4 doi: 10.2202/1544-6115.1175. Article32. [DOI] [PubMed] [Google Scholar]
- Schäfer J., Opgen-Rhein R., Strimmer K., Opgen-Rhein R., Strimmer K., Strimmer K. Reverse engineering genetic networks using the GeneNet package. R News. 2006;6:50–53. http://cran.r-project.org/doc/Rnews. [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Vingron M., Scholkopf B., Weigel D., Lohmann J.U., Scholkopf B., Weigel D., Lohmann J.U., Weigel D., Lohmann J.U., Lohmann J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Sokolov L.N., Dominguez-Solis J.R., Allary A.L., Buchanan B.B., Luan S., Dominguez-Solis J.R., Allary A.L., Buchanan B.B., Luan S., Allary A.L., Buchanan B.B., Luan S., Buchanan B.B., Luan S., Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. 2006;103:9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J., Williams L.A., Farmer L.M., Vierstra R.D., Callis J., Farmer L.M., Vierstra R.D., Callis J., Vierstra R.D., Callis J., Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic qcid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H., Horimoto K., Horimoto K. Inference of a genetic network by a combined approach of cluster analysis and graphical Gaussian modeling. Bioinformatics. 2002;18:287–297. doi: 10.1093/bioinformatics/18.2.287. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Young J., Berriz G.F., Brost R.L., Chang M., Berriz G.F., Brost R.L., Chang M., Brost R.L., Chang M., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wei H., Persson S., Mehta T., Srinivasasainagendra V., Chen L., Page G.P., Somerville C., Loraine A., Persson S., Mehta T., Srinivasasainagendra V., Chen L., Page G.P., Somerville C., Loraine A., Mehta T., Srinivasasainagendra V., Chen L., Page G.P., Somerville C., Loraine A., Srinivasasainagendra V., Chen L., Page G.P., Somerville C., Loraine A., Chen L., Page G.P., Somerville C., Loraine A., Page G.P., Somerville C., Loraine A., Somerville C., Loraine A., Loraine A. Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 2006;142:762–774. doi: 10.1104/pp.106.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker J. Graphical models in applied multivariate statistics. Wiley; New York: 1990. [Google Scholar]
- Wille A., Buhlmann P., Buhlmann P. Low-order conditional independence graphs for inferring genetic networks. Stat. Appl. Genet. Mol. Biol. 2006;5 doi: 10.2202/1544-6115.1170. Article1. [DOI] [PubMed] [Google Scholar]
- Wille A., Zimmermann P., Vranova E., Furholz A., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Zimmermann P., Vranova E., Furholz A., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Vranova E., Furholz A., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Furholz A., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Hennig L., Prelic A., von Rohr P., Thiele L., Prelic A., von Rohr P., Thiele L., von Rohr P., Thiele L., Thiele L., et al. Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. Genome Biol. 2004;5:R92. doi: 10.1186/gb-2004-5-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.J., Bowles D.J., Bowles D.J. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14:1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.K., Tegner J., Collins J.J., Tegner J., Collins J.J., Collins J.J. Reverse engineering gene networks using singular value decomposition and robust regression. Proc. Natl. Acad. Sci. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip K.Y., Yu H., Kim P.M., Schultz M., Gerstein M., Yu H., Kim P.M., Schultz M., Gerstein M., Kim P.M., Schultz M., Gerstein M., Schultz M., Gerstein M., Gerstein M. The tYNA platform for comparative interactomics: A web tool for managing, comparing and mining multiple networks. Bioinformatics. 2006;22:2968–2970. doi: 10.1093/bioinformatics/btl488. [DOI] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C., Xu C., Benning C., Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xia Y., Trifonov V., Gerstein M., Xia Y., Trifonov V., Gerstein M., Trifonov V., Gerstein M., Gerstein M. Design principles of molecular networks revealed by global comparisons and composite motifs. Genome Biol. 2006;7:R55. doi: 10.1186/gb-2006-7-7-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yugi K., Nakayama Y., Kojima S., Kitayama T., Tomita M., Nakayama Y., Kojima S., Kitayama T., Tomita M., Kojima S., Kitayama T., Tomita M., Kitayama T., Tomita M., Tomita M. A microarray data-based semi-kinetic method for predicting quantitative dynamics of genetic networks. BMC Bioinformatics. 2005;6:299. doi: 10.1186/1471-2105-6-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z.H., Demura T., Ye Z.H., Ye Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W., Hirsch-Hoffmann M., Hennig L., Gruissem W., Hennig L., Gruissem W., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]