Abstract
Rhomboids are a recently discovered family of widely distributed intramembrane serine proteases. They have diverse biological functions, including the regulation of growth factor signaling, mitochondrial fusion, and parasite invasion. Despite their existence in all branches of life, the sequence identity between rhomboids is low. We have combined BLAST-based database mining with functional and structural data to generate a comprehensive genomic analysis of eukaryotic rhomboid-like proteins. We show that robust membrane topology models are necessary to classify active rhomboid proteases unambiguously, and we define rules for distinguishing predicted active proteases from the larger evolutionary group of rhomboid-like proteins. This leads to a revision of estimates of numbers of proteolytically active rhomboids. We identify three groups of eukaryotic rhomboid-like proteins: true active rhomboids, a tightly clustered group of novel inactive rhomboids that we name the iRhoms, and a small number of other inactive rhomboid-like proteins. The active proteases are themselves subdivided into secretase and PARL-type (mitochondrial) subfamilies; these have distinct transmembrane topologies. This enhanced genomic analysis leads to conclusions about rhomboid enzyme function. It suggests that a given rhomboid can only cleave a single orientation of substrate, and that both products of rhomboid catalyzed intramembrane cleavage can be released from the membrane. Our phylogeny predictions also have evolutionary implications: Despite the complex classification of rhomboids, our data suggest that a rhomboid-type intramembrane protease may have been present in the last eukaryotic common ancestor.
Intramembrane proteolysis has over the last few years become recognized as an important cellular regulatory mechanism. Intramembrane proteases fall into three mechanistic classes, the S2P metalloproteases, the GxGD-type aspartyl proteases, including presenilin/γ-secretase and SPP, and the rhomboid serine proteases (for reviews, see Brown et al. 2000; Weihofen and Martoglio 2003; Wolfe and Kopan 2004). The rhomboid gene was first discovered in Drosophila, where it was named after an embryonic mutant phenotype (Mayer and Nusslein-Volhard 1988). More recently, Drosophila Rhomboid-1 was shown to be the founding member of a class of polytopic membrane proteins conserved throughout evolution (Wasserman et al. 2000; Koonin et al. 2003). Genetic and cell biological analysis revealed that rhomboids are intramembrane serine proteases (Urban et al. 2001). Drosophila Rhomboid-1 cleaves membrane-tethered growth factor precursors, releasing the active form and triggering their secretion; thereby, it is the primary activator of epidermal growth factor receptor (EGFR) signaling (Lee et al. 2001; Urban et al. 2002a). The Caenorhabditis elegans rhomboid ROM1 has similarly been implicated in EGFR control (Dutt et al. 2004).
In other eukaryotic species, much less is known about the role of intramembrane proteolysis by rhomboids, but there is evidence for significant functions in a variety of contexts. For example, in the apicomplexan parasites Plasmodium falciparum and Toxoplasma gondii, rhomboids are involved in the shedding of adhesion molecules and have been implicated in host cell invasion (Brossier et al. 2005; Dowse et al. 2005; Baker et al. 2006; O’Donnell et al. 2006). In the yeast Saccharomyces cerevisiae, Drosophila, and mammals, a subclass of rhomboids located in the inner mitochondrial membrane has recently been the focus of attention (Esser et al. 2002; Herlan et al. 2003; McQuibban et al. 2003). In S. cerevisiae, the mitochondrial rhomboid Pcp1 (or Rbd1) controls mitochondrial membrane fusion by cleaving the dynamin-like GTPase Mgm1 (Herlan et al. 2003, 2004; McQuibban et al. 2003). Pcp1/Rbd1 is conserved across eukaryotes, and related but not identical functions have been shown for the orthologs in Drosophila (Rhomboid-7) and mice (PARL) (Cipolat et al. 2006; McQuibban et al. 2006). Finally, two putative substrates (thrombomodulin and ephrin-B3) for mammalian nonmitochondrial rhomboids were identified by candidate testing, although their physiological significance remains unclear (Lohi et al. 2004; Pascall and Brown 2004).
There has been much recent progress in the molecular understanding of rhomboid function, and how these enzymes perform the unusual cleavage of peptide bonds in the hydrophobic plane of the cellular membrane. Rhomboid activity has been reconstituted in vitro, enabling mechanistic questions to be addressed (Lemberg et al. 2005; Maegawa et al. 2005; Urban and Wolfe 2005). Complementary to this functional analysis, high-resolution structures of the Escherichia coli rhomboid GlpG have recently provided insight into its architecture (Wang et al. 2006; Wu et al. 2006; Ben-Shem et al. 2007; Lemieux et al. 2007). Although much remains to be resolved, these studies allow predictions about how one class of rhomboids act, revealing a dyad between a conserved serine and histidine in their catalytic center, with subsidiary functions in other domains (Lemberg et al. 2005; Wang et al. 2006). The molecular structure function prediction is, however, hampered by the diversity of the rhomboid family.
Many genes have been annotated as rhomboid proteases by sequence similarity algorithms such as PSI-BLAST and by hidden Markov models (Wasserman et al. 2000; Bateman et al. 2002; Koonin et al. 2003; Brossier et al. 2005; Dowse and Soldati 2005), but false positives are found. Although it has been stated that the rhomboids are uniquely conserved among polytopic membrane proteins (Koonin et al. 2003), sequence similarity over the entire length of distant homologs is actually quite low and the limited constraints of the rhomboid protease domain hampers the classification of active rhomboid proteases. In this article, we have exploited recent understanding of rhomboid structure and mechanism to enhance BLAST-based predictions. From this we derive a new stringent and function-based definition of rhomboids that depends on multiple sources of evidence, enabling comprehensive and accurate annotation of related sequences. As well as providing the first robust classification of rhomboids, we report a novel subfamily of highly conserved inactive rhomboid-like proteins, which we name iRhoms. This functionally enhanced genomic analysis also leads to mechanistic and evolutionary conclusions about rhomboid enzymes. Notably, we propose that rhomboids can only cleave substrates in a single membrane orientation but can release both N- and C-terminal protein domains from substrates.
Results and Discussion
The minimum consensus sequence for rhomboid proteases
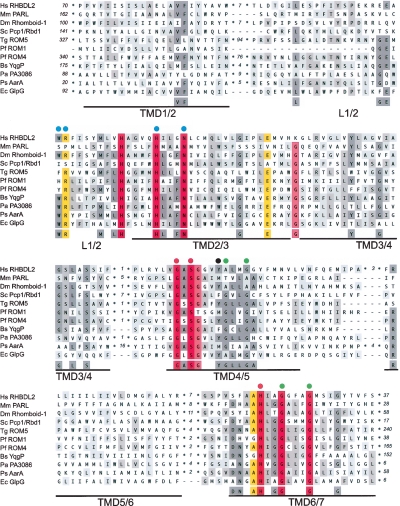
We aligned the sequences of the subset of predicted rhomboids that have been functionally studied in mutagenesis experiments to determine the minimum sequence requirements for the active enzymes. Alignment of the full-length proteins by automatic methods, including ClustalW (Thompson et al. 1994), T-Coffee (Notredame et al. 2000), and MUSCLE (Edgar 2004), is unsatisfactory due to the heterogeneity of tails and sequence insertions (data not shown). Multiple sequence alignment of just the conserved membrane-integral portion shows that although all transmembrane domains (TMDs) can be aligned, substantial conservation is only observed in a few regions, specifically the active site formed by the serine protease motif (GxSx in TMD4 and H in TMD6) and a domain of unknown function (in the L1 loop and TMD2) with a prominent tryptophan-arginine motif (WR) (Fig. 1; Urban et al. 2001; Lemberg et al. 2005). Recent crystal structures of the E. coli rhomboid GlpG confirm that these residues have structural and functional significance (for details, see Fig. 1; Wang et al. 2006).
Figure 1.
Functionally based rhomboid protease consensus. Conserved membrane integral portion of rhomboids that have been used in mutagenesis experiments (Urban et al. 2001, 2002b; Esser et al. 2002; McQuibban et al. 2003; Brossier et al. 2005; Lemberg et al. 2005; Maegawa et al. 2005; Urban and Wolfe 2005; Baker et al. 2006; Cipolat et al. 2006; O’Donnell et al. 2006; Stevenson et al. 2007) were aligned. For human (Homo sapiens, Hs) RHBDL2, Drosophila melanogaster (Dm) Rhomboid-1, Toxoplasma gondii (Tg) ROM5, Plasmodium falciparum (Pf) ROM1 and ROM4, Escherichia coli (Ec) GlpG, Providencia stuartii (Ps) AarA, Pseudomonas aeruginosa (Pa) rhomboid PA3086, and Bacillus subtilis (Bs) YqgP TMD1 to TMD6 are shown; for mouse (Mus musculus, Mm) PARL and Saccharomyces cerevisiae (Sc) Pcp1/Rbd1, the topologically equivalent TMD2 to TMD7 were aligned. For accession numbers see below. Alignment was performed by ClustalW and corrected manually to remove gaps in predicted TMDs and loops extending the sequence of the E. coli GlpG rhomboid. The more variable N- and C-terminal domains (including additional TMDs) were excluded from the alignment (number of amino acids is indicated). TMD predictions were based on the structure of the E. coli rhomboid GlpG and are underlined (Wang et al. 2006). The L1 loop (L2 for PARL-type rhomboids) containing widely conserved and functional important residues is indicated; invariant residues forming the active site (GxSx and H) are labeled by red dots; residues affecting the activity only in certain rhomboids or experimental conditions are highlighted by blue dots; position of a tyrosine residue of the E. coli GlpG rhomboid (Y205) suggested to be involved in positioning of the catalytic histidine (H254) (Wang et al. 2006) is labeled by a black dot; small residues such as glycines (G) and alanines (A) in TMD4/6 and TMD6/7 that allow tight helix packing are labeled by green dots (Ben-Shem et al. 2007). The background color reflects the degree of identity/similarity of sequence alignment (100%, red; 90%–99%, light-red; 80%–89%, yellow; 50%–79%, dark gray; 30%–49%, light gray). For accession numbers for the eukaryotic rhomboids, see Figure 3 and Table S1. The accession number for E. coli GlpG is Swiss-Prot:P09391; P. stuartii AarA is Swiss-Prot:P46116; P. aeruginosa PA3086 is Swiss-Prot:Q9HZC2; and B. subtilis YqgP is Swiss-Prot:P54493.
This alignment emphasizes that the rhomboid protease consensus is very restricted. Notably, similar sequence motifs are found in unrelated polytopic membrane proteins. For instance, a GxSx-sequence similar to the rhomboid active site consensus is common in TMD5 of the Sec61/SecY superfamily (Van den Berg et al. 2004), a coincidence with no expected functional implication. The limited protease consensus, plus the existence of many apparently proteolytically inactive rhomboid-like proteins, makes it challenging to classify the entire family of active rhomboid proteases by simple sequence comparison; the BLAST E-value threshold has to be set low, and consequently, genes that lack key rhomboid protease characteristics are hit (for examples, see Table S1). To improve the efficiency of classifying active rhomboid proteases, the context of the conserved motifs and the topology of the protein must be taken into account (see below). Similar conclusions were also reached in the case of G-protein-coupled receptors (Hedman et al. 2002).
Refining rhomboid topology
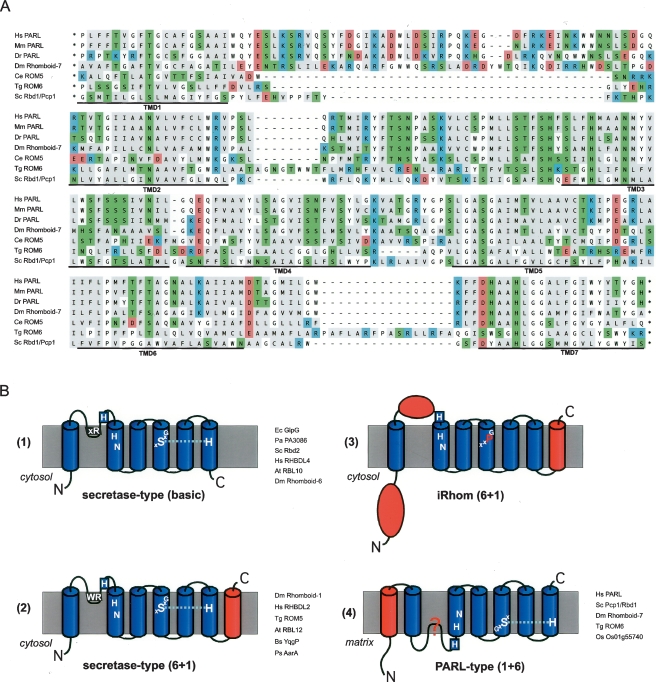
The need to position conserved rhomboid sequences in the context of overall TMD topology highlights the need to predict rhomboid TMDs with precision. Koonin et al. (2003) have proposed that rhomboids adopt three different topologies: bacterial and archaeal rhomboid having a basic six TMD-core, most eukaryotic rhomboids having a seventh TMD fused to the C terminus (6 + 1), and a subfamily of eukaryotic rhomboids (named after the human PARL and subsequently shown to be mitochondrial) (Esser et al. 2002; Herlan et al. 2003; McQuibban et al. 2003) with a seventh TMD fused to the N terminus (1 + 6) (Koonin et al. 2003). Confusion arises, however, for the experimentally well-studied PARL homolog in yeast, Pcp1/Rbd1, and the predicted T. gondii ortholog, ROM6, in which six TMDs have been proposed (Koonin et al. 2003; McQuibban et al. 2003; Dowse and Soldati 2005). If true, this would suggest that topology has not been conserved within the PARL subfamily, in turn suggesting that specific topology may not be fundamental to rhomboid function. We therefore decided to re-examine the topology of PARL and its orthologs from mouse, zebrafish, Drosophila melanogaster, C. elegans, T. gondii, and S. cerevisiae.
TMD prediction, particularly in polytopic membrane proteins, is imprecise so we compared the results of four TMD-prediction programs. This has been shown to be a valid way of judging the reliability of predicted topology models (Nilsson et al. 2000; Friedmann et al. 2004). Although these algorithms were designed for proteins in the secretory pathway and the mechanism of import of mitochondrial membrane proteins is less well understood, it is expected that translocation-mediated recognition of TMDs is based on similar principles, justifying this approach (von Heijne 2006). Not all the algorithms predict all TMDs, but combining these results according to a “major-vote principle” (Nilsson et al. 2000) and superimposing the six TMD-core on the known structure of GlpG, increases the quality of the topology model (see Fig. 1) and supports a universal seven TMD structure for PARL-type rhomboids (Fig. 2A; Table S2). Within this framework, the few TMDs that are not predicted by any program, such as TMD2 of C. elegans PARL (ROM5), can nevertheless be clearly aligned; an aspartate (D), a charged residue not common in TMDs, explains the prediction failure. This comparative analysis predicts an extra TMD in S. cerevisiae Pcp1/Rbd1 and T. gondii ROM6, making them similar to other members of the PARL subfamily; this has implications for rhomboid function (see below).
Figure 2.
Rhomboid topology. (A) Multiple-sequence alignment of the membrane integral portion of PARL-type rhomboids from human (Homo sapiens, Hs), mouse (Mus musculus, Mm), zebrafish (Danio rerio, Dr), Drosophila melanogaster (Dm; Rhomboid-7), Caenorhabditis elegans (Ce; named ROM5 by automated annotation), Toxoplasma gondii (Tg; ROM6), and Saccharomyces cerevisiae (Sc; Pcp1/Rbd1). Assuming the overall protein architecture is conserved, we manually corrected gaps in predicted TMDs of the ClustalW-based alignment. Typically for rhomboids, the TMDs have a high content of polar amino acids, which occur predominantly in conserved positions. In the alignment, the functional characteristics of the amino acids are indicated by background color (acidic, red; strong basic, blue; polar and weak basic, green; and hydrophobic, gray). (B) Topology models for different rhomboid proteases and catalytically inert iRhoms; extra domains fused to the basic six TMD rhomboid core are highlighted in red; the key conserved residues and the L1 structure extending sidewise in the membrane are indicated (Urban et al. 2001; Lemberg et al. 2005; Wang et al. 2006); examples of bacterial and eukaryotic rhomboids are listed. (For accession numbers, see Table S1; Figs. 1, 3.)
A new classification of rhomboid topologies
By modifying previous rhomboid topology models (Urban et al. 2001; Koonin et al. 2003; Maegawa et al. 2005), we now suggest four different topological classes for rhomboid-like proteins (Fig. 2B). The basic class of a six-TMD core is found in E. coli GlpG and some eukaryotic rhomboids such as S. cerevisiae Rbd2 (YPL246C) (Daley et al. 2005; Maegawa et al. 2005; Kim et al. 2006). The next class, with Drosophila Rhomboid-1 as its most studied member, has a putative extra TMD fused to the C terminus and a variable N-terminal domain (Urban et al. 2001). In contrast with a previous proposal (Koonin et al. 2003), we note that this topology is not unique to eukaryotes: Many bacterial rhomboids are predicted to have a 6 + 1 TMD structure (Fig. 2B). The third class is characterized by a large globular domain inserted into the L1 loop and variations in the active site (see below). Note that all these three classes can have additional globular domains, fused either to the N or C termini (Table S3). Finally, the PARL-subfamily has a predicted extra TMD fused to the N terminus of the rhomboid core, thereby changing the position of the catalytic residues to TMD5 and TMD7 (instead of TMD4 and TMD6 in other rhomboids) (Fig. 2A); PARLs also have long N-terminal extensions (Jeyaraju et al. 2006). Taken together, this clearly shows that substantial diversification between different rhomboid proteases has occurred, and it remains to be addressed how extra TMDs affect the structure and function of more complex rhomboids.
How many rhomboid proteases are there in key species?
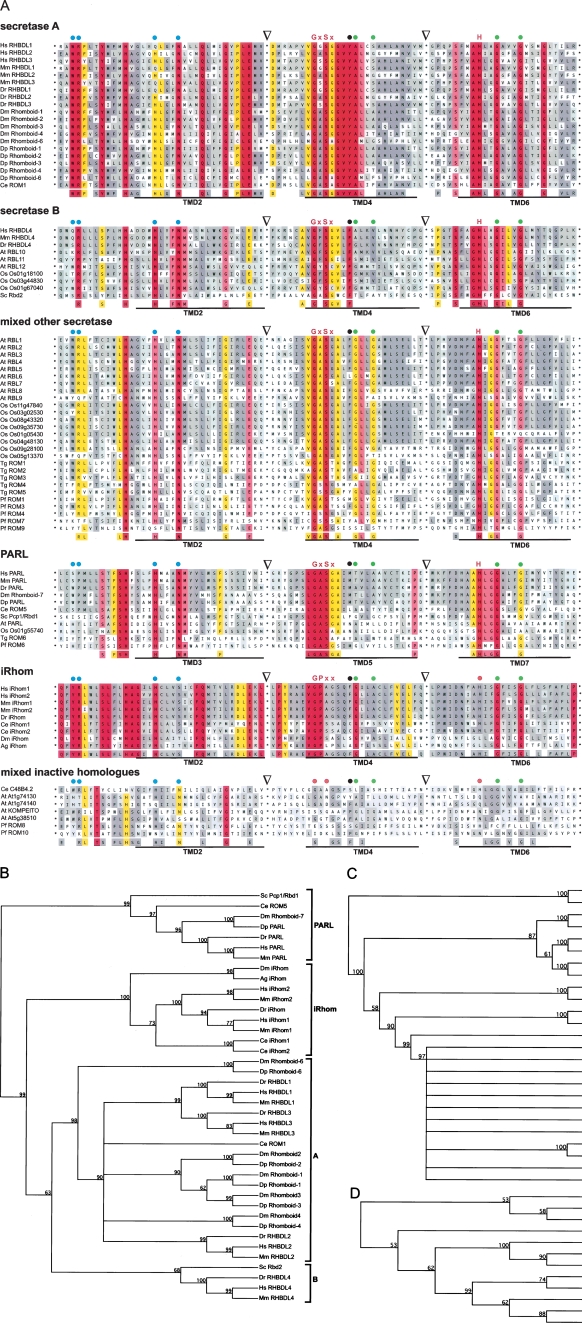
In order to generate a complete list of active rhomboid proteases for significant model organisms and to remove falsely annotated genes, we have exploited our new definitions of rhomboids. We propose defining as “rhomboids” only genes that are predicted to encode catalytically active proteases; homologs that do not share the protease consensus are more broadly defined as “rhomboid-like genes” (see below). In evolutionary terms, we are thus distinguishing the evolutionarily coherent rhomboid-like family from the functionally active rhomboid proteases. The rhomboid-like family is operationally defined as genes identified by sequence homology; the rhomboid proteases are a subset that includes only genes with all necessary features for predicted proteolytic activity. The steps in this process were as follows: (1) BLAST-based homology search using the core domain of unambiguous rhomboid proteases (as listed in Fig. 1; for details, see Methods). We searched against the nonredundant NCBI protein database (by PSI-BLAST) and the Ensembl and MIPS databases (by BLAST) and analyzed sequences obtained for rhomboid characteristics. (2) We constructed manually adjusted topology models (as in Fig. 2). (3) We examined candidates to determine whether the minimal rhomboid-protease consensus (GxSx and H) fits the six TMD protease core (i.e., do the catalytic residues lie in a topologically appropriate position?). (4) Finally, we looked for the presence of additional conserved features, such as the residues characteristic of L1/TMD2 (see Fig. 1). In order not to lose any more distantly related but bona fide rhomboids, the last step was not applied absolutely. A complete list of the rhomboid proteases thus defined in humans, mouse, zebrafish, Drosophila, C. elegans, S. cerevisiae, P. falciparum, T. gondii, Arabidopsis, and rice (Oryza sativa) is given in Figure 3A. Revising previous suggestions, we find five putative rhomboid proteases in humans, mice, and zebrafish (Danio rerio); six in Drosophila, six in P. falciparum; two in C. elegans; 13 in Arabidopsis; and 12 in rice (O. sativa). In agreement with previous reports, we find six rhomboid homologs in T. gondii and two in S. cerevisiae. This stringent approach has allowed us to remove genes that had been previously suggested to be rhomboid proteases by BLAST-based sequence comparison and Pfam automated gene annotation, which relies on hidden Markov models (for details, see Table S1); some of these are closely related inactive homologs that lack only key catalytic residues (Fig. 3A; see discussion below). Note that we cannot rule out splice variants of some of the inactive rhomboid-like proteins found in poorly annotated genome sequences, and these could in principle include the complete protease consensus. Apart from this caveat, we are confident that all rhomboid proteases in these species have now been identified.
Figure 3.
Phylogeny of eukaryotic rhomboids. Phylogenetic and functionally based analysis of rhomboid proteases and catalytically inactive homologs from human (Homo sapiens, Hs), mouse (Mus musculus, Mm), zebrafish (Danio rerio, Dr), Drosophila melanogaster (Dm), Drosophila pseudoobscura (Dp), Caenorhabditis elegans (Ce), Saccharomyces cerevisiae (Sc), Toxoplasma gondii (Tg), Plasmodium falciparum (Pf), Arabidopsis thaliana (At), and rice (Oryza sativum, Os). For accession numbers see below. (A) Multiple-sequence alignment of the conserved region according to structure-based TMD prediction (see Fig. 1). The sequences are classified into secretase-type (A, B, and other), PARL-type, iRhoms, and mixed inactive homologs. For secretase rhomboids, iRhoms and inactive homologs the C-terminal portion of L1, TMD2, TMD4, and TMD6 were used for the alignment; for PARL and its orthologs the topological equivalent portion of L2, TMD3, TMD5, and TMD7 are shown; the junctions of artificial splices are indicated by triangles. The degree of similarity is color-indicated as in Figure 1; for rhomboid proteases the key catalytic residues (GxSx and H) are highlighted (for iRhoms, the conserved GPxx instead); other functionally and structurally important positions are highlighted as described in Figure 1; TMDs are underlined. For a quantitative Two-Sample Logo comparison of the different subgroups, see Figure S1. Phylogenetic tree of rhomboid proteases and iRhoms in animals and yeast (B), plants (C), and apicomplexan parasites (D). Dendrograms were constructed by UPGMA analysis of the alignment in Figure 3A. Clades are indicated by square brackets; bootstrap values that represent the statistical significance are indicated in the internal nodes; orthology relationships of Arabidopsis and rice rhomboids (Tripathi and Sowdhamini 2006) not visible in the tree structure are indicated by braces.
Accession numbers: For human, mouse, and Arabidopsis rhomboids, see Table S1; for details of the rice genes, see MIPS plant genome database (http://mips.gsf.de/projects/plants/). The accession number for zebrafish (D. rerio, Dr) RHBDL1 is Ensembl:ENSDARP00000082440, Dr RHBDL2 is Swiss-Prot:Q7ZUN9, Dr RHBDL3 is Swiss-Prot:Q566N3, Dr RHBDL4 is Swiss-Prot:Q568J3, and Dr PARL is Ensembl:ENSDARP00000057438; D. melanogaster (Dm) Rhomboid-1 is Swiss-Prot:P20350, Dm Rhomboid-2 is Swiss-Prot:Q86P37, Dm Rhomboid-3 is Swiss-Prot:Q9W0F8, Dm Rhomboid-4 is Swiss-Prot:Q9VYW6, Dm Rhomboid-6 is Swiss-Prot:Q86BL6, and Dm Rhomboid-7 (PARL) is Swiss-Prot:A1Z8R8; D. pseudoobscura (Dp) Rhomboid-1 is GenBank:EAL31292, Dp Rhomboid-2 is GenBank:EAL31289, Dp Rhomboid-3 is GenBank:EAL31296, Dp Rhomboid-4 is GenBank:EAL32611, Dp Rhomboid-6 is GenBank:EAL33827, and Dp PARL is GenBank:EAL25960; C. elegans (Ce) ROM1 is Swiss-Prot:Q19821 and Ce ROM5 (PARL) is GenBank:NP_491125; S. cerevisiae (Sc) Rbd2 is Swiss-Prot:Q12270 and Sc Pcp1/Rbd1 (PARL) is Swiss-Prot:P53259; T. gondii (Tg) ROM1 is Swiss-Prot:Q695U0, Tg ROM2 is Swiss-Prot:Q695T9, Tg ROM3 is Swiss-Prot:Q6IUY1, Tg ROM4 is Swiss-Prot:Q695T8, Tg ROM5 is Swiss-Prot:Q6GV23, and Tg ROM6 (PARL) is Swiss-Prot:Q2PP52; P. falciparum (Pf) ROM1 is GenBank:AAN35734, Pf ROM3 is GenBank:CAD51095, Pf ROM4 is GenBank:CAD51434, Pf ROM6 (PARL) is GenBank:CAD52576, Pf ROM7 is GenBank:CAD52703, and Pf ROM9 is GenBank:CAD51515. Note that there is no ROM2 and ROM5 annotated in P. falciparum (for details, see Dowse and Soldati 2005). For the accession numbers for the iRhoms, see Figure 5. The accession numbers for the inactive rhomboid homologs are Swiss-Prot:P34356 for Ce C48B4.2, GenBank:AAN36722 for Pf ROM8, and GenBank: CAG25001 for Pf ROM10; for the Arabidopsis proteins, see Table S1.
Rhomboid nomenclature
In conjunction with this genome-wide analysis, we propose some rationalization of rhomboid nomenclature (see Fig. 3A; Table S1) to avoid future confusion. We propose keeping established names of genes that have already been published, with the exception that running numbers in the name should be based on their appearance in the literature, which leads to a few alterations. Based on functional differences, we further suggest distinguishing PARL-type and secretase-type rhomboids. Since all species analyzed so far have only one copy of the PARL subfamily, the scope for confusion is not great, so we suggest that previously used names such as Drosophila Rhomboid-7 and S. cerevisiae Pcp1 be retained, as long as reference is made to these being of the PARL subfamily. Finally, we propose redefining the tightly clustered inactive subfamily (see below) as iRhoms instead of true rhomboids.
Phylogenetic relationship of eukaryotic rhomboid homologs
Having established a complete list of rhomboid proteases and putative inactive rhomboid-like proteins for various eukaryotes, we next questioned their phylogenetic relationships. We were prompted to revisit this by the observation that the two rhomboids in S. cerevisiae, Pcp1/Rbd1 and Rbd2, localize to mitochondria and secretory pathway respectively (Huh et al. 2003; McQuibban et al. 2003) yet had both been placed in the PARL subfamily (Koonin et al. 2003), which is now known to be mitochondrial. We wondered whether by using stringent alignments of functionally important regions of rhomboids, we could develop a phylogenetic tree that reflected the current understanding of rhomboids more fully, including the known subcellular localization (see Methods). Bootstrap analysis of our consensus tree shows that indeed all PARL-type rhomboids fall into one clade, but now places the second yeast rhomboid, Rbd2, in a different clade (Fig. 3B). This analysis also separated the non-PARL rhomboids into many subgroups, indicating a substantial diversification. To enable a better comparison between more closely related species, we analyzed parasites and plants separately, because they have very divergent rhomboids (see below). As shown in Figure 3B, this simplified phylogenetic tree shows four major clades: the PARL-type rhomboids; a major clade consisting of bona fide rhomboids (secretase-type A); a second clade of secretase rhomboids (B-type); and, finally, a clade of more distantly related rhomboids that lack catalytic residues; we have termed this last group the iRhoms and discuss them below. The sequence distinctions between the different rhomboid groups was analyzed and quantified using Two Sample Logo (Vacic et al. 2006; Fig. S1).
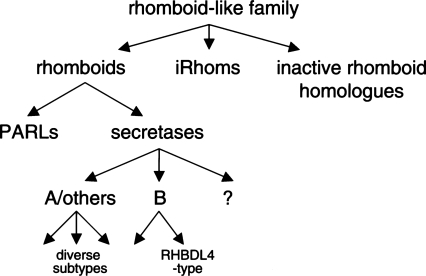
A few rhomboid-like proteins did not fit into any of these groups (Fig. 3A): By virtue of having mutated core residues, they are predicted to be catalytically inactive, but they do not cluster with the iRhoms (data not shown). These include, for example, C. elegans C48B4.2 (formerly ROM2 by automated annotation) and AT1G74130, AT1G74140, AT5G38510, and KOMPEITO from Arabidopsis. These do not form a coherent phylogenetic group (data not shown), and we believe them to be relatively recent mutations of active rhomboids (e.g., At1g74130 and At1g74140, which are not found in other plants, are presumably derived from Arabidopsis PARL (At1g18600)) (Tripathi and Sowdhamini 2006). We refer to these as inactive rhomboid-like genes but do not further classify them. Figure 4 provides a summary of the proposed classification of the rhomboid-like family. We now outline some features of the rhomboid-like groups and subfamilies and discuss the implications of this tree.
Figure 4.
Summary of classification of rhomboid-like proteins. For details, see text. The secretase rhomboids that do not cluster clearly with the A or B class may represent very divergent A-type or possibly additional, as yet unrecognized, classes.
PARL-type rhomboids
Members of this subfamily all have the 1 + 6 TMD topology discussed above and lack features typical of other rhomboids such as a characteristic arginine residue (R) in the L1/2 and a conserved glutamate residue (E) at the C-terminal junction of TMD2/3 (Figs. 3A, S1). Note that the WR motif found in some PARLs is located at a topologically distinct site within the predicted TMD2 (Fig. 2A), suggesting that it is not equivalent to the highly conserved and functionally important WR motif in the non-PARL rhomboids. Despite these differences, PARL-type rhomboids have all the hallmarks of the serine protease catalytic dyad, although the GxSx and histidine are shifted to TMDs 5 and 7, respectively (Fig. 3A). The biological significance of PARLs belonging to a distinct subfamily is supported by their high degree of overall sequence similarity (Table S4), their identical topology (Fig. 2; Table S2), and their predicted mitochondrial localization (Esser et al. 2002; Herlan et al. 2003, 2004; McQuibban et al. 2003, 2006; Cipolat et al. 2006). Furthermore, the substrate of PARL-type rhomboids in S. cerevisiae, Drosophila, and mouse appears to have been conserved (Herlan et al. 2003; McQuibban et al. 2003, 2006; Cipolat et al. 2006), suggesting that their function is also related. The branching within the PARL subgroup reflects the phylogenetic species tree. This adds weight to the validity of the model and suggests that PARL-type rhomboids may have derived from a common ancestor.
Secretase-type rhomboids
The secretase subfamily is so called because all its experimentally characterized members are located in the secretory pathway; it contains the majority of eukaryotic rhomboids. Although the sequence similarity within this subfamily is quite high, significant differences exist, and we find these proteins split into two clades (Figs. 3, S1). Secretase-A rhomboids have a 6 + 1 TMD topology described above, while secretase-B rhomboids have the six TMD core only (Fig. 2B). Note, however, that we find one exception in each class: Drosophila Rhomboid-6 has six TMDs, and Arabidopsis RBL12 and its rice ortholog Os01g67040 are predicted to have 6 + 1. These could represent annotation errors, but they may imply that the TMD topology distinction between the secretase-A and -B rhomboids is not absolute. Another notable distinction between the A- and B-classes is that the WR-motif in the L1 loop is strictly conserved in the A-class, whereas, with the exception of the more distant plant rhomboids RBL12 and Os01g67040, the B-class has only the conserved arginine (Figs. 3A, S1). The functional importance of this motif has been highlighted by genetic, biochemical, and structural studies, although its exact function remains unclear (Urban et al. 2001; Lemberg et al. 2005; Wang et al. 2006).
There are also clear distinctions between the A- and B-class secretase rhomboids in the sequence around the catalytic serine: There is a highly conserved GxSxGVYA sequence in the A-class compared with a slightly less rigid consensus of GxSxxxF in the B-class (Figs. 3A, S1). The tyrosine (Y) and phenylalanine (F) residues, which are implicated in the stabilization of the histidine within the serine protease catalytic dyad (Wang et al. 2006), also allow a clear distinction between A- and B-class (see Fig. S1). Another interesting variation is observed in the first x-position of the GxSx-motif of all vertebrate secretase rhomboids accessible by the Ensembl genome browser: There is a glycine (G) in RHBDL1 orthologs, an alanine (A) in RHBDL2 orthologs, a serine (S) in RHBDL3 orthologs, and a phenylalanine (F) in RHBDL4 orthologs (Fig. 3A; data not shown). We speculate that this position influences the activity or substrate specificity, as has been observed for a residue neighboring the active site of the intramembrane protease presenilin/γ-secretase (Yamasaki et al. 2006).
There has been much diversification within the secretase-A class of vertebrate rhomboids, but significant relationships can nevertheless be inferred (Fig. 3B). All Drosophila secretase rhomboids (Rhomboids-1, -2, -3, -4, and -6) fall into the secretase-A class. Consistent with their common function in EGFR control, Rhomboids-1, -2, and -3 are the most closely related; Rhomboid-4 has a minor role in EGFR control (M. Freeman, unpubl.) and is more divergent. Rhomboid-6 is the most divergent Drosophila secretase rhomboid and interestingly is the only one with no detectable function in EGFR control (M. Freeman, unpubl.).
The secretase-B rhomboids represent a previously unrecognized class. It contains S. cerevisiae Rbd2 and a group of orthologous rhomboids from human, mouse, and zebrafish (Fig. 3B). These orthologs are the founding members of a subclass of secretase-B rhomboids, which we name after mammalian RHBDL4 (Fig. 4; Table S1). RHBDL4-like rhomboids are found in all chordate genomes annotated by Ensembl and in Arabidopsis and rice (Fig. 3C; Table S5; data not shown). Despite the prediction of mitochondrial targeting (TargetP and MitoPred; for details, see Methods), immunofluorescence analysis in mammalian tissue culture cells reveals that RHBDL4 is localized in the secretory pathway (M.K. Lemberg and M. Freeman, unpubl.). Based on these results, we suggest that the RHBDL4-like rhomboids are a distinct subclass of rhomboids within the secretase-B class.
The wide distribution of RHBDL4 orthologs in evolution, combined with their distant relationship to yeast Rbd2, the only secretase-type rhomboid in yeast, suggests that the B subclass may be ancient. The observation that its members have only the core six TMDs is also consistent with them resembling an ancestral precursor, as is the appearance of some A-like characteristics in yeast Rbd2 such as the GASG active site motif (Fig. 3A). It is attractive to speculate that the more complex eukaryotic rhomboids may have derived from such a simple rhomboid (although this ancient form appears to have been lost in nematodes and insects). If true, this would make rhomboids a rare case where topology appears to have evolved by attachment of nonhomologous TMDs, instead of by the more typical internal gene duplication or non-covalent oligomerization (von Heijne 2006).
Catalytically inert iRhoms
As has been noted previously (Koonin et al. 2003; Freeman 2004; Nakagawa et al. 2005), there are a significant number of rhomboid-like proteins that lack key catalytic residues (Figs. 3A, S1). Because the overall fold is expected to be retained, as indicated by the strict conservation of small amino acid residues crucial for helix packing in the E. coli rhomboid GlpG (see Fig. 1; Ben-Shem et al. 2007), serine and histidine residues in other positions are topological distinct and are not therefore expected to act as surrogates (Fig. 3A). Consistent with this, we have not detected proteolytic activity for any of these rhomboid-like proteins we have tested (M.K. Lemberg and M. Freeman, unpubl.). More generally, putatively inactive homologs have been identified in many protease families, but their functions remain mysterious. Our phylogenetic alignment clusters most of the inactive rhomboids into a discrete clade, suggesting a functional relationship between them. Members of this new group, which we name iRhoms (for inactive rhomboids), have a characteristic large loop inserted between TMDs 1 and 2 (Fig. 2, class 3). iRhoms occur in all animal genomes accessible by the Ensembl database (Fig. 5; data not shown). Importantly, the clear phylogenetic clustering of iRhoms implies that they are not simply an unrelated set of pseudogenes or evolutionary remnants of active rhomboids but instead a true orthologous group (Fig. 3B; Table S6).
Figure 5.
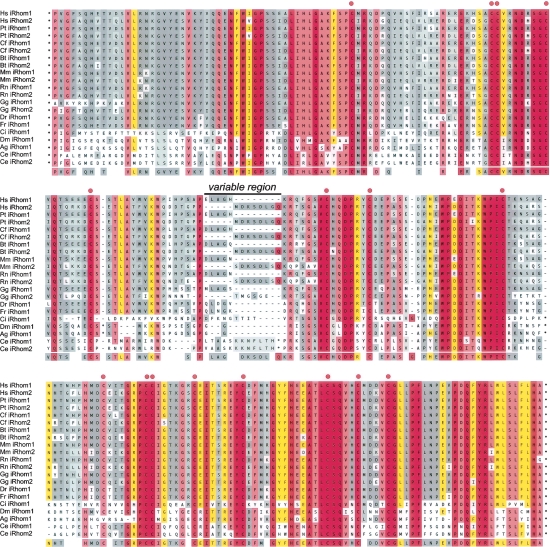
Conserved iRhom homology domain (IRHD). Genes encoding iRhoms were found in all animal genomes accessible by the Ensembl genome browser. ClustalW-based multiple sequence alignment of the extremely conserved luminal domain between TMD1 and TMD2 (for accession numbers, see below). Two paralogs were found in vertebrates, excluding fish (D. rerio and T. rubripes), and in C. elegans. Variable inserted sequences from C. intestalis (three residues), D. melanogaster (10 and six residues), A. gambiae (Ag) (two and six residues), C. elegans iRhom1 (eight residues), and C. elegans iRhom2 (27 residues) are hidden as indicated by an asterisk; position of the conserved cysteine (C) is indicated by red dots. The region showing the highest degree in variation between the vertebrate orthologs iRhom1 and iRhom2 is marked; degree of conservation is colored as in Figure 1. Note that A. gambiae iRhom appears to lack one of the 16 cysteine residues.
Accession Numbers: For human and mouse iRhoms, see Table S1. Due to the current coverage of the D. pseudoobscura genome project, no iRhom is currently annotated, but a clear ortholog was found in all insect genomes accessible by the Ensembl genome browser (e.g., Anopheles gambiae (Ag); Ensembl:ENSANGP00000021207). Note that the gene for Drosophila melanogaster (Dm) (Swiss-Prot:Q76NQ1) (Rhomboid-5) is ambiguously predicted. Comparison with predicted exon-intron structure of Ag iRhom suggests that the related Dm iRhom mRNA starts with the sequence (GenBank: AA699010) (M.K. Lemberg and M. Freeman, unpubl.). The Swiss-Prot entry (Swiss-Prot:Q8IH64) that is falsely annotated as Rhomboid-5 consists of a fragment of Ribophorin I (Swiss-Prot:Q76NQ0). The accession number for zebrafish (Danio rerio, Dr) iRhom is Swiss-Prot:Q6GMF8, Takifugu rubripes (Fr) iRhom is Ensembl: SINFRUP00000147713, and Ciona intestalis (Ci) iRhom is Ensembl:ENSCINT00000006045. For the other species, the accession numbers of iRhom1 and iRhom2, respectively, are: Pan troglodytes (Pt), Ensembl:ENSPTRP00000012854 and Ensembl:ENSPTRP00000016460; Canis familiaris (Cf), GenBank:XP_547222.1 and Ensembl:ENSCAFP00000007595; Bos taurus (Bt), Ensembl:ENSBTAP00000026388 and Ensembl:ENSBTAP00000004463; Rattus norvegicus (Rn), GenBank:NP_001025205.1 and GenBank:XP_221133.3; Gallus gallus (Gg), Ensembl:ENSGALP00000012018 and Ensembl: ENSGALP00000003004; and Caenorhabditis elegans (Ce), GenBank:NP_503013 [ROM3 by automated annotation] and GenBank:NP_001041013 [ROM4 by automated annotation].
Different members of the iRhom group are, however, missing different catalytic residues. Some have the serine but not the histidine; others have the histidine but not the serine; some lack both (Figs. 3A; S1). In the most anomalous cases, for example, C. elegans iRhom1 and iRhom2 (formerly ROM3 and ROM4 by automated annotation), both catalytic residues are present (Fig. 3A). In contrast to this relatively low conservation of catalytic residues, all iRhoms have a conserved proline (P) in the first x-position of the GxSx rhomboid catalytic motif. Although the role of the residue in this position is unknown, its proximity to the catalytic serine suggests that it might influence enzyme activity. Furthermore, the peptide backbone may contribute to the conformation of the active site, a hypothesis that is consistent with the invariant nature of the glycine within the GxSx rhomboid protease motif (Urban et al. 2001; Lemberg et al. 2005; Wang et al. 2006; Ben-Shem et al. 2007). Because of the distinct chemical nature of prolines, this active site conformation is likely to be structurally disrupted. Consistent with this hypothesis, a mutant of Drosophila Rhomboid-1 with a proline at this position (Rho1-A216P) has no significant proteolytic activity against its substrates Spitz and Gurken (M.K. Lemberg and M. Freeman, unpubl.). The universal presence of this proline leads us to propose that all members of this clade are not active rhomboid intramembrane proteases, including C. elegans iRhom1 and iRhom2, which formerly would have been predicted to be active proteases. Once initially mutated to inactivity, probably by the addition of proline to the active site, the other catalytic residues would no longer be under strong selective pressure, providing an explanation for their variable conservation.
The iRhoms are considerably larger than other rhomboids, with very long N-terminal domains preceding the membrane-integral domain and the expanded L1 loop between TMD1 and TMD2 (240–270 amino acids compared with 32 amino acids for GlpG). The crystal structure of GlpG predicts that the L1 loop is embedded in the outer leaflet of the membrane (Fig. 2B; Wang et al. 2006). As shown in Figure 5, the sequence of this extended L1 domain is highly conserved in all iRhoms, and we have termed it the IRHD for iRhom homology domain. Strikingly, 16 cysteine residues are conserved, and we suggest that these form disulphide bridges that would stabilize a globular fold. Some sequences between these cysteines are of variable length and sequence and could form exposed loops. In vertebrates with more than one iRhom, these variable sequences are diagnostic for the subtype, called iRhom1 and iRhom2, respectively (Fig. 5; Table S1).
Plant and apicomplexan parasite rhomboids
Rhomboids in apicomplexan parasites, including the malaria parasite Plasmodium, have been the focus of intense study recently because of evidence that they are essential in parasite invasion (O’Donnell et al. 2006). Other studies have focused on the large numbers of rhomboids in the genomes of the plants Arabidopsis and rice (O. sativa) (Koonin et al. 2003; Kanaoka et al. 2005; Garcia-Lorenzo et al. 2006; Tripathi and Sowdhamini 2006). Most of these parasite and plant rhomboids are quite distant from other eukaryotic rhomboids, so we have carried out a phylogenetic analysis on them separately.
In contrast with previous reports (Garcia-Lorenzo et al. 2006; Tripathi and Sowdhamini 2006), our analysis of all Arabidopsis and rice rhomboids revealed five clades (Fig. 3C). The PARL orthologs form the first subfamily. The second clade corresponds to the secretase-B class. Within this, Arabidopsis RBL10 and rice Os01g18100 are clear orthologs of vertebrate RHBDL4 (Table S5; data not shown). As in other eukaryotes, cellular localization algorithms predict these rhomboids to be mitochondrial (for details, see Methods), but as argued above, functional evidence suggests that they are in fact located in the secretory pathway. All remaining plant rhomboids divide into three rather poorly defined clades, none of which obviously clusters with the secretase-A rhomboids or iRhoms (Fig. 3C). The differences between these three clades are minor, and the physiological relevance of this grouping will have to be evaluated. Nevertheless, initial evidence for functional diversification between them can be drawn from their predicted subcellular localization. RBL8 (D-clade) is predicted to localize to chloroplasts, and RBL9 (C-clade) is predicted to have dual localization to mitochondria and chloroplasts; in contrast all members of the E-clade are predicted to localize to the secretory pathway (for details, see Methods; data not shown).
Finally, we studied the phylogeny of rhomboids from the apicomplexan parasites Toxoplasma and Plasmodium. Consistent with previous reports, we observed four major clades representing the secretase rhomboids and PARL (Brossier et al. 2005; Dowse and Soldati 2005). Surprisingly, Plasmodium has two additional putative rhomboids, of which one clusters in the PARL subfamily (Fig. 3D). This rhomboid, called ROM9, was not included in the published phylogeny and is predicted to be mitochondrial. However, the predicted 6 + 1 topology with a large insertion into the L1 loop (Fig. 2B) suggests that, even if it is confirmed to be mitochondrial, ROM9 is not functionally related to PARL. Rhomboids in the other clades and Plasmodium ROM7 are predicted to be in the secretory pathway.
It is unclear why the plant and parasite rhomboids do not fall clearly into the same subfamilies as other eukaryotic rhomboids. Presumably they are either genuinely functionally distinct subfamilies or, in fact, are members of the same subfamilies but are too divergent for this to be readily apparent. Indeed, most plant and parasite rhomboids share characteristics of both subgroups: from the A-clade they have the 6 + 1 TMD topology and sequence elements, such as the WR-motif (Figs. 2B, 3A); from the B-clade they have the characteristic phenylalanine (F) in TMD4 (Fig. 3A, labeled with blue and black dots, respectively). Functional and structural studies have highlighted both these sequence motifs as important (for details, see Fig. 1; Urban et al. 2001; Lemberg et al. 2005; Wang et al. 2006), suggesting that these mixed forms may represent evolutionary intermediates between an ancestral rhomboid (of the B-type) and the potentially more recently evolved rhomboids of the higher eukaryotes (A-clade).
On the relationship between prokaryotic and eukaryotic rhomboids
Very little is known about the function of bacterial rhomboid proteases, although recently the rhomboid from Providencia stuartii, was shown to activate the TatA protein transporter (Stevenson et al. 2007). The sequences of bacterial rhomboids are very diverse, and their comprehensive analysis is beyond the scope of this paper. Nevertheless, the approach developed in this study can be used to classify bacterial rhomboids broadly. The majority have the basic six TMD architecture, which leads us to speculate that the proposed ancestral eukaryotic rhomboid may have derived from a bacterial rhomboid. In contrast, P. stuartii rhomboid AarA and Bacillus subtilis rhomboid YqgP have a 1 + 6 TMD architecture (Fig. 2B), suggesting that, like the eukaryotic rhomboids, they may have evolved by gene fusion events. Bacterial rhomboids can also be classified by characteristic sequence motifs. All rhomboids have the catalytic GxSx and histidine residues in TMDs 4 and 6, implying that they use the conserved rhomboid serine protease dyad (Lemberg et al. 2005; Wang et al. 2006). Like the eukaryotic secretase rhomboids, the first x-position of the rhomboid active site motif shows substantial variation. As in eukaryotes, alanine (A) is by far the most common residue of rhomboids currently in the NCBI database; leucine (L), as found in E. coli rhomboid GlpG (Fig. 1), is the second most common; residues less frequently found are methionine, serine, phenylalanine, and valine. Full phylogenetic analysis of the prokaryotic rhomboids remains a future challenge but is likely to provide important functional and evolutionary insights.
Functional implications of the new rhomboid classification
The identification of an extra TMD in all members of the PARL subfamily has caused us to re-evaluate aspects of the published experimental literature and turns out to have important mechanistic consequences for proteolysis by rhomboids. The additional TMD shifts the serine protease active site residues from TMD4 and TMD6 in other rhomboids to TMD5 and TMD7 (Fig. 6A). The presence of an N-terminal mitochondrial targeting sequence, which is predicted to be directed to the mitochondrial matrix (topologically equivalent to the cytoplasm) (Schatz and Dobberstein 1996), coupled with the 1 + 6 TMD structure, suggests that the PARL active site has the opposite orientation within the membrane to other rhomboids. In secretase rhomboids the catalytic GxSx and histidine are located in TMDs 4 and 6, which both are of out-to-in orientation. In contrast, these catalytic motifs in PARLs occur in in-to-out TMDs (Fig. 6A). Significantly, there is a corresponding inversion of substrate orientation: PARL substrates have an Nin/Cout topology, but secretase rhomboids cleave type I membrane proteins (Nout/Cin). This inversion of the active sites of PARLs, has not been apparent until now because of the failure to detect all the TMDs in S. cerevisiae PARL (Pcp1/Rbd1) (see above and Table S2). The striking correlation between enzyme and substrate inversion suggests the possibility that all rhomboids may only cleave one substrate orientation, a conclusion supported by almost all functional evidence. A very recent publication, however, challenges this view (Tsruya et al. 2007), and further functional work is needed to resolve this question.
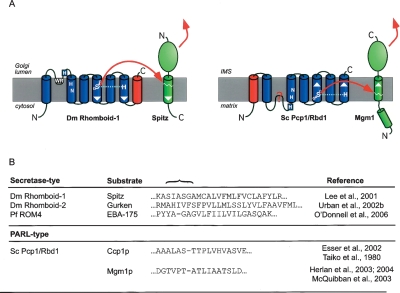
Figure 6.
Secretase- and PARL-type rhomboids have active sites with opposite orientations. (A) Topology model of Drosophila Rhomboid-1 (Urban et al. 2001) and S. cerevisiae PARL (Pcp1/Rbd1). Cleavage by Drosophila Rhomboid-1 releases the N-terminal portion of the membrane tethered growth factor Spitz into the Golgi lumen, thereby allowing its secretion to trigger EGFR signaling in neighboring cells (Lee et al. 2001). In contrast, S. cerevisiae PARL, Pcp1/Rbd1, cleaves its substrate Mgm1 to release the C-terminal portion into the intermembrane space (IMS) (Herlan et al. 2004). Topology model as in Figure 2B; extra TMDs fused to the C terminus of Drosophila Rhomboid-1 and the N terminus of PARL (Pcp1/Rbd1) are colored red. The orientation of the active site TMDs and substrate TMD is indicated by white arrowheads. (B) Examples of substrate TMDs of secretase- and PARL-type rhomboids. Cleavage site region is indicated by a brace. Experimentally determined cleavage sites are indicated in the sequences by dashes. Note that the substrates for S. cerevisiae PARL (Pcp1/Rbd1) are cleaved in secondary non-typical short TMDs (Takio et al. 1980; Herlan et al. 2003, 2004).
Examination of the active sites and substrates of PARL and secretase rhomboids also suggests another important mechanistic conclusion. The PARL active sites are predicted to lie close to the matrix side of the membrane (topologically equivalent to the cytoplasm), but the released fragment of the substrate is the intermembrane space (IMS) domain (Fig. 6A). That is, the cleaved fragment with the long TMD remnant is released (Fig. 6B). On the other hand, the active site of secretase type rhomboids is close to the other side of the membrane—the luminal or extracellular side, which is topologically equivalent to the IMS; the released fragment of all known substrates of these rhomboids is the side with the short TMD remnant. Therefore both halves of rhomboid cleaved substrates can be released from the membrane. This raises the intriguing possibility that in some cases, rhomboid cleavage may lead to bidirectional signaling, for example, simultaneously releasing substrate domains into the cytoplasm and the lumen/extracellular space.
Conclusions
Although primary sequence comparison (e.g., by PSI-BLAST or by the use of hidden Markov models) is a powerful approach to identify rhomboid-like proteins, a topological prediction of the TMD structure and the analysis of the rhomboid protease consensus enhances the gene annotation and allows discrimination between rhomboid proteases, inactive homologs and unrelated false positives.
We define four topological classes of rhomboids by virtue of their number and position of TMDs, their orientation in the membrane, and the existence of characteristic extramembrane domains. Although the overall function of this protease core is expected to be conserved, the structural and functional implication of these extra TMDs remains an important question to be addressed.
We define true rhomboids as being active intramembrane serine proteases (and those that are predicted to be active by virtue of their sequence). There are numerous rhomboid-like proteins that are missing catalytically essential active site residues. We propose that these not be called rhomboids, despite being rhomboid-like. There is one tightly clustering orthologous group of inactive rhomboids that we name the iRhoms (for inactive rhomboids). The iRhom family all contain a large globular domain inserted into the L1 loop, which we have called the IRHD. Other rhomboid-like proteins that lack catalytic residues but do not cluster with the iRhoms are scattered across evolution so are not classified as a coherent clade.
Our analysis allows us to predict for the first time the number of active rhomboids in sequenced genomes. We therefore revise the number in several species, including humans. This reduces the total number of intramembrane proteases for mouse and human to 13 (five rhomboids, one S2P, and seven GxGD-type), instead of 16 as previously suggested (Overall and Blobel 2007).
We find four major phylogenetic clades of eukaryotic rhomboid-like proteins: secretase-type, which are divided into A and B classes; PARLs, the mitochondrial subfamily; and finally iRhoms (which we no longer define as true rhomboids). Note, however, that rhomboids from plants and apicomplexan parasites are too divergent to incorporate clearly into these four clades.
This genomic analysis suggests new areas of study and leads to functional conclusions. For example, in the organism in which rhomboids have been most studied, Drosophila, Rhomboid-6 stands out as the most divergent of the secretase rhomboids. Analysis of this gene has not been reported but would now be an interesting focus. More substantially, the topology that we report for all PARL-type rhomboids is consistent with two mechanistic conclusions. The first is that a given rhomboid may only cleave one orientation of substrate TMD. The second is that both products of a rhomboid-catalyzed transmembrane cleavage can leave the membrane, raising the possibility of bidirectional signaling by rhomboids.
Finally, the revised phylogeny of rhomboids, based on functional and structural data that have only recently become available, suggests the existence of a previously unrecognized rhomboid subgroup represented in yeast, plants and vertebrates (secretase-B). These rhomboids have the most basic six TMD domain architecture, which we predict to resemble an ancestral template for all eukaryotic rhomboids. It was previously proposed that rhomboids have spread through eukaryotes by several independent horizontal gene transfer events (Koonin et al. 2003). On the basis of the proposed simpler phylogenetic relationship between all eukaryotic rhomboids (see Fig. 3B), we believe that a model of primarily vertical evolution from an ancestral gene present in the last common eukaryotic ancestor is now the more parsimonious conclusion. Since the putative ancestral eukaryotic form resembles the most common form of the prokaryotic rhomboids, it is also attractive to postulate the existence of a single rhomboid ancestor in the last common universal ancestor of all organisms. We note, however, that this remains speculative until the phylogeny of prokaryotic and archaeal rhomboids have been further characterized.
Methods
Sequence data
Rhomboid sequences were retrieved by BLAST and PSI-BLAST search (Altschul et al. 1997) from the NCBI database (nonredundant protein sequences; restricted to eukaryotic sequences) (http://www.ncbi.nlm.nih.gov/BLAST/), from the Ensembl genome browser (http://www.ensembl.org/), and the MIPS plant genome database (http://mips.gsf.de/projects/plants/). Except where otherwise stated, we used default parameters. For the iterative homology search by PSI-BLAST, we used a cut-off E-value of 0.01.
TMD prediction and comparative topology analysis
Rhomboid topology models were constructed by superimposing TMD predictions from four different prediction algorithms on a ClustalW multiple-sequence alignment of homologs and orthologs (using MacVector7.2.2) (Thompson et al. 1994). To ensure optimum results, the alignments were also performed with T-Coffee (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi) (Notredame et al. 2000) and MUSCLE (http://www.drive5.com/muscle/) (Edgar 2004). Manual adjustments were also introduced as appropriate. Where possible, precise TMD boundaries were based on a comparison with structural information taken from the E. coli rhomboid GlpG (Wang et al. 2006). As prediction algorithms, we used TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), HMMTOP version 2.0 (http://www.enzim.hu/hmmtop/index.html), PSORT II (psort.nibb.ac.jp/form2.html), and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
Multiple-sequence alignment and phylogenetic analysis
We obtained 84 sequences for rhomboid proteases and rhomboid-like proteins. Based on our topology model, we artificially spliced together the conserved regions (C-terminal 13 amino acids of L1, TMD2, TMD4, and TMD6 for secretase-type rhomboids; C-terminal 13 amino acids of L2, TMD3, TMD5, and TMD7 for PARL-type rhomboids). In total, 86 amino acids were aligned, and a phylogeny tree was constructed based on the UPGMA analysis using MacVector7.2.2 software. To test the support of individual clades, 1000 bootstrap replicas were performed. Differences between aligned subgroups were determined by Two Sample Logo analysis (http://www.twosamplelogo.org/) (Vacic et al. 2006); two sample t-test with the Bonferroni correction revealed statistical significant variations between the subgroup tested (positive sample) and the unrelated sequences (negative sample). Related rhomboid sequences were analyzed with the EMBOSS pairwise alignment algorithm (http://www.ebi.ac.uk/emboss/align/), MEME-MAST search tools (http://meme.sdsc.edu/meme/intro.html) (Bailey and Gribskov 1998), and the MPsrch protein database query implementing a Smith-Waterman algorithm (http://www.ebi.ac.uk/MPsrch/).
Prediction of subcellular localization and protein search for conserved protein domains
Sequences were analyzed by TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/), ChloroP (http://www.cbs.dtu.dk/services/ChloroP/), MITOPRED (http://bioinformatics.albany.edu/~mitopred/), PSORT II (http://psort.nibb.ac.jp/form2.html), and rps-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Acknowledgments
We thank Markus Zettl, Kvido Strisovsky, and Colin Adrain for their helpful comments on the manuscript. We thank Madan Babu for advice with the phylogenetic analysis, helpful discussion, and critical reading of the manuscript. M.K.L. was supported by an EMBO Longterm Fellowship and by a fellowship from the Swiss National Science Foundation.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6425307
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M., Gribskov M. Methods and statistics for combining motif match scores. J. Comput. Biol. 1998;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- Baker R.P., Wijetilaka R., Urban S., Wijetilaka R., Urban S., Urban S. Two plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS. Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L., Howe K.L., Marshall M., Sonnhammer E.L., Marshall M., Sonnhammer E.L., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., Fass D., Bibi E., Fass D., Bibi E., Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossier F., Jewett T.J., Sibley L.D., Urban S., Jewett T.J., Sibley L.D., Urban S., Sibley L.D., Urban S., Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc. Natl. Acad. Sci. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.S., Ye J., Rawson R.B., Goldstein J.L., Ye J., Rawson R.B., Goldstein J.L., Rawson R.B., Goldstein J.L., Goldstein J.L. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Craessaerts K., Metzger K., Frezza C., Annaert W., D’Adamio L., Metzger K., Frezza C., Annaert W., D’Adamio L., Frezza C., Annaert W., D’Adamio L., Annaert W., D’Adamio L., D’Adamio L., et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Daley D.O., Rapp M., Granseth E., Melen K., Drew D., von Heijne G., Rapp M., Granseth E., Melen K., Drew D., von Heijne G., Granseth E., Melen K., Drew D., von Heijne G., Melen K., Drew D., von Heijne G., Drew D., von Heijne G., von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- Dowse T.J., Soldati D., Soldati D. Rhomboid-like proteins in Apicomplexa: Phylogeny and nomenclature. Trends Parasitol. 2005;21:254–258. doi: 10.1016/j.pt.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Dowse T.J., Pascall J.C., Brown K.D., Soldati D., Pascall J.C., Brown K.D., Soldati D., Brown K.D., Soldati D., Soldati D. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. Int. J. Parasitol. 2005;35:747–756. doi: 10.1016/j.ijpara.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Dutt A., Canevascini S., Froehli-Hoier E., Hajnal A., Canevascini S., Froehli-Hoier E., Hajnal A., Froehli-Hoier E., Hajnal A., Hajnal A. EGF signal propagation during C. elegans vulval development mediated by ROM-1 rhomboid. PLoS Biol. 2004;2:e334. doi: 10.1371/journal.pbio.0020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E., Tursun B., Ingenhoven M., Michaelis G., Pratje E., Ingenhoven M., Michaelis G., Pratje E., Michaelis G., Pratje E., Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Freeman M. Proteolysis within the membrane: Rhomboids revealed. Nat. Rev. Mol. Cell Biol. 2004;5:188–197. doi: 10.1038/nrm1334. [DOI] [PubMed] [Google Scholar]
- Friedmann E., Lemberg M.K., Weihofen A., Dev K.K., Dengler U., Rovelli G., Martoglio B., Lemberg M.K., Weihofen A., Dev K.K., Dengler U., Rovelli G., Martoglio B., Weihofen A., Dev K.K., Dengler U., Rovelli G., Martoglio B., Dev K.K., Dengler U., Rovelli G., Martoglio B., Dengler U., Rovelli G., Martoglio B., Rovelli G., Martoglio B., Martoglio B. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J. Biol. Chem. 2004;279:50790–50798. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- Garcia-Lorenzo M., Sjodin A., Jansson S., Funk C., Sjodin A., Jansson S., Funk C., Jansson S., Funk C., Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman M., Deloof H., Von Heijne G., Elofsson A., Deloof H., Von Heijne G., Elofsson A., Von Heijne G., Elofsson A., Elofsson A. Improved detection of homologous membrane proteins by inclusion of information from topology predictions. Protein Sci. 2002;11:652–658. doi: 10.1110/ps.39402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A.S., Vogel F., Bornhovd C., Neupert W., Reichert A.S., Bornhovd C., Neupert W., Reichert A.S., Neupert W., Reichert A.S., Reichert A.S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Herlan M., Bornhovd C., Hell K., Neupert W., Reichert A.S., Bornhovd C., Hell K., Neupert W., Reichert A.S., Hell K., Neupert W., Reichert A.S., Neupert W., Reichert A.S., Reichert A.S. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 2004;165:167–173. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Howson R.W., Weissman J.S., O’Shea E.K., Weissman J.S., O’Shea E.K., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jeyaraju D.V., Xu L., Letellier M.C., Bandaru S., Zunino R., Berg E.A., McBride H.M., Pellegrini L., Xu L., Letellier M.C., Bandaru S., Zunino R., Berg E.A., McBride H.M., Pellegrini L., Letellier M.C., Bandaru S., Zunino R., Berg E.A., McBride H.M., Pellegrini L., Bandaru S., Zunino R., Berg E.A., McBride H.M., Pellegrini L., Zunino R., Berg E.A., McBride H.M., Pellegrini L., Berg E.A., McBride H.M., Pellegrini L., McBride H.M., Pellegrini L., Pellegrini L. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc. Natl. Acad. Sci. 2006;103:18562–18567. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M.M., Urban S., Freeman M., Okada K., Urban S., Freeman M., Okada K., Freeman M., Okada K., Okada K. An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 2005;579:5723–5728. doi: 10.1016/j.febslet.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Kim H., Melen K., Osterberg M., von Heijne G., Melen K., Osterberg M., von Heijne G., Osterberg M., von Heijne G., von Heijne G. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc. Natl. Acad. Sci. 2006;103:11142–11147. doi: 10.1073/pnas.0604075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Rogozin I.B., Davidovic L., Letellier M.C., Pellegrini L., Makarova K.S., Rogozin I.B., Davidovic L., Letellier M.C., Pellegrini L., Rogozin I.B., Davidovic L., Letellier M.C., Pellegrini L., Davidovic L., Letellier M.C., Pellegrini L., Letellier M.C., Pellegrini L., Pellegrini L. The rhomboids: A nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.R., Urban S., Garvey C.F., Freeman M., Urban S., Garvey C.F., Freeman M., Garvey C.F., Freeman M., Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Lemberg M.K., Menendez J., Misik A., Garcia M., Koth C.M., Freeman M., Menendez J., Misik A., Garcia M., Koth C.M., Freeman M., Misik A., Garcia M., Koth C.M., Freeman M., Garcia M., Koth C.M., Freeman M., Koth C.M., Freeman M., Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux M.J., Fischer S.J., Cherney M.M., Bateman K.S., James M.N., Fischer S.J., Cherney M.M., Bateman K.S., James M.N., Cherney M.M., Bateman K.S., James M.N., Bateman K.S., James M.N., James M.N. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O., Urban S., Freeman M., Urban S., Freeman M., Freeman M. Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by mammalian rhomboids. Curr. Biol. 2004;14:236–241. doi: 10.1016/j.cub.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Maegawa S., Ito K., Akiyama Y., Ito K., Akiyama Y., Akiyama Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- Mayer U., Nusslein-Volhard C., Nusslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes & Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- McQuibban G.A., Saurya S., Freeman M., Saurya S., Freeman M., Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- McQuibban G.A., Lee J.R., Zheng L., Juusola M., Freeman M., Lee J.R., Zheng L., Juusola M., Freeman M., Zheng L., Juusola M., Freeman M., Juusola M., Freeman M., Freeman M. Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. 2006;16:982–989. doi: 10.1016/j.cub.2006.03.062. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Guichard A., Castro C.P., Xiao Y., Rizen M., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Guichard A., Castro C.P., Xiao Y., Rizen M., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Castro C.P., Xiao Y., Rizen M., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Xiao Y., Rizen M., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Rizen M., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Zhang H.Z., Hu D., Bang A., Helms J., Bier E., Hu D., Bang A., Helms J., Bier E., Bang A., Helms J., Bier E., Helms J., Bier E., Bier E., et al. Characterization of a human rhomboid homolog, p100hRho/RHBDF1, which interacts with TGF-alpha family ligands. Dev. Dyn. 2005;233:1315–1331. doi: 10.1002/dvdy.20450. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Persson B., von Heijne G., Persson B., von Heijne G., von Heijne G. Consensus predictions of membrane protein topology. FEBS Lett. 2000;486:267–269. doi: 10.1016/s0014-5793(00)02321-8. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J., Higgins D.G., Heringa J., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- O’Donnell R.A., Hackett F., Howell S.A., Treeck M., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Hackett F., Howell S.A., Treeck M., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Howell S.A., Treeck M., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Treeck M., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Krnajski Z., Withers-Martinez C., Gilberger T.W., Blackman M.J., Withers-Martinez C., Gilberger T.W., Blackman M.J., Gilberger T.W., Blackman M.J., Blackman M.J. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall C.M., Blobel C.P., Blobel C.P. In search of partners: Linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 2007;8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- Pascall J.C., Brown K.D., Brown K.D. Intramembrane cleavage of ephrinB3 by the human rhomboid family protease, RHBDL2. Biochem. Biophys. Res. Commun. 2004;317:244–252. doi: 10.1016/j.bbrc.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Stevenson L.G., Strisovsky K., Clemmer K.M., Bhatt S., Freeman M., Rather P.N., Strisovsky K., Clemmer K.M., Bhatt S., Freeman M., Rather P.N., Clemmer K.M., Bhatt S., Freeman M., Rather P.N., Bhatt S., Freeman M., Rather P.N., Freeman M., Rather P.N., Rather P.N. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc. Natl. Acad. Sci. 2007;104:1003–1008. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takio K., Titani K., Ericsson L.H., Yonetani T., Titani K., Ericsson L.H., Yonetani T., Ericsson L.H., Yonetani T., Yonetani T. Primary structure of yeast cytochrome c peroxidase. II. The complete amino acid sequence. Arch. Biochem. Biophys. 1980;203:615–629. doi: 10.1016/0003-9861(80)90219-2. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L.P., Sowdhamini R., Sowdhamini R. Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics. 2006;7:200. doi: 10.1186/1471-2164-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsruya R., Wojtalla A., Carmon S., Yogev S., Reich A., Bibi E., Merdes G., Schejter E., Shilo B.Z., Wojtalla A., Carmon S., Yogev S., Reich A., Bibi E., Merdes G., Schejter E., Shilo B.Z., Carmon S., Yogev S., Reich A., Bibi E., Merdes G., Schejter E., Shilo B.Z., Yogev S., Reich A., Bibi E., Merdes G., Schejter E., Shilo B.Z., Reich A., Bibi E., Merdes G., Schejter E., Shilo B.Z., Bibi E., Merdes G., Schejter E., Shilo B.Z., Merdes G., Schejter E., Shilo B.Z., Schejter E., Shilo B.Z., Shilo B.Z. Rhomboid cleaves Star to regulate the levels of secreted Spitz. EMBO J. 2007;26:1211–1220. doi: 10.1038/sj.emboj.7601581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Wolfe M.S., Wolfe M.S. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Lee J.R., Freeman M., Lee J.R., Freeman M., Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Urban S., Lee J.R., Freeman M., Lee J.R., Freeman M., Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 2002a;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Schlieper D., Freeman M., Schlieper D., Freeman M., Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr. Biol. 2002b;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- Vacic V., Iakoucheva L.M., Radivojac P., Iakoucheva L.M., Radivojac P., Radivojac P. Two Sample Logo: A graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W.M.J., Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A., Clemons W.M.J., Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A., Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A., Hartmann E., Harrison S.C., Rapoport T.A., Harrison S.C., Rapoport T.A., Rapoport T.A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Ha Y., Zhang Y., Ha Y., Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- Wasserman J.D., Urban S., Freeman M., Urban S., Freeman M., Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signalling. Genes & Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- Weihofen A., Martoglio B., Martoglio B. Intramembrane-cleaving proteases: Controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 2003;13:71–78. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- Wolfe M.S., Kopan R., Kopan R. Intramembrane proteolysis: Theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Wu Z., Yan N., Feng L., Oberstein A., Yan H., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Yan N., Feng L., Oberstein A., Yan H., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Feng L., Oberstein A., Yan H., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Oberstein A., Yan H., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Yan H., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Baker R.P., Gu L., Jeffrey P.D., Urban S., Shi Y., Gu L., Jeffrey P.D., Urban S., Shi Y., Jeffrey P.D., Urban S., Shi Y., Urban S., Shi Y., Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat. Struct. Mol. Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- Yamasaki A., Eimer S., Okochi M., Smialowska A., Kaether C., Baumeister R., Haass C., Steiner H., Eimer S., Okochi M., Smialowska A., Kaether C., Baumeister R., Haass C., Steiner H., Okochi M., Smialowska A., Kaether C., Baumeister R., Haass C., Steiner H., Smialowska A., Kaether C., Baumeister R., Haass C., Steiner H., Kaether C., Baumeister R., Haass C., Steiner H., Baumeister R., Haass C., Steiner H., Haass C., Steiner H., Steiner H. The GxGD motif of presenilin contributes to catalytic function and substrate identification of gamma-secretase. J. Neurosci. 2006;26:3821–3828. doi: 10.1523/JNEUROSCI.5354-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]