Abstract
The goals of the human genome project did not include sequencing of the heterochromatic regions. We describe here an initial sequence of 1.1 Mb of the short arm of human chromosome 21 (HSA21p), estimated to be 10% of 21p. This region contains extensive euchromatic-like sequence and includes on average one transcript every 100 kb. These transcripts show multiple inter- and intrachromosomal copies, and extensive copy number and sequence variability. The sequencing of the “heterochromatic” regions of the human genome is likely to reveal many additional functional elements and provide important evolutionary information.
The sequencing of the human genome focused explicitly on the euchromatic regions of the genome, and all remaining large gaps (>300 kb) coincide with regions considered to be heterochromatic (Lander et al. 2001; Venter et al. 2001; IHGSC 2004). These consist of centromeres, telomeres, the secondary constrictions on 1q, 9q and 16q, distal Yq and the short (p) arms of the acrocentric chromosomes (13, 14, 15, 21, and 22). In total these regions represent a significant proportion of the genome, estimated at ∼200 Mb (∼6.5%) (Morton 1991; IHGSC 2004).
The sequences and annotation of the long arms of acrocentric chromosomes have been reported but there is very little data on the short (p) arms. Cytogenetic data show that the p arms contain large heterochromatic regions (Craig-Holmes and Shaw 1971; Verma et al. 1977). Molecular evidence suggests that they are composed mainly of satellite and other repeat families, including satellites I (Kalitsis et al. 1993), II (Hollis and Hindley 1988), III (Choo et al. 1990), β (Waye and Willard 1989), and repeats ChAB4 (Cserpán et al. 2002), 724 (Kurnit et al. 1986), and D4Z4-like (Lyle et al. 1995). These repeats have a complex pattern and are often organized in subfamilies shared between different acrocentric chromosomes and the heterochromatin of other chromosomes, for example D4Z4 (Lyle et al. 1995). The p arms encode the ribosomal (RNR) genes but may encode other genes (Tapparel et al. 2003).
While the function of acrocentric short arm repeat regions is unknown, there is evidence that they are involved in diverse processes, including disease. For example, transcription of satellite III sequences has a role in nuclear stress granules (Valgardsdottir et al. 2005). The most common chromosomal rearrangements in humans are Robertsonian translocations (ROBs; ∼1 in 1000 births), which involve exchanges between acrocentric p arms. Three to five percent of ROBs are associated with phenotypic abnormalities (Warburton 1991).
The sequence of HSA21p would be not only important for understanding chromosome 21 genetics and disease (Antonarakis et al. 2004), but a necessary step in the characterization of these unexplored regions of the genome and toward the complete sequence of the human genome. However, the fact that this region of the genome is homologous to other regions, highly repetitive, and polymorphic presents certain technical challenges. Here we present 1.1 Mb of 21p sequence, and identify and characterize transcripts from the region.
Results and Discussion
We prepared a BAC library from the somatic cell hybrid WAV17 (Slate et al. 1978), which contains only human chromosome 21 (HSA21), derived from a single HSA21, in a mouse background. This ensures that derived non-mouse non-HSA21q BACs map to 21p, and that we do not sequence multiple 21p alleles. To ensure the integrity of WAV17, we karyotyped the line and used FISH to confirm the human chromosomal content (Supplemental Fig. 1). We also genotyped WAV17 for six HSA21 microsatellite markers and observed only one allele in each case, confirming the monoallelic composition of HSA21 in WAV17 (P < 1.5 × 10−5).
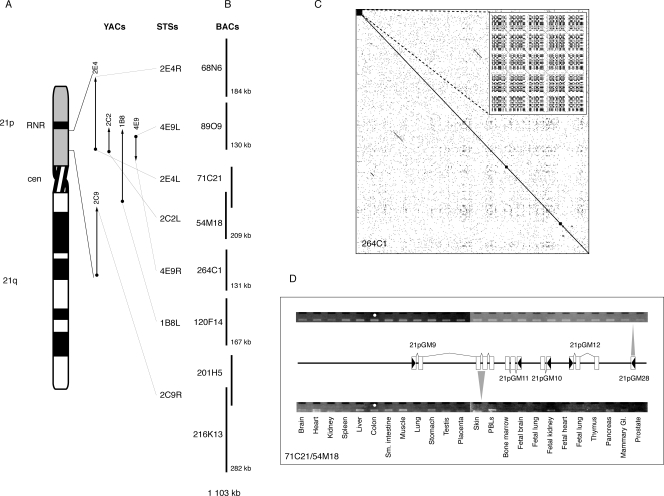
A BAC library (CHORI-507) was prepared from MboI partially digested WAV17 DNA (Supplemental Fig. 2). 21p clones were identified by screening human-positive clones with sequence-tagged sites (STSs) known to map to 21p (Fig. 1A). A total of 96 positive clones were identified and binned according to location (Supplemental Tables 1, 2). Eight of these clones were selected for sequencing, based on their predicted location, to cover as wide a region as possible (Fig. 1B). The end sequences of these clones did not match either mouse or HSA21q sequence, indicating that they are derived from 21p. To confirm this, we tested seven BACs by FISH to normal human lymphoblastoid cells and each hybridized to 21p (Supplemental Fig. 3). Signals were also detected on the p arms of other acrocentric chromosomes and other genomic locations as expected. The total sequence length of these eight clones is 1,251,403 bp (Supplemental Table 3). Aligning the sequences identified two partial overlaps with 100% sequence identity (71C21/54M18 and 201H5/216K13), giving a total nonredundant length of 1,103,574 bp (Fig. 1B). This is estimated to be ∼10% of 21p (Morton 1991; IHGSC 2004). The order and orientation of the BACs in Figure 1 are based on the YAC contig-derived STSs used to identify them.
Figure 1.
Isolation and analysis of 21p BACs. (A) Location of YACs (Wang et al. 1999) and seven STS probes used for screening the CHORI-507 library. (B) Eight BACs identified with 21p STSs from the CHORI-507 library were sequenced. The order shown is based on the order of the STS probes. (C) Dotplot of BAC 264C1 shows that there are no large regions of satellite or highly repetitive sequence. (Inset) Magnification of the first 1500 bp shows that there is a small region of satellite III at one end of the clone. The pattern of satellite III as a variable number of (ATTCC) repeats separated by a ∼10-bp consensus (Prosser et al. 1986) can be seen. (D) Schematic of the five gene models identified within BACs 71C21/54M18. RT-PCR expression patterns of 21pGM9 and 21pGM28 are shown. White boxes represent exons; black arrowheads indicate transcription direction.
Self- and pairwise-dotplots indicated few satellite sequences in these clones (Fig. 1C; Supplemental Fig. 4). Identifying all repetitive elements with RepeatMasker (http://www.repeatmasker.org) showed that the 21p sequence appears to have a repeat content similar to euchromatic regions (Supplemental Table 4). To test this, we compared 21p with the euchromatic portion of the human genome (Lander et al. 2001). The results show that, whilst SINE (fewer, χ2 P < 0.001) and LTR (more, χ2 P < 0.001) compositions are different, LINEs and DNA elements are not significantly different (Supplemental Table 4). The GC content of the 21p sequence is 39.7% (BAC range 34%–44%), very similar to the genome average of 41% (Lander et al. 2001).
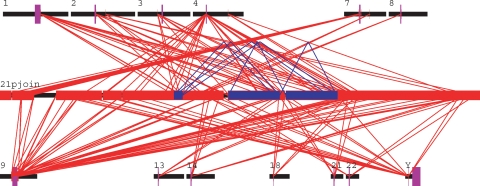
A segmental duplication analysis (sequences >90% and >1 kb in length) of the 21p BAC sequences against the entire human genome (both hg17 and hg18; WGAC method45; Supplemental Table 5) shows that 95.2% (1.05 Mb) of the sequenced 21p region consists of segmental duplications. A total of 278 alignments were identified and are distributed primarily among pericentromeric and subtelomeric regions (Fig. 2). The majority of duplications (249/278, 90%) are interchromosomal as opposed to intrachromosomal. A hundred and seventy (61%) map within 5 Mb of human centromeres, 18 are subtelomeric (2 Mb from telomere), and 90 (32%) map to neither centromeres nor telomeres, including 49 (9%), which map to random chromosomal (unmapped) sequence. Thus, most (61%) of the interchromosomal duplications map to the pericentromeric regions of human chromosomes, particularly chromosomes 1, 2, 4, 7, and 9. The sequence identity spectrum of 21p segmental duplications is skewed toward longer (average 30 kb) and higher identity alignments (average 96.7%) when compared to the genome, pericentromeric or subtelomeric averages. Finally, even though there are few intrachromosomal duplications, several appear to be palindromic in structure similar to what has been observed on the Y chromosome (Skaletsky et al. 2003). Whilst it is clear that 21p largely consists of regions duplicated elsewhere in the genome, given the paucity of sequence data for these regions it is difficult to interpret the exact nature and distribution of the duplications.
Figure 2.
Intrachromosomal and interchromosomal segmental duplications on 21p. 21p sequence is shown in the center (21pjoin) with other chromosomal regions shown above and below. Intrachromosomal (blue) and interchromosomal (red) segmental duplications (>90% identity, >10 kb) between 21p and the human genome (hg18) are depicted. Purple bars indicate the position of centromeres on each chromosome.
A prime motivation for sequencing the human genome is the identification of genes, and the observation that the 21p sequences described here are similar to euchromatic sequences in terms of the repeat content raised the possibility that 21p may also encode genes. We used a combination of ab initio and experimentally based prediction methods to identify 26 gene models (21pGMs). We tested the gene models for expression by RT-PCR in a panel of 24 human tissues; 38% (10/26) were positive by RT-PCR in at least one tissue (Fig. 1D; Supplemental Table 6). For each positive GM, at least one PCR product was sequenced and only those found to be identical to 21p sequence were considered to be positive.
The majority (25/26, 96%) of GMs have open reading frames (ORFs) ranging in length from 120 to 2532 bp (28–843 amino acids). The ORFs were translated and searched by BLAST against protein databases and translated human and other genomes; 10/26 (38%) have significant homology with known human proteins, and these are all conserved in at least one other species (Supplemental Table 7). In addition, 3 (12%) 21pGMs have a CpG island close to their predicted 5′ end, and two of these are expressed. This is low compared to ∼50% of genes in the genome. However, since there are many more predicted CpG islands in the BAC sequences, and in many cases we do not know the location of the start site of the gene, this number is an underestimate of predictions with CpG islands.
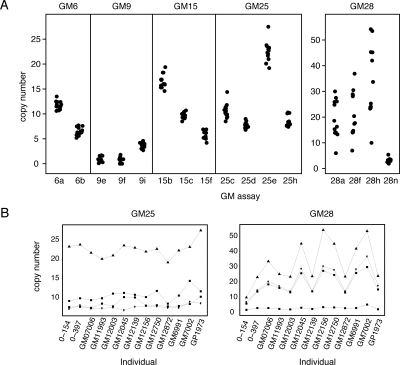
Since sequences on 21p are also found elsewhere in the human genome, five gene models were selected for copy number analysis (21pGM6, 21pGM9, 21pGM15, 21pGM25, 21pGM28). We designed two to four assays per gene model (Supplemental Table 9) and determined the number of copies per genome with quantitative real-time PCR (qPCR) in 13 individuals. 21pGMs are present in multiple copies with considerable inter-individual variation in copy number (Fig. 3A; Supplemental Table 8), ranging from 1 to 27 copies per haploid genome for most of the gene models, and 21pGM28 with 10–54 copies.
Figure 3.
Copy number polymorphism of five gene models in 13 unrelated individuals. (A) Each gene model was tested with two to four assays: X-axis, gene model assay; Y-axis, copy number. Each dot represents an individual. Note the different Y-axis scale for the left and right panels. (B) Graphical representation of the relationship between four different assays for 21pGM25 and 21pGM28 in the 13 individuals.
Noticeably, different assays for a particular gene model often gave different copy number results. For example, 21pGM25 shows no obvious correlation between the copy number of four assays among different individuals (Fig. 3B). For the gene models tested only 21pGM28 showed high correlation between assays in different individuals (Spearman rho 0.71–0.95, P < 0.001; Fig. 3B). Although all the primer sets were selected based on their high efficiency (0.95–1.05), we cannot exclude that the variations between different assays might reflect a technical difference in the qPCR. On the other hand, the extensive nucleotide variability present in the gene models (see below) suggests that the presence of partial deletions/duplications and SNPs might produce such a variable pattern in gene copy number at different loci. Taken together the copy number polymorphism data suggest a complex pattern of partial gene sequence repeats spread over multiple loci in the genome.
To identify if a 21pGM was expressed from a single locus or multiple loci, and to analyze the consequences of the variation on the predicted ORFs, we analyzed RT-PCR sequence variants for gene models 21pGM6, 21pGM9, 21pGM15, 21pGM25, and 21pGM28 from two sources: first, somatic hybrid cell lines carrying single human acrocentric chromosomes to detect the variability within a single chromosome; second, in tissues to detect the genome-wide variability of expressed 21pGMs.
A total of 454 clones were sequenced, aligned, and grouped to identify the minimal number of expressed loci (Supplemental Table 9). This identified both nucleotide variation and alternative splicing (Supplemental Fig. 5). Each gene model has multiple cDNA groups showing that there are a large number of loci expressing similar transcripts. For 21pGM15, 21pGM25, and 21pGM28 one group matched the original gene prediction (Supplemental Table 8); however, for 21pGM6 and 21pGM9 there was no identical match to the 21p prediction. Many of the nucleotide variants introduce a protein truncation codon (PTC) into the ORF compared to the original gene model (Supplemental Table 8). For example, all 21pGM6 variants have a PTC indicating that these loci are probably not translated. For 21pGM9 and 21pGM15, there are variants which extend the ORF giving longer homology with known proteins.
The analysis from monochromosomal somatic cell hybrids shows that the acrocentrics have multiple expressed copies of a 21pGM variant. For example, 21pGM15-related sequences are expressed from at least 11, 7, 8, and 6 loci on HSA13, HSA15, HSA21, and HSA22, respectively. In addition, some variants are present on multiple acrocentric chromosomes. For example, loci identical to 21pGM15 are expressed from HSA13, HSA15, HSA21, and HSA22 in monochromosomal somatic cell hybrids.
The 21p sequence we describe here is similar to the euchromatic portion of the genome (IHGSC 2004) in terms of repeat and transcript content. The data suggest that 21p and other unsequenced regions of the genome contain euchromatic-like sequence. Gene predictions identified 26 gene models, 38% of which were validated by RT-PCR, a confirmed transcript every ∼100 kb, similar to the gene density of the euchromatic portion of the human genome (IHGSC 2004). Comparison of gene prediction confirmation rates with similar approaches for known and novel genes in euchromatic sequence (Harrow et al. 2006) suggests that multiple genes map to heterochromatic regions, but that some of the 21pGMs may correspond to nonexpressed pseudogenes. The situation is similar in the Drosophila and Arabidopsis genomes, where euchromatic-like sequences and functional protein coding genes are embedded within heterochromatic regions (The Arabidopsis Genome Initiative 2000; Hoskins et al. 2002).
A question remains, however: Is the considerable amount of transcription which we observe from functional genes or pseudogenes? It is worth noting that we only looked for spliced genes. Whilst some of the transcripts we observed are obviously pseudogenes because they have PTCs which would truncate the predicted protein (identified by homology), some of the gene models have the characteristics of functional genes, that is, long ORFs (up to 2 kb), correct splicing, homology with known proteins, conserved in multiple species, and tissue-specific expression. 5′ and 3′ RACE will be required to determine the complete structure of these predicted genes. To gain insight into potentially functional genes it will also be necessary to produce antibodies to the putatively expressed proteins to determine their presence in cells and tissues. It is clear, however, that there are unexpected, extensive levels of transcription from 21p and, therefore, we would argue, from other heterochromatic regions of the genome.
The observation of extensive copy number polymorphisms and sequence variation of the transcripts reveals a complex underlying genomic structure. Functional copies of these gene models may be located in chromosomal regions outside of the short arms. The number of functional copies for each gene model is unknown but this may also vary among different individuals. A further unanswered question relates to the minimum copy number required for normal function of the transcript.
The complex duplication architecture of the 21p sequence is reminiscent of what has been seen for human pericentromeric regions (Guy et al. 2003; Grunau et al. 2006). Indeed, She et al. (2004) showed that 35% of pericentromeric regions contain euchromatic-like sequences originating from duplication of euchromatic gene-containing segments of DNA. In this initial analysis we only looked at sequences in proximal 21p (centromere to the ribosomal genes; Fig. 1); almost nothing is known about the distal region (ribosomal to telomere).
The repetitive nature and structural features of HSA21p pose technical challenges to studying the region. Wang et al. (1999) constructed a YAC contig of the proximal part of HSA21p, between the RNR gene cluster and the centromere. This map provides the best physical map of an acrocentric short arm to date and provides a framework for further studies of 21p since it provides locations for STS markers. However, this contig is not suitable as a physical map for sequencing since it excludes the distal portion of the chromosome, and the YACs were isolated from a whole genome library so the chromosome specificity of these regions are not certain. There are two advantages in our strategy of constructing a BAC library from WAV17. First, since 21p is known to be homologous to many regions of the genome, this ensures that any human BACs isolated from the library CHORI-507 which do not map to 21q are derived from 21p. Second, this avoids sequencing multiple alleles with a different repeat content. A limitation of our method, however, is that HSA21p contains many satellite and other repeat sequences, which biases the distribution of restriction enzyme sites. Therefore, the library CHORI-507 will not represent some regions of HSA21p. Countering this potential limitation, however, the method of preparation we used will have depleted many satellite sequences and consequently enriched for euchromatic-like sequences. Since these are potentially the most interesting regions, at least initially, this BAC library therefore provides an important resource for further sequencing of 21p. A library prepared from sheared DNA will be required for completion of the 21p sequence (Osoegawa et al. 2007).
We show that a large gap in the genome, which is known to include heterochromatin and has homology with other heterochromatic regions of the genome, contains sequences which have the properties of euchromatic sequence, including a similar transcript density. This strengthens the case for completing more of the human genome sequence since the data indicate that some of the remaining gaps in the genome do not consist only of repetitive sequence and may harbor functional elements such as genes.
Methods
WAV17
The somatic cell hybrid WAV17 (Slate et al. 1978) was obtained from Coriell (http://ccr.coriell.org/nigms; GM08854) and cultured according to the recommended protocols. The monoallelic composition of HSA21 in WAV17 was assessed by genotyping with markers D21S11, D21S1270, D21S1435, D21S1411, D21S226, and IFNAR1.
Preparation of BAC library CHORI-507
The CHORI-507 BAC library was prepared as described (Osoegawa et al. 1998). Detailed methods are described in Supplemental materials.
Screening BAC library CHORI-507
To identify human clones, CHORI-507 was screened with 32P-labeled α-satellite and Alu probes, and total human genomic DNA. This resulted in 2400 (1.09%) positive clones after screening using α-satellite and Alu probes, and 2752 (1.24%) positives with total human genomic DNA. This is consistent with the fact that the HSA21 content of WAV17 is ∼1%–3% in an aneuploid mouse background. The nonredundant positive clones (3208) were re-arrayed into 384-well plates and gridded on nylon membrane for screening with 21p-specific probes. To identify specific 21p BACs, overgo probes were designed to STSs corresponding to the YAC ends 2E4R, 2E4L, 4E9L, 4E9R, 1B8L, 2C2R, 2C9R from Wang et al. (1999). The library clone size distribution is given in Supplemental Figure 2, and a summary of the library screens is given in Supplemental Tables 1, 2.
FISH
Cell lines were grown in RPMI 1640 with Glutamax I medium (Invitrogen) supplemented with 10% fetal calf serum and a 1% penicillin/streptomycin mix. Cultures were exposed to colcemid (0.1 μg/mL; Invitrogen) for 1.5 h at 37°C and harvested according to routine cytogenetic protocols. G-banding was performed following standard cytogenetic methods. The following probes were used in FISH to check the integrity of HSA21 in the WAV17 cell line: HSA21 paint (Vysis), cosmid c55A10 (Chen et al. 1999), D21Z1 (Maratou et al. 1999), human and mouse Cot-1 DNA (Invitrogen). Interphase nuclei and metaphase spreads were counterstained with DAPI (Vysis) diluted in Vectashield antifade (Vector Labs). BAC and cosmid DNA was labeled with either a Biotin or Digoxigenin-Nick Translation kit (Roche) and detected with anti-dig fluorescein (green) or avidin (red). Cells were viewed under a Nikon fluorescence microscope equipped with appropriate filter combinations. Monochromatic images were captured and superimposed using the Applied Imaging automated imaging system.
BAC sequencing
Detailed methods are described in Supplemental materials.
Gene predictions
EST sequences from dbEST (Boguski et al. 1993) were aligned to the HSA21p sequence using the program EXONERATE (Slater and Birney 2005). We considered only spliced alignments with sequence identity of 90% or higher and at least 88% coverage, and discarded those for which there existed a better spliced alignment elsewhere in the genome. Selected EST alignments were divided into four groups according to the properties of the alignment: best, the best alignment for this EST is in HSA21p sequence; pseudo, the best alignment for this EST is in euchromatin, but this is not spliced, indicating a possible processed pseudogene in the euchromatin; paralog, the coverage of the EST alignment is 100% but the percentage identity is not as high as the best match in euchromatin, hence a potential paralogue; random, there is a better alignment for this EST in an unassembled euchromatin contig. From these alignments we calculated the set of unique exon–exon pairs (75 in total).
Using RepeatMasker (http://www.repeatmasker.org) without the low-complexity filter (option -nolow), we masked the HSA21p sequences and performed in silico gene prediction using the two ab initio gene prediction programs, GeneID (Parra et al. 2000) and GENSCAN (Burge and Karlin 1997), and the comparative gene predictor SGP (Parra et al. 2003), in this case using mouse genome sequence (version mm5) as reference. We thus obtained three sets of predictions which we merged together into a single set of predicted exon–exon pairs removing duplicates. We further removed those exon–exon pairs that overlap with any of the EST exon–exon pairs calculated before. We obtained a set of 182 unique exon–exon pairs obtained by ab initio and comparative gene prediction and not occurring in the EST-predicted set. We classified the 182 exon–exon pairs into those being predicted by all three gene predictors, by two of them, and those that are specific to one gene predictor. This classification, the coordinates, and the sequences of these predictions are available through the on-line Supplemental Material Web site at http://genome.imim.es/datasets/hsa21p.
Exon–exon pairs were assembled into 26 gene models (21pGMs) by aligning them with the genomic sequence and combining exon pairs conforming to the splice-site consensus sequence.
Sequence analysis
For general sequence analysis, programs within the EMBOSS package were used (Rice et al. 2000). BLAST searches were carried out at Ensembl (http://www.ensembl.org). Segmental duplications were detected using a BLAST-based detection scheme (WGAC) 45 to identify all pairwise similarities representing duplicated regions (≥1 kb and ≥90% identity) within the finished BAC sequence of chromosome 21p arm. The 21p BAC sequences were manually joined in their natural order and compared to all other chromosomes in the NCBI genome assembly (hg17 and 18). Satellite repeats were detected using RepeatMasker (version 2002/05/15) on sensitive settings (www.repeatmasker.org) (Smit et al. 1996). The program PARASIGHT (J. Bailey, unpubl.) was used to generate images of pairwise alignments.
RT-PCR
Gene models were tested in 24 human cDNAs (brain, heart, kidney, spleen, liver, colon, small intestine, muscle, lung, stomach, testis, placenta, skin, PBLs, bone marrow, fetal brain, fetal liver, fetal kidney, fetal heart, fetal lung, thymus, pancreas, mammary glands, prostate) as described (Reymond et al. 2002). All amplicons spanned introns.
Gene copy number
Relative DNA copy number was determined as described (Lyle et al. 2006) with some modifications. Detailed methods are described in Supplemental Material. DNAs from a total of 13 individuals were used: eight unrelated individuals and mother and son from CEPH families (Coriell), and three unrelated caucasian individuals from the University of Geneva DNA bank. For each gene prediction a minimum of two assays was designed. Seven assays were used for normalization of input DNA. Control assays were designed in regions that are present in one (AKAP1) and two copies (HBG2) per haploid genome. Assay sequences are available in Supplemental Table 10.
Quantitative PCR reactions were performed using PowerSYBR Green PCR master Mix (Applied Biosystems) and each DNA sample was amplified in three replicates. Fluorescence data were collected using the ABI Prism 7900 Sequence Detection System (Applied Biosystems) and data analysis was performed with Qbase (Hellemans et al. 2007).
Polymorphisms
RNA was obtained from human brain, testis, heart, spleen, thymus, and lung (Ambion) and from somatic cell hybrids carrying human chromosome 13, 14, 15, 21, and 22 (GM10898, GM10479, GM11418, GM08854, GM10888; Coriell). Each predicted gene model was tested with the appropriate primer set (Supplemental Table 10) designed to span the gene model. Detailed methods are described in Supplemental Material.
Acknowledgments
We thank Dr. Jane Hewitt for comments on the manuscript and Dr. Mike Morris for cosmid c55A10 DNA. This work was supported by grants from the Swiss National Science Foundation (R.L., A.R., and S.E.A.), the NCCR Frontiers in Genetics (S.E.A.), the European Commission (BioSapiens NoE to R.G. and S.E.A. and AnEUploidy IP to A.R. and S.E.A.), Blanceflor-Boncompagni Ludovisi Foundation (P.P.) and ChildCare foundations (S.E.A.), a bilateral scientific and technological cooperation project between the University of Antwerp and Shanghai Chinese National Human Genome Centre (C.V.B. and S.-Y.W.), and NIH grant HG002385 (E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org and at genome.imim.es/datasets/hsa21p. The BAC sequence data from this study have been submitted to the EMBL Nucleotide Sequence Database under accession nos. CR381572, CR381670, CR392039, CR381535, CR381653, CR382332, CR382285, and CR382287.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6675307
References
- Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S., Dermitzakis E.T., Reymond A., Deutsch S., Reymond A., Deutsch S., Deutsch S. Chromosome 21 and down syndrome: From genomics to pathophysiology. Nat. Rev. Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Boguski M.S., Lowe T.M.J., Tolstoshev C.M., Lowe T.M.J., Tolstoshev C.M., Tolstoshev C.M. dbEST— Database for “expressed sequence tags.”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chen H., Rossier C., Morris M.A., Scott H.S., Gos A., Bairoch A., Antonarakis S.E., Rossier C., Morris M.A., Scott H.S., Gos A., Bairoch A., Antonarakis S.E., Morris M.A., Scott H.S., Gos A., Bairoch A., Antonarakis S.E., Scott H.S., Gos A., Bairoch A., Antonarakis S.E., Gos A., Bairoch A., Antonarakis S.E., Bairoch A., Antonarakis S.E., Antonarakis S.E. A testis-specific gene, TPTE, encodes a putative transmembrane tyrosine phosphatase and maps to the pericentromeric region of human chromosomes 21 and 13, and to chromosomes 15, 22, and Y. Hum. Genet. 1999;105:399–409. doi: 10.1007/s004390051122. [DOI] [PubMed] [Google Scholar]
- Choo K.H., Earle E., McQuillan C., Earle E., McQuillan C., McQuillan C. A homologous subfamily of satellite III DNA on human chromosomes 14 and 22. Nucleic Acids Res. 1990;18:5641–5648. doi: 10.1093/nar/18.19.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig-Holmes A.P., Shaw M.W., Shaw M.W. Polymorphism of human constitutive heterochromatin. Science. 1971;174:702–704. doi: 10.1126/science.174.4010.702. [DOI] [PubMed] [Google Scholar]
- Cserpán I., Katona R., Praznovszky T., Novák E., Rózsavölgyi M., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Katona R., Praznovszky T., Novák E., Rózsavölgyi M., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Praznovszky T., Novák E., Rózsavölgyi M., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Novák E., Rózsavölgyi M., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Rózsavölgyi M., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Csonka E., Mórocz M., Fodor K., Hadlaczky G., Mórocz M., Fodor K., Hadlaczky G., Fodor K., Hadlaczky G., Hadlaczky G. The chAB4 and NF1-related long-range multisequence DNA families are contiguous in the centromeric heterochromatin of several human chromosomes. Nucleic Acids Res. 2002;30:2899–2905. doi: 10.1093/nar/gkf382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C., Buard J., Brun M.E., De Sario A., Buard J., Brun M.E., De Sario A., Brun M.E., De Sario A., De Sario A. Mapping of the juxtacentromeric heterochromatin-euchromatin frontier of human chromosome 21. Genome Res. 2006;16:1198–1207. doi: 10.1101/gr.5440306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J., Hearn T., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Hearn T., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Mudge J., Viggiano L., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Viggiano L., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Koczan D., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Thiesen H.-J., Bailey J.A., Horvath J.E., Eichler E.E., Bailey J.A., Horvath J.E., Eichler E.E., Horvath J.E., Eichler E.E., Eichler E.E., et al. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10p. Genome Res. 2003;13:159–172. doi: 10.1101/gr.644503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Reymond A., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chen C.-K., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Lagarde J., Gilbert J.G.R., Storey R., Swarbreck D., Gilbert J.G.R., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J., Mortier G., De Paepe A., Speleman F., Vandesompele J., De Paepe A., Speleman F., Vandesompele J., Speleman F., Vandesompele J., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis M., Hindley J., Hindley J. Satellite II DNA of human lymphocytes: Tandem repeats of a simple sequence element. Nucleic Acids Res. 1988;16:363. doi: 10.1093/nar/16.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R.A., Smith C.D., Carlson J.W., Carvalho A.B., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Smith C.D., Carlson J.W., Carvalho A.B., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Carlson J.W., Carvalho A.B., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Carvalho A.B., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Mungall C.J., Sullivan B.A., Sutton G.G., Sullivan B.A., Sutton G.G., Sutton G.G., et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:1–16. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IHGSC (International Human Genome Sequencing Consortium) Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Vissel B., Shaffer L.G., Choo K.H.A., Earle E., Vissel B., Shaffer L.G., Choo K.H.A., Vissel B., Shaffer L.G., Choo K.H.A., Shaffer L.G., Choo K.H.A., Choo K.H.A. A chromosome 13-specific human satellite I DNA subfamily with minor presence on chromosome 21: Further studies on Robertsonian translocations. Genomics. 1993;16:104–112. doi: 10.1006/geno.1993.1147. [DOI] [PubMed] [Google Scholar]
- Kurnit D.M., Roy S., Stewart G.D., Schwedock J., Neve R.L., Bruns G.A.P., Van Keuren M.L., Patterson D., Roy S., Stewart G.D., Schwedock J., Neve R.L., Bruns G.A.P., Van Keuren M.L., Patterson D., Stewart G.D., Schwedock J., Neve R.L., Bruns G.A.P., Van Keuren M.L., Patterson D., Schwedock J., Neve R.L., Bruns G.A.P., Van Keuren M.L., Patterson D., Neve R.L., Bruns G.A.P., Van Keuren M.L., Patterson D., Bruns G.A.P., Van Keuren M.L., Patterson D., Van Keuren M.L., Patterson D., Patterson D. The 724 family of DNA sequences is interspersed about the pericentromeric regions of human acrocentric chromosomes. Cytogenet. Cell Genet. 1986;43:109–116. doi: 10.1159/000132305. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lyle R., Wright T.J., Clark L.N., Hewitt J.E., Wright T.J., Clark L.N., Hewitt J.E., Clark L.N., Hewitt J.E., Hewitt J.E. The FSHD associated repeat, D4Z4, is a member of a dispersed family of homeobox containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- Lyle R., Radhakrishna U., Blouin J.-L., Gagos S., Everman D.B., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Radhakrishna U., Blouin J.-L., Gagos S., Everman D.B., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Blouin J.-L., Gagos S., Everman D.B., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Gagos S., Everman D.B., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Everman D.B., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Gehrig C., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Delozier-Blanchet C., Solanki J.V., Patel U.C., Nath S.K., Solanki J.V., Patel U.C., Nath S.K., Patel U.C., Nath S.K., Nath S.K., et al. Split-hand/split-foot malformation 3 (SHFM3) at 10q24, development of rapid diagnostic methods and gene expression from the region. Am. J. Med. Genet. A. 2006;140:1384–1395. doi: 10.1002/ajmg.a.31247. [DOI] [PubMed] [Google Scholar]
- Maratou K., Siddique Y., Kessling A.M., Davies G.E., Siddique Y., Kessling A.M., Davies G.E., Kessling A.M., Davies G.E., Davies G.E. Novel methodology for the detection of chromosome 21-specific α-satellite DNA sequences. Genomics. 1999;57:429–432. doi: 10.1006/geno.1999.5786. [DOI] [PubMed] [Google Scholar]
- Morton N.E. Parameters of the human genome. Proc. Natl. Acad. Sci. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K., Woon P.Y., Zhao B., Frengen E., Tateno M., Catanese J.J., de Jong P.J., Woon P.Y., Zhao B., Frengen E., Tateno M., Catanese J.J., de Jong P.J., Zhao B., Frengen E., Tateno M., Catanese J.J., de Jong P.J., Frengen E., Tateno M., Catanese J.J., de Jong P.J., Tateno M., Catanese J.J., de Jong P.J., Catanese J.J., de Jong P.J., de Jong P.J. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Osoegawa K., Vessere G.M., Li Shu C., Hoskins R.A., Abad J.P., de Pablos B., Villasante A., de Jong P.J., Vessere G.M., Li Shu C., Hoskins R.A., Abad J.P., de Pablos B., Villasante A., de Jong P.J., Li Shu C., Hoskins R.A., Abad J.P., de Pablos B., Villasante A., de Jong P.J., Hoskins R.A., Abad J.P., de Pablos B., Villasante A., de Jong P.J., Abad J.P., de Pablos B., Villasante A., de Jong P.J., de Pablos B., Villasante A., de Jong P.J., Villasante A., de Jong P.J., de Jong P.J. BAC clones generated from sheared DNA. Genomics. 2007;89:291–299. doi: 10.1016/j.ygeno.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Blanco E., Guigo R., Blanco E., Guigo R., Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Wiehe T., Fickett J.W., Guigo R., Fickett J.W., Guigo R., Guigo R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J., Frommer M., Paul C., Vincent P.C., Frommer M., Paul C., Vincent P.C., Paul C., Vincent P.C., Vincent P.C. Sequence relationships of three human satellite DNAs. J. Mol. Biol. 1986;187:145–155. doi: 10.1016/0022-2836(86)90224-x. [DOI] [PubMed] [Google Scholar]
- Reymond A., Marigo V., Yayiaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Marigo V., Yayiaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Yayiaoglu M.B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Scamuffa N., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Caccioppoli C., Dermitzakis E.T., Lyle R., Banfi S., Dermitzakis E.T., Lyle R., Banfi S., Lyle R., Banfi S., Banfi S., et al. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420:582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- She X., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Sutton G., Halpern A.L., Eichler E.E., Halpern A.L., Eichler E.E., Eichler E.E. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Repping S., Pyntikova T., Ali J., Bieri T., Pyntikova T., Ali J., Bieri T., Ali J., Bieri T., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Slate D.L., Shulman L., Lawrence J.B., Revel M., Ruddle F.H., Shulman L., Lawrence J.B., Revel M., Ruddle F.H., Lawrence J.B., Revel M., Ruddle F.H., Revel M., Ruddle F.H., Ruddle F.H. Presence of human chromosome 21 alone is sufficient for hybrid cell sensitivity to human interferon. J. Virol. 1978;25:319–325. doi: 10.1128/jvi.25.1.319-325.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G.S., Birney E., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F.A., Hubley R., Green P., Hubley R., Green P., Green P. 1996 RepeatMasker Open-3.0. http://www.repeatmasker.org/ [Google Scholar]
- Tapparel C., Reymond A., Girardet C., Guillou L., Lyle R., Lamon C., Hutter P., Antonarakis S.E., Reymond A., Girardet C., Guillou L., Lyle R., Lamon C., Hutter P., Antonarakis S.E., Girardet C., Guillou L., Lyle R., Lamon C., Hutter P., Antonarakis S.E., Guillou L., Lyle R., Lamon C., Hutter P., Antonarakis S.E., Lyle R., Lamon C., Hutter P., Antonarakis S.E., Lamon C., Hutter P., Antonarakis S.E., Hutter P., Antonarakis S.E., Antonarakis S.E. The TPTE gene family: Cellular expression, subcellular localization and alternative splicing. Gene. 2003;323:189–199. doi: 10.1016/j.gene.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G., Giordano M., Cobianchi F., Riva S., Biamonti G., Cobianchi F., Riva S., Biamonti G., Riva S., Biamonti G., Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verma R.S., Dosik H., Lubs H.A., Dosik H., Lubs H.A., Lubs H.A. Size variation polymorphisms of the short arm of human acrocentric chromosomes determined by R-banding by fluorescence using acridine orange (RFA) Hum. Genet. 1977;38:231–234. doi: 10.1007/BF00527408. [DOI] [PubMed] [Google Scholar]
- Wang S.Y., Cruts M., Del Favero J., Zhang Y., Tissir F., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Cruts M., Del Favero J., Zhang Y., Tissir F., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Del Favero J., Zhang Y., Tissir F., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Zhang Y., Tissir F., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Tissir F., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Potier M.C., Patterson D., Nizetic D., Bosch A., Chen H., Patterson D., Nizetic D., Bosch A., Chen H., Nizetic D., Bosch A., Chen H., Bosch A., Chen H., Chen H., et al. A high-resolution physical map of human chromosome 21p using yeast artificial chromosomes. Genome Res. 1999;9:1059–1073. doi: 10.1101/gr.9.11.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: Clinical significance and distribution of breakpoints. Am. J. Hum. Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Waye J.S., Willard H.F., Willard H.F. Human β satellite DNA: Genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl. Acad. Sci. 1989;86:6250–6254. doi: 10.1073/pnas.86.16.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]