Abstract
The latest revolution in the DNA sequencing field has been brought about by the development of automated sequencers that are capable of generating giga base pair data sets quickly and at low cost. Applications of such technologies seem to be limited to resequencing and transcript discovery, due to the shortness of the generated reads. In order to extend the fields of application to de novo sequencing, we developed the SHARCGS algorithm to assemble short-read (25–40-mer) data with high accuracy and speed. The efficiency of SHARCGS was tested on BAC inserts from three eukaryotic species, on two yeast chromosomes, and on two bacterial genomes (Haemophilus influenzae, Escherichia coli). We show that 30-mer-based BAC assemblies have N50 sizes >20 kbp for Drosophila and Arabidopsis and >4 kbp for human in simulations taking missing reads and wrong base calls into account. We assembled 949,974 contigs with length >50 bp, and only one single contig could not be aligned error-free against the reference sequences. We generated 36-mer reads for the genome of Helicobacter acinonychis on the Illumina 1G sequencing instrument and assembled 937 contigs covering 98% of the genome with an N50 size of 3.7 kbp. With the exception of five contigs that differ in 1–4 positions relative to the reference sequence, all contigs matched the genome error-free. Thus, SHARCGS is a suitable tool for fully exploiting novel sequencing technologies by assembling sequence contigs de novo with high confidence and by outperforming existing assembly algorithms in terms of speed and accuracy.
For almost 20 years, the Sanger technique (Sanger et al. 1977) has remained the gold standard for DNA sequence determination. Despite many improvements on the original technology that have boosted both read length and throughput, Sanger-based sequencing of genomes is still a costly enterprise (Bentley 2006). The sequencing field has experienced a major shift by the introduction of innovative sequencing technologies developed by 454 Life Sciences and Solexa/Illumina, each capable of generating sequence data at a fraction of the cost and much quicker when compared to the Sanger method (Margulies et al. 2005; Bentley 2006). Present equipment sold by 454 Life Sciences allows generation of 100 Mbp of sequence data with 200- to 300-bp reads within 8 h. The Illumina 1G sequencer generates up to 1.3 Gbp in 25- to 36-bp reads in a single 80-h run. The advent of this second generation of DNA sequencing technology raises the question whether, despite the shortness of reads, accurate assembly can be computed at an acceptable computational cost de novo.
For the assembly of sequence fragments of ∼500 bp in length, a variety of sophisticated assembly algorithms have been suggested, including PHRAP (Ewing and Green 1994), the TIGR assembler (Sutton et al. 1995), CAP3 (Huang and Madan 1999), the Celera assembler (Myers et al. 2000), ARACHNE (Batzoglou et al. 2002; Jaffe et al. 2003), Phusion (Mullikin and Ning 2003), and EULER (Pevzner et al. 2001). These algorithms proved useful on very different shotgun sequencing projects, such as the fruit fly genome (Adams et al. 2000), the draft of the pufferfish genome (Aparicio et al. 2002), and the mouse genome (Waterston et al. 2002). In sequencing projects that use Sanger technology, genomes are typically covered 6- to 10-fold. To assemble such data sets, the algorithms described above put great emphasis on the optimal exploitation of all reads. Issues like the correction of sequencing errors and the assembly of reads containing mismatches increase the complexity of these algorithms. Due to their complexity, existing assemblers are incapable of assembling very large numbers of reads, even on very large computers.
Given the low cost of sequence data generated by second-generation sequencing instruments, sequence fragments can be provided to cover a target sequence in excess of 100-fold. When the resulting huge numbers of very short reads are assembled, simplicity becomes a virtue for an assembly algorithm. One approach to assemble reads as short as 25 bases was reported by Warren et al. (2007). The described program SSAKE, however, generates misassemblies due to contig extension across repeat borders and due to difficulties in handling erroneous reads. Here, we describe SHARCGS (SHort-read Assembler based on Robust Contig extension for Genome Sequencing), which is capable of assembling millions of very short reads, copes with sequencing errors, and virtually never generates misassemblies.
In this paper, we first demonstrate the feasibility of assemblies using 25–40 base reads under idealized conditions, assuming that all possible reads of a given length are available and no sequencing errors are permitted. Next, we focus on a realistic scenario, showing how to cope with missing reads and how to detect reads that contain sequencing errors out of a large pool of reads. We calculate assemblies for simulated reads of sequenced BAC clones from Arabidopsis, Drosophila, and human, as well as simulated reads of microbial sequences. Finally, we generate a 36-mer read data set from Helicobacter acinonychis (genome size 1.55 Mbp) on the Illumina 1G sequencing instrument and use SHARCGS to assemble the genome based on these data.
Results and Discussion
Assembling simulated short read (25–40-mer) data under idealized assumptions
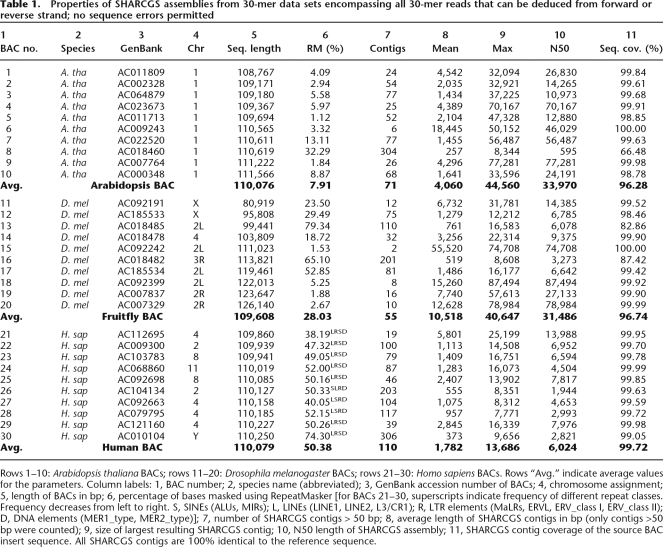
Assuming that all possible reads are present in the data set, no sequencing errors have occurred, and read length is fixed, the quality of the assembly solely depends on intrinsic properties of the sequence. We assembled simulated ideal 30-mer reads from a set of 60 BAC insert sequences with an average length of 110 kbp each, derived from Arabidopsis thaliana, Drosophila melanogaster, and Homo sapiens (Table 1 and Supplementary Table 1). We found between 1 (Drosophila, BAC supp20) and 520 (Human, BAC supp26) SHARCGS contigs per BAC. As a quality measure for the assembled SHARCGS contigs, we compared the N50 sizes of the assemblies from different BACs. Both Arabidopsis and Drosophila BACs had an average N50 length of >30 kbp, and human BACs were assembled to an average N50 length of 5–6 kbp.
Table 1.
Properties of SHARCGS assemblies from 30-mer data sets encompassing all 30-mer reads that can be deduced from forward or reverse strand; no sequence errors permitted
Rows 1–10: Arabidopsis thaliana BACs; rows 11–20: Drosophila melanogaster BACs; rows 21–30: Homo sapiens BACs. Rows “Avg.” indicate average values for the parameters. Column labels: 1, BAC number; 2, species name (abbreviated); 3, GenBank accession number of BACs; 4, chromosome assignment; 5, length of BACs in bp; 6, percentage of bases masked using RepeatMasker [for BACs 21–30, superscripts indicate frequency of different repeat classes. Frequency decreases from left to right. S, SINEs (ALUs, MIRs); L, LINEs (LINE1, LINE2, L3/CR1); R, LTR elements (MaLRs, ERVL, ERV_class I, ERV_class II); D, DNA elements (MER1_type, MER2_type)]; 7, number of SHARCGS contigs > 50 bp; 8, average length of SHARCGS contigs in bp (only contigs >50 bp were counted); 9, size of largest resulting SHARCGS contig; 10, N50 length of SHARCGS assembly; 11, SHARCGS contig coverage of the source BAC insert sequence. All SHARCGS contigs are 100% identical to the reference sequence.
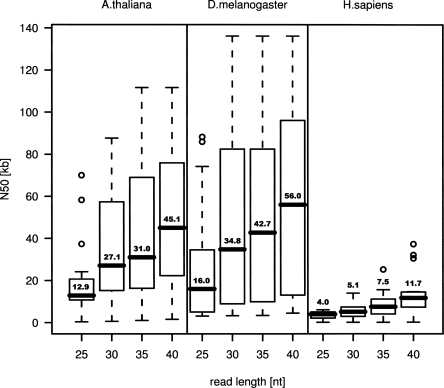
The results in Table 1 and Supplementary Table 1 highlight the quality optimum that can be achieved in 30-mer-based assemblies. However, the assembly can be improved by using longer reads. Figure 1 summarizes the N50 changes when contig assembly is carried out with 25–40-mer reads, respectively, calculated from assemblies with 20 BACs per species. There is a clear tendency toward larger N50. For instance, an increase of read length from 30 to 40 bases almost doubled N50 in all three species. Longer reads have a better performance in bridging regions of ambiguity or low complexity, resulting in larger sequence contigs.
Figure 1.
Distributions of N50 values in relation to different read length illustrated using box plots. An N50 length of x kbp indicates that 50% of the source sequence is covered by contigs of x kbp or larger. The data was collected from assemblies under perfect assumptions (all possible reads are present exactly once, no sequencing errors), based on 20 BACs with an average size of 110 kbp for each species. The boxes stretch from the first to the third quartile of the respective distribution, thus covering 50% of the corresponding data. The boxes are split with a line at the median. For sake of clarity, the median value is given for each box plot. Whiskers indicated with dashed lines are 1.5 times longer than the box but do not stretch beyond minima and maxima. They are used to define the outliers, data points outside the range of the whiskers, which are marked as small circles.
Repeat content and assembly quality
To determine the influence of the repeat content on the assembly quality, we analyzed the GenBank BAC sequences with RepeatMasker (www.repeatmasker.org). While human BACs, with the exception of Y chromosome derived clones and a few outliers, showed uniform repeat content (40%–55%), the Drosophila set showed high variance with some BACs being almost repeat-free (1% repeat content) and some containing >50% repetitive DNA. The data show that a higher fraction of repetitive regions results in shorter N50 lengths. We got the best SHARCGS assemblies in Drosophila for BACs with a repeat content of ∼5% or lower, resulting in N50 of >57 kbp. Repeats that occur only once per BAC, or which are very diverged, will essentially behave like single-copy sequences and will not affect assembly quality. To assess whether the number of low-complexity (LC) or simple repeats (SR), as identified by RepeatMasker, have an effect on the assembly quality, we counted the number of such sequence segments per BAC. Arabidopsis BACs contained 40 LC/SRs on average, BACs from Drosophila contained 44 LC/SRs, and human BACs contained 37 LC/SRs (numbers based on clones in Table 1). Since the N50 for human BAC assemblies are the lowest in our sample, we conclude that LC/SRs in this data set have no effect onto SHARCGS assembly. This is supported by closer inspection of the data: For instance, Drosophila BAC 13 (N50 of 6 kbp) contained 19 LC/SRs, while BAC 18 contained 61 LC/SRs and had an N50 of 87 kbp. With very few exceptions, SHARCGS contigs covered the inserts of BACs > 99%. Two outliers were BAC 13 and BAC 8. BAC 13 was exceptionally repeat-rich (79% repetitive DNA), while the insert of BAC 8 (repeat content 32%) contained 38 kbp of highly similar tandem repeats (Supplementary Fig. 1).
Assembling simulated short-read data with missing reads and sequencing errors
Under real conditions, the number of reads that start at a given position of the target sequence can vary so that more than one read or none at all are available. In addition, a certain chance for wrong base calls exists.
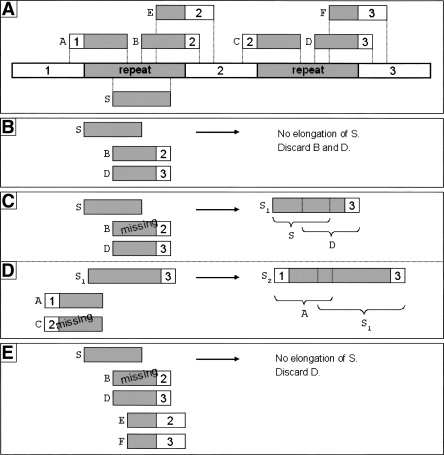
Missed positions (or “gaps”) result in decreased contig lengths, as one can expect breaks to occur whenever contig extension fails due to missing reads. The more serious problem might be wrong assembly if ambiguities (or “forks”) are not detected because of missing reads. This problem and its solution implemented in SHARCGS are illustrated in Figure 2. For each read R that is to be assembled onto a contig, SHARCGS inspects both the forward and reverse strand for ambiguities. Read R is only used to extend the contig, if other reads matching the contig with shorter overlaps do not lead to ambiguities. In this manner, gaps can be checked safely for ambiguities (see Methods for details).
Figure 2.
Detection of ambiguities during contig elongation despite missing read information. (A) Target sequence to assemble. Repetitive segments are shown in gray, unique sequence segments are shown in white and indicated with numbers. The positions of reads A–F and S are also shown in panel A. These reads are used to explain the principles of error-free contig extension in panels B–E. (B) Assembly under idealized assumptions (all reads present). Read S to be elongated to the right. An ambiguity is found, both reads B and D could extend read S; neither read D nor B will be assembled. (C, D) Assembly under real assumptions, but without robust contig extension. (C) Read S to be elongated to the right, read B is missing so that the ambiguity cannot be detected; read D would be assembled to S. (D) Naively built contig S1 will be elongated to the far right end (not shown), then the elongation to the left starts, one crucial read is missing again (read C) and the ambiguity cannot be detected; read A would be assembled to contig S1, wrongly connecting sequence segment 1 with sequence segment 3. (E) Assembly under real assumptions with SHARCGS’ robust contig extension. Different overlaps are checked before assembling D, and if any ambiguity is found for smaller overlaps the assembly of D will be rejected. Since both reads E and F could be assembled to the read S, read D will not be assembled to read S.
In a BAC-based sequencing project, sequencing from BAC pools increases efficiency in terms of time and money. The resulting data sets contain reads from more than a single clone. The sequencing technology itself can be used to discriminate between reads derived from individual clones by using indices. We assumed distinguishable, barcoded BACs, and we simulated reads for 1–16 different BACs per pool. Increasing the pool size implies that fewer reads are available per BAC, and thus the coverage per BAC is decreasing. Assuming 5 million 30-mer reads per sample, the coverage is 1363/n, with n indicating the number of BACs per pool (insert size 110 kbp per BAC).
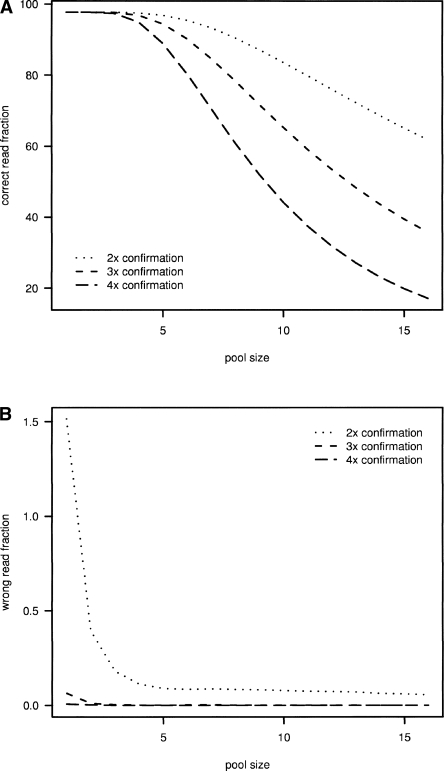
To avoid assembling faulty reads, we assumed that it is unlikely that the same error occurs multiple times in a data set of limited size. Thus, the correct reads for a certain base must outnumber reads with a wrong base call at this position. By comparing the read counts, we are able to determine a read data set that can be used for the assembly, e.g., reads that are present at least three times. The number of correct reads per BAC decreases with increasing complexity of the BAC pool, since fewer reads are available for the individual BAC (Fig. 3A). For example, assuming a sequencing error rate of 0.6%, threefold confirmation of a 30-mer read results in 94% of reads retained in a pool of 5 BACs (coverage ∼270-fold) and 65% of reads being retained in a pool of 10 BACs (coverage ∼135-fold), assuming average insert size of 110 kbp per BAC. Figure 3B indicates the proportion of false reads per BAC that remain in the data set after applying a two- to fourfold redundancy filtering, depending on pool size. Particularly for small pools, twofold redundancy is not sufficient and the data set contains many false reads. Conversely, three- to fourfold redundancy checks constitute reliable filters that ensure that pools of size two or larger will be essentially free of false reads (Fig. 3B). Since fourfold redundancy filtering leads to a rapid decrease of reads available for assembly, threefold filtering seems to be optimal (Fig. 3A). However, SHARCGS adapts this filtering criterion to the input data and combines contigs generated for up to three filter settings. Furthermore, a second filtering step described in more detail in the Methods section allows the use of reads which are confirmed only twice.
Figure 3.
Read filtering and selection prior to assembly to avoid inclusion of reads containing sequencing errors. (A) Proportion of correct reads that remain in the data set by applying two- to fourfold redundancy filtering (i.e., by counting reads present at least twice, threefold, or fourfold in the data set) depending on BAC pool size (average insert size per BAC 110 kbp). (B) Proportion of false reads that remain in the data set for assembly after application of two- to fourfold redundancy filtering. Data were simulated with 0.6% error rate.
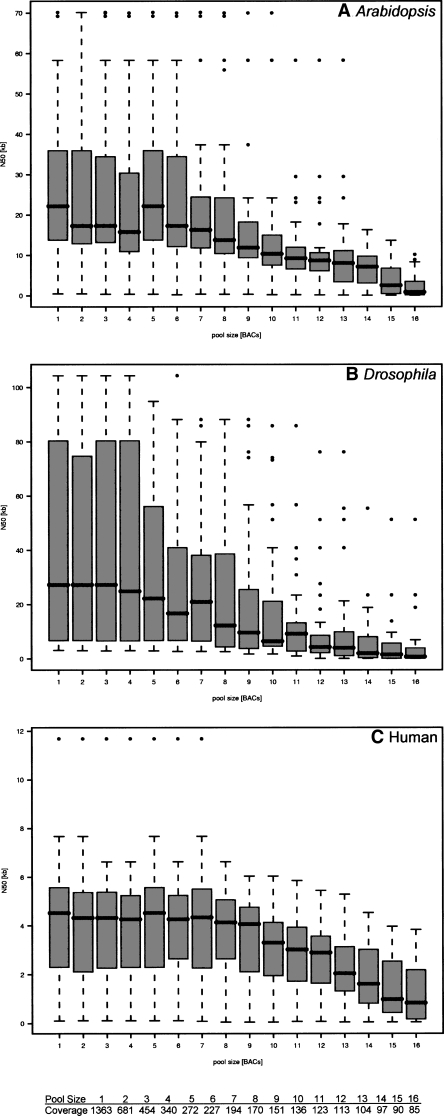
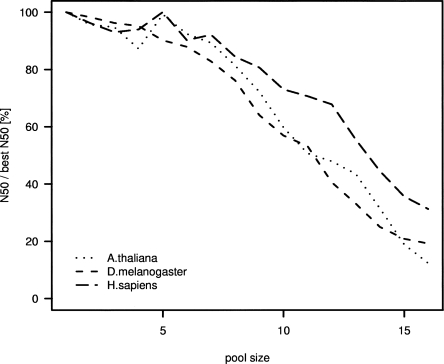
We applied all the features above and assumed a sequencing error rate of 0.6% to the assembly of the set of 60 BACs described earlier (Fig. 4A–C). The SHARCGS assembly took ∼15 min per BAC on a Dual Xeon 2.8-GHz 32-bit Linux machine with 4 GB of RAM. In the context of these assemblies we calculated 942,380 contigs > 50 bp, corresponding to 489.59 Mbp. All these contigs, with one single exception, could be aligned perfectly without mismatches or gaps against the BAC reference sequences. The only falsely assembled contig we observed was 57 bp in size and was derived from reads from a conserved 30-bp inverted repeat located within Drosophila BAC supp20 (GenBank AC099006). From these data, we can conclude that our algorithm is perfectly reliable with respect to assembling contigs de novo from short reads and that our filtering to remove reads containing sequencing errors is efficient. The box plots in Figure 4A–C illustrate the feasibility to assemble BACs from indexed pools. Noteworthy is the relatively constant average N50 that is observed for pooled clones, even though the variance is high because of sequence differences inherent in BACs (see Table 1, e.g., repeat content). In both Arabidopsis and Drosophila, 5–6 BACs can be pooled to still achieve an N50 at ∼20 kbp, and for human BACs we find an N50 of 4–5 kbp for pools of up to 9 BACs. With increased pool sizes, contigs become progressively shorter. This can be explained by redundancy checks becoming so stringent that sufficient reads are not retained for the assembly. Figure 5 shows this effect in comparison to the N50 sizes of assemblies based on simulated reads under ideal assumptions for different pool sizes.
Figure 4.
Dependency of N50 on BAC pool size for three different species (A–C). The data shown were collected from assemblies under realistic assumptions, with an error rate of 0.6% per base call and reads generated from randomly chosen positions. We mimic the BAC pool size by generating the proportional number of reads, i.e., for pool size n, we generate 5 million/n reads per BAC, corresponding to a coverage of 1363/n. BAC pool sizes (1–16) and corresponding coverages are indicated at the bottom of the figure. For each species we used 20 BACs with average length 110 kbp and simulated 30-mer data sets three times per BAC. Thus, for each pool size distribution, 60 assembly results are shown using box plots. Boxes stretch from the first to the third quartile of the respective distribution and cover 50% of the corresponding data. The median inside each box is indicated with a line. Whiskers (dashed lines) are 1.5 times longer than the box but do not stretch beyond minima and maxima and are used to define the outliers (small circles).
Figure 5.
Summary of decrease in N50 size due to pooling (for corresponding coverages, see Fig. 4). The plot compares N50 values for a given pool size with the optimal N50 achieved for pool size 1. The lines show the average fraction of N50 per pool size divided by the optimal (“best”) N50. For each pool size, we average over 20 BACs and three simulations each, using the same data as in Fig. 4.
Assembling simulated short-read data of microbial sequences
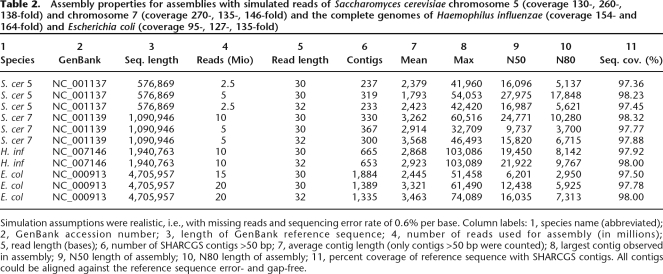
To further evaluate the prospects of SHARCGS, we assembled yeast chromosomes and bacterial genomes based on simulated 30- or 32-mer data sets under realistic assumptions as described above (missing reads, 0.6% sequencing error rate; Table 2). We chose Saccharomyces cerevisiae chromosomes V (0.58 Mbp) and VII (1.09 Mbp), as well as the genomes of Haemophilus influenzae (1.94 Mbp) and E. coli (4.71 Mbp). Depending on sequencing depth (coverage range 130- to 270-fold), we obtained an N50 of up to 28 kbp for yeast chromosome V and up to 18 kbp for yeast chromosome VII. The bacterial genomes (coverage range 95- to 164-fold) could be assembled with an N50 of 22 kbp (H. influenzae) and 16 kbp (E. coli). In all these assemblies the sequence coverage exceeded 97% and all contigs were error-free, as determined by matching them back against the reference sequences.
Table 2.
Assembly properties for assemblies with simulated reads of Saccharomyces cerevisiae chromosome 5 (coverage 130-, 260-, 138-fold) and chromosome 7 (coverage 270-, 135-, 146-fold) and the complete genomes of Haemophilus influenzae (coverage 154- and 164-fold) and Escherichia coli (coverage 95-, 127-, 135-fold)
Simulation assumptions were realistic, i.e., with missing reads and sequencing error rate of 0.6% per base. Column labels: 1, species name (abbreviated); 2, GenBank accession number; 3, length of GenBank reference sequence; 4, number of reads used for assembly (in millions); 5, read length (bases); 6, number of SHARCGS contigs >50 bp; 7, average contig length (only contigs >50 bp were counted); 8, largest contig observed in assembly; 9, N50 length of assembly; 10, N80 length of assembly; 11, percent coverage of reference sequence with SHARCGS contigs. All contigs could be aligned against the reference sequence error- and gap-free.
Assembling Illumina short-read data of the H. acinonychis genome
In order to evaluate SHARCGS’ performance on real data, we chose to sequence and assemble the genome of H. acinonychis, a bacterial species isolated from the stomach of large felines. The sequences of the H. acinonychis genome (1.55 Mbp) and of the H. acinonychis plasmid pHac1 (3661 bp) were determined recently with Sanger technology (Eppinger et al. 2006). The H. acinonychis genome contains 38% GC and 90% coding regions (Eppinger et al. 2006), and the total repeat content is 0.48% (7509 bp) according to RepeatMasker analysis. In detail, RepeatMasker found five reverse transcriptase homologs (340 bp, 0.02%), 14 small RNAs (932 bp, 0.06%), 10 simple repeats (760 bp, 0.05%), and 141 low-complexity regions (5479 bp, 0.35%). Based on the same DNA preparation that Eppinger and coworkers had used for Sanger sequencing of the H. acinonychis genome, we generated 12.3 million 36-mer reads on the Illumina 1G sequencing instrument (representing 272-fold coverage). To determine a per-base error rate, we matched these reads against the H. acinonychis sequence using Illumina’s ELAND software. ELAND reported 110,569 reads matching to the plasmid and 8,278,979 reads matching to the genome: 512,436 (4.2%) reads did not pass ELAND’s quality check based on the first 32 base calls of each read; 6,652,321 (54.1%) reads matched exactly to the reference sequences (H. acinonychis or plasmid); 1,475,601 (12.0%) contained one mismatch; 682,388 (5.6%) reads contained two mismatches; and 2,966,045 (24.1%) reads could not be matched to the reference sequences. This translates into a per-base error rate of at least 1.54% for the data set.
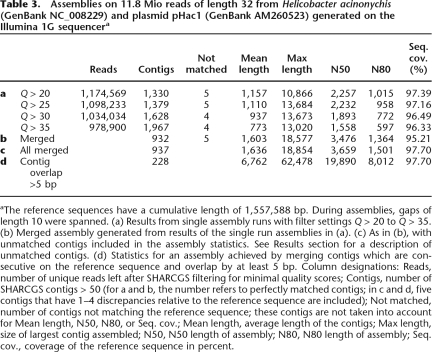
We removed reads containing unspecified bases from the complete data set of 12.3 million reads and trimmed the last four bases of each read. We assembled the remaining 11.8 million reads without taking ELAND results into account, thus relying exclusively on read filtering and assembly procedures implemented within SHARCGS. Different from the assemblies based on simulated reads described above, the filtering of Illumina read data relied on the utilization of cumulative base quality values (see Methods). We used four different base quality thresholds (Q > 20, Q > 25, Q > 30, and Q > 35) to generate four read data sets (Table 3a). For instance, the read data sets Q > 20 and Q > 35 contained 1,174,569 and 978,900 unique reads, respectively, to be left for contig assembly. Each read data set was assembled separately. Thereafter, we took advantage of SHARCGS’ feature to automatically merge contigs generated from different filter settings (Table 3b). The inclusion of reads that contain sequencing errors (reads are retained when applying weak filtering criteria) result in contig breaks, and missing reads (reads are removed when applying strong filtering criteria) result in contig breaks, too. However, runs with different filtering criteria result in contig breaks at different positions. Merging overlapping contigs generated in different assembly runs therefore improves the assembly quality (see Methods for details). In our case, assemblies from single runs contained 1303–1948 contigs with N50 between 1558 and 2257 bp. For the final H. acinonychis assembly we merged the contigs from these four assembly runs, resulting in an increased N50 size by more than 50% relative to the best single assembly. The final assembly of H. acinonychis contains 937 contigs that cover 98% of the genome with an N50 of 3659 bp: 932 of these contigs are error-free, and 5 contigs contain mismatches at 1–4 positions. In three of these five contigs, discrepancies between the SHARCGS assembly and the reference sequence are supported by 103, 117, or 198 Illumina reads, respectively. These high numbers of supporting reads suggest that the sequence based on Illumina reads is correct. The remaining two discrepancies are located at the ends of SHARCGS contigs.
Table 3.
Assemblies on 11.8 Mio reads of length 32 from Helicobacter acinonychis (GenBank NC_008229) and plasmid pHac1 (GenBank AM260523) generated on the Illumina 1G sequencera
aThe reference sequences have a cumulative length of 1,557,588 bp. During assemblies, gaps of length 10 were spanned. (a) Results from single assembly runs with filter settings Q > 20 to Q > 35. (b) Merged assembly generated from results of the single run assemblies in (a). (c) As in (b), with unmatched contigs included in the assembly statistics. See Results section for a description of unmatched contigs. (d) Statistics for an assembly achieved by merging contigs which are consecutive on the reference sequence and overlap by at least 5 bp. Column designations: Reads, number of unique reads left after SHARCGS filtering for minimal quality scores; Contigs, number of SHARCGS contigs > 50 (for a and b, the number refers to perfectly matched contigs; in c and d, five contigs that have 1–4 discrepancies relative to the reference sequence are included); Not matched, number of contigs not matching the reference sequence; these contigs are not taken into account for Mean length, N50, N80, or Seq. cov.; Mean length, average length of the contigs; Max length, size of largest contig assembled; N50, N50 length of assembly; N80, N80 length of assembly; Seq. cov., coverage of the reference sequence in percent.
The assembly described here was carried out entirely de novo, i.e., solely depending upon the read sequences and base quality values. We positioned the contigs on the reference genome and found that many contigs overlapped. By merging contigs overlapping more than five bases, our assembly collapsed into 228 contigs, increasing the N50 length 5.4-fold (Table 3d). This suggests that microread-based assemblies can be improved with modest additional effort, by carrying out a hybrid assembly together with low, e.g., one- or twofold coverage of the target genome in 454 or Sanger reads.
Comparative evaluation of SHARCGS
Warren et al. (2007) recently presented SSAKE, an algorithm to be used for assembling short-read sequences. To evaluate the performance of SHARCGS relative to SSAKE, we compared assembly results achieved with either algorithm. We chose eight BACs (three each for Drosophila and Arabidopsis and two for human) out of our 60-BAC test set, namely for each species the clone with the largest insert, as well as the clones with the largest and the minimal proportion of repetitive DNA. For human, BACs were selected from the autosomes, thereby excluding the Y chromosome, which has atypical genomic organization. The largest human clone was at the same time the clone with the highest repeat content.
Even though SSAKE does have a “stringent” mode implemented that should in principle avoid the extension of contigs across regions of ambiguity, in practice a large number of false joins were observed. In realistic simulations with missing reads and sequencing errors, 25% (on average, range 0%–58%) of SSAKE contigs could not be aligned to the BAC reference sequence. This indicates that the performance of SSAKE is particularly vulnerable to the presence of sequencing errors in a read data set. Considering only correct SSAKE contigs, an average BAC insert coverage of 76% was determined. In contrast, all contigs generated by SHARCGS could be perfectly aligned to the source sequences. The fraction of each BAC insert sequence covered with SHARCGS contigs was on average 93% (Supplementary Table 2). We conclude that the large proportion of unmatched contigs will seriously limit the utility of the Warren et al. (2007) algorithm as a tool for de novo assembly.
We have attempted a similar comparison with the Euler2 algorithm from Pevzner et al. (2001). Although we have tweaked the source code to make Euler2 accept millions of input reads, the algorithm did not finish a single assembly under realistic assumptions within one day on a 64-GB Opteron machine. The large amounts of input data apparently cause this assembler to generate exceedingly large A-Bruijn graphs with many millions of nodes. In particular when many sequencing errors render the graphs complex, Euler2 runs into performance problems. Under idealized assumptions, Euler2 was able to generate assemblies for five of our BACs. The resulting Euler2 assemblies, however, contained various wrong contigs of length up to 33 kbp. The correct contigs covered between 33% and 95% of the target sequence with N50 between 53 bp and 51 kbp. We conclude from these results that Euler2 is not well adapted to this particular assembly task.
Conclusions
There are multiple applications for microread-sequencing technologies, including genome resequencing, genotyping, transcript identification and profiling, transcription factor binding site analysis, and methylome characterization. SHARCGS extends the portfolio of applications to comprise de novo sequence assembly. Our evaluation of SHARCGS was based both on simulated reads and on reads generated on the Illumina 1G sequencer. We have shown that SHARCGS generates error-free contigs from short-read data containing as much as 1.5% of sequencing errors, as long as high coverage of the target genome is provided (200-fold or more). With these conditions, we assembled a bacterial genome with a size of 1.6 Mpb (H. acinonychis). Assuming that the accuracy of short-read sequencing technologies will improve, larger bacterial genomes are realistic targets, as shown for the 4.4-Mbp genome of E. coli on simulated reads with an error rate of 0.6%. To assemble more complex eukaryotic genomes, a BAC-wise approach may be considered. The choice of the BAC pool size depends on the repeat content of the genome.
In the present implementation of our algorithm, single-nucleotide polymorphisms would cause spurious contig breaks. If BACs from the same chromosomal region derived from different haplotypes were sequenced in one single pool, sequence variants may be discarded from the data set, unless indices were used.
Microread-based assemblies can easily be improved, e.g., by integration of reads from HTP pyrosequencing or conventional Sanger reads at low coverage. These modifications consolidate the assembly by bridging gaps and regions of ambiguity.
Methods
The SHARCGS algorithm consists of a filtering step to avoid reads with sequencing errors, an assembly step to generate contigs, and a final contig merging step including computation of quality measures.
Filtering for confirmed reads
SHARCGS obtains a large set of reads of equal length, optionally augmented by quality scores per base call, as its input. Thereby, it is adapted to cope with a substantial number of reads with sequencing errors. When the error rate per base call is 0.6%, about 16% of 30-mer reads contain errors. Pcorrect, the probability that a single read contains no sequencing errors can be estimated by the product of all its base calls being correct independently. Thus:
where Perror represents the error rate per base call and r the read length.
In order to assemble reliable contigs, we suggest removing unconfirmed low-quality reads from the assembly. We consider a read as being confirmed when the following two conditions hold: Reads are generated multiple times, and overlapping partners exist. Without quality scores, SHARCGS simply filters for reads generated at least n times, n being a parameter of this filtering step. When quality measures are available, the first filtering step is modified: SHARCGS then filters for reads having high minimal quality values. Quality values of all occurrences of a read on the same or the opposite strand are combined, similar to Phrap (Ewing and Green 1994), i.e., the maximum is used when a confirmation comes from the same strand, while the sum is used for confirmations from the opposite strand. The minimal quality threshold q is a parameter of this filtering variant. When reads are generated with very high coverage, some wrong reads may pass the first filtering step. For instance, when simulating 300,000 30-mer reads with an error rate of 0.6% per base call from a 90-kbp target sequence (i.e., coverage ∼100), we observe up to 500 wrong reads among those generated twice.
In simulations, the second criterion allows the removal of virtually all wrong reads left over after the first filtering step, at the expense of discarding very few correct ones. In this second filtering step, our algorithm removes reads that lack partners with perfectly matching overlap. The minimal overlap used to search for matching partners is a parameter of this filtering step, to be chosen larger than half of the read length. We consider reads to be confirmed, if at least one matching partner exists for each of its ends, thus if it is covered entirely at least twice. Reads containing sequencing errors are very unlikely to find partners on both sides.
After the filtering steps, SHARCGS generates reverse complements for all confirmed reads and keeps only one copy of each read, before starting the core assembly algorithm.
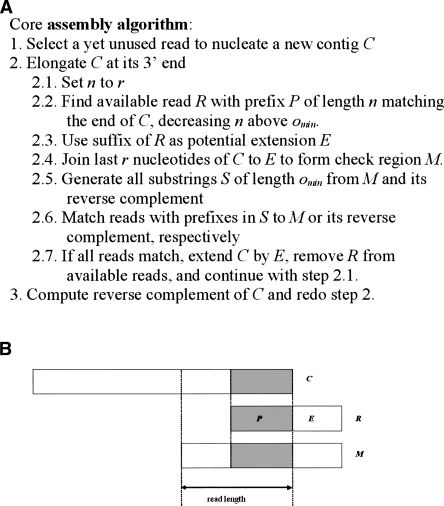
The core assembly algorithm
The core assembly algorithm is based on a contig extension scheme using a prefix-tree to look up potential extensions efficiently. Reads are used in turn to nucleate novel contigs. A contig is elongated at its end, as long as we find reads with a prefix of minimal length, which perfectly matches the end of the contig. The algorithm tries to extend the current contig by the second part of the matching read but first checks both strands for ambiguities. Such ambiguities occur when parts of the sequence are repeated. At the end of a repeat, the prefix of two reads will match to the repeated sequence, but their suffixes will match to different parts of the sequence that cannot be arranged unambiguously. In this case, the elongation of the contig is terminated. We search for ambiguities using prefixes of minimal length extracted from each of the last few positions of the putative contig’s end. Each of these prefixes is used to search for other reads which start with the same prefix, and all such reads must match perfectly to the end of the putative contig. By applying this approach, we are able to detect ambiguities although several reads in a row may be missing from the input data. After the elongation of a contig at its 3′ end has been terminated, we compute its reverse complement and try the elongation at the other end the same way. An illustration of the elongation step and the core assembly algorithm’s pseudocode is given in Figure 6.
Figure 6.
Description of the core assembly algorithm. (A) Pseudocode overview of the steps during assembly of a single contig. The parameter omin controls the stringency of the algorithm, and r denotes the read length. (B) Illustration of the elongation step. Contig C is to be elongated to the right. Read R is a candidate for elongation found in the data set of reads, because its prefix (gray) matches the end of C perfectly. The suffix of read R (white) is the potential extension E for contig C. The length of the check region M is the sum of read length r, and the length of the extension E. Substrings of M and its reverse complement are used to search for matching read prefixes in the data set. Only if all of these reads match M exactly is C extended by E.
Merging contigs from several assembly runs and generation of quality measures
Setting the filtering parameters to very stringent levels discards large amounts of data from the input. The resulting assembly has very short contigs, because reads from too many positions in the target sequence are unavailable. When including weakly confirmed reads in the assembly, it is more difficult to filter reads containing sequencing errors. Such reads, however, would stop the algorithm from assembling long contigs, since reads containing sequencing errors cause spurious ambiguities. The contig breaks due to confirmed reads with sequencing errors tolerated by weak filtering are unlikely to occur at the same positions as the contig breaks caused by missing reads after application of strong filtering criteria. This is why we run the core assembly algorithm automatically for weak, medium, and strong filter parameter settings. For each parameter setting, the core algorithm only generates contigs contained in the target sequence. SHARCGS attempts to merge contigs from different core assembly runs by finding exact overlaps at least as long as the read length.
If quality measures are supplied with the raw reads, SHARCGS computes quality measures for any position in all contigs. It adopts the paradigm described in the Phrap documentation (Ewing and Green 1994). For a given position x in a contig, SHARCGS finds all covering reads and selects the best quality measure observed for x. It does the same for the opposite strand and adds the two quality measures to obtain the final value.
Fine-tuning parameters
We have introduced two parameters for our algorithm: the confirmation level for filtering incorrect reads and the minimal overlap used in the second filtering step as well as in the contig elongation step. The proper setting of both is crucial for the assembler’s reliability, and thus we provide automated conservative settings as described below.
The optimal setting of both parameters depends on the length of the target sequence. We use the abundance of reads generated several times as a fingerprint of the target sequence length. We expect the number of reads generated x times to be distributed according to a binomial distribution with parameters k, the number of reads generated, and P, the probability to read correctly at a given position in the target sequence. The probability P is given by Pcorrect/n, where n is the length of the target sequence. In the input data, we can count the number of reads generated x times for any x. In a maximum likelihood approach, we determine the combination of error rate and target sequence length, which most probably causes the distribution of read confirmations observed in the input data. This estimation is accurate in simulated data (within 5% of the real sequence length).
According to the target sequence length n, we determine the confirmation levels for which n/2 (strong filtering) to n (weak filtering) reads pass the filter. At most we expect to observe one read per position in the target sequence, so we do not expect to have more than n correct reads. Moreover, we do not expect to be able to assemble input data with decent quality, if more than half of the reads are missing.
The minimal overlap with which the core assembly algorithm should check for ambiguities, as well as the minimal overlap used in the second filtering step to detect unreliable reads, depends on the lengths of gaps expected in the input data. In this context, we call gaps runs of consecutive positions for which no confirmed read is available. The probability Pg that a gap at least of length g starts at a given position depends on the number of available reads navail and the length of the target sequence n as follows:
where Pmiss is the probability of no confirmed read being available at a given position. The number of such gaps expected in the whole target sequence is n · Pg. We suggest setting the minimal overlap omin such that the expected number of gaps larger than r − omin is <1, r being the read length.
Implementation
We have implemented all steps of the described algorithm in Perl and tested the script under version 5.8.4 of this scripting language. The program runs from any Linux shell without installation of additional software or modules. Full documentation and a user introduction are included in the script and may be viewed by calling the program without any parameters. The assembly of a BAC of size 100 kbp typically takes <15 min on an Intel Xeon 2.8-GHz 32-bit Linux machine. The corresponding memory footprint of our program is smaller than 1 GB of RAM. The assembly of E. coli takes less than 10 h and uses 24 GB of RAM on an AMD Opteron 2.8-GHz 64-bit Linux machine. The filtering step for the assembly of H. acinonychis takes <1 h and uses 24 GB of RAM. The assemblies of H. acinonychis at four different Q levels and the final merging take 4 h in total and use 6 GB of RAM.
Evaluation
In order to evaluate the performance of our algorithm, we have simulated short reads from a number of sequences, computed contigs, and matched them back to the reference sequences. Since our method is particularly valuable to the de novo sequencing of BAC inserts, we chose three eukaryotic BAC libraries, i.e., the Clemson T-BAC library (Choi et al. 1995) for Arabidopsis thaliana, RPCI-98 (Hoskins et al. 2000) for Drosophila melanogaster, and RPCI-11 (Osoegawa et al. 2001) for Homo sapiens for the selection of sequenced BAC inserts as target sequences. We collected all sequenced BAC inserts of these libraries from the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov, version as of December 2006) and sorted the sequences by length. We chose 20 successive BAC insert sequences from each of the three data sets so that the average sequence length for each of the species was ∼110 kbp (112 kbp in A. thaliana, 109 kbp in D. melanogaster, and 110 kbp in H. sapiens). In order to mimic the real situation for sequencing even more closely, we added the sequence of the cloning vector pBACe3.6 to each of the test sequences and filtered all contigs matched to this vector before evaluating the assembly results.
Short-read generation in silico
We have implemented a program that simulates short reads from input target sequences. Parameters for this Perl script are the read length, the number of reads, and the average error rate per base call. The script generates reads from any position in the target sequence with equal probability and decides for each base call independently with a constant error probability whether it is generated correctly. When introducing an error, our simulation program decides with equal probability for one of the three possible substitution errors.
Preparation of fragment libraries and Illumina sequencing
Genomic DNA was fragmented by nebulization. Sheared fragments were processed according to the recommended Solexa/Illumina protocol (end repair, A-tailing, adapter ligation, size selection, and preamplification). Amplified material was loaded onto channels of the flow cells at 2 pM and 4 pM concentration. Sequencing was carried out by running 36 cycles on the 1G Illumina sequencing instrument. Image deconvolution and quality value calculation were performed using the Goat module (Firecrest v.1.8.28 and Bustard v.1.8.28 programs) of the Illumina pipeline v.0.2.2.3. Per-base error rates were calculated on the basis of alignments generated with ELAND (Gerald module v.1.27 of the Illumina pipeline).
Program and data availability
The programs and data generated in this work are available from our Web site http://sharcgs.molgen.mpg.de.
Acknowledgments
We thank Mark Achtman and Giovanna Morelli for providing DNA from H. acinonychis and Andrew Hufton for comments on the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6435207
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Batzoglou S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Berger B., Mesirov J.P., Lander E.S., Mesirov J.P., Lander E.S., Lander E.S. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Choi S.D., Creelman R., Mullet J., Wing R.A., Creelman R., Mullet J., Wing R.A., Mullet J., Wing R.A., Wing R.A. Construction and characterization of a bacterial artificial chromosome library from Arabidopsis thaliana. Weeds World. 1995;2:17–20. [Google Scholar]
- Eppinger M., Baar C., Linz B., Raddatz G., Lanz C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Baar C., Linz B., Raddatz G., Lanz C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Linz B., Raddatz G., Lanz C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Raddatz G., Lanz C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Lanz C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Keller H., Morelli G., Gressmann H., Achtman M., Schuster S.C., Morelli G., Gressmann H., Achtman M., Schuster S.C., Gressmann H., Achtman M., Schuster S.C., Achtman M., Schuster S.C., Schuster S.C. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120.eor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1994;8:186–194. [PubMed] [Google Scholar]
- Hoskins R.A., Nelson C.R., Berman B.P., Laverty T.R., George R.A., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Nelson C.R., Berman B.P., Laverty T.R., George R.A., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Berman B.P., Laverty T.R., George R.A., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Laverty T.R., George R.A., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., George R.A., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Ciesiolka L., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Naeemuddin M., Arenson A.D., Durbin J., David R.G., Arenson A.D., Durbin J., David R.G., Durbin J., David R.G., David R.G., et al. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–2274. doi: 10.1126/science.287.5461.2271. [DOI] [PubMed] [Google Scholar]
- Huang X., Madan A., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe D.B., Butler J., Gnerre S., Mauceli E., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S., Butler J., Gnerre S., Mauceli E., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S., Gnerre S., Mauceli E., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S., Mauceli E., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S., Mesirov J.P., Zody M.C., Lander E.S., Zody M.C., Lander E.S., Lander E.S. Whole-genome sequence assembly for mammalian genomes: ARACHNE 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullikin J.C., Ning Z., Ning Z. The Phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E.W., Sutton G.G., Delche A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Sutton G.G., Delche A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Delche A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., Mobarry C.M., Reinert K.H., Remington K.A., Reinert K.H., Remington K.A., Remington K.A., et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Osoegawa K., Mammoser A.G., Wu C., Frengen E., Zeng C., de Jong P.J., Mammoser A.G., Wu C., Frengen E., Zeng C., de Jong P.J., Wu C., Frengen E., Zeng C., de Jong P.J., Frengen E., Zeng C., de Jong P.J., Zeng C., de Jong P.J., de Jong P.J. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P.A., Tang H., Waterman M.S., Tang H., Waterman M.S., Waterman M.S. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R., Nicklen S., Coulson A.R., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G.G., White O., Adams M.D., Kerlavage A.R., White O., Adams M.D., Kerlavage A.R., Adams M.D., Kerlavage A.R., Kerlavage A.R. TIGR Assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1995;1:9–19. [Google Scholar]
- Warren R.L., Sutton G.G., Jones S.J., Holt R.A., Sutton G.G., Jones S.J., Holt R.A., Jones S.J., Holt R.A., Holt R.A. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]