Abstract
The fate of all aerobic organisms is dependent on the varying intracellular concentrations of NADH and NADPH. The former is the primary ingredient that fuels ATP production via oxidative phosphorylation, while the latter helps maintain the reductive environment necessary for this process and other cellular activities. In this study we demonstrate a metabolic network promoting NADPH production and limiting NADH synthesis as a consequence of an oxidative insult. The activity and expression of glucose-6-phosphate dehydrogenase, malic enzyme, and NADP+-isocitrate dehydrogenase, the main generators of NADPH, were markedly increased during oxidative challenge. On the other hand, numerous tricarboxylic acid cycle enzymes that supply the bulk of intracellular NADH were significantly downregulated. These metabolic pathways were further modulated by NAD+ kinase (NADK) and NADP+ phosphatase (NADPase), enzymes known to regulate the levels of NAD+ and NADP+. While in menadione-challenged cells, the former enzyme was upregulated, the phosphatase activity was markedly increased in control cells. Thus, NADK and NADPase play a pivotal role in controlling the cross talk between metabolic networks that produce NADH and NADPH and are integral components of the mechanism involved in fending off oxidative stress.
If an aerobic organism is to survive, it is essential that an adequate supply of NADPH is available. This nicotinamide nucleotide provides a reductive environment that enables the oxidative cell to nullify the reactive oxygen species (ROS) generated as a consequence of oxidative phosphorylation, a process key to the generation of ATP (9, 18, 22). All organisms that utilize oxygen as the terminal e− acceptor have evolved intricate molecular strategies that allow them to combat the inherent dangers associated with living in an aerobic environment (11, 26). Catalase, superoxide dismutase (SOD), and glutathione peroxidase are some of the enzymes that help decrease oxidative tension during aerobic respiration (4). However, the effectiveness of these proteins as the scavengers of ROS depends on the availability of NADPH. This nucleotide supplies the reductive power necessary to quell the oxidative potential of ROS. Hence, the production of this reducing agent is an integral part of the oxidative energy-generating machinery of all aerobic organisms. Production of ATP via oxidative phosphorylation cannot proceed effectively in the absence of a continual supply of NADPH (14, 31).
Glucose-6-phosphate dehydrogenase (G6PDH), NADP+-isocitrate dehydrogenase (ICDH-NADP+), malic enzyme (ME), 6-phosphogluconate dehydrogenase (6PGDH), and glutamate dehydrogenase-NADP+ (GDH-NADP+) are some of the important enzymes that enable aerobic cells to fulfill their requirement for NADPH (34). NADH, which is generated essentially during the catabolism of acetyl-coenzyme A via the tricarboxylic acid (TCA) cycle, is a potent prooxidant as its downstream metabolism mediated by complexes I, III, and IV produces the majority of the ROS generated in aerobic organisms (13). Although this metabolic network is provides the bulk of cellular energy, it can also create a highly oxidative environment that is harmful to the organisms. Hence, a fine balance has to be maintained between these two nicotinamide nucleotides if a cell is to function in an efficient manner. Thus, a normal functioning cell has to have adequate levels of NADPH and ATP and a small amount of NADH. On the other hand, a high concentration of NADH coupled with a low concentration of NADPH promotes an oxidative milieu, a condition conducive to cellular dysfunction and diseases (13, 36).
As part of our study to elucidate the metabolic adaptation evoked by environmental stress, the impact of menadione, a generator of superoxide (O•2−) in the metabolism of the soil microbe Pseudomonas fluorescens, has been examined. Here, the ability of this organism to survive in the presence of this toxicant by upregulating the production of NADPH and decreasing the synthesis of NADH is demonstrated. The participation of the TCA cycle and numerous NADPH-generating enzymes in combating the toxicity of menadione is discussed below. The intriguing influence of NAD+ kinase (NADK) and NADP+ phosphatase (NADPase), two critical modulators of metabolic pathways due to their abilities to regulate the availability of NAD+ and NADP+, is also explained.
MATERIALS AND METHODS
Microbial growth conditions and isolation of cellular fractions.
The bacterial strain P. fluorescens ATCC 13525 was obtained from the American Type Culture Collection and grown in a mineral medium containing (per liter of deionized water) 6.0 g Na2HPO4, 3.0 g KH2PO4, 0.8 g NH4Cl, 0.2 g MgSO4·7H2O, and 4 g citric acid. Trace elements were present at concentrations described previously (1). The pH was adjusted to 6.8 with dilute NaOH. The media were dispensed in 200-ml amounts into 500-ml Erlenmeyer flasks with foam plugs and autoclaved for 20 min at 121°C. Menadione (100 μM) was added after the medium was autoclaved. (Note that the concentration of menadione used in oxidative stress studies is usually 50 to 500 μM [35, 38, 40].) The media were then inoculated with 1 ml of stationary-phase cells grown in a medium not amended with menadione on an aerated gyratory water bath shaker (model 76; New Brunswick Scientific) at 26°C at 140 rpm. The Bradford protein assay was performed in triplicate in order to determine the amount of cells in a culture by measuring the solubilized cellular proteins, and bovine serum albumin was used as the standard (3). The cells were treated with 1 M NaOH in order to solubilize the protein. The bacterial cells were harvested after various growth intervals and suspended in a cell storage buffer (pH 7.3) consisting of 50 mM Tris-HCl, 5 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride. The cells were disrupted by sonication and centrifuged at 3,000 × g for 30 min at 4°C to remove intact bacteria. Centrifugation at 180,000 × g for 2 h resulted in a soluble cell extract (CFE) and a membrane CFE. The soluble fraction was further centrifuged at 180,000 × g for 1 h to obtain a membrane-free system. The CFE fractions were kept at 4°C for up to 5 days, and various enzymatic activities were monitored.
Monitoring NADPH-producing enzymes: activity and protein expression.
ICDH-NADP+, G6PDH, ME, GDH-NADP+, and 6PGDH were analyzed by monitoring NADPH production at 340 nm in triplicate (37). Isocitrate, glucose-6-phosphate, malate, glutamate, and 6-phosphogluconate, respectively, at concentrations ranging from 1 to 5 mM were used as the substrates. The concentration of the cofactor (NADP+) ranged from 0.1 to 0.5 mM. The reaction was performed in 25 mM Tris-HCl buffer (pH 7.3) with 5 mM MgCl2 and 0.1 mg protein·ml−1 of the soluble CFE for 15 min. For ICDH, ME, and GDH, the keto acids α-ketoglutarate (α-KG) and pyruvate were also quantitated with the aid of 2,4-dinitrophenylhydrazine at 450 nm (30). For ICDH activity, 4 mM malonate was added in order to inhibit isocitrate lyase (EC. 4.1.3.1). Controls consisted of the reaction mixtures without the CFE and substrates.
BN-PAGE, SDS-PAGE, and in-gel activity stains.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed as described by Schagger and von Jagow (33), with the following modifications. Sixty micrograms of proteins was prepared in BN buffer (50 mM bis-Tris, 500 mM ɛ-aminocaproic acid [pH 7.0]). Gels were electrophoresed under BN conditions; 80 V was used for the stacking gel, and the voltage was increased to 200 V when the proteins reached the separating gel. The blue cathode buffer (50 mM Tricine, 15 mM bis-Tris, and 0.02% Coomassie blue G-250 [pH 7.0] at 4°C) was changed to the colorless cathode buffer (50 mM Tricine and 15 mM bis-Tris [pH 7.0] at 4°C) once the running front reached halfway through the separating gel. Electrophoresis was stopped before the running front was out of the gel. The gels were placed subsequently in an equilibration buffer (25 mM Tris-HCl [pH 7.3], 5 mM MgCl2) for 15 min. Immediately after incubation, the gels were placed in the Tris buffer described above containing 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride (INT) (0.4 mg/ml), phenazine methosulfate (PMS) (0.2 mg/ml), and NADP+ (0.5 mM). For in-gel detection of ICDH-NADP+, 0.1 mM NADP+ was used to ensure specificity. Formazan precipitation was utilized to visualize the active enzymes. Isocitrate (2 mM), glucose-6-phosphate (2 mM), malate (5 mM), and glutamate (5 mM) were the substrates for ICDH, G6PDH, ME, and GDH, respectively. Following identification of the specific activity bands, two-dimensional (2D) BN-PAGE analyses were also performed under the conditions described previously (20). 2D sodium dodecyl sulfate (SDS)-PAGE using a discontinuous buffer system was performed according to the method of Laemmli (16). A 10% isocratic gel was used for these experiments. Samples were solubilized in 62.5 mM Tris-HCl (pH 6.8)-2% SDS-10% glycerol-2% 2-mercaptoethanol at 100°C for 5 min. Electrophoresis was carried out at a constant voltage of 200 V, and slab gels were fixed, stained, and destained according to standard procedures. The protein content was analyzed by Coomassie blue staining and subsequent densitometric analysis (SCION Corporation, Frederick, MD). Controls consisted of reaction mixtures without substrate and cofactors.
Analyses of NADH-generating enzymes.
NAD+-dependent ICDH (ICDH-NAD+), α-KG dehydrogenase (α-KGDH), NAD+-dependent GDH (GDH-NAD+), malate dehydrogenase (MDH), and pyruvate dehydrogenase (PDH) were analyzed by monitoring the formation of NADH at 340 nm in triplicate (37). Isocitrate, α-KG, glutamate, and pyruvate, respectively, were the substrates used. The cofactor was NAD+ (0.1 and 0.5 mM), and the reaction was performed in 25 mM Tris-HCl buffer (pH 7.3) with 5 mM MgCl2 and 0.1 mg ml−1 protein equivalent of the membrane CFE for 15 min. Coenzyme A was utilized as a cofactor for PDH and α-KGDH. For ICDH-NAD+, α-KGDH, GDH-NAD+, and PDH, the keto acids α-KG and pyruvate were also quantitated using 2,4-dinitrophenylhydrazine at 450 nm (30). Controls consisted of the reaction mixtures without the CFE and substrates. BN-PAGE was performed as described above with the following modification. Sixty micrograms of proteins from membrane CFE was prepared in BN buffer containing 1% maltoside. Following electrophoresis, gels were equilibrated for 15 min in Tris-HCl buffer and then placed in the same buffer containing INT (0.4 mg/ml), PMS (0.2 mg/ml), and NAD+ (0.5 mM). For in-gel detection of ICDH-NAD+, 0.1 mM NAD+ was used to ensure band specificity. Isocitrate (5 mM), α-KG (5 mM), glutamate (5 mM), malate (5 mM), and pyruvate (5 mM) were the substrates for ICDH-NAD+, α-KGDH, GDH-NAD+, MDH, and PDH, respectively. Following identification of specific enzymatic bands, 2D BN-PAGE and SDS-PAGE were also performed under the conditions described above.
In-gel activities of NADK, NADHK, and NADPase.
NADK was analyzed as previously described by Mailloux et al. (21). Soluble CFE proteins were electrophoresed as described above, and gels were incubated in equilibration buffer containing INT (0.4 mg/ml) and PMS (0.2 mg/ml) supplemented with NAD+ (0.5 mM), ATP (3 mM), isocitrate (5 mM), and ICDH-NADP+ (5 U). Following identification of specific enzymatic bands, 2D BN-PAGE and SDS-PAGE were also performed under the conditions described above. Controls consisted of reaction mixtures without substrate and cofactors. For detection of NADH kinase (NADHK), the gels were incubated with dichloroindophenol (16.7 μg/ml), NADH (0.5 mM), ATP (3 mM), oxidized glutathione (2 mM), INT (4 mg/ml), and glutathione reductase (5 U). NADPase activity was identified using native PAGE and enzyme-coupled assays. Sixty micrograms of soluble protein extract was electrophoresed using a Ponceau S cathode buffer (0.0125% [wt/vol] Ponseau S in cathode buffer) under native conditions. Reactions were carried out as described above in equilibration buffer containing 5 mM malate, 1 mM NADP+, 10 U of MDH, INT, and PMS. Following band visualization the reactions were stopped using a destaining solution and documented. Band specificity was ascertained using known the phosphatase inhibitors okadaic acid (7.5 μM) and sodium orthovanadate (1 mM) (28). Exclusion of MDH was used to ensure band specificity. To evaluate the regulation of NADK and NADPase during oxidative stress, a 10-mg equivalent to menadione-exposed cells was introduced into citrate control medium and subsequently cultured for 8 h. Similar experiments were performed with control cells transferred into menadione-containing medium. Following the 8-h incubation, cells were isolated, and the activities of NADK and NADPase were assessed as described above.
Enzymatic activities after various growth intervals.
Cells were harvested after various time intervals, and the cellular fractions were isolated. The ICDH-NADP+, G6PDH, and ME activities and protein contents in the soluble CFE were analyzed by BN-PAGE, 2D BN-PAGE, and 2D SDS-PAGE. The ICDH-NAD+, α-KGDH, and PDH activities and protein contents were analyzed in a similar manner with cells harvested after corresponding time intervals. In this case, membrane CFE was utilized.
The menadione-mediated regulation of enzyme activity and expression was achieved using recovery assays. Ten milligrams of protein equivalent of menadione-stressed cells was transferred to citrate (control) medium, and 10 mg of protein equivalent of control cells was transferred to menadione (100 μM)-supplemented medium. Following incubation for 8 h, cells were harvested and the cellular fractions were isolated and assayed to determine enzymatic activities and protein concentrations. For comparative studies of enzymatic activities, cells were harvested at the same growth phase (i.e., 25 h for control cultures and 30 h for menadione cultures).
Metabolite analysis.
The soluble CFE isolated from control and menadione-stressed cells were used to determine the NAD(P)+ levels. Cell extract treatments were performed at neutral pH to ensure accurate measurement of the nucleotides (39); 2 mg/ml of protein equivalent to soluble CFE was heated gently for 2 min to remove proteins and lipids. Following removal of the precipitate, the supernatant was analyzed. Similarly, NADK activity was determined by assessing the nucleotide levels by high-performance liquid chromatography (HPLC). For NADK activity, 2 mg/ml of soluble CFE was incubated for 30 min at 26°C in a reaction buffer (25 mM Tris, 5 mM MgCl2; pH 7.0) containing 1 mM NAD+ and 1 mM ATP (21). Peaks were quantified using the EMPOWER software. The HPLC analysis was carried out with a Waters Alliance HPLC equipped with a Waters model 2487 UV-Vis dual-wavelength detector and a C18 reverse-phase column (3.5 μm; amide cap; 4.6 mm by 150 mm [inside diameter]; Symmetry column; Phenomenex, Torrance, CA). A mobile phase consisting of 95% 20 mM KH2PO4 (pH 7) and 5% acetonitrile (dissolved in Millipore water) operating at a flow rate of 0.2 ml/min at the ambient temperature was utilized. The absorbance was monitored at 254 nm. A neutral pH was maintained to ensure accurate measurement of the nucleotides. 13C nuclear magnetic resonance (13C-NMR) analyses were performed using a Varian Gemini 2000 spectrometer operating at 50.38 MHz for 13C. Experiments were performed with a 5-mm dual probe (35° pulse; 1-s relaxation delay; 8 kilobytes of data; 2,000 scans). Chemical shifts were referenced to standard compounds under the same conditions. CFE (2 mg/ml) was incubated for 10 min in a phosphate reaction buffer containing 5 mM [2,4-13C]citrate. The reaction was stopped by heating at 95°C for 5 min. Following removal of the precipitate, the 13C-NMR was recorded.
Statistical analyses.
Data were expressed as means ± standard deviations. Statistical correlations of data were checked for significance using the Student t test (P ≥ 0.05). All experiments were performed twice and in triplicate.
RESULTS
Metabolite analysis.
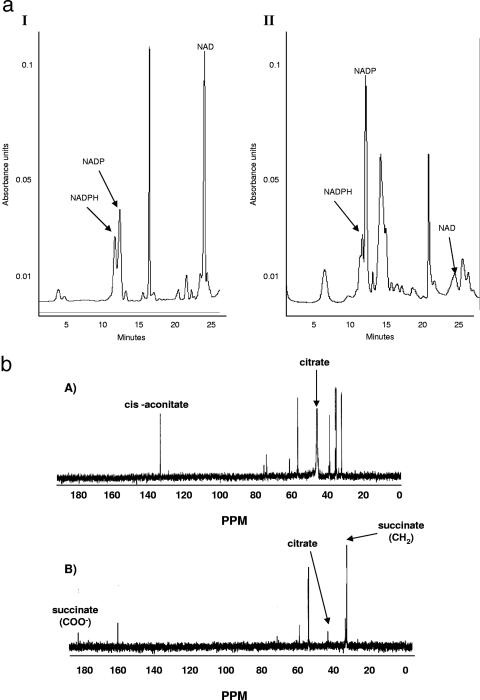
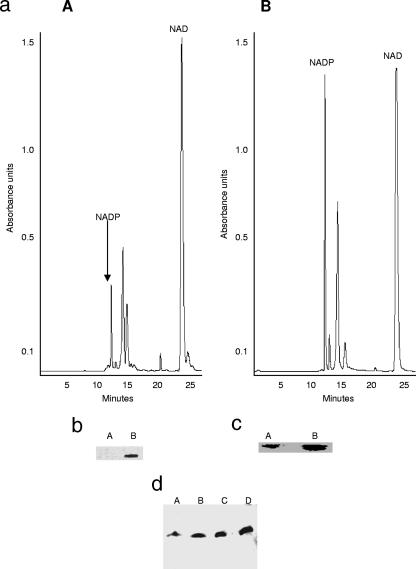
In the presence of 100 μM menadione P. fluorescens experienced a decreased growth rate. However, compared to the control cultures, in the stationary phase of growth, which was attained approximately 5 h later in the menadione-supplemented cultures, the biomass was similar. Thus, it appeared that the organism had adapted to this oxidative challenge. HPLC analyses of the nucleotides in the soluble CFE revealed marked variation in the nature of the species associated with the organism. In the cells isolated from the stressed cultures, peaks indicative of NADP+ were more abundant, while the levels of NAD+ were higher in the soluble CFE from the control cultures (Fig. 1). The identities of the peaks were further confirmed by spiking the samples with known standards. The occurrence of the disparate metabolic pathways was also confirmed by 13C-NMR studies. The disparity in the content of the distinct reducing factors prompted us to examine the metabolic changes that would trigger such a change. The main NADPH-generating enzymes were examined. Control cells were harvested at 25 h, while menadione-exposed cells were isolated at 30 h.
FIG. 1.
(a) HPLC analyses of soluble CFE from cells grown in control (panel I) and menadione (panel II) media. Cells were isolated at the same growth phase. The mobile phase was 95% 20 mM KH2PO4 buffer-5% acetonitrile, the flow rate was 0.2 ml·min−1, and UV detection was at 254 nm. (b) 13C-NMR analysis of citrate metabolism in P. fluorescens isolated from control (panel A) and menadione-stressed (panel B) cultures. CFE (2 mg/ml) was incubated in a phosphate reaction buffer containing 5 mM [2,4-13C]citrate for 10 min. Known standards were used as references for the spectrum.
Oxidative stress promotes NADPH production.
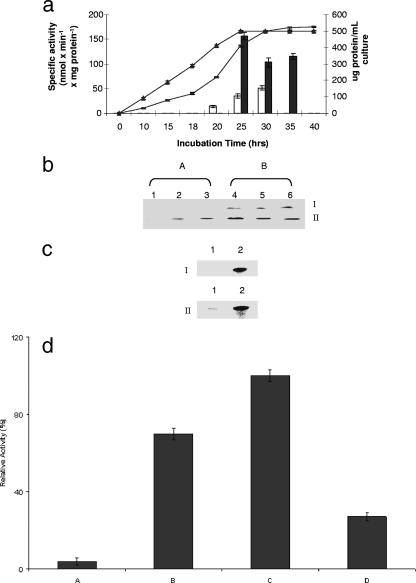
The level of ICDH-NADP+, an enzyme usually associated with the cytoplasmic fraction, was found to be at least threefold higher in the stressed cultures, while the level of G6PDH, a key component of the pentose phosphate pathway, was fourfold higher than the level in the control (Table 1). Similar increases in the microbe cultured in menadione-supplemented medium were observed for ME, GDH-NADP+, and 6PGDH, enzymes known to participate in the production of NADPH. A time profile analysis of ICDH-NADP+ indicated that there was a sharp increase in the activity of this enzyme at 25 h. A threefold increase was recorded (Fig. 2a). BN-PAGE analyses revealed the same pattern; however, two bands corresponding to ICDH-NADP+ activity were evident (Fig. 2b). It is important to note that these activity bands were observed with 0.01 to 0.1 mM NADP+. One of the bands was absent in the control even when the reaction mixture was incubated for longer periods. In an effort to detect the amount of protein associated with these activity bands, the bands were excised and analyzed by 2D SDS-PAGE. Coomassie blue staining revealed significant amounts of protein associated with the activity bands from the menadione cultures (Fig. 2c). No band was evident with band I in the control cultures, thus pointing to an ICDH-NADP+ isoenzyme only in the cells from the menadione medium. This activity was also confirmed by HPLC. The appearance of α-KG and the decrease in the isocitrate peaks in the soluble CFE from cells grown in the menadione cultures occurred within minutes. In the control CFE, only an isocitrate peak was observed (data not shown). To further elucidate the influence of menadione on the expression of ICDH-NADP+, cells grown in the control medium (25 h) were transferred to the menadione medium, while cells harvested from menadione cultures (30 h) were incubated in the control medium. There was a sharp increase in ICDH-NADP+ activity in the control cells exposed to menadione for only 8 h (Fig. 2d). The reverse trend was evident in the menadione culture cells incubated in the control medium. A sharp decrease in ICDH-NADP+ activity was evident. This clearly established the pivotal role that oxidative stress plays in the upregulation of this NADPH-generating enzyme.
TABLE 1.
Specific activities of NADPH-generating enzymes in cells harvested from menadione-containing and control media at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells)
| NADP+-dependent enzyme | Sp act (nmol min−1 mg protein−1)a
|
|
|---|---|---|
| Control | Menadione stressed | |
| ICDH-NADP+ | 35 ± 5 | 103 ± 3* |
| G6PDH | 16 ± 3 | 72 ± 6* |
| ME | 75 ± 11 | 130 ± 8* |
| GDH-NADP+ | 15 ± 2 | 36 ± 3* |
| 6PGDH | 5.1 ± 1.1 | 10 ± 1.2* |
The level of NADPH was monitored. The values are means ± standard deviations. An asterisk indicates that the P value is ≤0.05 (n = 3).
FIG. 2.
(a) ICDH-NADP+ activity in P. fluorescens after various growth intervals. ▴, cellular yield in control cultures; −, cellular yield in menadione-containing cultures; open bars, ICDH-NADP+ in soluble CFE from control cultures; solid bars, ICDH-NADP+ in soluble CFE from menadione cultures. The error bars indicate standard deviations (n = 3). (b) In-gel ICDH-NADP+ activity in P. fluorescens after different growth intervals. Lanes 1, 2, and 3, soluble CFE obtained from control cells after 15, 24, and 30 h of growth, respectively; lanes 4, 5, and 6, soluble CFE obtained from menadione-stressed cells after 25, 30, and 35 h of growth, respectively. (The time intervals corresponded to the same growth phases; note the appearance of isoenzyme I in the menadione cultures.) (c) Expression of ICDH-NADP+ in control and menadione-stressed P. fluorescens as determined by 2D BN-PAGE. Lanes 1 and 2 contained the soluble CFE from control and menadione-stressed cultures, respectively. For panel I band I was excised and loaded. For panel II, band II was excised and loaded. Activity bands corresponding to similar growth phases were excised and loaded. (d) Regulation of ICDH-NADP+ total activity in P. fluorescens. Bar A, control cells; bar B, control cells transferred to menadione medium; bar C, menadione-stressed cells; bar D, menadione-stressed cells transferred to control medium. One hundred percent corresponds to 107 nmol·min−1·mg protein−1. The error bars indicate standard deviations (n = 3).
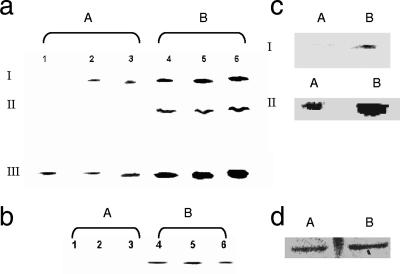
There was also a significant difference in the nature of G6PDH in the control and menadione-challenged cells. In the latter cells, three intense bands corresponding to the activity of this enzyme were evident, compared to two bands in the soluble CFE from the control cells (Fig. 3a). Band II was completely absent, while band I was evident after 24 h of incubation in the control cells. 2D BN-PAGE and SDS-PAGE revealed marked variation in the protein content associated with these bands in the CFE from the stressed cultures compared to the control (data not shown). In addition, an activity band corresponding to decarboxylation of malate to pyruvate by ME was prominent only in the soluble CFE from the stressed cells (Fig. 3b). 2D BN-PAGE analysis of the activity band indicated that there was a sharp increase in the expression of ME in the cultures subjected to oxidative stress (data not shown). The GDH-NADP+ activity in the soluble CFE isolated from the cells grown in menadione medium was significantly higher than that in the control (Fig. 3c). Analysis of the protein bands obtained following 2D SDS-PAGE by the SCION image software revealed a fourfold increase in protein associated with GDH-NADP+ activity in the soluble CFE from the menadione cultures. Quantification of the bands confirmed the observed increase in the activity of GDH-NADP+. The activity of NADHK was also markedly increased in the menadione-stressed cells (Fig. 3d). The purity of the CFE preparations was verified by monitoring the activity of G6PDH, a cytoplasmic enzyme, and succinate dehydrogenase (SDH), a membrane-bound enzyme. No in-gel activity was observed in the membrane CFE for G6PDH and succinate dehydrogenase in the soluble CFE (data not shown).
FIG. 3.
(a) BN-PAGE analysis of G6PDH activity in control and menadione-stressed cells after various growth intervals. Lanes 1, 2, and 3, soluble CFE from cells grown in a citrate medium (control) for 15, 24, and 30 h, respectively; lanes 4, 5, and 6, soluble CFE from cells grown in a menadione-stressed medium for 24, 30, and 35 h, respectively. I, II, and III indicate the positions of the isoenzymes of G6PDH. (b) BN-PAGE analysis of ME activity in control and menadione-stressed cells after various growth intervals. Lanes 1, 2, and 3, soluble CFE from cells grown in citrate medium (control) for 15, 24, and 30 h, respectively; lanes 4, 5, and 6, soluble CFE from cells grown in menadione medium for 25, 30, and 35 h, respectively. (c) Activity and expression of GDH-NADP+ in control and menadione-stressed cells. Lane A, control cells; lane B, menadione-stressed cells. For gel I in-gel activity staining was used, and for gel II Coomassie blue staining was used (the activity band in a one-dimensional BN-PAGE gel was excised and loaded onto a 2D BN-PAGE gel). Note that the concentration of NADP+ was 0.5 mM, and no activity band was observed with NAD+ (0.5 mM). Activity bands from gel I corresponding to the same growth phase were excised and loaded. Note that cells were isolated at the same growth interval (25 h for the control cells and 30 h for the menadione-stressed cells). (d) In-gel activity staining for NADHK in P. fluorescens grown in control (lane A) and menadione-stressed (lane B) cultures. Note that cells were isolated at the same growth interval (25 h for control cells and 30 h for menadione-stressed cells).
NADH-generating enzymes.
Examination of the NADH-forming enzymes revealed an entirely different activity profile. In this case, a marked decrease in NADH was evident in the menadione cultures. Critical enzymes of the TCA cycle that supply the reducing factors for oxidative phosphorylation were significantly downregulated in response to menadione stress. While a threefold decrease was observed for ICDH-NAD+, there was a sixfold decrease in α-KGDH activity in the membrane CFE from the menadione cultures (Table 2). Similar declines were observed with PDH and GDH-NAD+. No significant variation was observed with MDH.
TABLE 2.
Specific activities of NADH-generating enzymes in cells harvested from menadione-containing and control media at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells)a
| NAD+-dependent enzyme | Sp act (nmol min−1 mg protein−1)a
|
|
|---|---|---|
| Control | Menadione stressed | |
| ICDH-NAD+ | 55 ± 5 | 15 ± 3* |
| a-KGDH | 40 ± 4 | 7 ± 1* |
| GDH-NAD+ | 50 ± 7 | 12 ± 3* |
| PDH | 6 ± 0.3 | 2 ± 0.2 |
| MDH | 11 ± 1 | 13 ± 1* |
The level of NADH was monitored. The values are means ± standard deviations. An asterisk indicates that the P value is ≤0.05 (n = 3).
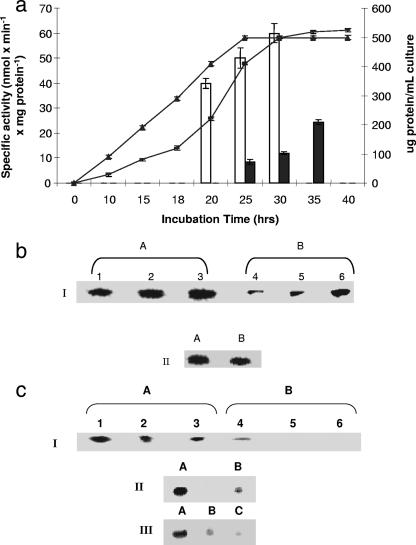
ICDH-NAD+ activity varied with the incubation time. A marked decrease in the menadione cultures was evident after 25 h of incubation (Fig. 4a and b). The protein content associated with the bands was also lower than the content in the control. The activity of α-KGDH, a pivotal metabolic enzyme that provides NADH and also essential precursors for amino acid synthesis, was significantly diminished in the membrane CFE isolated from the menadione cultures. Indeed, the diminished activity corresponded to decrease protein expression, as revealed by 2D electrophoretic analysis (Fig. 4c). The key role of menadione in modulating the activity of this enzyme was further shown by transferring control cells into the menadione medium. In this case, a sharp decline in α-KGDH activity was evident (Fig. 4c). In-gel activity as determined by BN-PAGE confirmed the significant changes observed by spectrophotometric analyses. HPLC studies provided further evidence for the decline in α-KGDH activity in the menadione-stressed cultures. Membrane CFE isolated from the stressed cells was unable to metabolize α-KG, while control membrane CFE decarboxylated α-KG to produce succinate, fumarate, and malate. Incubation of control membrane CFE with labeled citrate and NAD+ resulted in a faster reaction, with numerous peaks attributable to the metabolites of the TCA cycle. No such peaks were evident with the membrane CFE isolated from the menadione-grown cells (data not shown). A similar decrease in protein expression in the case of GDH-NAD+ was also evident (Fig. 5a). In addition, activity staining for PDH also revealed the diminished ability of this NAD+-dependent enzyme to metabolize pyruvate in the cells isolated from the menadione cultures (Fig. 5b).
FIG. 4.
(a) Time profile of P. fluorescens growth and ICDH-NAD+ activity. ▴, cellular yield in control cultures; −, cellular yield in menadione containing cultures; open bars, ICDH-NAD+ activity in soluble CFE from control cultures; solid bars, ICDH-NAD+ activity in soluble CFE from menadione cultures. The error bars indicate standard deviations (n = 3). (b) ICDH-NAD+ activity after different growth intervals and the influence of menadione on protein levels. For gel I, activity staining of membrane CFE for ICDH-NAD+ is shown. Lanes 1, 2, and 3, soluble CFE from cells grown in citrate medium (control) for 15, 24, and 30 h, respectively; lanes 4, 5, and 6, membrane CFE from cells grown in menadione-stressed medium for 25, 30, and 35 h, respectively. Note that the time intervals correspond to similar growth phases (lanes 1, 2, and 3, 20, 30, and 35 h; lanes 3, 4, and 5, 25, 30, and 35 h). For gel II, Coomassie blue staining of ICDH-NAD+ on a 2D BN-PAGE gel is shown (activity bands from gel I corresponding to the same growth phase were excised and loaded). (c) Influence of menadione on α-KGDH activity and protein levels. For gel I, activity staining of membrane CFE for α-KGDH on a BN-PAGE gel at different growth intervals is shown. Lanes 1, 2, and 3, membrane CFE from cells grown in citrate medium (control) for 15, 24, and 30 h, respectively: lanes 4, 5, and 6, membrane CFE from cells grown in a menadione-stressed medium for 24, 30, and 35 h, respectively. Note that the time intervals correspond to similar growth phases. For gel II, Coomassie blue staining of α-KGDH on a 2D BN-PAGE gel of the activity bands obtained by one-dimensional BN-PAGE activity staining of membrane CFE for α-KGDH is shown (activity bands from gel I corresponding to the same growth phase were excised and loaded). In gel III, regulation of α-KGDH activity (in-gel activity staining) is shown. Lane A, membrane CFE from control cells; lane B, membrane CFE from menadione-stressed cells; lane C, membrane CFE obtained following the transfer of control cells to menadione-stressed medium.
FIG. 5.
(a) Effect of menadione on activity and expression of GDH-NAD+. Activity and Coomassie blue staining on one-dimensional and 2D BN-PAGE gels are shown. For gel I, an in-gel activity stain for GDH-NAD+ was used; for gel II, Coomassie blue staining of GDH-NAD+ on a 2D BN-PAGE gel is shown. Lane A, control cells; lane B, menadione-stressed cells. Cells were isolated at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells). (b) BN-PAGE analysis of PDH activity in control and menadione-stressed cells. Lanes A and B contained membrane CFE from control and menadione-stressed cultures, respectively. Note that cells were isolated at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells).
NADK and NADPase.
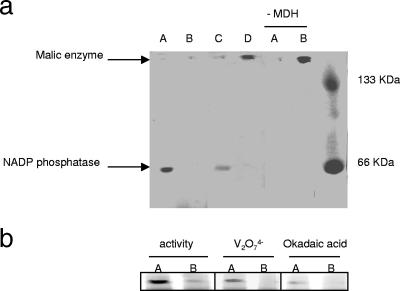
The modulation of the biosynthesis of NADPH and NADH prompted us to examine the precursors that feed these metabolic processes, namely, NADP+ and NAD+. One of the crucial enzymes in the homeostasis of these two reduced nucleotides is NADK. The regulation of this enzymatic activity can easily allow an organism to switch between a reductive state and an oxidative state. Incubation of the soluble CFE isolated from the menadione-challenged cells with NAD+ and ATP showed that there was a significant increase in the activity of NADK (Fig. 6a). A pronounced peak at 14.5 min attributable to NADP+ and a decreased NAD+ peak at 24 min were evident. The hydrolysis of ATP to ADP was also noticeable. These observations were further confirmed by assessing the in-gel activity of the NADK. The formation of NADP+ at the site of NADK activity was monitored with the aid of ICDH-NADP+ (Fig. 6b). A dark band was evident in CFE from the stressed cells, while the band resulting from the control CFE was barely evident. 2D BN-PAGE revealed more protein associated with the activity band obtained from menadione cultures (Fig. 6c). To further evaluate the notion that oxidative stress caused an increase in NADK, we introduced menadione-treated cells into medium containing citrate. The cells were incubated for 8 h. The menadione-treated cells exhibited virtually no change in NADK activity when they were exposed to citrate-containing medium (Fig. 6d). In contrast, control cells incubated in menadione-containing cultures displayed an increase in NADK activity. The NADPase activity was more pronounced in the control cultures than in the menadione cultures. Furthermore, exposure of menadione-treated cells to control medium resulted in an increase in the activity of NADPase (Fig. 7a). The activity bands corresponding to NADPase were confirmed using known phosphatase inhibitors (Fig. 7b). In addition, HPLC analysis of NADPase activity further confirmed the diminished ability of menadione-stressed cells to hydrolyze the phosphoester bond associated with NADP+ (data not shown). Thus, the production of NADP+ and the production of NADPH were markedly increased in the cells subjected to oxidative stress.
FIG. 6.
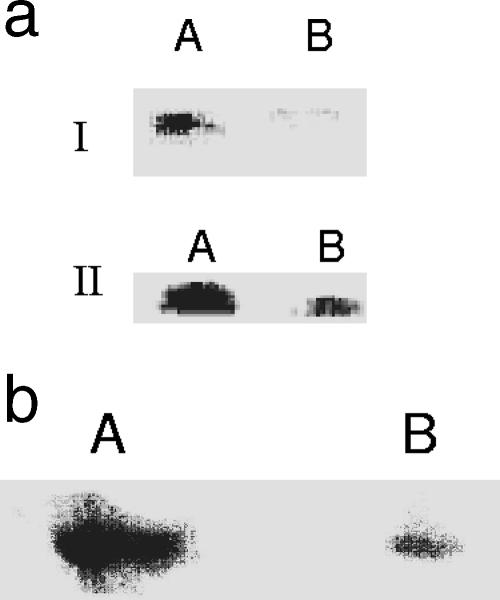
(a) HPLC analysis of NADK activity in soluble CFE from cells grown in control medium (panel A) and menadione-containing medium (panel B) isolated at same growth phase (25 h for control cells and 30 h for menadione-stressed cells). (b) In-gel activity staining of NADK in soluble CFE from control and menadione-stressed media. Lane A, soluble CFE from control culture; lane B, soluble CFE from menadione culture. Note that the cells were harvested at the same growth phase (25 h for the control culture and 30 h for the menadione culture) and no band was observed until addition of ATP to the incubation mixture). (c) 2D BN-PAGE analysis of NADK protein expression. Lanes A and B contained soluble control CFE and soluble menadione-stressed CFE, respectively. The activity bands from panel b were used for 2D analysis. (d) Menadione-mediated modulation of NADK activity: activity staining for NADK in P. fluorescens cells exposed to control conditions (lane A), P. fluorescens cells exposed to menadione stress (lane B), menadione-stressed cells exposed to control medium (lane C), and control cells exposed to menadione medium (lane D).
FIG. 7.
(a) In-gel detection of NADPase in P. fluorescens cells exposed to control conditions (lanes A), P. fluorescens cells exposed to menadione stress (lanes B), menadione-stressed cells exposed to control medium (lane C), and control cells exposed to menadione medium (lane D). Note that cells were isolated at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells). Reaction mixtures lacking MDH were used to evaluate the position of ME and ensure band specificity for NADPase. (b) Evaluation of the sensitivity of the NADPase activity band for known phosphatase inhibitors. P. fluorescens was isolated from control medium (lanes A) and menadione stress medium (lanes B) and subsequently tested for NADPase activity. Reaction mixtures containing either sodium orthovanadate (1 mM) or okadaic acid (7.5 μM), two known phosphatase inhibitors, were used to confirm band specificity. Note that cells were isolated at the same growth phase (25 h for control cells and 30 h for menadione-stressed cells).
DISCUSSION
NADH and NADPH are the two reduced nicotinamide nucleotides that constitute the basis of life. The former provides ATP in all aerobic organisms via the process of oxidative phosphorylation, while the latter helps nullify the oxidative environment triggered by the oxidation of NADH. Hence, their regulation is intricately linked to the survival and proliferation of all organisms that utilize oxygen as an e− sink (12, 29). An imbalance in the concentrations of these moieties leads to cellular dysfunction, diseases, and death. A reductive environment promoted by NADPH is essential for normal cellular activity. The data presented in this paper reveal the interplay between these two reducing factors during the oxidative stress triggered by menadione. There appears to be a metabolic shift that promotes the enhanced production of NADPH and diminished synthesis of NADH in menadione-challenged cells. This metabolic network is modulated by the availability of NAD+ and NADP+ via the activities of NADK and NADPase.
Superoxide is a potent oxidant and is known to oxidize Fe-S clusters in proteins and membranes and essential sulfydryl groups in vital enzymes. This toxicant, which is also part of normal cellular metabolism, is detoxified essentially with the aid of SOD and catalase (23, 27). The former dismutates O•2− into H2O2, while the latter neutralizes H2O2, producing H2O and O2. Glutathione peroxidase can also detoxify H2O2 with the participation of glutathione. Although a body of literature exists on the regulation of these enzymes in response to O•2− and other ROS, there is a dearth of information on the metabolic modules that are an integral part of this detoxification strategy in all aerobic organisms (6, 23, 27). These frontline enzymes in the defense against ROS are ineffective in the absence of a reductive moiety. NADPH is, in fact, the real force that mediates the functioning of these enzymes. Once the ROS have been detoxified, the enzymes have to be recharged directly or indirectly with the help of NADPH (19, 22, 34). Glutathione, which is required for the activity of glutathione peroxidase, has to be regenerated with the participation of glutathione reductase, an enzyme that utilizes NADPH as the cofactor (11, 26). Similarly, catalase is oxidized during ROS detoxification, and reduction of the Fe, an essential component of this heme enzyme, is effected with the aid of NADPH. Hence, understanding the homeostasis of NADPH is essential if one is to appreciate the global network involved in the detoxification of ROS. In this study, numerous key enzymes involved in NADPH production were upregulated in an effort to mitigate the oxidative stress imposed by menadione. Isoenzymes associated with ICDH-NADP+ and G6PDH were also evident. Hence, the creation of a reductive environment mediated by the enhanced formation of NADPH is instrumental to the survival of any organism in an oxidative environment. The role of NADPH in diminishing oxidative tension has been well documented (8, 25), and the importance of ICDH-NADP+ in creating a reductive environment has begun to emerge only recently (2, 15, 17, 24). The presence of this enzyme in the peroxisome has been linked to the ability of this organelle to neutralize H2O2 (7). It is important to note that in this instance NADHK also contributed to the NADPH budget during oxidative stress. However, enhanced production of NADPH without a decrease in the synthesis of NADH may not be very effective. Hence, it is essential to limit the production of NADH, a generator of ROS in oxidative organisms.
In this case, the microbe appeared to decrease the production of NADH, a prooxidant responsible for generation of most of the ROS as a consequence of ATP production via oxidative phosphorylation. Although a decrease in ATP production would ensue, the diminution of NADH synthesis would indeed help reduce the oxidative burden of the cell. ICDH-NAD+ and α-KGDH, two key producers of NADH in the TCA cycle, were severely impeded, as were PDH and GDH-NAD+. The reduction in NADH formation would decrease ROS production at the expense of the aerobic phosphorylation of ADP to ATP. This adaptation would confer on the cell the ability to survive in the presence of oxidative tension, albeit to the detriment of ATP synthesis. This is the first demonstration of a global strategy to enhance NADPH production and to decrease the activity in the TCA cycle, the main metabolic network responsible for NADH formation.
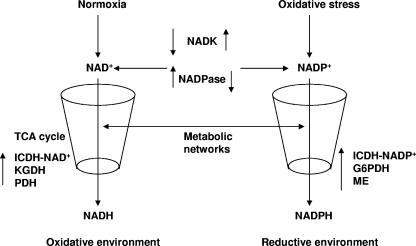
Although disparate metabolic networks are involved in the syntheses of NADH and NADPH, these processes can be regulated by enzymes that mediate the conversion of NAD+ into NADP+. NADK mediates the conversion of NAD+ to NADP+, and NADPase effects the transformation of NADP+ into NAD+. These enzymes, by modulating the intracellular concentrations of NAD+ and NADP+, can act as potent regulators of NADPH and NADH metabolic networks. The results of this study indeed point to instrumental roles of NADK and NADPase in combating oxidative stress. NADK was upregulated in the microbe exposed to menadione, while NADPase was downregulated. Thus, it is quite conceivable that the conversion of NAD+ to NADP+ decreases the availability of the former. This decrease in NAD+ has a negative influence on the TCA cycle, a metabolic network which is the main engine that supplies the reducing equivalents for the production of ATP via oxidative phosphorylation. The diminution in the level of NAD+ impedes NADH production and also lowers the oxidative stress of the cell since ROS production as a consequence of oxidative phosphorylation is reduced. Hence, NADK and NADPase may be powerful effectors of the metabolic state of the cell, influencing ATP production and intracellular oxidative tension. The ability of NADK to synthesize NADP+ has a positive influence on the metabolic modules involved in the production of NADPH, hence providing the reductive environment essential for neutralizing oxidative stress (5, 10, 32). The upregulation of NADK and the downregulation of NADPase may serve the dual purpose of diminishing oxidative stress by enhancing the production of NADPH and diminishing the formation of NADH. Hence, the modulation of NAD+ and NADP+ mediated by NADK and NADPase may dictate the metabolic routes that an organism follows during oxidative stress. Figure 8 depicts such a possible metabolic shift during oxidative stress.
FIG. 8.
Critical role of NADK and NADPase in NAD+ and NADP+ homeostasis. These two enzymes regulate the metabolic networks involved in NADPH and NADH homeostasis during oxidative stress.
In conclusion, this study shows that a global understanding of any biological process is critical if one is to appreciate how living systems function. Although ROS-detoxifying enzymes, such as catalase and SOD, are important in nullifying oxidative stress, the efficacy of these enzymes depends on the reductive environment promoted by NADPH. However, the production of NADPH is governed by the supply of NADP+, a process that is mediated by NADK and NADPase. The upregulation of the kinase and the downregulation of the phosphatase decrease the level of NAD+, a situation that effectively impedes metabolic networks that participate in the production of ROS. On the other hand, a concomitant increase in NADP+ promotes NADPH production, thus further decreasing the oxidative tension. This metabolic strategy may be essential if the organism is to survive in an oxidative environment. Hence, the flux of metabolites may be a critical factor determining the metabolic pathways that a cell may follow. This is the first demonstration of how NADK and NADPase may dictate the redox potential of a cell by modulating the levels of NAD+ and NADP+.
Acknowledgments
Funding for this project was generously provided by the Ontario Center of Excellence and Industry Canada.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Anderson, S., V. D. Appanna, J. Huang, and T. Viswanatha. 1992. A novel role for calcite in calcium homeostasis. FEBS Lett. 308:94-96. [DOI] [PubMed] [Google Scholar]
- 2.Beriault, R., D. Chenier, R. Singh, J. Middaugh, R. Mailloux, and V. Appanna. 2005. Detection and purification of glucose 6-phosphate dehydrogenase, malic enzyme, and NADP-dependent isocitrate dehydrogenase by blue native polyacrylamide gel electrophoresis. Electrophoresis 26:2892-2897. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brioukhanov, A. L., and A. I. Netrusov. 2004. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Moscow) 69:949-962. [DOI] [PubMed] [Google Scholar]
- 5.Chai, M. F., Q. J. Chen, R. An, Y. M. Chen, J. Chen, and X. C. Wang. 2005. NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 59:553-564. [DOI] [PubMed] [Google Scholar]
- 6.Christofidou-Solomidou, M., and V. R. Muzykantov. 2006. Antioxidant strategies in respiratory medicine. Treat. Respir. Med. 5:47-78. [DOI] [PubMed] [Google Scholar]
- 7.Corpas, F. J., J. B. Barroso, L. M. Sandalio, J. M. Palma, J. A. Lupianez, and L. A. del Rio. 1999. Peroxisomal NADP+-dependent isocitrate dehydrogenase. Characterization and activity regulation during natural senescence. Plant Physiol. 121:921-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Flores, M., M. A. Ibanez-Hernandez, R. E. Galvan, M. Gutierrez, G. Duran-Reyes, R. Medina-Navarro, D. Pascoe-Lira, C. Ortega-Camarillo, C. Vilar-Rojas, M. Cruz, and L. A. Baiza-Gutman. 2006. Glucose-6-phosphate dehydrogenase activity and NADPH/NADP+ ratio in liver and pancreas are dependent on the severity of hyperglycemia in rat. Life Sci. 78:2601-2607. [DOI] [PubMed] [Google Scholar]
- 9.Giro, M., N. Carrillo, and A. R. Krapp. 2006. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 152:1119-1128. [DOI] [PubMed] [Google Scholar]
- 10.Grose, J. H., L. Joss, S. F. Velick, and J. R. Roth. 2006. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc. Natl. Acad. Sci. USA 103:7601-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensley, K., K. A. Robinson, S. P. Gabbita, S. Salsman, and R. A. Floyd. 2000. Reactive oxygen species, cell signaling, and cell injury. Free Radic. Biol. Med. 28:1456-1462. [DOI] [PubMed] [Google Scholar]
- 12.Hogan, M. C., C. M. Stary, R. S. Balaban, and C. A. Combs. 2005. NAD(P)H fluorescence imaging of mitochondrial metabolism in contracting Xenopus skeletal muscle fibers: effect of oxygen availability. J. Appl. Physiol. 98:1420-1426. [DOI] [PubMed] [Google Scholar]
- 13.Jezek, P., and L. Hlavata. 2005. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int. J. Biochem. Cell Biol. 37:2478-2503. [DOI] [PubMed] [Google Scholar]
- 14.Jo, S. H., M. K. Son, H. J. Koh, S. M. Lee, I. H. Song, Y. O. Kim, Y. S. Lee, K. S. Jeong, W. B. Kim, J. W. Park, B. J. Song, and T. L. Huh. 2001. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 276:16168-16176. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. J., and J. W. Park. 2005. Oxalomalate, a competitive inhibitor of NADP+-dependent isocitrate dehydrogenase, regulates heat shock-induced apoptosis. Biochem. Biophys. Res. Commun. 337:685-691. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lee, J. H., E. S. Yang, and J. W. Park. 2003. Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol-induced liver injury. J. Biol. Chem. 278:51360-51371. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. M., H. J. Koh, D. C. Park, B. J. Song, T. L. Huh, and J. W. Park. 2002. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic. Biol. Med. 32:1185-1196. [DOI] [PubMed] [Google Scholar]
- 19.Maeng, O., Y. C. Kim, H. J. Shin, J. O. Lee, T. L. Huh, K. I. Kang, Y. S. Kim, S. G. Paik, and H. Lee. 2004. Cytosolic NADP(+)-dependent isocitrate dehydrogenase protects macrophages from LPS-induced nitric oxide and reactive oxygen species. Biochem. Biophys. Res. Commun. 317:558-564. [DOI] [PubMed] [Google Scholar]
- 20.Mailloux, R. J., R. Hamel, and V. D. Appanna. 2006. Aluminum toxicity elicits a dysfunctional TCA cycle and succinate accumulation in hepatocytes. J. Biochem. Mol. Toxicol. 20:198-208. [DOI] [PubMed] [Google Scholar]
- 21.Mailloux, R. J., R. Singh, and V. D. Appanna. 2006. In-gel activity staining of oxidized nicotinamide adenine dinucleotide kinase by blue native polyacrylamide gel electrophoresis. Anal. Biochem. 359:210-215. [DOI] [PubMed] [Google Scholar]
- 22.Marino, D., E. M. Gonzalez, P. Frendo, A. Puppo, and C. Arrese-Igor. 2007. NADPH recycling systems in oxidative stressed pea nodules: a key role for the NADP+-dependent isocitrate dehydrogenase. Planta 225:413-421. [DOI] [PubMed] [Google Scholar]
- 23.Melov, S., J. Ravenscroft, S. Malik, M. S. Gill, D. W. Walker, P. E. Clayton, D. C. Wallace, B. Malfroy, S. R. Doctrow, and G. J. Lithgow. 2000. Extension of life-span with superoxide dismutase/catalase mimetics. Science 289:1567-1569. [DOI] [PubMed] [Google Scholar]
- 24.Middaugh, J., R. Hamel, G. Jean-Baptiste, R. Beriault, D. Chenier, and V. D. Appanna. 2005. Aluminum triggers decreased aconitase activity via Fe-S cluster disruption and the overexpression of isocitrate dehydrogenase and isocitrate lyase: a metabolic network mediating cellular survival. J. Biol. Chem. 280:3159-3165. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen, P., R. T. Awwad, D. D. Smart, D. R. Spitz, and D. Gius. 2006. Thioredoxin reductase as a novel molecular target for cancer therapy. Cancer Lett. 236:164-174. [DOI] [PubMed] [Google Scholar]
- 26.Nordberg, J., and E. S. Arner. 2001. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31:1287-1312. [DOI] [PubMed] [Google Scholar]
- 27.Nordlund, A., and M. Oliveberg. 2006. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc. Natl. Acad. Sci. USA 103:10218-10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma, E., D. A. Ragozzino, S. Di Angelantonio, G. Spinelli, F. Trettel, A. Martinez-Torres, G. Torchia, A. Arcella, G. Di Gennaro, P. P. Quarato, V. Esposito, G. Cantore, R. Miledi, and F. Eusebi. 2004. Phosphatase inhibitors remove the run-down of gamma-aminobutyric acid type A receptors in the human epileptic brain. Proc. Natl. Acad. Sci. USA 101:10183-10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson, G. H., S. M. Knobel, P. Arkhammar, O. Thastrup, and D. W. Piston. 2000. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc. Natl. Acad. Sci. USA 97:5203-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanov, V., M. T. Merski, and R. P. Hausinger. 1999. Assays for allantoinase. Anal. Biochem. 268:49-53. [DOI] [PubMed] [Google Scholar]
- 31.Rydstrom, J. 2006. Mitochondrial transhydrogenase—a key enzyme in insulin secretion and, potentially, diabetes. Trends Biochem. Sci. 31:355-358. [DOI] [PubMed] [Google Scholar]
- 32.Sakuraba, H., R. Kawakami, and T. Ohshima. 2005. First archaeal inorganic polyphosphate/ATP-dependent NAD kinase, from hyperthermophilic archaeon Pyrococcus horikoshii: cloning, expression, and characterization. Appl. Environ. Microbiol. 71:4352-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 34.Singh, R., R. Beriault, J. Middaugh, R. Hamel, D. Chenier, V. D. Appanna, and S. Kalyuzhnyi. 2005. Aluminum-tolerant Pseudomonas fluorescens: ROS toxicity and enhanced NADPH production. Extremophiles 9:367-373. [DOI] [PubMed] [Google Scholar]
- 35.Sun, J. S., Y. H. Tsuang, W. C. Huang, L. T. Chen, Y. S. Hang, and F. J. Lu. 1997. Menadione-induced cytotoxicity to rat osteoblasts. Cell. Mol. Life Sci. 53:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace, D. C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wynn, J. P., A. Kendrick, A. A. Hamid, and C. Ratledge. 1997. Malic enzyme: a lipogenic enzyme in fungi. Biochem. Soc. Trans. 25:S669. [DOI] [PubMed] [Google Scholar]
- 38.Yan, G., Z. Hua, G. Du, and J. Chen. 2006. Adaptive response of Bacillus sp. F26 to hydrogen peroxide and menadione. Curr. Microbiol. 52:238-242. [DOI] [PubMed] [Google Scholar]
- 39.Yates, S. P., and A. R. Merrill. 2005. Characterization of oxidized nicotinamide adenine dinucleotide (NAD+) analogues using a high-pressure- liquid-chromatography-based NAD+-glycohydrolase assay and comparison with fluorescence-based measurements. Anal. Biochem. 340:41-51. [DOI] [PubMed] [Google Scholar]
- 40.Zadzinski, R., A. Fortuniak, T. Bilinski, M. Grey, and G. Bartosz. 1998. Menadione toxicity in Saccharomyces cerevisiae cells: activation by conjugation with glutathione. Biochem. Mol. Biol. Int. 44:747-759. [DOI] [PubMed] [Google Scholar]