Abstract
We applied a novel negative selection strategy called genomic array footprinting (GAF) to identify genes required for genetic transformation of the gram-positive bacterium Streptococcus pneumoniae. Genome-wide mariner transposon mutant libraries in S. pneumoniae strain R6 were challenged by transformation with an antibiotic resistance cassette and growth in the presence of the corresponding antibiotic. The GAF screen identified the enrichment of mutants in two genes, i.e., hexA and hexB, and the counterselection of mutants in 21 different genes during the challenge. Eight of the counterselected genes were known to be essential for pneumococcal transformation. Four other genes, i.e., radA, comGF, parB, and spr2011, have previously been linked to the competence regulon, and one, spr2014, was located adjacent to the essential competence gene comFA. Directed mutants of seven of the eight remaining genes, i.e., spr0459-spr0460, spr0777, spr0838, spr1259-spr1260, and spr1357, resulted in reduced, albeit modest, transformation rates. No connection to pneumococcal transformation could be made for the eighth gene, which encodes the response regulator RR03. We further demonstrated that the gene encoding the putative DNA repair protein RadA is required for efficient transformation with chromosomal markers, whereas transformation with replicating plasmid DNA was not significantly affected. The radA mutant also displayed an increased sensitivity to treatment with the DNA-damaging agent methyl methanesulfonate. Hence, RadA is considered to have a role in recombination of donor DNA and in DNA damage repair in S. pneumoniae.
The gram-positive bacterium Streptococcus pneumoniae is an important opportunistic human pathogen. In healthy carriers, it resides asymptomatically in the nasopharynx, but in susceptible individuals, mostly children, the elderly, and immunocompromised patients, the pathogen can spread to other organs to cause disease, such as pneumonia, otitis media, or meningitis. Aspects that hamper the development of therapeutic and prophylactic tools against S. pneumoniae are its immense genetic diversity and adaptability. Vaccine efficacy, for example, is limited by the over 90 different capsular serotypes of S. pneumoniae (28), and major surface proteins, such as PspA, exist in different immunoreactive families (29). A major cause for this variation is the ability of S. pneumoniae to take up DNA from its environment via natural genetic transformation.
Natural genetic transformation is a complex mechanism, occurring in various gram-positive organisms, which requires the assembly of a multicomponent DNA translocon in the cell membrane as well as the presence of multiple cytoplasmic factors involved in processing and integration of foreign DNA into the genome. In S. pneumoniae, competence for genetic transformation is regulated by a two-component signal transduction system (TCS) that is induced by a small heptadecapeptide pheromone known as the competence stimulating peptide (CSP) (27, 51). This small peptide is secreted into the medium, and its accumulation in exponentially growing pneumococcal cultures is sensed by the TCS in the cell envelope, which leads to the activation of a regulatory pathway that controls the expression of most other genes now known to be required for DNA uptake, binding, and processing (Fig. 1). In addition, competence development appears to be involved in several other aspects of the pneumococcal life cycle. Some of these, such as selective lysis of neighboring cells, named fratricide (23), may be important for acquiring DNA. In other cases, competence appears to be induced more as a general response to (antibiotic) stress (17, 56) or as a signal to initiate the formation of pneumococcal biofilms (48).
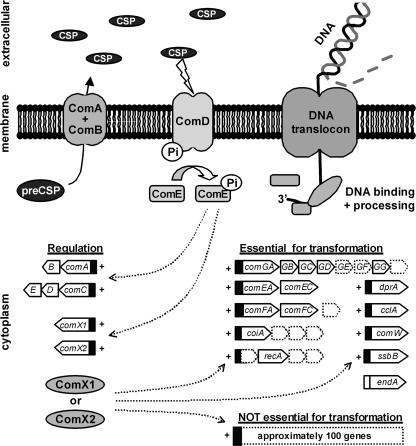
FIG. 1.
Regulation of genes essential for pneumococcal transformation. The precursor of CSP is the gene product of comC, which is processed and excreted into the medium by the ComAB ABC transporter. The accumulation of CSP in exponentially growing pneumococcal cultures is sensed by the ComD histidine kinase in the bacterial membrane. Subsequent phosphorylation of the cytoplasmic response regulator ComE leads to increased CSP production and to ComX1/X2 synthesis. Except for the endA gene, these alternative sigma factors control the expression of all genes required for DNA uptake and processing. In addition, they induce the expression of approximately 100 other genes that are not essential for pneumococcal transformation. Dotted arrows in the figure indicate activation of the promoters (black rectangles) by the transcriptional regulator ComE and alternative sigma factors ComX1 and ComX2. Genes in competence operons with no known role in pneumococcal transformation are shown as dotted boxes.
The application of various techniques has led to the identification of genes that are directly involved in development of pneumococcal competence and transformation. Already in the late 1970s, Morrison and Baker had identified 14 competence-related proteins by analysis of protein expression profiles (46). Research on pneumococcal transformation was boosted with the availability of synthetic CSP, enabling more reproducible transformation rates (26), and with the application of novel genetic tools, such as insertion duplication mutagenesis (40, 47), differential fluorescence induction (6), and other transcriptional reporter strategies (13, 52). Upon the release of pneumococcal genome sequences (21, 30, 62), transcriptome analyses using microarrays revealed the truly complex nature of the competence regulon, which drives the expression of at least 124 genes (18, 53). Interestingly, only 22 of the genes regulated by CSP, together with the endA gene, have thus far been shown to be indispensable for proper pneumococcal transformation.
Recently, novel technologies have been developed to identify conditionally essential genes of bacteria by using microarrays to track large transposon mutant libraries before and after a challenge of choice (4, 59). In these approaches, mutant-specific microarray probes are generated by PCR amplification of the DNA adjacent to the transposon insertion site or by in vitro transcription from promoters engineered at the ends of transposons. Microarray hybridization patterns of these probes reveal which mutants disappear during challenge and, consequently, which genes are conditionally essential. We developed a similar negative selection strategy, called genomic array footprinting (GAF), for use with S. pneumoniae and have successfully applied this technology to identify genes essential for survival during zinc stress (12). Given the pivotal role of competence development in the pneumococcal life cycle, we used GAF in an attempt to identify the full complement of genes specifically essential for genetic transformation. To this end, mariner transposon mutant libraries were challenged by transformation with an antibiotic resistance cassette and growth in the presence of the corresponding antibiotic. In this study, we show that many genes expected to be essential for pneumococcal transformation in our experimental setup could readily be identified by GAF. In addition, we discuss the identification of several genes that have previously not been associated with this process. Finally, we demonstrate that one of these genes, encoding the RadA DNA repair protein, is essential for efficient pneumococcal transformation with DNA that needs to be integrated into the chromosome.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae was routinely grown in GM17 broth (33) or on GM17 agar plates or Colombia agar (Oxoid) plates supplemented with 5% sheep blood (Biotrading) at 37°C and 5% CO2. For transformation, S. pneumoniae was grown as standing cultures in cCAT medium (10 g liter−1 Casamino Acids [Difco], 5 g liter−1 tryptone [Difco], 10 g liter−1 yeast extract [Difco], 5 g liter−1 NaCl, 16 mM K2PO4, 0.2% glucose, and 0.15 g liter−1 glutamine) or CTM medium (cCAT medium supplemented with 0.2% bovine serum albumin and 1 mM CaCl2), and transformants were selected on cCAT medium or blood agar plates. Escherichia coli was grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth in a shaking incubator. Lactococcus lactis was grown at 30°C on GM17 agar plates or as standing cultures in GM17 broth. When indicated, antibiotics were used at the following concentrations: spectinomycin, 150 μg ml−1; kanamycin, 500 μg ml−1; erythromycin, 0.25 μg ml−1; chloramphenicol, 2.5 μg ml−1 for S. pneumoniae and 5 μg ml−1 for L. lactis; and ampicillin, 100 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant featuresa | Reference |
|---|---|---|
| Strains | ||
| S. pneumoniae | ||
| R6 | Unencapsulated derivative of D39 | 30 |
| D39 (FP22) | D39 with cps gene locus replaced by Kmr cassette; template for Δcps PCR product | 49 |
| D39 ΔaliA | D39 with aliA gene replaced by Ermr cassette | 32 |
| TIGR4 | Wild-type strain; serotype 4 | 62 |
| TIGR4 (FP23) | TIGR4 with cps gene locus replaced by Kmr cassette | 49 |
| CP1250 | Unencapsulated derivative of Rx and D39 | 51 |
| E. coli | ||
| DH5α | Host for plasmids pR412T7 and pR410 | 24 |
| L. lactis | ||
| LL108 | Host for plasmid pNG8048E | 42 |
| Plasmids | ||
| pR412T7 | Donor for mariner transposon, conferring Spr; contains T7 promoter; Ampr | 12 |
| pR410 | Donor for mariner transposon, conferring Kmr; Ampr | 61 |
| pNG8048E | Replicating plasmid derived from pNZ8048; Ermr Cmr | 33 |
Kmr, kanamycin resistance; Ermr, erythromycin resistance; Spr, spectinomycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
General DNA techniques.
Chromosomal DNA was isolated from pneumococcal cultures by cetyltrimethylammonium bromide extraction as described previously (64). Plasmids were isolated from E. coli or L. lactis broth cultures by use of a HighSpeed Plasmid Midi kit (QIAGEN Benelux B.V.). For in vitro transposon mutagenesis (37), 1 μg of pneumococcal DNA was incubated in the presence of purified HimarC9 transposase (36), with 0.5 μg of plasmid pR412T7 (12) or pR410 (61) as the donor for mariner transposons conferring spectinomycin or kanamycin resistance, respectively. The resulting transposition products were repaired with T4 DNA polymerase and E. coli DNA ligase (25). The Δcps locus of S. pneumoniae strain D39 (FP22) was PCR amplified with primer pair FI4 and PE21 (49), using Taq DNA polymerase (Integro). PCR conditions were as follows: 93°C for 4 min; 30 cycles of 93°C for 30 s, 55°C for 1 min, and 72°C for 2 min; and 72°C for 4 min. Δcps PCR products were purified by ethanol precipitation and dissolved in H2O.
Genetic transformation of S. pneumoniae.
Preparation and transformation of precompetent S. pneumoniae cell stocks were performed essentially as described previously (44). Briefly, cCAT medium was inoculated with several colonies and grown to an optical density at 600 nm (OD600) of 0.25 to 0.3. After a 30-fold dilution of the culture in CTM medium, cells were grown to an OD600 of 0.1, pelleted, concentrated 10-fold in CTM-pH 7.8 (CTM medium adjusted to pH 7.8 with NaOH) containing 15% glycerol, and stored at −80°C. For transformation, precompetent cells were grown for 15 min at 37°C in a 10-fold volume of CTM-pH 7.8 supplemented with 100 ng/ml CSP-1, for strains R6 and CP1250, or CSP-2, for the TIGR4 strain. After the addition of DNA, cultures were incubated at 32°C for 30 min and then shifted to 37°C. One hour after the temperature shift, transformants were selected on solid medium supplemented with the proper antibiotics. DNA uptake efficiencies of wild-type and mutant strains were probed by transformation with 1 μg ml−1 chromosomal DNA or PCR product or 0.1 μg ml−1 plasmid DNA. At least three independent transformation experiments were performed, and transformation efficiencies are given as percentages of that of the wild-type strain or a 2,500-CFU mutant library.
Construction and challenge of transposon mutant libraries.
To obtain pneumococcal mariner transposon mutant libraries, CSP-induced precompetent S. pneumoniae R6 cells were transformed with 1 μg ml−1 of pR412T7-mutagenized R6 chromosomal DNA. After a 1-hour incubation at 37°C, precompetent stocks of these mutant libraries were prepared by diluting the transformation culture 100-fold in cCAT medium. Subsequent steps were performed as described above. To counterselect for nontransformants, cCAT and CTM media were supplemented with spectinomycin. The transposon mutant library was challenged for DNA uptake by transformation with 1 μg ml−1 Δcps PCR product. Transformants were selected by growth in cCAT medium supplemented with spectinomycin and kanamycin. For the kinetics experiment, in which the effect of growth time on the selection of genes required for DNA uptake was assessed, a single transformation culture was used. Samples were taken at approximately 15, 18, 21, and 24 generations of growth, while at the same times cultures were diluted eightfold in fresh medium. For the challenge and control experiments, at least three independent transformation experiments were performed, and the growth time was approximately 25 generations. Generation times were estimated by measuring the OD600 and were confirmed by counting the CFU of serially diluted samples on selective solid medium.
GAF.
The GAF technology was performed essentially as described previously (12). Briefly, chromosomal DNA extracts from the challenged and nonchallenged mutant libraries were digested with the AluI (New England Biolabs) and TaqI (Invitrogen) endonucleases, which cut the genome 9,985 and 4,832 times, respectively. The resulting DNA fragments were used as a template for an in vitro T7 RNA polymerase reaction (T7 MegaScript; Ambion). After removal of template DNA by DNase I treatment, fluorescent Cy3/Cy5-labeled dUTP nucleotides (GE Healthcare) were incorporated by reverse transcription, using Superscript III (Invitrogen). Fluorescently labeled DNA probes were combined and hybridized in Slidehyb buffer 1 (Ambion) to pneumococcal microarrays, which are superamine glass slides spotted with duplicates of amplicons representing 2,087 S. pneumoniae TIGR4 open reading frames (ORFs) and 184 ORFs unique to R6 (34). The microarrays were incubated for 16 h at 45°C and then washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; Invitrogen) containing 0.25% sodium dodecyl sulfate for 5 min, followed by two washes, with 1× SSC and 0.5× SSC, for 5 min each. Finally, the slides were dipped into H2O and dried by centrifugation.
Microarray analysis.
Dual-channel array images were acquired on a GenePix 4200AL microarray scanner and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). Spots were screened visually to identify those of low quality. Slide data were processed and normalized using MicroPreP as described previously (63). Net signal intensities were calculated using a grid-based background subtraction. Automatically and manually flagged spots and spots with low background-subtracted signal intensities (sum of Cy3 and Cy5 net signals, <700) were filtered out of all data sets prior to analysis. For the kinetics experiment, only genes with data available from all array experiments were analyzed. DNA microarray data for the challenge experiments with the full-genome transposon mutant library were obtained from three (control) or four (challenge) independent biological replicates and analyzed using Cyber-T Student's t test for paired data (43; http://visitor.ics.uci.edu/genex/cybert/index.shtml). The following selection criteria were applied: for the control experiment, a minimum of five reliable measurements, and for the challenge experiment, a minimum of seven reliable measurements; a Bayes P value of <0.001; and a false discovery rate of <0.05.
Construction of deletion mutants.
A megaprimer PCR method was employed to replace target genes in the genomes of S. pneumoniae strains R6, CP1250, and TIGR4 with the spectinomycin resistance cassette of plasmid pR412T7. Details of the primer sequences are available on request. The spectinomycin resistance cassette of plasmid pR412T7 was amplified using primers PBpR412_L and PBpR412_R. Flanking regions of ∼500 bp, containing less than 150 bp of the coding sequence of the target genes, were amplified by PCR, with chromosomal DNA as the template and the following PCR conditions: 93°C for 4 min; 35 cycles of 93°C for 30 s, 55°C for 1 min, and 68°C for 1.5 min; and 68°C for 4 min. For each flanking region, the primer closest to the target gene (extension plus _L2 or _R2) contained an additional sequence complementary to primer PBpR412_L or PBpR412_R. In a second PCR, the PCR products of the two flanking regions and the antibiotic resistance cassette were combined, leading to incorporation of the antibiotic resistance cassette between the two flanking regions of the target gene. The PCR conditions were as follows: 93°C for 4 min; 30 cycles of 93°C for 30 s, 55°C for 1 min, and 68°C for 3 min; and 68°C for 7 min. In both PCRs, Pwo DNA polymerase (Roche) was used, and the MgSO4 concentration was optimized for each reaction. DNA constructs were introduced into S. pneumoniae via transformation. Correct integration of the antibiotic resistance cassette into the target gene was validated by a PCR using Taq DNA polymerase and a combination of flanking primers (extension plus _L1 or _R1) with gene-specific primers (extension plus _C) or the cassette-specific primer PBMrTn7. PCR conditions were as follows: 93°C for 4 min; 35 cycles of 93°C for 30 s, 55°C for 1 min, and 72°C for 1.5 min; and 72°C for 4 min.
MMS sensitivity assay.
For methyl methanesulfonate (MMS) survival analysis, cells were grown in GM17 medium to the exponential stage (OD600 = 0.15 to 0.2) and then exposed to 0.1% MMS at 37°C for different times. To score the survival of cells, exposed cell cultures were serially diluted in phosphate-buffered saline and plated onto Columbia sheep blood agar plates for overnight growth. The experiment was repeated three times.
Microarray data accession numbers.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under GEO Series accession number GSE8362.
RESULTS
Identification of pneumococcal genes essential for efficient genetic transformation by GAF technology.
To identify genes essential for pneumococcal transformation using GAF technology, we transformed mariner transposon mutant libraries of the unencapsulated S. pneumoniae strain R6 with donor DNA containing an antibiotic resistance marker and counterselected the transformants for transformation-defective mutants by growth under selective pressure. Transformed mutant libraries that were grown without selective pressure were used as controls. The donor for antibiotic resistance was the PCR product of a capsule gene locus knockout (KO; Δcps), consisting of a kanamycin resistance cassette flanked by parts of the dexB and aliA genes, which replaces the entire cps locus during integration into the chromosome (49). Since capsule production in the R6 strain is already impaired, i.e., the first nine genes of the capsule gene cluster are absent or truncated (30), the number of mutant-specific growth defects as a result of the insertion was expected to be low. Transformation rates obtained with the Δcps PCR product were typically 100,000 per ml of competent cells, and we extrapolated that a mutant library should contain fewer than 5,000 unique transposon mutants per ml to give each mutant ample (20-fold) opportunity to acquire the antibiotic resistance cassette.
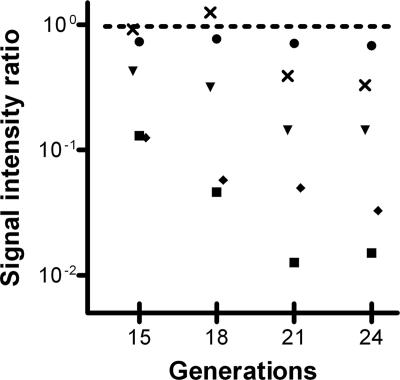
In our approach, chromosomal DNA from cultured mutant libraries was used to generate mutant-specific probes by consecutive endonuclease fragmentation and in vitro transcription of the T7 promoter sequences engineered at the ends of the mariner transposon. It was expected that DNA of nontransformed cells, which do not survive antibiotic selective pressure, could still affect the outcome of the GAF screen by interference with probe synthesis and subsequent microarray analysis. To study if we could reduce this background effect by increasing growth times, thus decreasing the relative abundance of DNA derived from nontransformed cells in the total DNA extract, we performed a kinetics experiment. A small mutant library of about 2,500 CFU was transformed and grown under selective pressure for about 15, 18, 21, or 24 generations before DNA extraction. GAF analysis using TaqI endonuclease for DNA fragmentation revealed that the ratios of signal intensities of genes known to be required for efficient pneumococcal transformation, such as coiA, comEC (celB), comGD (cglD), and comD, decreased with an increasing number of generations, reaching a plateau at 21 to 24 generations (Fig. 2). At the same time, the mean absolute ratio of all genes, excluding the 23 known to be involved in transformation (53) and the genes replaced by the Δcps PCR product, did not alter significantly over the different time points (Fig. 2). These results imply that, overall, the stoichiometry of the mutants within the challenged mutant library remained unaffected by increased growth times, whereas the outcome of the GAF screen improved considerably. During this analysis, we noticed several apparent false-positive results, i.e., genes with no known role in genetic transformation but localized adjacent to transformation-related genes such as coiA (data not shown). To address this issue, we repeated GAF on the 24th-generation samples, using AluI instead of TaqI endonuclease. Since AluI cuts the genome twice as often as TaqI, fewer DNA fragments will represent multiple genes. Indeed, genes adjacent to coiA were no longer picked up, while known competence genes were still reliably identified (data not shown). Consequently, to reduce the chance of selecting genes adjacent to transformation-related genes, the next set of experiments was performed using AluI.
FIG. 2.
Challenge kinetics of alterations in the relative abundance of mutants in comEC (▪), coiA (⧫), comGD (▾), and comD (×) genes in a 2,500-CFU mutant library. Challenged and nonchallenged mutant libraries were grown for about 15, 18, 21, or 24 generations, and signal intensity ratios for each gene were determined by GAF. The average absolute ratio (•) was calculated at different time points, excluding known competence genes and genes in the Δcps PCR product. The dotted line indicates a signal intensity ratio of 1.
To screen the entire genome of S. pneumoniae strain R6 for genes essential for genetic transformation, we constructed a mutant library of about 40,000 CFU. Since 2,043 genes have been predicted for the R6 strain (30), this amounts to almost 20-fold coverage of the genome, which can be considered close to saturation. After transformation with the Δcps PCR product, the mutant library was grown under selective antibiotic pressure for approximately 25 generations before extraction of chromosomal DNA and GAF analysis. As a control, we analyzed three replicates of mutant libraries that were transformed and grown without selective antibiotic pressure. This showed that the maximal signal intensities varied by a factor of <1.2 due to growth and labeling alone (data not shown). Although we expected mutants of each gene with a signal alteration above this value to have altered transformation efficiencies, we decided to use a signal intensity difference between the unchallenged and challenged libraries of 1.8-fold to ensure that changes in transformation efficiency were significant. After excluding the genes present on the cps locus, 23 genes met this criterion after challenge (Table 2). Interestingly, two genes were found to have increased signals, suggesting that the corresponding mutants had become more abundant during challenge due to increased transformation efficiencies compared to that of the wild-type strain. These genes code for the HexA and HexB mismatch repair proteins, which play a role during homologous recombination (15). In the absence of either HexA or HexB, mismatches between donor DNA and the genome are not recognized and excised, resulting in increased integration efficiencies (5, 16, 55). The signal decreases observed for the other 21 genes indicated that the corresponding mutants had become less abundant during challenge, implying that these genes are putatively essential for efficient pneumococcal transformation in our experimental setup. Eight of the observed genes, i.e., comGA (cglA), comGB (cglB), comGG (cglG), comD, coiA, comEC, comFA (cflA), and endA, have previously been described to be essential for efficient pneumococcal transformation (11, 27, 35, 40, 51, 52, 57). Interestingly, we also identified comGF (cglF), which is part of the comG operon and most likely encodes a component of the DNA translocon (40). However, the appearance of comGF in this GAF screen could also be the result of polar effects of a transposon insertion in this gene on the expression of the other genes in this operon. Alternatively, loss of the comGF signal could be the consequence of the disappearance of mutants in comGG, as these two genes share an AluI fragment. The same holds true for spr2014, which shares an AluI restriction fragment with comFA. Interestingly, several genes we selected, namely radA (spr0025), spr2011, and parB (spr2046), have previously been described to be part of the competence regulon. Although expression of these genes was found to be upregulated during competence development, no substantial effect of their mutation on transformation has been seen (53). Most likely, the spr2011 gene was identified because it shares an AluI fragment with the comFC (cflB) gene (spr2012), which is essential for pneumococcal transformation (53). The comFC gene itself was not identified in our analysis. This suggests that the probe generated by the transposon insertion in the comFC gene, which also anneals to the spr2011-specific amplicon, anneals less efficiently to the comFC-specific amplicon itself. The parB gene is in the same operon as the htrA gene (spr2045), encoding the serine protease HtrA with a putative general chaperone function (22). It has previously been shown that overexpression of this gene can lead to decreased competence development and transformation rates (60). Possibly, transposon mutants in parB affect htrA expression levels and, indirectly, transformation rates. For these reasons, the relationship between pneumococcal transformation and the spr2011 and parB genes was not studied further in this work. For eight genes that were found in the challenge, no connection with competence development and/or genetic transformation could be made based on present knowledge; these included genes coding for the response regulator RR03 (spr0334), a putative ABC transporter system (spr0459 and spr0460), a putative inositol monophosphatase (spr1260), and four hypothetical proteins. Orthologs of the hypothetical ORFs in the TIGR4 genome are annotated as encoding putative methylases (spr0838 and spr1259), a cell division protein (spr0838), and permeases (spr1357 and spr0777).
TABLE 2.
Genes identified by GAF as being involved in pneumococcal transformation
| Gene | Common namea | Functiona | Essential for competenceb | Reference | Ratioc |
|---|---|---|---|---|---|
| Negatively selected | |||||
| spr0025 | radA | DNA repair | No | 53 | 0.36 |
| spr0344 | rr03 | Response regulator | No | 38 | 0.50 |
| spr0459 | ABC transporter, ATP-binding protein | − | 0.49 | ||
| spr0460 | ABC transporter, permease | − | 0.49 | ||
| spr0777 | Putative permease | − | 0.50 | ||
| spr0838 | Tetrapyrrole methylase/cell division protein | − | 0.45 | ||
| spr0857 | comEC (celB) | DNA pore | Yes | 52 | 0.14 |
| spr0881 | coiA | DNA processing | Yes | 40 | 0.17 |
| spr1259 | NOL1/NOP2/sun family, RNA methylase | − | 0.30 | ||
| spr1260 | Inositol monophosphatase | − | 0.55 | ||
| spr1357 | Putative permease | − | 0.36 | ||
| spr1779 | endA | Endonuclease, degradation of donor strand | Yes | 35, 57 | 0.54 |
| spr1858 | comGG (cglG) | Cell wall channel | Yes | 53 | 0.44 |
| spr1859 | comGF (cglF) | Cell wall channel | − | 0.35 | |
| spr1863 | comGB (cglB) | ComGC-GG export | Yes | 52 | 0.16 |
| spr1864 | comGA (cglA) | ComGC-GG export | Yes | 52 | 0.11 |
| spr2011 | Ribosomal subunit interface protein | No | 53 | 0.48 | |
| spr2013 | comFA (cflA) | Putative helicase/DNA translocase | Yes | 40 | 0.51 |
| spr2014 | Hypothetical protein | − | 0.46 | ||
| spr2042 | comD | Sensor for CSP | Yes | 27, 51 | 0.17 |
| spr2046 | parB | Chromosome segregation protein | No | 53 | 0.41 |
| Positively selected | |||||
| spr0160 | hexB | DNA mismatch repair | No | 55 | 3.4 |
| spr1888 | hexA | DNA mismatch recognition | No | 5, 16 | 4.5 |
Names and functions of genes were derived from the NCBI (http://www.ncbi.nlm.nih.gov/) and KEGG (http://www.kegg.com/) websites.
Competence data were obtained from the cited references. Yes, essential; no, not essential; −, unknown.
Ratios of signal intensities of genes derived from microarray data (intensity when challenged/intensity when not challenged).
Phenotypic validation of genes identified during GAF.
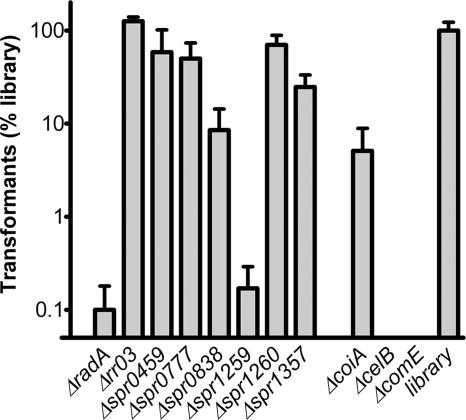
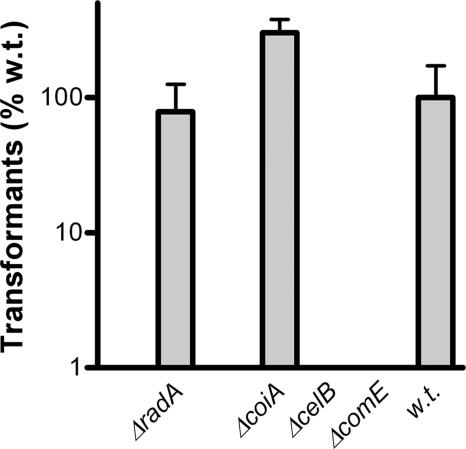
To examine whether the eight putative transformation genes picked up by GAF and the radA gene are indeed required for efficient pneumococcal transformation, directed KO mutants in S. pneumoniae strain R6 were generated by allelic replacement with a spectinomycin resistance cassette. Since the spr0459 and spr0460 genes are likely to encode one ABC transporter system, a single mutant disrupting both genes was constructed. KO mutants for genes encoding elements of competence regulation (comE), the DNA translocon (comEC), and DNA processing (coiA) were included as positive controls. Transformation efficiencies of the mutant strains were determined by using the Δcps PCR product as the donor of foreign DNA and were compared to the efficiency of a mutant library (Fig. 3). As expected, genetic transformation was completely abolished for both the comE and comEC mutants and was reduced to approximately 5% of the mutant library level for the coiA mutant. Of the eight mutants that were tested, only three, i.e., the radA, spr1259, and spr0838 mutants, had transformation efficiencies of 10% or lower, and they were considered to be affected in genetic transformation in our experimental setup. For four other mutants, i.e., the spr0459 spr0460, spr0777, spr1260, and spr1357 mutants, the transformation efficiency had not decreased below 10%, and the effects of these genes on competence could be considered modest, at best. Most likely, the spr1260 gene was identified because it shares an AluI restriction fragment with the spr1259 gene. For the Δrr03 mutant, no decrease in transformation efficiency was observed. To rule out the possibility that altered growth rates of the mutants that displayed no or modest effects on transformation efficiency caused them to disappear from the mutant library, we performed growth experiments. However, no clear-cut growth defects of the mutants were observed before and after transformation with the Δcps PCR product (data not shown). Furthermore, we tested whether these mutants had more prominent alterations in their transformation efficiencies when they were transformed in mixed cultures of mutant and wild-type strains, a situation which is more similar to the experimental setup of the GAF screen itself. However, this was not the case (data not shown). Hence, the appearance of the spr0459-spr0460, spr0777, and spr1357 genes in the GAF screen was most likely the result of the modestly decreased transformation efficiencies of their mutants. It remains unclear why the rr03 gene was picked up in this GAF screen.
FIG. 3.
Transformation of S. pneumoniae R6 mutant strains and a 2,500-CFU mutant library with the Δcps PCR product. Transformation efficiencies are given as percentages relative to that of the 2,500-CFU mutant library. Data were obtained from three independent experiments and are presented as means ± standard deviations.
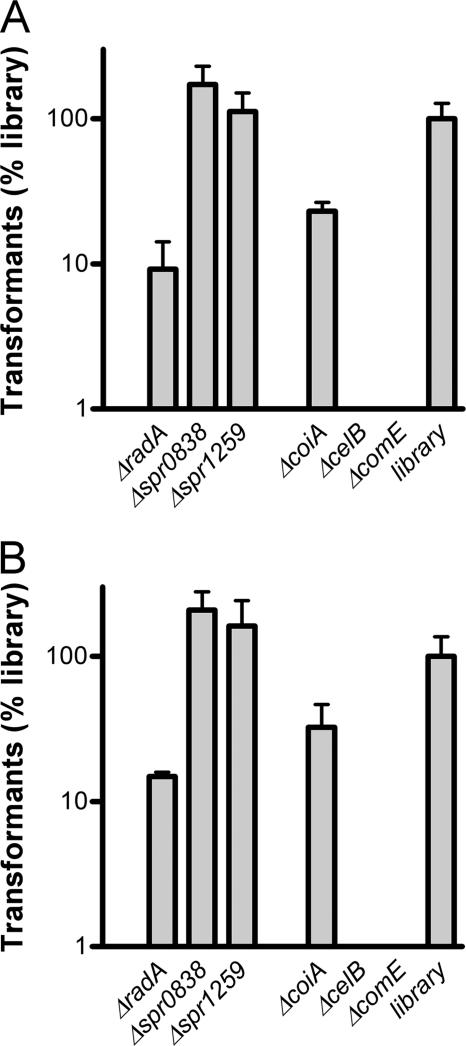
To probe if the transformation defects in the radA, spr0838, and spr1259 mutants were Δcps PCR product specific, we performed two additional transformation experiments, one with in vitro transposition products of R6 chromosomal DNA with the mariner transposon from plasmid pR410 (kanamycin resistance) and one with chromosomal DNA of an ΔaliA strain (erythromycin resistance). The first type of donor DNA was expected to give random genomic insertions, ruling out any gene-specific effects on transformation rates, whereas the second has a different antibiotic marker in a gene that is not required for in vitro growth. Interestingly, with both types of donor DNA, the spr0838 and spr1259 mutants no longer displayed reductions of their transformation rates (Fig. 4A and B), making a general role of these genes in pneumococcal transformation highly unlikely. Possibly, the deletion of the remnants of the R6 capsule genes caused some phenotypic change in connection with these two genes. For the radA mutant, the transformation rate increased slightly, to 10% of the mutant library level, which is still twofold lower than that of the coiA mutant with these types of donor DNA. This suggests that the radA gene is required for efficient pneumococcal transformation in general.
FIG. 4.
Transformation of S. pneumoniae R6 mutant strains and a 2,500-CFU mutant library with pR410-mutagenized R6 DNA (A) or D39 ΔaliA chromosomal DNA (B). Transformation efficiencies are given as percentages relative to that of the 2,500-CFU mutant library. Data were obtained from three independent experiments and are presented as means ± standard deviations.
Function of RadA in pneumococcal competence.
For both E. coli and Bacillus subtilis, RadA has been shown to be involved in genomic recombination of donor DNA during transformation (8, 14). To probe if this is also the case for pneumococcal RadA, we performed transformation experiments with plasmid pNG8048E, which does not need to recombine with the genome for replication. For the comE and comEC mutants, which are both essential for uptake of DNA, transformation with plasmid DNA was completely abolished (Fig. 5). Disruption of the coiA gene, which is required for efficient recombination of internalized donor DNA with the pneumococcal genome, had no negative impact on transformation with the replicating plasmid. Also, the radA mutant had transformation rates close to the wild-type level with this type of donor DNA. This indicates that RadA is required for efficient integration of donor DNA into the chromosome but not for efficient uptake of replicating plasmid DNA into the cell.
FIG. 5.
Transformation of S. pneumoniae R6 mutant and wild-type strains with replicating plasmid pNG8048E. Transformation efficiencies are given as percentages relative to that of the wild-type (w.t.) strain. Data were obtained from three independent experiments and are presented as means ± standard deviations.
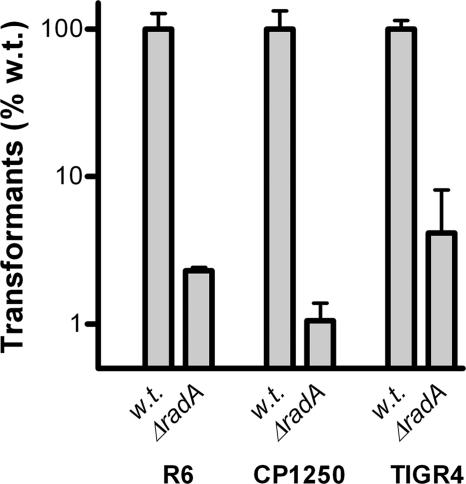
In a previous study, radA was found to be the third gene of a putative CSP-induced operon composed of six genes (53). In that study, disruption of the last five genes of this putative operon in the Rx derivative strain CP1250 was found to have no impact on pneumococcal transformation with integrative DNA (53). To assess if this apparent inconsistency with our findings was strain specific, we constructed radA mutants in the TIGR4 and CP1250 backgrounds to probe their transformation efficiencies. Since the CP1250 strain contains a type 3 capsule gene cluster (inactive) (58), which is one of the few capsule types that is not flanked by the aliA gene (10), it was not possible to efficiently transform the CP1250 wild-type strain with the Δcps PCR product. To overcome this constraint, we used chromosomal DNA from an unencapsulated D39 (FP22) strain as donor DNA for transformation of the R6 and CP1250 strains and DNA of an unencapsulated TIGR4 (FP23) strain (49) for TIGR4 transformation. Similar to the R6 radA mutant, the TIGR4 and CP1250 radA mutant strains were almost completely deficient for transformation (Fig. 6). This demonstrates that the requirement for the radA gene for pneumococcal transformation with integrative DNA is conserved at least between the R6, CP1250, and TIGR4 strains. We also disrupted the last five genes of the putative radA operon (annotated as four genes for R6) in the CP1250 and R6 strains to probe if these mutants would behave differently from the single radA gene KO strains. Interestingly, both the R6 and CP1250 spr0024-spr0027 mutant strains had severe growth problems in the CTM medium that we used for transformation assays, and consequently, their transformation efficiencies could not be determined.
FIG. 6.
Transformation of S. pneumoniae R6, CP1250, and TIGR4 wild-type and ΔradA mutant strains with chromosomal DNA from the D39 Δcps and TIGR4 Δcps strains. Transformation efficiencies are given as percentages relative to those of the wild-type (w.t.) strains. Data were derived from three independent experiments and are presented as means ± standard deviations.
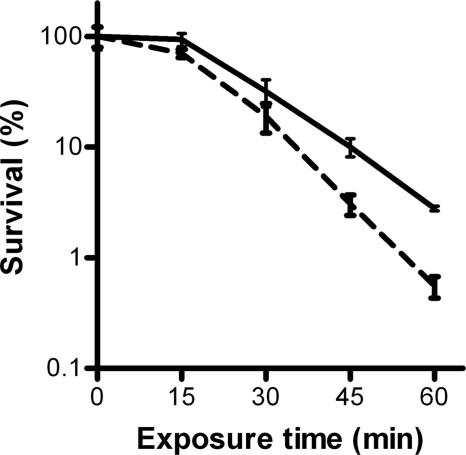
In addition to a role in recombination of DNA during transformation, RadA has also been described to contribute to DNA damage repair in E. coli and B. subtilis (8, 14). To assess if the product of the identified gene has a similar function in S. pneumoniae, we compared survival rates of the radA mutant and the wild-type R6 strain after treatment with MMS. The radA mutant displayed an increased sensitivity to treatment with the MMS mutagen (Fig. 7), which is indicative of a role of RadA in DNA repair. No effect of the radA mutation on sensitivity to ionizing radiation (UV light) was observed (not shown).
FIG. 7.
MMS sensitivity assay results. Exponentially growing cultures of S. pneumoniae R6 wild-type (solid line) and radA mutant (dotted line) strains were incubated at 37°C with 0.1% MMS, and survival of the cells was determined at the indicated time points. Survival rates are given as percentages relative to the number of cells present before the addition of MMS. Data were derived from three independent experiments and are presented as means ± standard deviations.
DISCUSSION
With the availability of genome sequences, classical methods to solve biological issues are rapidly being complemented with novel genome-wide approaches. Recently, we described a novel genomic approach in which microarrays are used to track transposon mutants within genome-wide mutant libraries of S. pneumoniae (12). In the present study, we applied this technology (GAF) to the complex biological process of pneumococcal competence by addressing which genes are essential for transformation.
In a mutant library of 40,000 CFU, we identified 21 genes whose mutants disappeared during a challenge for DNA uptake. Only eight genes found in our screen have been described previously as being essential for pneumococcal transformation. The remainder of the 23 genes known to be required for this process (53) were not identified directly by applying GAF. In part, this was a consequence of our experimental design. The addition of synthetic CSP obviates the need for the CSP synthesis and efflux pathway, encoded by the comAB and comC genes (26, 31). Furthermore, small differences in growth due to transposon insertions might result in a loss of mutants while generating a mutant library. For example, mutants in the recA gene, which is required for DNA recombination into the genome, are impaired in growth and were therefore less likely to be present in our initial library (40). Certain aspects of the competence regulon itself and characteristics of single mutants may affect the outcome of GAF as well. Two copies of the gene encoding the ComX alternative sigma factor, comX1 and comX2, are present on the R6 genome, and single mutants are not affected in transformation efficiency (41). On the other hand, a number of mutants will not show up due to technological limitations of the GAF procedure itself. For instance, transposon insertions in short genes that have multiple AluI restriction sites, such as comGC (cglC), are unlikely to generate GAF probes long enough for efficient hybridization to the corresponding amplicon on the array (12). Moreover, several genes, such as comW, comGD, comEA, comFC, and ssbB, are located on AluI fragments that also carry adjacent genes that are not essential for competence, resulting in reduced alterations in signal intensity. This phenomenon was exemplified by the identification of comGD in the 2,500-CFU mutant library of the kinetics experiment but not in the 40,000-CFU mutant library of the genome-wide GAF screen. Interestingly, other studies have also reported limitations in using pneumococcal mutant libraries for the identification of competence genes. While utilizing insertion duplication mutagenesis, Lee and coauthors (40) failed to identify mutants in the comCDE and several other transformation-specific loci, whereas they expected their 20,000-CFU mutant library to be saturating as well. Moreover, three large-scale signature-tagged mutagenesis screens did not identify mutants in capsule genes to be essential for virulence in various models of pneumococcal disease (25, 39, 54), even though unencapsulated mutants are avirulent. This suggests that the construction and maintenance of pneumococcal mutant libraries that represent mutants in most genes are difficult.
In addition to known competence genes, we identified 13 genes previously not shown to be required for pneumococcal transformation. As anticipated, some of them were adjacent to genes known to be essential for transformation, i.e., comGF, spr2011, and spr2014, or near genes that could affect competence development and transformation indirectly, i.e., parB. However, most of the mutants in the nine remaining genes, which were clustered in seven loci, were indeed at least slightly impaired in efficient pneumococcal transformation in our experimental setup. Interestingly, most genes whose KO mutants had slightly decreased transformation efficiencies, i.e., spr0459-spr0460, spr0777, and spr1357, encode parts of putative ABC transporters and/or permeases. This class of proteins is especially recognized for a role in the import of a variety of substrates and might therefore be necessary to feed the process of competence development. Also, other permeases have been shown to influence competence. A mutant of the AliB oligopeptide transporter is almost completely defective in competence development (1, 50), although its transformation in the presence of synthetic CSP remains unaffected (2). Several metal ion importers were shown to be important for transformation as well (19, 20). Further study on the substrate specificities of the permeases identified in this work should help to reveal their role in pneumococcal transformation.
Only the appearance of the rr03 gene in the GAF screen remains unexplained. A role for this gene in competence is not supported by earlier studies (38) or by our own experiments. It is also unlikely that polar effects of rr03 mutants on adjacent genes resulted in its identification. The ORF upstream of rr03 codes for its cognate histidine kinase, HK03, whose fate is connected to that of the response regulator itself, and the three ORFs downstream of rr03, i.e., spr0345 to spr0347, have been annotated as encoding truncations of a putative DNA alkylation repair enzyme, AlkD. In fact, a mutant disrupting the spr0345 to spr0347 genes was unaffected in pneumococcal transformation (our unpublished observation). The only link between the rr03 gene and competence was shown in a ΔciaR strain, where rr03 expression was induced upon addition of CSP (18). Interestingly, there appears to be a delicate balance between the expression of TCSs, e.g., comDE and ciaRH, and competence development (23, 45). Since the mariner transposons we used in this study do not have a transcriptional stop, it is not unlikely that altered expression of elements in TCS03 by mariner transposons, not their deletion, affects competence as well. Consequently, the rr03 gene could appear in our GAF screen, whereas our KO mutants remained unaffected in competence.
The capsule-specific decrease in transformation efficiency of the Δspr0838 and Δspr1259 strains cannot be explained by our current knowledge of the corresponding genes. The product of the spr0838 gene is homologous to the E. coli FtsK cell division protein (9) and the B. subtilis SpoEIII sporulation factor (65). The spr1259 gene encodes a hypothetical protein with homology to bacterial rRNA methylases, such as YebU in E. coli (3). Although FtsK and YebU are both implicated in cell growth (3, 9), the pneumococcal spr0838 and spr1259 mutants had no growth defects (our unpublished data). Therefore, we speculated that some aspects of the transformation process affect the outcome of transformation with the Δcsp PCR product. Interesting in this respect is the recent finding that some cps genes are not only required for the production of capsular polysaccharides but also considered to have a role in pneumococcal growth in vitro, which is manifested in a prolonged lag phase of pneumococcal growth (7). It has been suggested that this is a consequence of alteration in the levels of metabolic products, which affects the ability of the pneumococcus to respond and/or adapt to environmental changes. Consequently, deletion of the remnants of the R6 capsule gene cluster might result in phenotypic changes that decrease the recovery rate of each cell after it has integrated the Δcsp PCR product. Although this is expected to affect all mutants within the library, these effects might, for unknown reasons, be especially profound in the Δspr0838 and Δspr1259 strains.
The most interesting gene we identified in this study was the one coding for the RadA DNA repair protein. The radA gene is part of a CSP-induced operon that has been identified in two independent microarray studies (18, 53). In one of these studies, deletion of the last five genes of the putative operon, including radA, from S. pneumoniae CP1250 had no effect on the transformation efficiency (53). This is rather surprising, as the R6 strain in which we constructed our radA mutant has the same ancestor as this Rx derivative, namely, D39 (49). Also, the annotation of the radA gene is puzzling; for the TIGR4 strain, radA has been annotated as a pseudogene, as it has a premature stop codon that comprises its third triplet codon. The R6 radA gene has an identical nucleotide sequence, but the start codon is annotated about 200 bp downstream of the annotated TIGR4 start codon. The translation product of this putative ORF would yield an amino acid sequence homologous to those of RadA proteins in other species, which are especially conserved at the C terminus (data not shown). Despite these contradictory data, the radA mutants that we constructed in the S. pneumoniae R6, TIGR4, and also CP1250 backgrounds unambiguously showed defects in chromosomal transformation. Because we failed to grow the R6 and CP1250 spr0024-spr0027 mutants in transformation media, we could not confirm or refute the wild-type transformation phenotype of the original CP1250 spr0024-spr0027 mutant described by Peterson and coworkers (53). The presented features of the pneumococcal radA mutants are comparable to the effects of disruption of radA homologs in other species (8, 14). Moreover, the importance of radA in the pneumococcal life cycle is manifested in other studies, where mutations in this gene reveal a distinct phenotype. For instance, radA is one of the few targets found in two different signature-tagged mutagenesis studies that employed a pneumonia model of pneumococcal disease (25, 54). Although the role of radA in pneumococcal disease is most likely not related to genetic transformation, its DNA repair function might be essential for S. pneumoniae during host infection. Altogether, these data suggest that the radA gene in S. pneumoniae is an active gene that is required at least for efficient chromosomal transformation and DNA damage repair.
Although it was designed as a negative selection strategy, the GAF technology also proved to be suitable for the positive selection of genes required for pneumococcal transformation. Mutants in genes encoding the HexA and HexB mismatch repair proteins were previously shown to positively affect pneumococcal transformation rates for donor DNA that contained one or several mismatches (5, 16, 55). Notably, HexA and HexB recognize point mismatches only in the homologous part of the donor DNA, not on insertional elements like the antibiotic resistance cassette of the Δcps PCR product that we used in our GAF screen. Alignment of the nucleotide sequences of the homologous dexB and aliA parts of the Δcps PCR product (49) with the R6 genome (30) revealed several mismatches in the dexB part of the Δcps PCR product (data not shown), thus explaining the increased transformation rates of the hexA- and hexB-deficient mutants in the GAF screen.
In conclusion, our work shows the value of the application of novel technologies, such as GAF, to examine well-studied biological systems, such as pneumococcal competence and transformation. Not only did it allow verification of the reliability of the GAF technology itself, but it also led to new insights into the use and interpretation of the experimental outcome of this technique. Moreover, the application of GAF resulted in the identification of novel factors that are important for pneumococcal transformation, which will increase our understanding of the pathways underlying this process.
Acknowledgments
We thank Francesco Ianelli (University of Siena, Siena, Italy) for sharing the D39 (FP22) and TIGR4 (FP23) strains; Marc Prudhomme, Jean-Pierre Claverys (both from CNRS, Toulouse, France), and Andrew Camilli (Tufts University School of Medicine, Boston, MA) for kindly providing plasmid pR410, a source of HimarC9 transposase, and the CP1250 strain; and Donald A. Morrison (University of Illinois at Chicago) for providing CSP and advice. We are also grateful to Jean-Pierre Claverys for critically reading the manuscript. Furthermore, we thank Colin Logie (Radboud University, Nijmegen, The Netherlands) for assistance with the MMS sensitivity assay.
This work was supported by IOP Genomic grant IGE3002 of SenterNovem, supported by the Dutch Ministry of Economic Affairs.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Alloing, G., P. de Philip, and J. P. Claverys. 1994. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J. Mol. Biol. 241:44-58. [DOI] [PubMed] [Google Scholar]
- 2.Alloing, G., C. Granadel, D. A. Morrison, and J. P. Claverys. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, N. M., and S. Douthwaite. 2006. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J. Mol. Biol. 359:777-786. [DOI] [PubMed] [Google Scholar]
- 4.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 5.Balganesh, T. S., and S. A. Lacks. 1985. Heteroduplex DNA mismatch repair system of Streptococcus pneumoniae: cloning and expression of the hexA gene. J. Bacteriol. 162:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed] [Google Scholar]
- 7.Battig, P., and K. Muhlemann. 2007. Capsule genes of Streptococcus pneumoniae influence growth in vitro. FEMS Immunol. Med. Microbiol. 50:324-329. [DOI] [PubMed] [Google Scholar]
- 8.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berge, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 12.Bijlsma, J. J., P. Burghout, T. G. Kloosterman, H. J. Bootsma, A. de Jong, P. W. Hermans, and O. P. Kuipers. 2007. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl. Environ. Microbiol. 73:1514-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 14.Carrasco, B., S. Fernandez, K. Asai, N. Ogasawara, and J. C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics 266:899-906. [DOI] [PubMed] [Google Scholar]
- 15.Claverys, J. P., and S. A. Lacks. 1986. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol. Rev. 50:133-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claverys, J. P., H. Prats, H. Vasseghi, and M. Gherardi. 1984. Identification of Streptococcus pneumoniae mismatch repair genes by an additive transformation approach. Mol. Gen. Genet. 196:91-96. [DOI] [PubMed] [Google Scholar]
- 17.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451-475. [DOI] [PubMed] [Google Scholar]
- 18.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 19.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 20.Dintilhac, A., and J. P. Claverys. 1997. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res. Microbiol. 148:119-131. [DOI] [PubMed] [Google Scholar]
- 21.Dopazo, J., A. Mendoza, J. Herrero, F. Caldara, Y. Humbert, L. Friedli, M. Guerrier, E. Grand-Schenk, C. Gandin, M. de Francesco, A. Polissi, G. Buell, G. Feger, E. Garcia, M. Peitsch, and J. F. Garcia-Bustos. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99-125. [DOI] [PubMed] [Google Scholar]
- 22.Gasc, A. M., P. Giammarinaro, S. Richter, and M. Sicard. 1998. Organization around the dnaA gene of Streptococcus pneumoniae. Microbiology 144:433-439. [DOI] [PubMed] [Google Scholar]
- 23.Guiral, S., T. J. Mitchell, B. Martin, and J. P. Claverys. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. USA 102:8710-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 25.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 26.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 28.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui, F. M., and D. A. Morrison. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr, A. R., P. V. Adrian, S. Estevão, R. de Groot, G. Alloing, J. P. Claverys, T. J. Mitchell, and P. W. Hermans. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 72:3902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloosterman, T. G., J. J. Bijlsma, J. Kok, and O. P. Kuipers. 2006. To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152:351-359. [DOI] [PubMed] [Google Scholar]
- 34.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 35.Lacks, S., B. Greenberg, and M. Neuberger. 1975. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 123:222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 38.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 39.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 40.Lee, M. S., B. A. Dougherty, A. C. Madeo, and D. A. Morrison. 1999. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl. Environ. Microbiol. 65:1883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 43.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 44.Martin, B., P. Garcia, M. P. Castanie, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367-379. [DOI] [PubMed] [Google Scholar]
- 45.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison, D. A., and M. F. Baker. 1979. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature 282:215-217. [DOI] [PubMed] [Google Scholar]
- 47.Morrison, D. A., M. C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J. Bacteriol. 159:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oggioni, M. R., C. Trappetti, A. Kadioglu, M. Cassone, F. Iannelli, S. Ricci, P. W. Andrew, and G. Pozzi. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 61:1196-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce, B. J., F. Iannelli, and G. Pozzi. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153:243-247. [DOI] [PubMed] [Google Scholar]
- 50.Pearce, B. J., A. M. Naughton, and H. R. Masure. 1994. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol. Microbiol. 12:881-892. [DOI] [PubMed] [Google Scholar]
- 51.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 52.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 54.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prats, H., B. Martin, and J. P. Claverys. 1985. The hexB mismatch repair gene of Streptococcus pneumoniae: characterisation, cloning and identification of the product. Mol. Gen. Genet. 200:482-489. [DOI] [PubMed] [Google Scholar]
- 56.Prudhomme, M., L. Attaiech, G. Sanchez, B. Martin, and J. P. Claverys. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89-92. [DOI] [PubMed] [Google Scholar]
- 57.Puyet, A., B. Greenberg, and S. A. Lacks. 1990. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J. Mol. Biol. 213:727-738. [DOI] [PubMed] [Google Scholar]
- 58.Ravin, A. W. 1959. Reciprocal capsular transformations of pneumococci. J. Bacteriol. 77:296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebert, M. E., K. P. Patel, M. Plotnick, and J. N. Weiser. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187:3969-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 63.van Hijum, S. A., A. de Jong, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 65.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]