Abstract
Chlamydia species are bacterial pathogens that affect over 140 million individuals worldwide. Ocular infection by Chlamydia trachomatis is the leading cause of preventable blindness, and urogenital tract infection by Chlamydia causes sexually transmitted disease. As obligate intracellular organisms, Chlamydia species have evolved mechanisms to evade the host immune system, including the degradation of the transcription factors regulatory factor X5 and upstream stimulation factor 1, which are required for the expression of major histocompatibility complex molecules I and II by CPAF and cleavage of p65 of the NF-κB pathway by the encoded CT441 protein. Here, we report the characterization of CT441 as a tail-specific protease. CT441 contains a PDZ domain of protein-protein interactions and a Ser/Lys dyad catalytic unit. Mutation at either Ser455 or Lys481 in the active site ablated CT441 activity of p65 cleavage. In addition, we found that the production of CT441 Tsp, which was detected at the middle and late stages of an infection, correlated with p65 cleavage activity. In addition to high homology, human and mouse p65 proteins also contain an identical C-terminal tail of 22 amino acid (aa) residues. However, only human p65 was susceptible to cleavage. Using molecular biology approaches, we mapped the p65 cleavage site(s) to a region that differs from that of mouse p65 by 6 aa residues. Additionally, the substitution of T352 with a proline inhibited p65 cleavage. Together, the study demonstrates that CT441 is a tail-specific protease that is capable of interfering with the NF-κB pathway of host antimicrobial and inflammatory responses.
The carboxyl-terminal processing proteases (Ctp), including the bacterial tail-specific protease (Tsp), are a group of endoproteases of posttranslational protein modification, maturation, and disassembly or degradation. The Ctp proteases have been found in Archaea, plant chloroplasts, bacteria (reviewed in reference 20), and viruses (5). A well-characterized Ctp is the P1D protease that contains a PDZ domain of protein-protein interaction and a domain of the S41B family peptidase (15, 20). P1D catalyzes the C-terminal processing of the D1 protein of photosystem II, an essential event for the consequent water oxidation and the generation of oxygen molecules in oxygenic photosynthetic organisms. Tail-specific proteases have been identified from bacterial pathogens of medical importance, including Borrelia, Chlamydia, Escherichia, Helicobacter, Pseudomonas, Salmonella, Shigella, Vibrio, and Yersinia. Limited examples indicate that these proteases play a critical role in bacterial protein modifications. For example, the inactivation of Tsp of Borrelia burgdorferi, the etiologic agent of Lyme borreliosis, affected borrelial protein synthesis and outer membrane protein P13 processing (19). Escherichia coli Tsp has been implicated in the processing of penicillin binding protein 3 (8). In general, the physiological functions of most bacterial Tsps are less well understood.
Chlamydiae are parasitic bacterial pathogens that infect the epithelium of the eye, causing the most preventable blindness in the world, and the urogenital tract, which is the most common cause of sexually transmitted disease in the United States (3). The annotated Chlamydia genome has two Tsps, CT441 and CT858 (25). The biological functions of CT441 and CT858 Tsps in Chlamydia replication are unknown; however, both proteins have been found to target host proteins to interfere with host cellular processes. CT858 degrades regulatory factor X5 (RFX5) and upstream stimulation factor 1 (USF-1), transcription factors required for the expression of the major histocompatibility complex molecules of antigen presentation (27). We found that CT441 Tsp was responsible for chlamydial protease activity that cleaves the p65 protein, an important regulator of the NF-κB pathway of inflammatory response. Degradation of RFX5 or cleavage of the p65 protein can suppress the host immune response against microbial infection.
The PDZ domain-containing Tsps are composed of a Ser/Lys dyad catalytic unit, which was first described by Slilaty and Little (23; reviewed in reference 20) and further defined from crystal structure studies of the P1D protein (15). Sequence analysis indicates that chlamydial CT441 Tsp and homologous proteins of Chlamydia serovars and biovars contain a conserved PDZ domain and a Ser/Lys catalytic unit. CT441 protease shows an overall identity of approximately 28% with P1D and bacterial Tsps, and the sequence identity around the active site of Ser and Lys is close to 70% among CT441 and many defined Tsp proteases of this group. To understand the functional roles of chlamydial Tsp in pathogen-host interactions, we performed mutagenesis analysis of cloned CT441 and identified the amino acid residues essential for p65 cleavage activity. Here, we report the characterization of CT441 as a PDZ domain-containing Tsp associated with p65 cleavage.
MATERIALS AND METHODS
Cell lines and bacterial strains.
HeLa 229 cells were obtained from the ATCC (Manassas, VA). HEK293T cells, which allow for episomal replication of transfected plasmids containing the simian virus 40 origin of replication, were cultured in Dulbecco's modified Eagle's medium (high glucose; Invitrogen, Carlsbad, CA) supplemented with nonessential amino acids, sodium pyruvate, l-glutamine, 20 μg/ml gentamicin, and 10% heat-inactivated defined fetal bovine serum (HyClone, Logan, UT). Chlamydia trachomatis serovar D and lymphogranuloma venereum (LGV2) were obtained from H. Caldwell of Rocky Mountain Laboratories of the NIH and G. Zhong of the University of Texas Health Science Center at San Antonio, respectively, and C. trachomatis of murine pneumonitis Nigg (MoPn) was obtained from L. M. de la Maza of the University of California Irvine. The bacteria were propagated in HeLa 229 cells, and the titers were determined as previously described (2, 18).
Antibodies.
Both rabbit and mouse anti-human p65, anti-Myc, and anti-Erk2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz). Monoclonal antibodies to chlamydial major outer membrane protein (MOMP) and to cysteine-rich protein 60 (CRP60) were kindly provided by L. M. de la Maza. Antibody to the hemagglutinin (HA) tag was obtained from Sigma (St. Louis, MO). Rabbit anti-CT441 polyclonal antibody was raised against a recombinant CT441 (amino acids [aa] 352 to 490) using a protocol approved by the IACUC. This antibody recognizes a single protein band of 70 kDa from Chlamydia-infected cell lysates and the purified elementary body of C. trachomatis.
Sequence analysis and molecular modeling.
A position-specific iterated and pattern-hit initiated BLAST (PSI-BLAST and PHI-BLAST) search was performed using the precursor sequence of CT441 (GenBank accession number NP_219953) as a query against the nonredundant peptide database. The region of aa 194 to 527 of CT441 was aligned with P1D (Protein Data Bank accession number 1FC7), and a comparative modeling was obtained using the MODELLER modeling package (21).
Infection assays.
Monolayers of HEK293T cells, or cells as specified, were infected with LGV2 at 1 inclusion-forming unit (IFU)/cell for various times or as indicated. In parallel experiments, an antibiotic agent was included throughout the infection process. The infected cells or control samples were lysed with a buffer containing 143 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), and a cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN). Soluble proteins were separated by SDS-polyacrylamide gel electrophoresis. The proteins were transferred onto a Immobilon-P polyvinylidene difluoride membrane (Millipore, Minneapolis, MN). Protein expression and p65 cleavage were detected with specific antibodies, followed by horseradish-conjugated secondary antibody and an ECL reagent kit (Pierce, Rockford, IL).
Molecular cloning and transfection.
Constructs for the expression of HA-tagged CT441, CT823, and CT858 as mature peptides were described previously (14). A standard PCR protocol was used to generate CT441 S455A and K481T. Briefly, the codon for S455 (TCG) was changed to GCA (S445A mutation), which effectively introduced a Pst1 site at this area. Similarly, the codon for K481 (AAA) was changed to ACC (K481T mutation). By introducing a silent mutation corresponding to G480 (GGA to GGT), a Kpn1 restriction site was created. The ligation of two corresponding PCR fragments into the pRK5HA vector (BamHI/HindIII) led to pRK5HA/CT441(S455A) and pRK5HA/CT441(K481T), respectively. Similarly, Myc-tagged p65 mutants were prepared using the pRK5Myc vector (BamHI/EcoRI site) for mammalian expression. All intended mutations were validated with automated DNA sequence analysis.
For protein expression and in vivo activity assays, 293T cells were transiently transfected with an expression plasmid or an empty vector using Fugene-6 (Roche). The cells were lysed 24 h posttransfection, and protein expression and cleavage of the endogenous p65 protein were detected by Western blotting analyses. Alternatively, the cell lysates were used as substrates in in vitro protein cleavage assays as described previously (14).
RESULTS
Chlamydial CT441 as a PDZ domain-containing tail-specific protease.
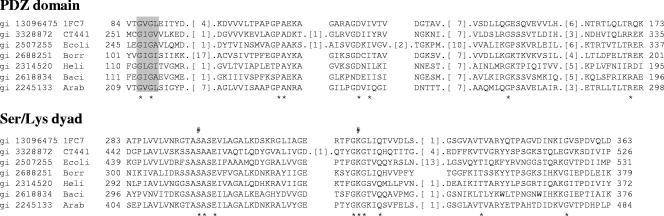
A search of the conserved-domain database of protein sequences using CT441 (GenBank accession number NP_219953) as the query revealed the protein as being a PDZ-Ctp/Tsp protease that contains a PDZ domain and a catalytic unit composed of a serine residue and a lysine residue (Fig. 1). CT441 shares an overall identity of approximately 28% with archaeal P1D (Protein Data Bank accession number 1FC7), and residues around the Ser/Lys active site have an overall identity of 50%. The PDZ domain was aligned to aa 251 through 335, and the catalytic unit was between aa 324 and 526, consistent with the previously proposed domain requirement of PDZ-Tsp proteins (1).
FIG. 1.
Multiple alignment of the conserved PDZ and the Ser/Lys dyad catalytic unit of carboxyl-terminal proteases and tail-specific proteases. The alignment was constructed using the conserved-domain BLAST and adjusted manually to include representative proteins of different species. Fragments of the D1 C-terminal processing protease (P1D) (Protein Data Bank accession number 1FC7) sequence of Scenedesmus obliquus around the PDZ and the catalytic unit are shown for comparison. The GenBank gene identifiers are shown on the left of each sequence. Species name abbreviations are as follows: 1FC7, S. obliquus P1D Ctp; CT441, C. trachomatis D CT441; Ecoli, E. coli; Borr, B. burgdorferi; Heli, Helicobacter pylori; Baci, Bacillus subtilis; Arab, Arabidopsis thaliana. The GLGF motifs of the PDZ domain are shaded, and the conserved Ser and Lys residues of Ser/Lys dyad are marked with #. * indicates identical residues.
Characterization of active site of CT441.
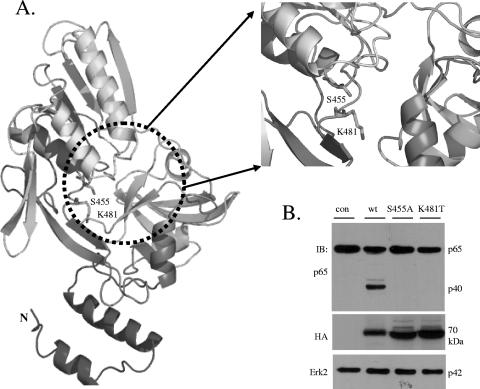
The structure of P1D (PDB accession number 1FC7) was therefore used as a structural template to generate a model for residues 194 to 527 of CT441 Tsp by employing the MODELLER program (21) (Fig. 2A). The PDZ domain of CT441 is formed by five-stranded antiparallel β-sheets with a single-turn helix inserted between the second and third β-sheets. The residues that form the highly conserved GLGF (Gly-Leu-Gly-Phe) loop of carboxylate binding (4, 9, 12) were conserved (residues 253 to 256, GIGV, in CT441). The three domains form a shallow cleft that resembles those observed in P1D and the hepatitis C virus NS3 protease. The Ser455 and Lys481 residues conserved in PDZ-Tsp situated in the middle of the cleft and the side chains of Ser455 and Lys481 are in a position for hydrogen bond formation. Together, these results indicate that CT441 is a PDZ domain-containing Tsp that may utilize a Ser/Lys dyad for catalytic activity.
FIG. 2.
Comparative modeling of CT441 Tsp against Protein Data Bank accession number 1FC7/P1D Ctp and verification of the active site of CT441 with mutagenesis. (A) Residues 194 to 527 of the CT441 precursor protein were aligned with the 336 residues of the C-terminal processing protease of photosystem II (P1D) (Protein Data Bank accession number 1FC7) using the MODELLER comparative modeling package (21). A model representing the PDZ domain and the catalytic unit of CT441 was obtained by satisfaction of spatial restraints. The Ser455 and Lys481 residues of the active site are shown as sticks. (B) Mutation of serine 455 or lysine 481 ablated p65 cleavage activity of CT441. wt CT441 or point mutations of S455A or K481T were expressed in 293T cells as N-terminal HA-tagged proteins by transient transfection. Protein expression and p65 cleavage were determined by immunoblotting (IB) analysis. The expression of Erk2 was used as a loading control.
To define the catalytic unit, we generated CT441 mutants (Ser455Ala and Lys481Thr) using standard protocols of molecular biology and investigated whether mutations at the catalytic unit affected p65 cleavage activity of CT441. To this end, a cDNA encoding the mature peptide of CT441 or the corresponding mutants was subcloned into a mammalian expression vector for N-terminal HA-tagged protein expression. Monolayers of 293T cells were then transfected for the expression of wild-type (wt) CT441 or the S455A or K481T mutant. CT441 expression and protein cleavage of endogenous p65 were detected by immunoblotting analysis using a specific antibody against the HA tag or the N terminus of p65, respectively. The expression of wt CT441 Tsp resulted in the detection of a p65 cleavage product of approximately 40 kDa. Mutation at either the Ser455 or Lys481 position ablated the p65 cleavage activity of CT441 (Fig. 2B), indicating that CT441 is a Ser/Lys dyad-containing protease responsible for p65 cleavage.
It is worth noting that the Chlamydia genome encodes several putative serine proteases that contain a leader peptide, including CT441, CT823, and CT858. CT823 is predicted to have a trypsin-like protease domain followed by a PDZ domain at the C-terminal region. We also noticed that CT858, initially annotated as being a putative Tsp, does not contain a PDZ domain and a Lys residue in the conserved Ser/Lys catalytic unit of PDZ-Ctp proteases. Therefore, both CT823 and CT858 proteins were expected to exhibit a different substrate specificity from that of CT441 even though the substrate for CT823 is unknown. Indeed, neither protein displayed p65 cleavage activity (14). Instead, CT858 was reported to degrade proteins, including the RFX5 and USF-1 transcription factors.
Detection of CT441 Tsp during Chlamydia infection.
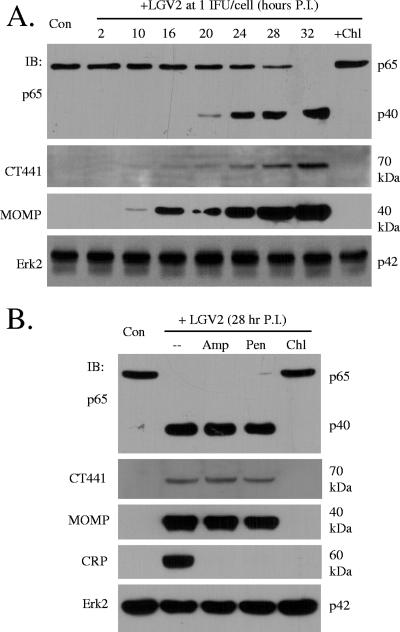
CT441 is predicted to be a σ28-regulated gene (26), suggesting that its expression is developmental cycle dependent. To this end, HeLa 229 cells were infected with Chlamydia LGV2 at a multiplicity of infection of 1 IFU/cell for various times, and CT441 expression was then detected by immunoblotting analysis to establish a time course of CT441 production. A protein of 70 kDa was detected approximately 10 to 16 h postinfection, and the expression persisted thereafter (Fig. 3A). Consistent with the notion of CT441 as a p65 cleavage protease, significant p65 cleavage was not detected until approximately 20 h postinfection (Fig. 3A), trailing CT441 expression.
FIG. 3.
CT441 expression and p65 cleavage during Chlamydia infection. Monolayers of HeLa 229 cells were infected with Chlamydia LGV2 at 1 IFU/cell for various times (A) or with 1 IFU for 28 h (B). In parallel experiments, an antibiotic agent was included at the beginning of an infection. The cells were lysed, and protein expression and p65 cleavage were detected by immunoblotting (IB) analyses using specific antibodies. Chl, chloramphenicol (60 μg/ml); Pen, penicillin (100 μg/ml); Amp, ampicillin (100 μg/ml); P.I., postinfection. Erk2 expression was used as a loading control.
Additionally, the p65 cleavage activity was dependent on bacterial replication and hence CT441 expression, since treatment with chloramphenicol, an agent that inhibits Chlamydia growth, prevented p65 cleavage (Fig. 3B). Treatment with penicillin or ampicillin had no effect on CT441 expression or p65 cleavage activity (Fig. 3B) since β-lactams such as penicillin were previously reported to inhibit Chlamydia maturation but not its growth (16, 17), which was verified by their inhibition of the expression of CRP60, a late gene product, but not MOMP, a middle and late gene product (Fig. 3B).
It is interesting that Tsps, β-lactam acylases, and bacterial type I leader peptidases use the Ser/Lys dyad for catalytic activity. An E. coli leader peptidase was reported to be sensitive to certain β-lactams (13, 20), although there seem to be no structural similarities among these proteases (15). We found no evidence to implicate CT441 Tsp in processing leader peptide-containing proteins since no precursor MOMP from β-lactam-treated samples was detected (Fig. 3B).
Identification of substrate cleavage site.
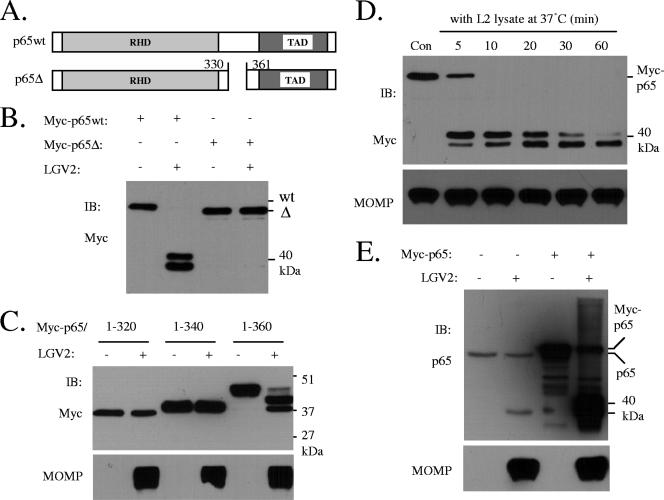
PDZ domains of Tsps recognize nonpolar amino acid residues with a carboxylate at the C terminus (24). The cleavage of p65 by CT441 Tsp was unlikely to be modulated by the C terminus of the p65 protein since the human p65 protein and the cleavage-resistant mouse p65 protein are identical at their C termini (last 22 aa residues) (not shown). A growing body of evidence shows PDZ domains can also recognize internal peptides that can form an unusual structure, a stable β-hairpin, also known as a β-finger (10). The human p65 protein seems not to have the consensus sequences for β-finger formation. We therefore attempted to characterize the cleavage site using Edman degradation. Since no conclusive results were obtained using the C-terminus-derived cleavage product (p22) of the p65 protein from those studies, we instead used deletion mutants to investigate p65 cleavage. An N-terminal Myc-tagged p65 (Myc-p65) or its deletion mutants were expressed in 293T cells, and total cell lysates were then incubated with a bacterial lysate in an in vitro cleavage assay (14). p65 cleavage was determined by immunoblotting analysis with an anti-Myc antibody. Incubation of Myc-p65 with LGV2 lysate resulted in the detection of two N-terminus-derived cleavage products, whereas the deletion of aa 331 to 360 (Myc-p65Δ) prevented p65 cleavage (Fig. 4B), suggesting that the cleavage site(s) resides in this region. This conclusion was also supported by the following evidence. We found that only a Myc protein containing the first 360 aa residues (aa 1 to 360), but not aa 1 to 320 or 1 to 340 of p65, was cleaved (Fig. 4C), further limiting the cleavage site(s) to a range of approximately 20 to 25 aa residues.
FIG. 4.
p65 deletion mutants that are resistant to cleavage. (A) Schematic drawing of p65 protein and a deletion mutant (deletion of aa 331 to 360 of p65). RHD, Rel-homologous domain; TAD, transactivation domain. (B) The p65 cleavage site(s) resides between amino acid residues 331 and 359. wt or mutant p65 proteins were expressed in 293T cells as N-terminal Myc-tagged proteins. The cell lysates were prepared from those samples and used as substrates for in vitro protease assays by incubation with a preparation of soluble chlamydial proteins (1 μg per reaction) (14). A deletion mutant (Δ331-360, Myc-p65Δ) was resistant to Chlamydia cleavage. (C) Chlamydia cleaves a p65 mutant containing the first 360 aa residues (residues 1 to 360), but not aa 1 to 320 or 1 to 340. (D) The initial cleavage product of Myc-p65 was subject to reprocessing. Con, control. (E) Comparison of relative motilities of the p40 cleavage product of endogenous p65 and the two 40-kDa products derived from Myc-p65 cleavage. Monolayers of 293T cells were transfected with an empty vector or pRK5Myc/p65. The cells remained uninfected or infected with LGV2 at 1 IFU/cell for 28 h. Protein cleavage of endogenous p65 or transiently expressed Myc-p65 in these samples was assayed by immunoblotting (IB) analysis using an N-terminus-specific p65 antibody.
Cleavage of the endogenous p65 protein led to the detection of one protein band of 40 kDa (p40) using an N-terminus-specific antibody (refer to Fig. 3A and B). However, two such protein bands were detected from the cleavage of overexpressed Myc-p65 (Fig. 4B and D). The slower-migrating band seemed to be subjected to reprocessing since prolonged incubation decreased its detection, which coincided with increased detection of the faster-migrating protein band (Fig. 4D, top). Indeed, the p40 cleavage product displayed a motility that fell within the range of these two proteins (Fig. 4E), suggesting that the slower-migrating band is the primary product of Myc-p65 cleavage. The reasons why and how two N-terminus-derived products were detected from the cleavage of Myc-p65 remain uninvestigated.
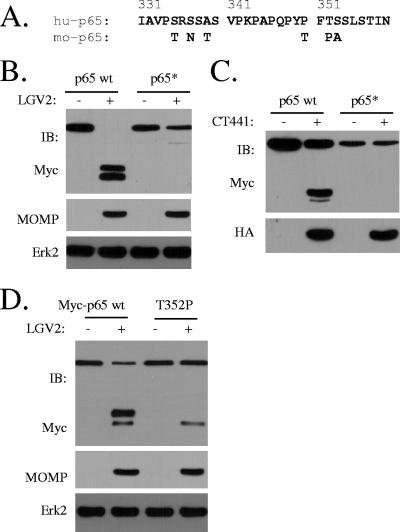
We noticed that within the cleavage region of p65, human p65 and the cleavage-resistant murine p65 differ by 6 aa residues (Fig. 5A). We therefore prepared a mutant that has all corresponding amino acid residues of human p65 replaced with the counterparts of mouse p65 and found that this mutant (Myc-p65*) was completely resistant to Chlamydia cleavage (Fig. 5B). In addition, we found that Myc-p65* was also resistant to CT441 cleavage (Fig. 5C), indicating that CT441 was responsible for chlamydial p65 cleavage activity. A peptidic bond between F351 and T352 resembles a scissile bond of serine proteases. Therefore, the T352 residue was replaced with a proline residue, since the substitution of a P′ residue with a proline tends to alter the substrate specificity of serine proteases (7). The T352P substitution prevented the production of the slower-migrating protein band, although the production of the faster-migrating protein band was not affected by T352P (Fig. 5D), suggesting that F351-T352 is a putative cleavage site. Mutants containing a single point mutation at the other sites did not affect p65 cleavage (data not shown). Since only one protein band was detected on SDS-polyacrylamide gels from the cleavage of endogenous p65, which migrated between the two cleavage products of a Myc-tagged p65 (Fig. 5E), we deduced that residue 351/352 is the site of p65 cleavage.
FIG. 5.
Identification of p65 cleavage site by CT441 Tsp. (A) Sequence alignment of the cleavage region of the human (hu-p65) and the cleavage-resistant mouse (mo-p65) p65 proteins. Within the region of cleavage, human and mouse p65 differ by 6 aa residues. (B) A mutant (Myc-p65*) that was prepared by the replacement of the corresponding amino acid residues within the cleavage region of the human p65 protein with the mouse p65 protein was resistant to Chlamydia and CT441 cleavage in 293T cells. Monolayers of 293T cells were transfected for Myc-p65 or Myc-p65* expression. The cells were then infected with LGV2 for 28 h. Protein expression and cleavage of Myc-p65 wt and Myc-p65* were detected with an anti-Myc antibody. MOMP expression was included as a measure of Chlamydia infection. (C) In a parallel experiment, the cells were cotransfected with HA-CT441 and Myc-p65 or Myc-p65*. CT441 expression and p65 protein cleavage were detected with anti-HA and anti-Myc antibodies, respectively. (D) A T352P mutation prevents the primary band production by Chlamydia cleavage. A peptidic bond between a phenylalanine and a nonproline residue resembles a scissile bond of serine proteases. Therefore, a T352P mutant was prepared and tested for Chlamydia cleavage. Note the absence of the slower-migrating protein band in the T352P lane. IB, immunoblot.
DISCUSSION
Using biochemical and molecular biological approaches, we showed previously that a chlamydial protein, CT441, has p65/RelA cleavage activity (14). p65 is a subunit of the NF-κB complex that regulates gene expression of the immediate-early response to pathogen infections (11, 22). We now provide evidence to show that CT441 protease activity is essential for p65 cleavage. CT441 belongs to a family of PDZ domain-containing tail-specific proteases that use a Ser/Lys catalytic unit for protease activity. Mutation of the nucleophilic serine 455 or lysine 481, a general base of Ser/Lys dyad serine proteases, rendered CT441 inactive. Consistent with its role in p65 cleavage in Chlamydia infection, we found that the expression of CT441 preceded p65 cleavage activity in infected cells. In addition, the substitution of T352 with a proline residue blocked p65 cleavage, indicating that F351-T352 is a primary site of p65 subjected to protease cleavage, since a proline residue that follows a phenylalanine at the P1 position is generally resistant to serine protease activity (6, 7).
The p65 protein and the NF-κB pathway of inflammatory response are well conserved among different species. The mouse and human p65 proteins share an overall identity of 88%. However, we found that mouse p65 was resistant to both Chlamydia and CT441 cleavage. The PDZ domain of Tsp engages the C terminus of a substrate with β-sheet interactions and hydrogen bonding through the highly conserved GLGF motif loop (4). Therefore, the species-specific cleavage of human p65 is likely sequence dependent since the last 22 aa at the C termini of mouse and human p65 proteins are identical. Indeed, a mutant of human p65 that has corresponding amino acid residues replaced with the counterparts of mouse p65 was completely resistant to cleavage by Chlamydia infection or CT441 overexpression (Fig. 4B).
Chlamydiae are obligate intracellular bacterial pathogens with a parasitic lifestyle. The organisms have evolved diverse mechanisms to secure a favorable habitat for progeny production. Concomitantly, the host mounts an antimicrobial response by the activation of innate and acquired arms of immune response. Zhong and colleagues observed that a chlamydial Tsp (CPAF or CT858) has the ability to degrade RFX5 and USF-1 proteins (27), transcription factors required for major histocompatibility complex molecule expression. We found that chlamydial CT441 cleaves p65 of the NF-κB pathway (14) to convert a regulator of the inflammatory response to a dominant negative inhibitor of the same pathway and, hence, to effectively suppress immune response. These studies demonstrated that Chlamydia has the ability to utilize the encoded proteases to regulate the host immune response. Therefore, the characterization and identification of chlamydial virulence factors shed light on our understanding of mechanisms that Chlamydia applies for immune evasion. These studies may also uncover novel targets for antivirulence agents.
Acknowledgments
We thank Patty Rutledge for administrative assistance.
This work was partially supported by a K22 grant from the NIAID, NIH.
This is TSRI manuscript 18766-IMM.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Beebe, K. D., J. Shin, J. Peng, C. Chaudhury, J. Khera, and D. Pei. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149-3155. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell, H. D., and L. J. Perry. 1982. Neutralization of Chlamydia trachomatis infectivity with antibodies to the major outer membrane protein. Infect. Immun. 38:745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2003. Summary of notifiable diseases—United States, 2003. Morb. Mortal. Wkly. Rep. 52:16-18. [Google Scholar]
- 4.Doyle, D. A., A. Lee, J. Lewis, E. Kim, M. Sheng, and R. MacKinnon. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85:1067-1076. [DOI] [PubMed] [Google Scholar]
- 5.Feldman, A. R., J. Lee, B. Delmas, and M. Paetzel. 2006. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J. Mol. Biol. 358:1378-1389. [DOI] [PubMed] [Google Scholar]
- 6.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 8.Hara, H., Y. Yamamoto, A. Higashitani, H. Suzuki, and Y. Nishimura. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J. Bacteriol. 173:4799-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison, S. C. 1996. Peptide-surface association: the case of PDZ and PTB domains. Cell 86:341-343. [DOI] [PubMed] [Google Scholar]
- 10.Hillier, B. J., K. S. Christopherson, K. E. Prehoda, D. S. Bredt, and W. A. Lim. 1999. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284:812-815. [PubMed] [Google Scholar]
- 11.Karin, M., and F. R. Greten. 2005. NF-κB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5:749-759. [DOI] [PubMed] [Google Scholar]
- 12.Kim, J. S., M. Groll, H. J. Musiol, R. Behrendt, M. Kaiser, L. Moroder, R. Huber, and H. Brandstetter. 2002. Navigation inside a protease: substrate selection and product exit in the tricorn protease from Thermoplasma acidophilum. J. Mol. Biol. 324:1041-1050. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, D., J. Weidner, P. Griffin, S. K. Shah, and W. B. Knight. 1994. Determination of the kinetic parameters of Escherichia coli leader peptidase activity using a continuous assay: the pH dependence and time-dependent inhibition by beta-lactams are consistent with a novel serine protease mechanism. Biochemistry 33:8347-8354. [DOI] [PubMed] [Google Scholar]
- 14.Lad, S. P., J. Li, J. da Silva Correia, Q. Pan, S. Gadwal, R. J. Ulevitch, and E. Li. 2007. Cleavage of p65/RelA of the NF-κB pathway by Chlamydia. Proc. Natl. Acad. Sci. USA 104:2933-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao, D. I., J. Qian, D. A. Chisholm, D. B. Jordan, and B. A. Diner. 2000. Crystal structures of the photosystem II D1 C-terminal processing protease. Nat. Struct. Biol. 7:749-753. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto, A., and G. P. Manire. 1970. Electron microscopic observations on the fine structure of cell walls of Chlamydia psittaci. J. Bacteriol. 104:1332-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molder, J. W. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis. 2:87-99. [PubMed] [Google Scholar]
- 18.Mukhopadhyay, S., A. P. Clark, E. D. Sullivan, R. D. Miller, and J. T. Summersgill. 2004. Detailed protocol for purification of Chlamydia pneumoniae elementary bodies. J. Clin. Microbiol. 42:3288-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostberg, Y., J. A. Carroll, M. Pinne, J. G. Krum, P. Rosa, and S. Bergstrom. 2004. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J. Bacteriol. 186:2074-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paetzel, M., and R. E. Dalbey. 1997. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem. Sci. 22:28-31. [DOI] [PubMed] [Google Scholar]
- 21.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 22.Santoro, M. G., A. Rossi, and C. Amici. 2003. NF-κB and virus infection: who controls whom. EMBO J. 22:2552-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slilaty, S. N., and J. W. Little. 1987. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl. Acad. Sci. USA 84:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73-77. [DOI] [PubMed] [Google Scholar]
- 25.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 26.Yu, H. H., E. G. Di Russo, M. A. Rounds, and M. Tan. 2006. Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli σ28 RNA polymerase. J. Bacteriol. 188:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]