Abstract
Genetic screens based on the use of MudJ-generated lac fusions permitted the identification of novel genes regulated by the Rcs signal transduction system in Salmonella enterica serovar Typhimurium. Besides genes that are also found in the Escherichia coli genome, our screens identified Salmonella-specific genes regulated by RcsB, including bapA, siiE, srfA, and srfB. Here we show that the srfABC operon is negatively regulated by RcsB and by PhoP. In vivo studies using mutants with constitutive activation of the Rcs and/or PhoPQ system suggested that there is an overlap between these regulatory systems in the control of Salmonella virulence.
Two-component systems are signal transduction devices found in all domains of life, and they are especially widespread in bacteria (91). These systems regulate diverse responses, including nutrient acquisition, energy metabolism, adaptation to environmental cues, complex developmental pathways, and host-pathogen interactions. Two-component systems are typically composed of a transmembrane sensor protein and a cytoplasmic transcriptional regulator. The transmembrane component harbors at least two domains: an input domain that senses the environmental stimulus and a cytoplasmic transmitter with histidine kinase activity that transforms the external stimulus into a cellular signal by autophosphorylation at a conserved histidine residue. The phosphorylated histidine is the source for phosphorylation of a conserved aspartic acid residue in the receiver domain of the transcriptional regulator. The phosphorylated transcription factor then mediates the cellular response, usually by differential expression of target genes. In a number of two-component systems, two histidine and two aspartic acid residues are present in four signaling domains that can be combined in several ways (91). On the basis of experimental evidence and protein sequence similarities, Escherichia coli and Salmonella enterica are thought to encode over 30 different two-component systems.
S. enterica can cause diseases ranging from gastroenteritis to typhoid fever in humans and other animals (73). Many virulence traits of S. enterica can be attributed to the presence of Salmonella pathogenicity islands (SPIs). The larger SPIs, SPI-1 and SPI-2, encode type III secretion systems (T3SS) that in S. enterica confer the ability to invade nonphagocytic cells and to survive and proliferate within the phagosome (45, 52, 90, 94). The PhoPQ two-component system has long been known as a master regulator of virulence in S. enterica. Both its inactivation (by null mutations in phoP or phoQ) and its hyperactivation (by a gain-of-function mutation in phoQ) result in strong virulence attenuation in mice (30, 34, 60, 61). Mg2+ and Ca2+ have been identified as the physiological signals detected by the sensor kinase PhoQ. The prevalent model suggests that Salmonella identifies its environment within the host by monitoring Mg2+ levels via the PhoQ protein (40). A low Mg2+ concentration is an indication of an intracellular environment and leads to activation of the PhoPQ system. In turn, a high Mg2+ concentration serves as a hallmark of an extracellular environment and leads to inactivation of the system. Accordingly, PhoP-activated genes (pag genes) are turned on inside host cells, whereas PhoP-repressed genes (prg genes) are expressed outside host cells. Only a fraction of PhoP-regulated genes are involved in Salmonella virulence. These genes appear to have been acquired by horizontal gene transfer (40). Other two-component systems that contribute to the regulation of Salmonella virulence are PmrA-PmrB, RcsC-RcsD-RcsB, OmpR-EnvZ, SsrA-SsrB, and SirA-BarA (for a review, see reference 3). Certain interactions between these regulatory systems have been shown. For instance, a subset of PhoP-activated genes are regulated via PmrA-PmrB (41, 78), while PhoP directly regulates SsrB-SsrA, a regulatory system whose activation is necessary for expression of the SPI-2-encoded T3SS (7).
The Rcs system was initially characterized in E. coli as a regulator of colanic acid capsule synthesis (9, 39, 82). The sensor protein RcsC, a hybrid histidine kinase, the intermediate phosphotransmitter RcsD (previously called YojN) (16, 84), and the transcriptional activator RcsB are the main components of the system, which also includes a second transcriptional activator, RcsA (83). RcsC has positive and negative regulatory effects on Rcs-regulated genes, and genetic data support the hypothesis that this protein has both kinase and phosphatase activities (18, 35, 56). RcsF is another component of the system and was originally proposed to be involved in RcsB phosphorylation (36). Recently, Majdalani et al. have shown that signaling proceeds through an ordered cascade, RcsF → RcsC → RcsD → RcsB (55). RcsF, rather than playing a role in RcsB phosphorylation, is critically involved in signal transduction from the cell surface to RcsC (55). The signals that activate the Rcs phosphorelay are largely unknown. Sledjeski and Gottesman (77) showed that osmotic upshift is an environmental signal that strongly but transiently induces colanic acid synthesis in E. coli in an rcsC- and rcsB-dependent manner. More recently, Hagiwara et al. (44) found that a combination of low temperature (20°C) and 0.4% glucose (or low temperature and zinc) is an effective stimulus for the Rcs activation. These authors also showed that, as far as signal transduction in response to glucose and zinc is concerned, the rcsF gene is also an essential component of the Rcs signaling system (44). A comprehensive review of the Rcs system has recently been published (54). In addition, recent evidence suggests that RcsB can receive a signal directly by accepting the phosphoryl group from acetyl phosphate (32).
Besides the transient or moderate effects exerted by environmental signals, mutations in some genes or overexpression of other genes can lead to permanent activation of the Rcs signal transduction pathway. Some examples in E. coli are mutations in mdoH (26) or pgsA (76) and overexpression of djlA (50). Overproduction of RcsB also induces capsule synthesis that results in mucoidy (9). This is in agreement with the view that overproduction of the response regulator mimics the physiological phosphorylation response (1, 21, 48). In S. enterica serovar Typhimurium, mutations in the essential gene igaA result in mucoidy and reduced motility (10) and cause overgrowth in certain eukaryotic cell types (11).
The Rcs system has been shown to participate in additional physiological processes, including synthesis of flagella (10, 31), cell division control (12), regulation of invasion proteins, flagellin, and Vi antigen in S. enterica serovar Typhi (2, 89), synthesis of the E. coli outer membrane protein OsmC (23), expression of the E. coli tolQRA operon (19), and resistance to chlorpromazine-induced stress (20). Synthesis of certain exopolysaccharides in Erwinia amylovora and Klebsiella pneumoniae (5, 65) is also regulated through Rcs signaling. Proteus mirabilis (4, 43) and E. coli also use this system to regulate swarming (16, 46).
A role for the Rcs system in the control of Salmonella virulence has recently been described. Modest attenuation of Salmonella virulence at late stages of infection in mice was shown for RcsC− mutants (24). A more pronounced effect on acute infection has been reported for mutations in igaA (25) or rcsC (35, 64) that hyperactivate the Rcs system. This effect is partially suppressed by mutations that prevent colanic acid capsule synthesis (wca mutations) (35, 64), suggesting that overproduction of capsule is one of the causes of attenuation in these mutants. The fact that suppression by wca mutations is only partial suggests that additional Rcs-regulated genes may be involved in Salmonella virulence.
Several global searches of genes regulated by the Rcs system have been carried out recently in E. coli (29, 44, 67). However, the limited overlap in the genes identified by these studies suggests that other members of the E. coli Rcs regulon remain to be identified. To our knowledge, systematic searches for members of the Rcs regulon have not been carried out in S. enterica. The evolutionary relatedness between E. coli and S. enterica anticipates a high degree of overlap between the two Rcs regulons. However, differences between the E. coli and Salmonella genomes and the involvement of Rcs in Salmonella virulence also suggest that specific genes might exist in S. enterica. In this work, we describe several new members of the Rcs regulon in S. enterica serovar Typhimurium and confirm the existence of Salmonella-specific genes regulated by RcsB. Among these genes are srfA and srfB, which are part of the putative operon srfABC. We show that RcsB and PhoP negatively regulate this operon. Interestingly, we provide evidence for functional overlap between these regulatory systems in the control of Salmonella virulence.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and strain construction.
S. enterica serovar Typhimurium strains used in this study are described in Table 1. Unless otherwise indicated, the strains were derived from the mouse-virulent strain ATCC 14028. Transductional crosses using phage P22 HT 105/1 int201 (74) were used for strain construction (57). To obtain phage-free isolates, transductants were purified by streaking on green plates (15). Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5.
TABLE 1.
Strains of S. enterica serovar Typhimurium used in this study
| Straina | Description | Source or reference |
|---|---|---|
| ATCC 14028 | Wild type | ATCC |
| 55130 | pho-24 (PhoP constitutive) | E. A. Groisman |
| SV4439 | rcsC52::MudQ | 10 |
| SV4514 | gmm::MudQ | This study |
| SV4530 | igaA1 | 25 |
| SV4535 | igaA3::CmryojN::KIXX | Laboratory stock |
| SV4573 | igaA3::Cmr/pNG1166 | This study |
| SV4608 | trg::MudJ | This study |
| SV4676 | srfB::MudJ | This study |
| SV4757 | rcsC54 (Rcs constitutive) | 35 |
| SV4773 | igaA5 | 25 |
| SV4918 | yjbH::MudJ | This study |
| SV4919 | siiE::MudJ | This study |
| SV4920 | melB::MudJ | This study |
| SV4921 | narH::MudJ | This study |
| SV4922 | dcuB::MudJ | This study |
| SV4923 | STM2176::MudJ | This study |
| SV4924 | yhhJ::MudJ | This study |
| SV5049 | ΔrcsB::Cmr | This study |
| SV5090 | gmm::MudQ/pIZ1589 | This study |
| SV5091 | srfA::MudJ | This study |
| SV5092 | yiaD::MudJ | This study |
| SV5093 | ΔrcsA::Cmr | This study |
| SV5094 | bapA::MudJ | This study |
| SV5095 | PSLT071::MudJ | This study |
| SV5106 | STM1491::MudJ | This study |
| SV5190 | srfA::3×FLAG | This study |
| SV5191 | srfB::3×FLAG | This study |
| SV5192 | srfC::3×FLAG | This study |
| SV5303 | prgH::lacZ | J. López-Garrido |
| SV5373 | ΔhilA | J. López-Garrido |
| SV5452 | ΔssrB::Cmr | This study |
| SV5470 | ssaV::lacZ | This study |
Derivatives of some of the strains were used as indicated in the text.
Construction of S. enterica mutants by gene targeting.
Disruption and replacement of rcsA, rcsB, ssrB, or ssaV with a chloramphenicol resistance gene were performed as described previously (22). Briefly, the chloramphenicol resistance gene from plasmid pKD3 was PCR amplified with primers RcsA-P1 and RcsA-P2 for rcsA, with primers RcsB-P1 and RcsB-P2 for rcsB, with primers SsrB-P1 and SsrB-P2 for ssrB, and with primers SsaV-P1 and SsaV-P2 for ssaV. The sequences of the primers used are shown in Table 2. The PCR product was used to transform the wild-type strain carrying the Red recombinase expression plasmid pKD46. When necessary, the antibiotic resistance cassette introduced by the gene-targeting procedure was eliminated by recombination with plasmid pCP20 (22).
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| RcsA-P1 | CAATTCGGCCCTGCGCTTTCAACCACTCGCGTACCAGAAAGTGTAGGCTGGAGCTGCTTC |
| RcsA-P2 | GAGGTACATTGCCAGTCCGGATGTCTCAGCGCATGTTAACCATATGAATATCCTCCTTAG |
| RcsB-P1 | ATTATTGCCGATGACCACCCGATTGTACTGTTCGGTATTCGTGTAGGCTGGAGCTGCTTC |
| RcsB-P2 | AGAGAGATAGTTGAGCAGCGCGATATCATTCTCTACGCCCCATATGAATATCCTCCTTAG |
| SsrB-P1 | AATATGACCAATGCTTAATACCATCGGACGCCCCTGGTTAGTGTAGGCTGGAGCTGCTTC |
| SsrB-P2 | TACTTAATATTATCTTAATTTTCGCGAGGGCAGCAAAATGCATATGAATA TCCTCCTTAG |
| SsaV-P1 | CATCATCGACAAATAAAATTTCTGGAGTCGCAATGCGTTCGTGTAGGCTGGAGCTGCTTC |
| SsaV-P2 | CAATTCATTCTTCATTGTCCGCCAACTCCTCTTCGCTAAGCATATGAATATCCTCCTTAG |
| SrfA-P1Flag | CCACGCGGCAATTCCGTTGACGTTTAAAAAGATAGGTGCCGACTACAAAGACCATGACGG |
| SrfA-P2Flag | GACGCTCTGTTTGTAATCACACAGATTGACCAACATAAAACATATGAATATCCTCCTTAG |
| SrfB-P1Flag | CAGCCACTACTGGATAGATAGTGGGAGTGTATACCTGAAAGACTACAAAGACCATGACGG |
| SrfB-P2Flag | CCCACTCGATAACAGCCTGGGTGGTGTTTAACGGTTTTGCATATGAATATCCTCCTTAG |
| SrfC-P1Flag | CCGACGTAGACAGAGCGCAATTAATTGCCCTGATAGCCGACTACAAAGACCATGACGG |
| SrfC-P2Flag | GATAAGTTTCCCCGTCGCATTGCTGTTTTCGCTTTTCTGACATATGAATATCCTCCTTAG |
| RcsB5' | AGCGGAATTCAGGAGGAATACATGAACAATATGAACG |
| RcsB3' | GTGAAAGCTTGTCGACAAGCGATTTATTCTTTGTCTG |
| MuL | CGAATAATCCAATGTCCTCC |
Construction of an ssaV::lac fusion in the Salmonella chromosome.
The FRT site generated by excision of the antibiotic resistance cassette (22) was used to integrate plasmid pCE36 to generate a transcriptional lac fusion in ssaV (27).
Chromosomal gene epitope tagging.
Addition of a 3×FLAG epitope tag at the 3′ ends of the srfA, srfB, and srfC genes was carried out as described previously (88) using primers SrfA-P1Flag, SrfA-P2Flag, SrfB-P1Flag, SrfB-P2Flag, SrfC-P1Flag, and SrfC-P2Flag (Table 2).
Media and chemicals.
The standard culture medium for S. enterica was Luria-Bertani (LB) broth. Solid LB medium contained 1.5% (final concentration) agar. Antibiotics were used at the following concentrations: kanamycin, 50 μg ml−1; chloramphenicol, 20 μg ml−1; and ampicillin, 100 μg ml−1. For some experiments 40 mg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 0.2% glucose or arabinose were added to LB medium. Motility assays were carried out in LB medium prepared without yeast extract (37). Solid motility medium contained agar at a final concentration of 0.25%. For SPI-1-inducing conditions, Salmonella strains were grown overnight at 37°C in LB medium containing 0.3 M NaCl in static conditions. For SPI-2-inducing conditions, cells from overnight cultures in LB medium were washed and diluted 1:100 with minimal medium at pH 5.8 (LPM) containing 80 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8), 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 0.1% Casamino Acids, 38 mM glycerol, 337.5 μM K2HPO4-KH2PO4 (pH 7.4), and 8 μM MgCl2 and incubated overnight at 37°C with shaking.
DNA amplification with PCR.
Amplification reactions were carried out with a Perkin Elmer Gene-Amp 2400 PCR system (Perkin Elmer Cetus, Foster City, CA). The final volume of reaction mixtures was 50 to 100 μl, and the final concentration of MgCl2 was 1 mM. Reagents were used at the following concentrations: deoxynucleoside triphosphates, 200 μM; primers, 1 μM; and Taq polymerase (Expand high-fidelity PCR system; Roche Diagnostics SL), 1 U per reaction mixture. The thermal program included the following steps: (i) initial denaturation for 2 min at 94°C; (ii) 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 to 3 min; and (iii) final incubation at 72°C for 7 min to complete extension.
Plasmids.
Plasmid pBAD18 (Apr) is a member of the pBAD series of vectors and permits tight regulation of cloned genes via the arabinose-inducible PBAD promoter (42). Plasmid pNG1166 is a pBAD18 derivative which carries the igaA gene under control of the PBAD promoter (10). Plasmid pIZ1589 was constructed as follows. Genomic DNA from strain ATCC 14028 was PCR amplified using primers RcsB5′ and RcsB3′ (Table 2), which introduce EcoRI and HindIII sites, respectively. The amplified DNA fragment was digested with EcoRI and HindIII for oriented cloning of the rcsB gene on pBAD18. Ligation mixtures were used to transform E. coli DH5α, with selection of Apr transformants on LB medium-ampicillin plates. One of the transformants was the source of pIZ1589, which carries the rcsB gene under the control of PBAD.
Mutagenesis with MudJ.
We employed the cis-complementation procedure of Hughes and Roth (49), in which a defective MudJ element is cotransduced with a Mud1 element that transiently provides transposition functions. Mud1 is the specialized transducing phage Mud1 (Ap Lac cts62) (13). MudI1734[KmLac] (14) is a transposition-deficient Mu derivative that generates operon fusions upon insertion; this element was renamed MudJ by Hughes and Roth (49).
Cloning and molecular characterization of MudJ inserts.
Genomic DNA from each MudJ-carrying isolate was digested with BamHI or XhoI and ligated with T4 DNA ligase to BamHI- or XhoI-digested pBluescript SKII. The ligation mixtures were transformed into E. coli DH5α, and MudJ-containing transformants were selected on LB medium plates supplemented with kanamycin. The DNA sequence of the fusion junctions and the flanking DNA was obtained by sequencing with an automated DNA sequencer (Sistemas Genómicos, Valencia, Spain) using primer MuL (87).
Sequence analysis.
Sequence analysis was performed with molecular biology algorithms from the National Center for Biotechnology Information at www.ncbi.nlm.nih.gov and the European Bioinformatics Institute at www.ebi.ac.uk.
β-Galactosidase assays.
Levels of β-galactosidase activity were determined for exponential- and stationary-phase cultures in LB medium as described previously (59), using the CHCl3-sodium dodecyl sulfate (SDS) permeabilization procedure.
Motility assays.
Liquid cultures were prepared in motility medium and incubated at 37°C with shaking. At the mid-exponential stage of growth, 5 μl of a culture was spotted in the center of a motility agar plate. The plate was incubated at 37°C. The diameter of the bacterial growth halo was measured every hour.
Western blotting and antibodies.
Salmonella strains were grown under SPI-1- or SPI-2-inducing conditions. The bacteria were then pelleted by centrifugation and resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. Proteins were separated by SDS-PAGE on 10% acrylamide gels and electrophoretically transferred to nitrocellulose filters for Western blot analysis using anti-Flag M2 monoclonal antibodies (1:10,000; Sigma). Goat anti-mouse horseradish peroxidase-conjugated antibodies (Bio-Rad) were used as secondary antibodies.
Mouse mixed infections and determination of CIs and COIs.
Eight-week-old female BALB/c mice (Charles River Laboratories, Santa Perpetua de Mogoda, Spain) were subjected to mixed infections. Groups of three or four animals were inoculated with two strains at a ratio of 1:1. Bacteria were grown overnight at 37°C in LB medium with shaking, diluted into fresh medium (1:100), and grown until the optical density at 600 nm was 0.3 to 0.6. Intraperitoneal inoculation was performed with 0.2 ml of saline containing 105 CFU. Bacteria were recovered from spleens 48 h after inoculation, and the CFU were enumerated on LB medium and on selective medium. A competitive index (CI) for each mutant was calculated by dividing the ratio of the mutant to the wild-type strain in the output (bacteria recovered from the host after infection) by the ratio of these strains in the input (initial inoculum) (6, 33, 85). The “cancelled-out” competitive index (COI) is the CI for mixed infections of double mutants with corresponding single mutant strains and was determined by dividing the ratio of a double mutant strain to the corresponding single mutant in the output by the ratio of these strains in the input (6).
Statistical analysis.
The CI or COI was expressed as the mean of at least three independent infections ± standard error. Student's t test was used to analyze every COI with two null hypotheses: (i) the mean COI is not significantly different from 1 and (ii) the mean COI is not significantly different from the CI of the corresponding single mutant. P values of ≤0.05 were considered significant.
RESULTS
Genetic screens for identification of Salmonella genes regulated by the Rcs system.
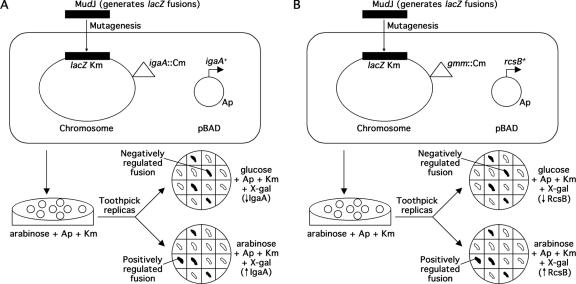
The initial goal of this work was identification of new genes regulated by the Rcs system in Salmonella. Given the involvement of the Rcs system in virulence, we were particularly interested in identifying genes that were Salmonella specific. For this purpose, we generated random transcriptional lac fusions in the Salmonella chromosome and compared their expression under strong activation conditions with their expression with a lack of activation of the Rcs system. Two different, independent screens were devised (Fig. 1).
FIG. 1.
Identification of IgaA- and RcsB-regulated genes. (A) Diagram of the genetic screen for identification of IgaA-regulated genes in S. enterica serovar Typhimurium. A plasmid carrying a wild-type igaA allele under the control of an arabinose-inducible promoter (PBAD) was introduced into a strain harboring a null igaA mutation. igaA is expressed in the presence of arabinose as the sole carbon source but not in the presence of glucose. lac transcriptional fusions were generated using MudJ. The expression pattern of 10,000 independent fusions was monitored in glucose- and arabinose-containing LB medium plates supplemented with X-Gal. (B) Diagram of the genetic screen carried out to identify RcsB-regulated genes. A plasmid with a copy of the wild-type rcsB gene under PBAD control was introduced into a strain harboring a null gmm mutation. rcsB is expressed in arabinose medium but not in glucose medium. lac transcriptional fusions were generated, as described above, with MudJ. The expression pattern of 20,000 independent fusions was monitored in glucose- and arabinose-containing LB medium plates supplemented with X-Gal. The gmm mutation was introduced to prevent mucoidy, thereby facilitating comparison of colony colors in different media. Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol.
(i) Identification of IgaA-regulated genes.
The product of igaA has a negative effect on the activation of the Rcs system. Null mutations in igaA are lethal. Mutants with some point mutations, like igaA1 and igaA5, are viable but show mucoidy and a lack of motility. rcsC, rcsD, and rcsB mutations suppress these phenotypes, suggesting that they are due to overactivation of the Rcs system (10, 25). Therefore, genes regulated by RcsB are expected to be regulated by IgaA in the opposite way. To carry out the first screen, we constructed strain SV4573 (Table 1), in which the null mutation igaA3::Cmr is complemented by a plasmid-borne, wild-type igaA allele expressed from an arabinose-dependent promoter (10, 25). In solid LB medium with arabinose, igaA is expressed and the strain forms nonmucoid colonies. In LB medium with glucose, the strain shows the phenotypes associated with a lack of igaA. Although null mutations in igaA are lethal (10, 25), we observed that patching of colonies growing in LB medium arabinose onto LB medium containing glucose allowed residual growth that was useful for the purpose of the screen. Mucoidy was observed in the presence of glucose, confirming the occurrence of reduced igaA expression and concomitant Rcs activation. The procedure is summarized in Fig. 1A.
For detection of IgaA-regulated loci, strain SV4573 was mutagenized with MudJ to generate transcriptional lacZ fusions. Ten thousand independent isolates carrying MudJ inserts were patched in grids onto LB medium containing glucose and onto LB medium containing arabinose in the presence of the chromogenic indicator X-Gal. Color differences between LB medium containing glucose and LB medium containing arabinose suggested that there was regulation of the fusion by igaA. Initially, 114 fusions were found to be differentially regulated. Reconstruction experiments and comparison of the β-galactosidase activities of the fusions in a wild-type background with the activities in an igaA5 background (25) allowed us to eliminate fusions whose phenotype was due to secondary mutations, insertion in the plasmid, or direct arabinose regulation. Finally, nine fusions were chosen for further study.
(ii) Identification of RcsB-regulated genes.
Plasmid pIZ1589, containing rcsB under PBAD promoter control, was transformed into wild-type strain ATCC 14028 (Table 1). Transformants carrying pIZ1589 were nonmucoid in glucose (when rcsB was not expressed) and mucoid in arabinose (when rcsB was expressed). This is in agreement with studies of E. coli indicating that overexpression of rcsB mimics overactivation of the system (9). Our second screen for Rcs-regulated genes is summarized in Fig. 1B. Plasmid pIZ1589 was introduced into strain SV4514 (gmm::MudQ), yielding strain SV5090 (Table 1). The gmm gene, also called wcaH, is necessary for production of colanic acid capsule. The advantage of this screen is that the strain is nonmucoid in both glucose and arabinose media, making color comparisons easier. Twenty thousand MudJ-carrying isolates were analyzed in this second screen, and 17 fusions were chosen for further study.
To ascertain whether the MudJ insertions provided by the screens could be ascribed to previously known Rcs-regulated genes, two tests were carried out: (i) the abilities of the different MudJ insertions to suppress the mucoid phenotype of the igaA5 mutant were analyzed and (ii) the insertions were transduced into the wild type (strain ATCC 14028) and transductants were tested for motility. Four insertions obtained in the first screen were found to be suppressors of mucoidy, and transductional linkage analysis revealed that these insertions were linked to gmm (not shown). Three additional insertions, one from the first screen and two from the second, caused a loss of motility in an otherwise wild-type background, suggesting that they affected flagellar genes. Putative insertions in genes involved in colanic acid synthesis or motility were not characterized further.
Characterization of novel members of the Rcs regulon.
Insertions that were suppressors of neither mucoidy nor motility were cloned and sequenced. To identify the loci where the MudJ element had inserted, chromosomal fragments containing the MudJ Kmr gene were cloned in pBluescript SKII. DNA sequencing was performed using the MuL primer (87). Genes positively regulated by RcsB (or negatively regulated by IgaA) and genes negatively regulated by RcsB (or positively regulated by IgaA) were identified. Data for these genes and their products are shown in Table 3.
TABLE 3.
IgaA- and RcsB-regulated fusions
| Insertion(s)a | Gene | Induction ratiob | Protein function and/or features | Putative transcriptional unit | Reference(s) |
|---|---|---|---|---|---|
| i36 | trg | −129.6 | Methyl-accepting chemotaxis protein | trg | 53, 81 |
| i59, i70 | srfB | −35.4 | Unknown | srfABC | 93 |
| i74, r50 | bapA | 5.4 | Large protein with repeats | bapABCD | 51 |
| r20, r44 | yjbH | 70.3 | Outer membrane lipoprotein | yjbEFGH | 29 |
| r52 | narH | −12.5 | β-Subunit of nitrate reductase A | narGHJI | 8, 79, 80 |
| r55, r56 | siiE | −3.3 | Large secreted protein coded in SPI-4 | siiABCDEF | 63, 92 |
| r71 | dcuB | −1.9 | Anaerobic C4-dicarboxylate transporter | dcuBfumB | 29, 38 |
| r85 | STM2176 | −1.9 | Glutathione S-transferase | STM2179- STM2178- STM2177- STM2176- STM2175 | 58 |
| r97 | melB | −1.9 | Melibiose uptake | melAB | 62, 66, 70, 75 |
| r111 | yhhJ | −3.3 | Transporter, ABC superfamily | yhiHyhhJ | 72 |
| r142, r171 | srfA | −9.5 | Similar to nuclear antigens | srfABC | 93 |
| r150 | PSLT071 | 4.5 | Unknown | PSLT071 | 68 |
| r153 | yiaD | 17.7 | Outer membrane lipoprotein | yiaD | 29 |
| r172 | STM1491 | 7.4 | ABC-type proline/glycine betaine transport, ATPase component | STM1494- STM1493- STM1492- STM1491 | 58 |
Insertions i36 to i74 were obtained in the first screen (with igaA expressed from a pBAD18 derivative), and insertions r20 to r172 arose from the second screen (with rcsB expressed from a pBAD18 derivative).
Induction ratios were determined as follows: igaA5/igaA+ for genes positively regulated by RcsB (positive values) and igaA+/igaA5 for genes negatively regulated by RcsB (negative values).
Null mutations in rcsC or rcsB suppress mucoidy and the lack of motility of an IgaA− mutant (10). Therefore, these mutations are expected to suppress the positive or negative effect of the igaA mutation on the expression of all genes found in our screens. Triple mutants bearing one of the MudJ insertions obtained in the screens, as well as the igaA5 mutation and a null rcsB or rcsC allele, were constructed. The β-galactosidase activities of these mutants indicated that all the genes studied were indeed regulated by RcsB and RcsC (Table 4). The only exception was STM2176, whose regulation appears to be RcsC independent.
TABLE 4.
β-Galactosidase activities of the Rcs-regulated fusions in different genetic backgrounds
| Insertion in gene | β-Galactosidase activity (Miller units)a
|
||||
|---|---|---|---|---|---|
| Wild type | igaA5 | igaA5 rcsA | igaA5 rcsB | igaA5 rcsC | |
| dcuB | 1,818 ± 36 | 974 ± 33 | 919 ± 21 | 1,941 ± 12 | 1,922 ± 53 |
| melB | 15 ± 0.9 | 8 ± 0.4 | 12 ± 0.3 | 18 ± 0.4 | 16 ± 0.8 |
| narH | 138 ± 25 | 11 ± 5 | 6 ± 2 | 69 ± 14 | 76 ± 16 |
| siiE | 22 ± 2 | 6 ± 0.1 | 6 ± 0.2 | 66 ± 4 | 68 ± 8 |
| srfA | 38 ± 0.3 | 4 ± 0.1 | 2 ± 0.1 | 86 ± 26 | 26 ± 0.3 |
| srfB | 248 ± 10 | 7 ± 0 | 7 ± 1.1 | 233 ± 0.8 | 225 ± 29 |
| trg | 648 ± 28 | 5 ± 0 | 5 ± 0.6 | 557 ± 72 | 588 ± 39 |
| yhhJ | 44 ± 3 | 12 ± 0.1 | 11 ± 0.4 | 49 ± 3 | 36 ± 0.1 |
| yiaD | 9 ± 0 | 159 ± 10 | 166 ± 0.9 | 8 ± 0 | 8 ± 0.3 |
| yjbH | 11 ± 0.1 | 773 ± 69 | 159 ± 27 | 7 ± 0.1 | 6 ± 0.2 |
| STM1491 | 57 ± 1.3 | 420 ± 36 | 512 ± 12 | 56 ± 13 | 84 ± 4 |
| STM2176 | 15 ± 0.9 | 8 ± 0.1 | 8 ± 0.3 | 15 ± 1 | 9 ± 1 |
| bapAb | 68 ± 5 | 369 ± 9 | 228 ± 3 | 45 ± 2 | 98 ± 3 |
| PSLT071 | 2 ± 0.3 | 9 ± 0.8 | 10 ± 0.2 | 2 ± 0 | 2 ± 0 |
β-Galactosidase activities were determined using stationary-phase cultures in LB medium. The data are the means ± standard deviations of two independent experiments. Similar results were obtained when the assays were performed with exponential-phase cultures.
Activities are variable from one experiment to another with this insertion.
RcsA is another component of the Rcs system known to participate in the expression of some but not all the genes regulated by RcsB (10). In a similar way, an rcsA mutation partially suppressed overexpression of yjbH in an igaA5 mutant but failed to suppress overexpression of STM1491 and yiaD or to restore expression of other genes in an igaA5 background (Table 4). Altogether, these results suggest that for the RcsB regulated genes found in this work, RcsA regulates only some of the genes that are positively regulated by the Rcs system.
srfABC operon is negatively regulated by PhoP.
Among the transcriptional units regulated by RcsB uncovered by our screens, the putative srfABC operon is of special interest. srfB was originally identified in a screen to find SsrB-regulated genes outside SPI-2, although the reported regulation by SsrB was only marginal (93). srfABC was proposed to constitute a multigene horizontal acquisition based on (i) a G+C content significantly higher than the genome average G+C content and (ii) the lack of E. coli homologues (93). A BLAST search revealed the presence of this putative operon in other Enterobacteriaceae, including Enterobacter sp. and Yersinia sp. Interestingly, the plant pathogen Pseudomonas syringae has an srfC ortholog coding for a protein that is secreted through a T3SS (69). Therefore, we decided to further explore the pattern of srfABC expression. Introduction of a 3×FLAG epitope at the 3′ end of srfA, srfB, or srfC permitted detection of C-terminally tagged proteins by Western blotting against the FLAG epitope. All products were the expected size (Fig. 2). Western blotting with anti-FLAG antibody showed that the igaA5 mutation inhibits synthesis of SrfA, SrfB, and SrfC and that this inhibition requires both RcsB and RcsC (Fig. 2).
FIG. 2.
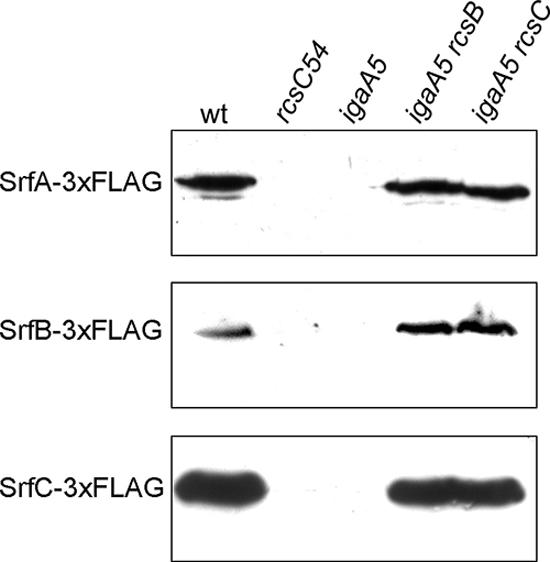
Regulation of srfA, sfrB, and srfC by RcsB at the protein level. Extracts from ATCC 14028 (wild-type strain) derivatives expressing 3×FLAG-tagged SrfA, SrfB, or SrfC were resolved by 10% SDS-PAGE. The same amount of protein was loaded in each lane. Immunoblotting was performed with a monoclonal anti-FLAG antibody. rcsC54 and igaA5 are mutants with constitutive activation of the Rcs system. wt, wild type.
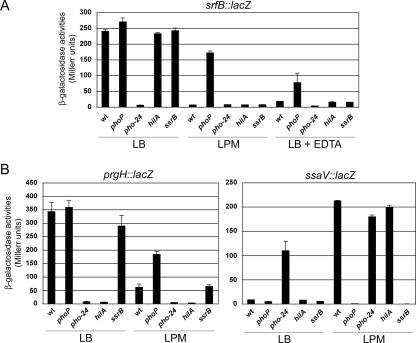
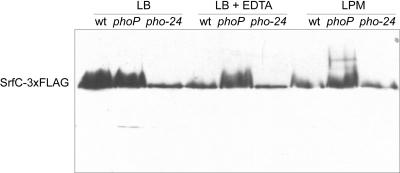
Next, we examined the srfABC expression pattern in media that optimize invasivity (SPI-1-inducing conditions) or that imitate the intracellular milieu (SPI-2-inducing conditions). We took advantage of the lacZ transcriptional fusion created by the MudJ insertion in srfB to measure the levels of transcription under SPI-1-inducing conditions (LB medium) and SPI-2-inducing conditions (LPM) in different genetic backgrounds, including wild type, phoP, pho-24 (a mutation that causes constitutive activation of the PhoPQ system), hilA, and ssrB (Fig. 3A). As controls, the same culture conditions and genetic backgrounds were used to monitor expression of the SPI-1 gene prgH and the SPI-2 gene ssaV. As expected, a prgH::lac transcriptional fusion was expressed under SPI-1-inducing conditions and was subjected to HilA positive regulation and PhoP negative regulation (Fig. 3B). In turn, an ssaV::lac transcriptional fusion was expressed under SPI-2-inducing conditions, and its expression was dependent on PhoP and SsrB (Fig. 3B). As shown in Fig. 3A, the srfABC operon is expressed in LB medium and repressed in LPM, and a phoP null mutation leads to expression of the operon even in LPM. These results provide evidence that the srfABC operon is expressed under SPI-1-inducing conditions and is repressed under SPI-2-inducing conditions in a PhoP-dependent manner. In support of this conclusion, expression of the operon is also repressed in a pho-24 background under SPI-1-inducing conditions and when LB medium is supplemented with EDTA, an Mg2+ chelator. In contrast, EDTA does not prevent transcription of the operon in a PhoP− background. The results obtained with the srfB::lacZ transcriptional fusion were confirmed at the protein level by detecting epitope-tagged SrfC by Western blotting (Fig. 4).
FIG. 3.
Transcriptional repression of srfABC by PhoP. Expression levels of srfB (A) and prgH and ssaV (B) were monitored with lacZ transcriptional fusions. Strains carrying the indicated mutations were cultured in the following media: LB medium for SPI-1 induction; LPM for SPI-2 induction; and LB medium containing EDTA for Mg2+ chelation. The β-galactosidase activities shown were measured in stationary-phase cultures, but similar results were obtained in exponential cultures. The data represent the averages and standard deviations from two experiments. wt, wild type.
FIG. 4.
Regulation of SrfC-3×FLAG levels by PhoP. Strains carrying the indicated mutations were cultured in LB medium (for SPI-1 induction), LPM (for SPI-2 induction), and LB medium containing EDTA (for Mg2+ chelation). Protein extracts were resolved by 10% SDS-PAGE. The same amount of protein was loaded in each lane. Immunoblotting was performed with a monoclonal anti-FLAG antibody. wt, wild type.
In vivo analysis reveals a partial overlap between the RcsB and PhoP regulons.
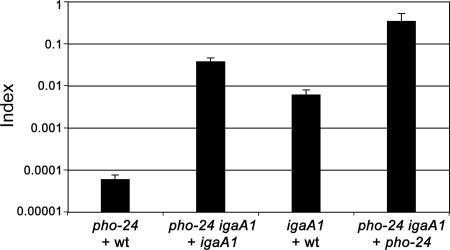
Regulation of the srfABC operon by RcsB and PhoP suggests the possibility that there is a genetic interaction between these systems. Since both systems regulate virulence in Salmonella, we investigated their hypothetical interaction in the mouse model of infection, taking advantage of a method that has been used previously for in vivo genetic analysis. BALB/c mice were infected intraperitoneally with a mixture of single and double mutants carrying igaA1 and pho-24 mutations, and a COI (6) was calculated. The COI (igaA1 pho-24 versus igaA1) was statistically different from the CI (pho-24 versus wild type) but also statistically different from 1 (Fig. 5). Similarly, the reciprocal COI (igaA1 pho-24 versus pho-24) was significantly different from 1 and from the CI (igaA1 versus wild type) (Fig. 5). These results suggest that there is a partial overlap between these regulatory systems in the control of certain virulence functions.
FIG. 5.
Overlap of PhoPQ and Rcs systems in the control of virulence: analysis of the pho-24 igaA1 double mutant in mixed infections with either the pho-24 or igaA1 single mutant. The indices represented are CIs (single mutant versus wild type) and COIs (double mutant versus single mutant). The strains used in each mixed infection are represented by the relevant mutations, as indicated under the bars. COIs were compared to 1.0 and to the CI relevant in each case. COIs are significantly different from the corresponding CIs and from 1.0 (P < 0.05). wt, wild type.
In support of this conclusion, we compared the expression of three representative genes (prgH [an SPI-1 gene], ssaV [an SPI-2 gene], and gmm [a capsule gene]) in PhoP-constitutive and Rcs-constitutive backgrounds. Note that overproduction of the colanic acid capsule is a relevant factor in the attenuation of Rcs constitutive mutants (35). Data in Table 5 show that both the PhoPQ and Rcs systems transcriptionally repress prgH. This result is in agreement with a previous report suggesting that the Rcs system negatively controls Salmonella invasion of epithelial cells (64). In contrast, ssaV is activated by PhoP but not by RcsB, and gmm is activated by RcsB but not by PhoP (Table 5). Altogether, these results support the view that there is partial overlap between Rcs and PhoPQ in the control of Salmonella virulence.
TABLE 5.
Partial overlap of Rcs and/or PhoPQ in the regulation of genes relevant for Salmonella virulence
| lac fusion in gene | β-Galactosidase activity (Miller units)a
|
||
|---|---|---|---|
| Wild type | igaA5 | pho-24 | |
| prgH | 120 ± 4 | 1 ± 0 | 3 ± 0 |
| ssaV | 7 ± 0.1 | 7 ± 0.2 | 103 ± 13 |
| gmm | 2 ± 0.1 | 901 ± 26 | 2 ± 0.1 |
β-Galactosidase activities were determined using stationary-phase cultures in LB medium. The data are the means ± standard deviations of two independent experiments. Similar results were obtained when the assays were performed with exponential-phase cultures.
DISCUSSION
The Rcs system is present in enteric and nonenteric bacteria, including E. coli, S. enterica, Vibrio cholerae, K. pneumoniae, E. amylovora, P. mirabilis, and Pseudomonas aeruginosa. From its initial description as a regulator of capsule synthesis (39), a diversity of roles have been assigned to the Rcs signaling system (54). Of special interest is the involvement of the Rcs system in Salmonella virulence (25, 35, 64).
Here, we identified RcsB-regulated genes in S. enterica serovar Typhimurium by comparing the expression of MudJ-generated fusions with a basal level of Rcs activation and the expression of MudJ-generated fusions with strong activation. The latter was achieved by using either igaA mutations or rcsB overexpression. The results of our screens overlap only partially with those obtained for E. coli (29, 44, 67). This can be explained by differences between E. coli and S. enterica and also by the diverse methodologies employed (macroarray, microarray, transcriptional fusions) and the disparate conditions used for activation of the Rcs system (growth at 20°C in medium with glucose and ZnCl2, DjlA overproduction, igaA mutations, rcsB overexpression).
Data in Table 4 confirm that all the genes identified in our screens are indeed regulated by RcsB. Several interesting details should also be noted. (i) STM2176 appears to be RcsC independent, in contrast to the remaining genes. (ii) rcsB and rcsC null mutations not only suppress repression of siiE by igaA5 but also increase siiE expression on their own. (iii) A similar effect is exerted on srfA by an rcsB mutation (but not by an rcsC mutation). These results suggest that under our experimental conditions, RcsB is not completely inactive in a wild-type background. These data could also indicate an RcsC-independent role for acetyl phosphate or another unknown donor in RcsB phosphorylation (28, 54).
Our screens unveiled genes already known to be members of the Salmonella Rcs regulon, including genes for colanic acid capsule synthesis, flagella, and chemotaxis (10). These genes served to validate our methodology but were not studied further. A second group included genes that have homologues in E. coli. None of them had previously been identified as a member of the Salmonella Rcs regulon, and only two, yiaD and yjbH, have been shown to be part of the E. coli Rcs regulon. E. coli yiaD and yjbH were found to be regulated by RcsC in a screen of transcriptional fusions generated by λplacMu53 (29). In the same study, macroarray analysis identified yjbE, yjbF, and yjbG, the three open reading frames upstream of yjbH, as members of the RcsC regulon (29). It has recently been shown that the yjb operon is involved in production of exopolysaccharides (28). Based on similarities to known genes, yiaD, yjbH, and yjbF are predicted to encode lipoproteins. Interestingly, many of the genes identified as members of the E. coli Rcs regulon in another study (44) are predicted to encode cell envelope-associated proteins, including three putative lipoproteins (OsmB, YajI, and YggG). All these genes are positively regulated by the system, as are yiaD and yjbH. This is in agreement with the involvement of the Rcs system not only in the production of capsule but also in the control of surface-related processes, such as swarming (4, 43, 46, 84) and biofilm formation (29). The remaining genes in this second group are negatively regulated by the system. The list includes dcuB and narH, suggesting that the Rcs system may also play a role in the control of anaerobic respiration. Interestingly FlhD and FlhC, which are negatively regulated by RcsB, have been shown to regulate a number of E. coli genes involved in anaerobic respiration (71), but dcuB and narH were not on the list. In addition, our data suggest that FlhD and FlhC do not regulate these genes in S. enterica (data not shown).
In previous work we showed that rcsC mutants with constitutive activation of the Rcs system are severely attenuated for virulence in intraperitoneally inoculated BALB/c mice and that overexpression of the colanic acid capsule is one of the factors responsible for attenuation (35). However, we also presented evidence that there are additional factors (35). Hence, we were especially interested in Salmonella genes regulated by the Rcs system that are not present in related bacteria. The Salmonella-specific genes revealed by our genetic screens are bapA, siiE, srfA, and srfB. The product of bapA has recently been described as a cell surface protein required for biofilm formation in S. enterica serovar Enteritidis (51). Interestingly, the Rcs system is involved in biofilm formation in E. coli (29). siiE is located in SPI-4, a Salmonella-specific chromosomal segment (92) predicted to be an operon containing six open reading frames which encode a secreted protein (SiiE) and components of a type I secretion system (58). Conclusive evidence that this island is required for intestinal but not systemic infection in mice was recently provided (63).
srfA and srfB, together with srfC, appear to form a 7-kb operon that is part of a putative horizontally acquired DNA segment that was described as regulated by SsrB (93), a regulator encoded within SPI-2 that is required for SPI-2-encoded T3SS expression (17, 47). Here, we show that the srfABC operon is expressed in SPI-1-inducing conditions but not in SPI-2-inducing conditions. In fact, our experiments show that in addition to being repressed by RcsB, srfABC is repressed by PhoP. In contrast, we provide evidence that srfABC is not under the control of SsrB (Fig. 3 and 4).
The previously reported regulation of some PhoP-activated genes by RcsB suggested the existence of a regulatory circuit between the PhoPQ and Rcs networks (86). However, in vivo studies demonstrated that attenuation of virulence caused by activation of the Rcs system is not related to loss of function of the PhoPQ system (25). These experiments thus failed to show any overlap between the two systems for the control of virulence. However, we show that the sfrABC operon is repressed by both RcsB and PhoP, providing examples of PhoP-repressed genes (prg genes) that are simultaneously repressed by RcsB. On the basis of the latter results, we investigated the effect of a mutation that causes constitutive activation of the Rcs system (igaA1) and a mutation that causes constitutive activation of the PhoPQ system (pho-24) in the mouse model. If virulence genes repressed by both systems exist, an epistatic effect should be detected in infections with double mutants. The main advantage of using igaA1 (CI, 0.006) in the virulence experiments instead of other more attenuated igaA mutations like igaA5 (CI, 0.0000036) (25) is that it can be expected to permit detection of complete additivity with pho-24 (CI, 0.00006), since with this system we are able to detect CI values as low as 0.0000004 but lower values are not accurately measured (25, 35; data not shown). The actual results (Fig. 5) are intermediate between additivity and epistasis and suggest that there is partial overlap between the Rcs and PhoPQ systems in the control of Salmonella virulence. This interpretation is strengthened by the existence of virulence genes that are repressed simultaneously by both systems, including prgH (Table 5) and other SPI-1 genes (64), whereas other genes are under the control of one system but not the other. Examples of the latter class include an SPI-2 gene (ssaV) and a capsule gene (gmm) (Table 5). Further investigations can be expected to reveal more precisely the overlap between the systems and to identify coregulated genes important for the systemic phase of Salmonella infection.
Acknowledgments
We are grateful to F. García-del Portillo for helpful discussions and to E. A. Groisman and J. López-Garrido for the generous gift of some strains.
This work was supported by grants BIO2004-04355-CO2-02 (to J.C.) and SAF2004-00227 (to F.R.-M.) from the Ministry of Education and Science of Spain and the European Regional Development Fund. C.G.-C. is a Ph.D. student supported by a predoctoral grant from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Aguilar, P. S., A. M. Hernandez-Arriaga, L. E. Cybulski, A. C. Erazo, and D. de Mendoza. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. [DOI] [PubMed] [Google Scholar]
- 3.Beier, D., and R. Gross. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9:143-152. [DOI] [PubMed] [Google Scholar]
- 4.Belas, R., R. Schneider, and M. Melch. 1998. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J. Bacteriol. 180:6126-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bereswill, S., and K. Geider. 1997. Characterization of the rcsB gene from Erwinia amylovora and its influence on exopolysaccharide synthesis and virulence of the fire blight pathogen. J. Bacteriol. 179:1354-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuzón, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. [DOI] [PubMed] [Google Scholar]
- 8.Blasco, F., C. Iobbi, G. Giordano, M. Chippaux, and V. Bonnefoy. 1989. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol. Gen. Genet. 218:249-256. [DOI] [PubMed] [Google Scholar]
- 9.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cano, D. A., G. Domínguez-Bernal, A. Tierrez, F. García-del Portillo, and J. Casadesús. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cano, D. A., M. Martínez-Moya, M. G. Pucciarelli, E. A. Groisman, J. Casadesús, and F. García-del Portillo. 2001. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69:6463-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballès, F., C. Bertrand, J. P. Bouché, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. [DOI] [PubMed] [Google Scholar]
- 13.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 16.Chen, M. H., S. Takeda, H. Yamada, Y. Ishii, T. Yamashino, and T. Mizuno. 2001. Characterization of the RcsC→YojN→RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65:2364-2367. [DOI] [PubMed] [Google Scholar]
- 17.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 18.Clarke, D. J., S. A. Joyce, C. M. Toutain, A. Jacq, and I. B. Holland. 2002. Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12. J. Bacteriol. 184:1204-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clavel, T., J. C. Lazzaroni, A. Vianney, and R. Portalier. 1996. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol. Microbiol. 19:19-25. [DOI] [PubMed] [Google Scholar]
- 20.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotter, P. D., C. M. Guinane, and C. Hill. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 25.Domínguez-Bernal, G., M. G. Pucciarelli, F. Ramos-Morales, M. García-Quintanilla, D. A. Cano, J. Casadesús, and F. García-del Portillo. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53:1437-1449. [DOI] [PubMed] [Google Scholar]
- 26.Ebel, W., G. J. Vaughn, H. R. Peters, and J. E. Trempy. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J. Bacteriol. 179:6858-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 28.Ferrières, L., S. N. Aslam, R. M. Cooper, and D. J. Clarke. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070-1080. [DOI] [PubMed] [Google Scholar]
- 29.Ferrières, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 30.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 32.Fredericks, C. E., S. Shibata, S. Aizawa, S. A. Reimann, and A. J. Wolfe. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734-747. [DOI] [PubMed] [Google Scholar]
- 33.Freter, R., P. C. O'Brien, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun. 34:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galán, J. E., and R. Curtiss III. 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog 6:433-443. [DOI] [PubMed] [Google Scholar]
- 35.García-Calderón, C. B., M. García-Quintanilla, J. Casadesús, and F. Ramos-Morales. 2005. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151:579-588. [DOI] [PubMed] [Google Scholar]
- 36.Gervais, F. G., and G. R. Drapeau. 1992. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J. Bacteriol. 174:8016-8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillen, K. L., and K. T. Hughes. 1991. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 173:2301-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golby, P., D. J. Kelly, J. R. Guest, and S. C. Andrews. 1998. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 180:6586-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gygi, D., M. M. Rahman, H. C. Lai, R. Carlson, J. Guard-Petter, and C. Hughes. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167-1175. [DOI] [PubMed] [Google Scholar]
- 44.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 46.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. USA 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 48.Hirakawa, H., K. Nishino, T. Hirata, and A. Yamaguchi. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185:1851-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes, K. T., and J. R. Roth. 1984. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J. Bacteriol. 159:130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelley, W. L., and C. Georgopoulos. 1997. Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol. Microbiol. 25:913-931. [DOI] [PubMed] [Google Scholar]
- 51.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penadés, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322-1339. [DOI] [PubMed] [Google Scholar]
- 52.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 53.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 54.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 55.Majdalani, N., M. Heck, V. Stout, and S. Gottesman. 2005. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 187:6770-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 57.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones & Barlett, Boston, MA.
- 58.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 59.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mizushima, K., S. Awakihara, M. Kuroda, T. Ishikawa, M. Tsuda, and T. Tsuchiya. 1992. Cloning and sequencing of the melB gene encoding the melibiose permease of Salmonella typhimurium LT2. Mol. Gen. Genet. 234:74-80. [DOI] [PubMed] [Google Scholar]
- 63.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 64.Mouslim, C., M. Delgado, and E. A. Groisman. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386-395. [DOI] [PubMed] [Google Scholar]
- 65.Nassif, X., N. Honore, T. Vasselon, S. T. Cole, and P. J. Sansonetti. 1989. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 3:1349-1359. [DOI] [PubMed] [Google Scholar]
- 66.Okada, T., K. Ueyama, S. Niiya, H. Kanazawa, M. Futai, and T. Tsuchiya. 1981. Role of inducer exclusion in preferential utilization of glucose over melibiose in diauxic growth of Escherichia coli. J. Bacteriol. 146:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 68.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 69.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prestidge, L. S., and A. B. Pardee. 1965. A second permease for methyl-thio-beta-d-galactoside in Escherichia coli. Biochim. Biophys. Acta 100:591-593. [DOI] [PubMed] [Google Scholar]
- 71.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 73.Scherer, C. A., and S. I. Miller. 2001. Molecular pathogenesis of salmonellae, p. 265-333. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, CA.
- 74.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 75.Schmitt, R. 1968. Analysis of melibiose mutants deficient in alpha-galactosidase and thiomethylgalactoside permease II in Escherichia coli K-12. J. Bacteriol. 96:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shiba, Y., Y. Yokoyama, Y. Aono, T. Kiuchi, J. Kusaka, K. Matsumoto, and H. Hara. 2004. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186:6526-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stewart, V. 1988. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52:190-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stewart, V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 82.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stout, V., A. Torres-Cabassa, M. R. Maurizi, D. Gutnick, and S. Gottesman. 1991. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173:1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 85.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tierrez, A., and F. García-del Portillo. 2004. The Salmonella membrane protein IgaA modulates the activity of the RcsC-YojN-RcsB and PhoP-PhoQ regulons. J. Bacteriol. 186:7481-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Torreblanca, J., S. Marqués, and J. Casadesús. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98:15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Virlogeux, I., H. Waxin, C. Ecobichon, J. O. Lee, and M. Y. Popoff. 1996. Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J. Bacteriol. 178:1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 91.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 92.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 94.Zhou, D., and J. Galán. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]