Abstract
Enzymes of the ATP-independent Deg serine endopeptidase family are very flexible with regard to their substrate specificity. Some family members cleave only one substrate, while others act as general proteases on unfolded substrates. The proteolytic activity of Deg proteases is regulated by PDZ protein interaction domains. Here we characterized the HhoA protease from Synechocystis sp. strain PCC 6803 in vitro using several recombinant protein constructs. The proteolytic activity of HhoA was found to increase with temperature and basic pH and was stimulated by the addition of Mg2+ or Ca2+. We found that the single PDZ domain of HhoA played a critical role in regulating protease activity and in the assembly of a hexameric complex. Deletion of the PDZ domain strongly reduced proteolysis of a sterically challenging resorufin-labeled casein substrate, but unlabeled β-casein was still degraded. Reconstitution of the purified HhoA with total membrane proteins isolated from Synechocystis sp. wild-type strain PCC 6803 and a ΔhhoA mutant resulted in specific degradation of selected proteins at elevated temperatures. We concluded that a single PDZ domain of HhoA plays a critical role in defining the protease activity and oligomerization state, combining the functions that are attributed to two PDZ domains in the homologous DegP protease from Escherichia coli. Based on this first enzymatic study of a Deg protease from cyanobacteria, we propose a general role for HhoA in the quality control of extracytoplasmic proteins, including membrane proteins, in Synechocystis sp. strain PCC 6803.
Proteolysis is an essential process in every living cell and is involved in protein quality control (45), as well as in regulation of diverse cellular events (11). In the cyanobacterium Synechocystis sp. strain PCC 6803, which is a widely used model system for studying photosynthesis and acclimation to abiotic stresses, 77 proteases are encoded in the genome (http://merops.sanger.ac.uk/). Three genes encode ATP-independent serine endopeptidases belonging to the Deg (HtrA) family (14, 19, 21, 40). The protease domain of these enzymes harbors the hallmark catalytic triad consisting of His, Asp, and Ser responsible for the proteolytic activity and may additionally confer a chaperone function (16, 42). Deg proteases usually contain one or two protein-protein interaction domains of the PDZ type C terminal of the protease domain (7). Studies of Escherichia coli DegP (also designated HtrA), E. coli DegS (also designated HhoB), and human HtrA2 showed that PDZ domains regulate the proteolytic activity (16, 18, 27, 38, 42, 46), are involved in the formation of homooligomeric complexes (16, 18, 38), and play a role in substrate recognition (7, 16, 23, 35).
In prokaryotes, Deg proteases are usually located in the periplasm and are implicated in the response to a variety of stresses, such as heat (3, 30, 33), oxidative stress (3, 47), and high-light stress (3, 39). Deg proteases are also essential for virulence in several pathogenic bacteria (33, 47). An intriguing feature of this protease family is its functional diversity; the enzymes range from general proteases to highly specific regulatory enzymes. E. coli DegP prevents aggregation of denatured proteins in the periplasm by acting as a chaperone at lower temperatures and as a general protease for unfolded substrates at higher temperatures (7, 23, 32, 42). DegP has further been implicated in limited proteolysis during the maturation of proteins secreted into the periplasm (5, 7). The homologous DegS protease in the same organism is a highly specific enzyme responsible for the primary cleavage inducing a proteolytic signal transduction cascade. Interaction of misfolded outer membrane porins with the PDZ domain activates DegS, which cleaves its only known substrate, the plasma membrane protein RseA in a periplasmic loop (44, 46). This is the first step of a proteolytic cascade which degrades the RseA protein and thus triggers the σE-dependent heat shock response (2, 7, 8).
The crystal structures of E. coli DegS (46) and human HtrA2 (27), both of which contain only one PDZ domain, showed the association of monomer subunits into homotrimeric complexes by interactions of the protease domains. In human HtrA2, the PDZ domains formed a lid which prevented access to the catalytic centers of the protease domain, and deletion of the PDZ domain enhanced proteolysis of β-casein (28). In E. coli DegS the PDZ domains extended sideways, forming a funnel-shaped structure with open access to the proteolytic center, which was present in an inactive conformation (46). Specific interactions with the PDZ domains are necessary to induce a conformational shift to activate the protease, and consequently, deletion of the PDZ domain yielded an inactive DegS variant. In contrast to these two homologues, E. coli DegP contains two PDZ domains, designated PDZ1 and PDZ2, and showed further assembly of two trimers into a cage-like hexameric structure (23). This hexamer formation was attributed to an extended flexible loop in the N-terminal regions of the protease domains, designated the LA loop, which showed complex interactions with the opposing trimer (7, 23). Two different conformations of the hexamer were observed in the crystal: a closed conformation where the PDZ2 domains interacted with the PDZ1 and PDZ2 domains of the opposite trimer and an open conformation where the PDZ domains extended sideways and the PDZ2 domains were too flexible to be resolved. Thus, it was suggested that the PDZ domains guarded the lateral entrance to the protease domains and coupled substrate binding and translocation into the DegP hexamer (23). Both forms of crystallized DegP were present in proteolytically inactive conformations where both substrate binding and catalysis were prevented (23). Recently, detailed deletion studies demonstrated that, in addition to the LA loop, both PDZ domains were necessary for hexamer formation (16, 18). Interestingly, only the PDZ1 domain was necessary for proteolysis but not for the formation of the hexameric complex.
In contrast to the detailed information reported for the localization, structure, and function of E. coli DegP and DegS, less is known about the role of the three Deg proteases in Synechocystis sp. strain PCC 6803. Care should be taken when the Deg/HtrA proteases of these two organisms are compared. Even though Synechocystis sp. strain PCC 6803 HtrA (htrA, slr1204), HhoA (hhoA, sll1679), and HhoB (hhoB, sll1427) have been designated like the E. coli proteases (19), the enzymes are not orthologous in the two organisms. This means that Synechocystis sp. strain PCC 6803 proteases are more similar to each other than to any Deg proteases from E. coli having the same designations and do not necessarily share the same protein structure and function (14, 17, 21). For instance, in contrast to the E. coli enzymes, all three Deg proteases in Synechocystis sp. strain PCC 6803 contain only one PDZ domain.
Proteome analyses demonstrated that HhoA from Synechocystis sp. strain PCC 6803 is a soluble protein in the periplasm (10) which associates with the plasma membrane (12). However, HhoA might also be located in the thylakoid lumen, because protein sorting in cyanobacteria is poorly understood and there are no methods for reliable separation of the periplasm from the thylakoid lumen (41). HhoB contains a predicted signal sequence for translocation to the periplasm or into the thylakoid lumen (17, 21), but this protease has not yet been identified in any proteome study. HtrA contains a predicted transmembrane segment at its N terminus and has been found in the outer membrane (13). Triple mutants of Synechocystis sp. strain PCC 6803 in which all three Deg proteases were inactivated exhibited a dramatic growth defect when they were exposed to high temperatures or high light intensities (3, 39). Based on these studies, it was proposed that Synechocystis sp. strain PCC 6803 Deg proteases protect the extracytoplasmic compartments from the effects of heat and light stress and from oxidative damage caused by reactive oxygen species which are generated by photosynthetic electron transport (3). Furthermore, HtrA, HhoA, and HhoB were suggested to have at least partially overlapping functions, because no severe phenotype was found in any of the double mutants (3). In contrast to this report, an earlier study indicated that a ΔhhoA insertion mutant was more sensitive to heat stress than the wild type or a ΔhhoB mutant, suggesting different physiological functions (40).
The very different mechanisms and functions of the E. coli enzymes, together with the unresolved function of the Deg proteases in Synechocystis sp. strain PCC 6803, encouraged us to characterize one of these enzymes, HhoA, in biochemical terms in vitro. We demonstrate here that mature HhoA protease, expressed as a soluble recombinant His-tagged fusion protein, acted as a general protease against unfolded model substrates. The proteolytic activity of HhoA increased with temperature and pH and was stimulated by addition of Mg2+ and Ca2+. We show that the PDZ domain of HhoA is essential for the proteolytic activity against certain substrates. Furthermore, recombinant HhoA formed a hexameric complex in solution, with two trimeric units assembled into a hexamer depending on the presence of the PDZ domain. Finally, we show that HhoA degraded only a limited set of proteins when it was added to isolated Synechocystis sp. strain PCC 6803 membrane fractions. Thus, our data demonstrate a dual role for the single PDZ domain in HhoA and support a role for this protease in protein quality control in the extracytoplasmic space, including membrane proteins.
MATERIALS AND METHODS
Strains and culture conditions.
A glucose-tolerant strain of Synechocystis sp. PCC 6803, designated WT, and a deletion mutant in which the HhoA-encoding gene sll1679 was interrupted by a cassette conferring kanamycin resistance, designated ΔhhoA (I. Adamska, P. F. Huesgen, and C. Funk, unpublished data), were cultured in BG-11 medium (36) buffered to pH 8.0 with 20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) using a photon flux density of 170 μmol photons m−2 s−1 at 30°C with 1.5% CO2.
Plasmids.
Genomic DNA was isolated from Synechocystis sp. strain PCC 6803 using a kit (Roche Diagnostics GmbH, Mannheim, Germany). The hhoA gene (sll1679) was amplified by PCR from genomic DNA with gene-specific primers (Operon Biotechnologies, Cologne, Germany) and ligated into the pET151-D/TOPO expression vector using a Champion directional cloning kit (Invitrogen GmbH, Karlsruhe, Germany) according to the manufacturer's instructions. A similar cloning strategy was used for engineering HhoA deletion constructs using following primers: for full-length HhoA, 5′-CAC CAT GAA ATA TCC CAC TTG GTT ACG-3′ and 5′-TTA ACT GGT GGG ATT ACG AAG TTG-3′; for mature HhoAΔN34, 5′-CAC CGC GGA CGA TTT GCC CCC G-3′ and 5′-TTA ACT GGT GGG ATT ACG AAG TTG-3′; and for HhoAPD lacking the PDZ domain, 5′-CAC CGC GGA CGA TTT GCC CCC G-3′ and 5′-GAG TTA TCC CCC CGC AGC GAG GGT-3′. To obtain plasmids for expression of proteolytically inactive mature HhoA (HhoAS237AΔN34) and HhoAPD (HhoAS237APD), the codon for Ser237 in the active site was changed to an Ala codon using primers 5′-CGG AAT GCC GGG GGC CCG TTG C-3′ and 5′-CAA CGG GCC CCC GGC ATT GCC G-3′ and a QuikChange II point mutagenesis kit (Stratagene Europe, Amsterdam, The Netherlands) according to the manufacturer's instructions. These primers also introduced an additional ApaI restriction site through a silent point mutation to facilitate distinction between the two plasmids. The inserted sequences and orientation in the plasmids were confirmed by DNA sequencing (GATC Biotech AG, Konstanz, Germany).
Expression and purification of recombinant protein.
E. coli BL21(DE3)Star Oneshot chemocompetent cells (Invitrogen GmbH, Karlsruhe, Germany) transformed with the expression plasmids were grown in Luria-Bertani medium containing 100 μg mg−1 ampicilin at 19°C to an optical density at 600 nm of 0.4 to 0.6. Expression of the protease constructs was induced by addition of 0.1 mM isopropyl-1-thio-d-galactoside (IPTG). Cultures were then grown overnight at 19°C and harvested by centrifugation at 5,000 × g for 10 min at 4°C. For purification of the HhoA constructs, cell pellets corresponding to 1-liter cultures were resuspended in 9 ml buffer A (50 mM HEPES, 300 mM NaCl; adjusted to pH 8.0) and 1 ml buffer B (50 mM HEPES, 300 mM NaCl, 500 mM imidazole; adjusted to pH 8.0). Cells were lysed on ice by 10 10-s sonication cycles with intervening 20-s cooling periods. Unbroken cells, insoluble cell debris, and inclusion bodies were pelleted by centrifugation for 1 h at 23,000 × g and 4°C. The supernatant was sterile filtered and purified by nickel affinity chromatography (HisTrap; GE Healthcare Europe GmbH, Munich, Germany) using an Ákta purifier fast protein liquid chromatography system (GE Healthcare Europe GmbH, Munich, Germany). A typical elution protocol included two washing steps with 10 ml buffer A containing 75 mM imidazole and 10 ml buffer A containing 100 mM imidazole before the purified protein was eluted with buffer B. The fractions were used directly for activity assays or desalted using a HiTrap desalting column (GE Healthcare Europe GmbH, Munich, Germany). Size exclusion chromatography was performed with a prepacked Superdex 200 column (GE Healthcare Europe GmbH, Munich, Germany). Dynamic light scattering of concentrated size exclusion chromatography elution fractions containing from 0.5 to 2 mg ml−1 protein was measured with a DynaPro instrument (Protein Solutions, Piscataway, NJ), and data were acquired with the Dynamics software, version 6 (Protein Solutions, Piscataway, NJ).
Protease activity assays.
To assay the proteolytic activity of the purified proteins, elution fractions containing 100 pmol purified protein as determined with the Bio-Rad protein assay (Bio-Rad Laboratories GmbH, Munich, Germany) were incubated with 8 μg resorufin-labeled casein (Roche Diagnostics GmbH, Mannheim, Germany) in 50 mM HEPES (pH 8.0) (adjusted at 20°C) and 20 mM CaCl2 at 40°C for 2 h unless indicated otherwise. As alternative substrates, 10 μg β-casein and 10 μg bovine serum albumin (BSA) (both from Sigma-Aldrich Chemie GmbH, Schnelldorf, Germany) were used.
Protease reconstitution assay.
Total membrane proteins containing 200 μg chlorophyll were prepared by rupturing preparations with glass beads essentially as described previously (22). The chlorophyll concentrations of cells or total membrane fractions were determined in methanol as described previously (29). One hundred picomoles of purified HhoAΔN34 was added to membrane fractions containing 50 μg chlorophyll a in 50 mM Tris (pH 7.5) supplemented with 20 mM CaCl2 and incubated for 2 h at the temperature indicated below. The bands of interest were excised and identified by mass spectrometry using a commercial service (Proteome Factory AG, Berlin, Germany).
Protein analysis.
Proteins were solubilized in LDS buffer and separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) as described previously (25). The gels were stained with Coomassie brilliant blue R250 (37).
Bioinformatics.
Sequence analysis was performed using SMART (26), TargetP (9), and SignalP (34) programs. For engineering of the truncated constructs, the secondary structure of HhoA was analyzed with PsiPred v2.5 (4) and compared to previously published structure and sequence alignments (7, 17).
RESULTS
Engineering and heterologous expression of full-length HhoA and deletion constructs.
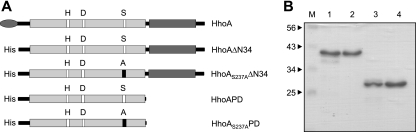
Analysis of the primary structure of HhoA from Synechocystis sp. strain PCC 6803 (encoded by the sll1679 gene) showed that this protein contains a putative N-terminal signal peptide, a protease domain of the trypsin type, and a single C-terminal PDZ domain (Fig. 1A). To assess the importance of conserved domains for proteolytic activity and oligomeric complex formation, we expressed full-length HhoA protein and various deletion constructs in E. coli. Our attempts to express full-length HhoA in a native form failed due to its insolubility and accumulation in inclusion bodies (data not shown). Deletion of the predicted 34-amino-acid periplasmic signal peptide overcame this problem and yielded the soluble HhoAΔN34 construct having a molecular mass of 41.4 kDa when it was expressed at 19°C (Fig. 1B, lane 1). Purified HhoAΔN34 was prone to slow self-degradation, as revealed by the accumulation of lower-molecular-mass bands that were observed in the elution fraction separated on a Coomassie blue-stained SDS gel (Fig. 1B, lane 1). In contrast, the HhoAS237AΔN34 construct, in which Ser237 of the catalytic triad is replaced by Ala (Fig. 1A), was autoproteolytically inactive since a single band at 41.4 kDa was visible in a Coomassie blue-stained SDS gel (Fig. 1B, lane 2). To investigate the role of the PDZ domain, HhoAPD (the suffix PD indicates the protease domain) and HhoAS237APD constructs having a molecular mass of 29.3 kDa were engineered. In both constructs the C-terminal PDZ domain was deleted by truncation after Gly282 (Fig. 1A), and in the latter construct the catalytic Ser237 was replaced by an Ala. Also in this case replacement of the catalytic Ser237 abolished the autoproteolysis compared with the HhoAPD construct (Fig. 1B, compare lanes 3 and 4).
FIG. 1.
Engineering and purification of HhoA deletion constructs. (A) Domain structure of the full-length HhoA protease and truncated constructs. Gray oval, putative signal peptides; light gray boxes, protease domains of the S1B subfamily of serine proteases, with the amino acid residues of the catalytic triad indicated; dark gray boxes, PDZ domains; His, six-His tag and linker introduced by the expression vector. (B) Ni2+ affinity-purified recombinant proteins (4 μg) separated by SDS-PAGE and visualized by Coomassie blue staining. Lane 1, HhoAΔN34; lane 2, HhoAS237AΔN34; lane 3, HhoAPD; lane 4, HhoAS237APD; lane M, markers.
PDZ domain modulates HhoA activity against unfolded protein substrates.
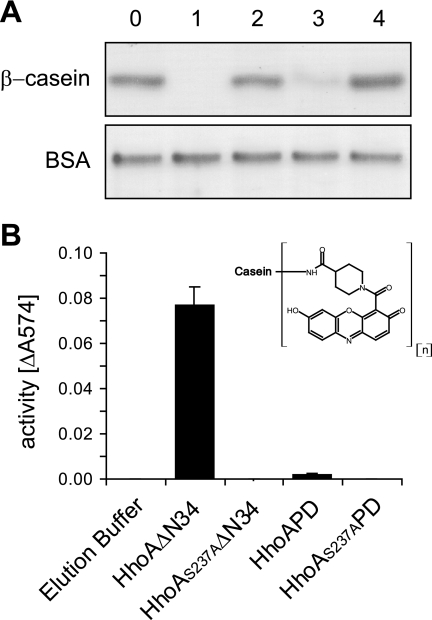
Purified HhoA deletion constructs were tested for their proteolytic activity against three model substrates, including β-casein, resorufin-labeled casein as a chromogenic substrate, and BSA. Elution fractions containing HhoAΔN34 readily degraded the naturally unfolded model substrates β-casein and resorufin-labeled casein but not the globular substrate BSA (Fig. 2A and B). Fractions containing purified HhoAPD were active against β-casein but not against resorufin-labeled casein or BSA. As expected, purified HhoAS237AΔN34, HhoAS237APD, or the elution buffer used as a control showed no proteolytic activity against the three substrates tested (Fig. 2A and B).
FIG. 2.
Proteolytic activities of purified HhoA deletion constructs with various model substrates. In all assays 100 pmol purified HhoA protein was incubated with the substrate for 2 h at 40°C in 50 mM HEPES (pH 8.0) and 20 mM MgCl2. (A) Degradation assays with β-casein and BSA visualized in a Coomassie blue-stained SDS-PAGE gel. A representative gel for three independent replicates is shown. Lane 0, elution buffer; lane 1, HhoAΔN34; lane 2, HhoAS237AΔN34; lane 3, HhoAPD; lane 4, HhoAS237APD. (B) Proteolytic activity against resorufin-labeled casein as determined by absorption at 574 nm. The chemical structure of the resorufin label is shown in the inset. The values are means ± standard deviations (n = 4).
PDZ domain mediates formation of a homohexameric HhoA complex.
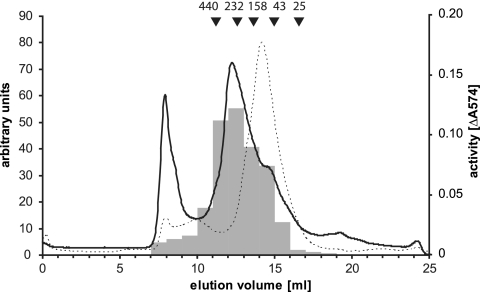
In order to investigate whether Synechocystis sp. strain PCC 6803 HhoA forms an oligomeric complex and to estimate the size of such a complex, HhoAΔN34 was subjected to analytical size exclusion chromatography. Two main peaks of absorption at 280 nm were observed during the elution (Fig. 3). While the first peak contained high-molecular-mass protein aggregates which could not be solubilized, the majority of HhoAΔN34 was eluted as a broad peak at an apparent molecular mass of approximately 245 kDa as calculated from a calibration curve obtained with marker proteins (Fig. 3). This indicated that there was formation of a complex composed of six 41.4-kDa monomeric subunits. Consistent with the observation that the purified HhoAΔN34 fraction contained some self-degradation fragments (Fig. 1B, lane 1), peak 2 was asymmetric and showed some tailing, as well as smaller peaks containing polypeptides with lower apparent molecular masses (Fig. 3). The distribution of the proteolytic activity within the collected fractions assayed with resorufin-labeled casein followed the elution profile of the main peak (Fig. 3). Size exclusion chromatography of the proteolytically inactive form HhoAS237AΔN34 resulted in a more symmetric main peak at roughly the same elution volume (data not shown). The more homogeneous HhoAS237AΔN34 complex was further investigated by dynamic light scattering. The concentrated size exclusion chromatography elution fractions were a monodisperse solution and contained particles with an apparent Stokes radius of 5.7 nm, corresponding to a molecular mass of 228 kDa (data not shown). Analytical size exclusion chromatography of the PDZ domain deletion construct HhoAS237APD revealed a complex with an apparent molecular mass of 80 kDa, which is somewhat smaller than the theoretical molecular mass of 88.2 kDa expected for a trimer formation (Fig. 3). Subsequent analysis of the elution fraction with dynamic light scattering showed that the protein solution was monodisperse and contained particles with a radius of 4.2 nm, corresponding to a molecular mass of 95 kDa (data not shown). This is consistent with a trimer form of HhoAS237APD.
FIG. 3.
Analysis of HhoA complex formation. The elution diagrams for size exclusion chromatography of recombinant HhoAΔN34 (solid line) and HhoAS237APD (dotted line) are shown. For HhoAΔN34, the proteolytic activity of 10 μl of selected fractions against resorufin-labeled casein is shown (gray columns and right axis).
Biochemical characterization of HhoA activity.
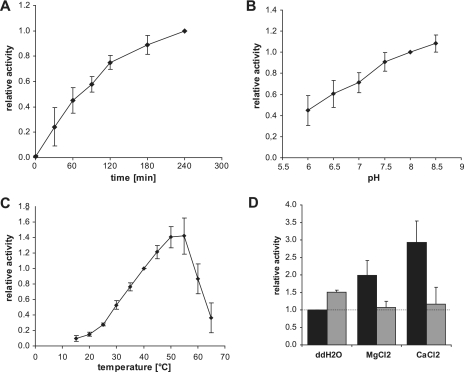
We further characterized the proteolytic activity of HhoAΔN34 with resorufin-labeled casein using different temperatures, buffer conditions, and salt concentrations. First, we tested the degradation kinetics of resorufin-labeled casein to determine when the amount of substrate becomes limiting (Fig. 4A). The assay proceeded in a linear manner for the first 2 h, and therefore we used this incubation time in the subsequent assays. The results obtained were expressed as relative activities normalized to the proteolytic activity of a reference point within the series because the observed proteolytic activity of 100 pmol protease varied somewhat for each independent protein preparation. The pH preference profile showed that HhoAΔN34 was active at all pH values tested, although the activity was twofold higher at pH 8.0 than at pH 6.0 (Fig. 4B). The effect of temperature was more pronounced than the effect of the pH value. At a low temperature, 15°C, HhoA was barely active, and the activity increased gradually with increasing temperature from 25°C up to 50°C and decreased rapidly at temperatures over 55°C, probably due to denaturation of the enzyme (Fig. 4C). We tested the requirement for metal ions for HhoA protease activity. Addition of the divalent cations Mg2+ and Ca2+ stimulated the HhoA activity two- and threefold, respectively (Fig. 4D). This effect could be reversed by addition of an equimolar concentration of EDTA (Fig. 4D). Weak stimulation of the proteolytic activity was also observed when 20 mM EDTA was added to the control reaction mixture, which may be attributed to capture of Ni2+ ions that were eluted with the protein from the Ni2+ affinity chromatography column.
FIG. 4.
Characterization of HhoA protease activity. For all assays 100 pmol purified HhoAΔN34 was incubated with 8 μg resorufin-labeled casein, and the experiment was repeated with at least three independent protein purifications. (A) Degradation kinetics at 40°C in 50 mM HEPES (pH 8.0) and 20 mM CaCl2. The values are means ± standard deviations (n = 4), normalized to the activity after 4 h. (B) Effect of pH. HhoAΔN34 was incubated in 50 mM morpholineethanesulfonic acid (MES) (pH 6.0 to 6.5) or 50 mM HEPES (pH 7.0 to 8.5) supplemented with 20 mM MgCl2 for 2 h at 40°C. The values are means ± standard deviations (n = 7), normalized to the activity at pH 8.0. (C) Effect of temperature. HhoAΔN34 was incubated in 50 mM HEPES (pH 8.0) and 20 mM CaCl2 for 2 h. The values are means ± standard deviations (n = 6) normalized to the activity at 40°C. (D) Effect of divalent cations. HhoAΔN34 was incubated for 2 h at 40°C in 50 mM HEPES (pH 8.0) supplemented with 20 mM MgCl2 or CaCl2 (black bars) and with 20 mM EDTA (gray bars). The values are means ± standard deviations (n = 6 for black bars and n = 3 for gray bars). ddH2O, double-distilled water.
Identification of HhoA substrates.
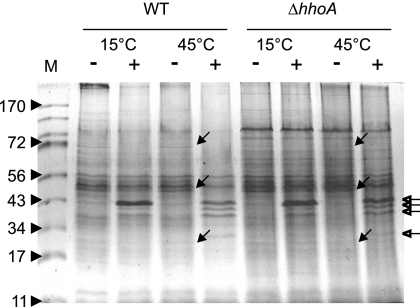
In order to test the substrate specificity of HhoA, we reconstituted purified recombinant HhoAΔN34 with total membrane proteins isolated from Synechocystis strain WT or the deletion mutant ΔhhoA prior to incubation of assay mixtures at two different temperatures. As a control, mock reactions were performed under the same conditions. After separation of proteins by SDS-PAGE, we observed at least three prominent bands at apparent molecular masses of approximately 70, 50, and 22 kDa, as judged from a Coomassie blue-stained gel, which were specifically degraded in samples with added HhoAΔN34 but not in control samples (Fig. 5). The degradation of similar proteins in membrane fractions of WT and ΔhhoA confirmed that the added recombinant HhoA mediated this proteolysis. Unfortunately, our attempt to identify these proteins by mass spectrometry failed.
FIG. 5.
Activity of HhoAΔN34 against isolated total membrane proteins. Isolated total membranes of WT or ΔhhoA mutant cells were incubated without (lanes −) or with (lanes +) recombinant HhoAΔN34 at 15 and 45°C in 50 mM Tris (pH 7.5) supplemented with 20 mM CaCl2, separated by SDS-PAGE, and visualized by Coomassie blue staining. The open arrows indicate bands of added HhoAΔN34 and its degradation fragments, and the filled arrows indicate protein bands specifically degraded by HhoAΔN34. Lane M contained markers.
DISCUSSION
This work provides the first biochemical characterization of the cyanobacterial protease HhoA. We showed that purified recombinant mature Synechocystis sp. strain PCC 6803 HhoA formed proteolytically active homohexameric complexes and degraded unfolded model substrates in vitro. Furthermore, we demonstrated that HhoA degraded selected membrane proteins at elevated temperatures in vitro. This is consistent with the earlier proposed function in protein quality control of the extracytoplasmic compartments, as suggested by studies of Synechocystis sp. strain PCC 6803 mutants (3).
PDZ domain of HhoA is essential for the formation of a homohexameric complex.
A similar function in protein quality control has been described for the well-studied homolog DegP from E. coli (7). However, a comparison of structural and biochemical properties of Synechocystis sp. strain PCC 6803 HhoA with E. coli DegP properties showed some remarkable differences. To date, all published structures of Deg proteases with one PDZ domain have shown trimerization involving interactions of the protease domains (27, 46). Only E. coli DegP, which contains two PDZ domains, showed dimerization of two trimers to form a cage-like hexameric structure (23). HhoA from Synechocystis sp. strain PCC 6803 was proposed to act as a trimer because it did not contain an extended LA loop as judged by protein sequence alignments and modeled best onto a trimeric template structure (17). However, our experimental data demonstrated that recombinant HhoA formed a proteolytically active hexameric complex (Fig. 3) and that the PDZ domain was essential for the dimerization of two trimers. The deletion construct of HhoA lacking the PDZ domain assembled only into a trimer. A similar behavior was also reported for E. coli DegP, which failed to assemble into a hexamer after deletion of either the LA loop or the PDZ2 domain (18). In the same study, the authors demonstrated that also the PDZ1 domain was necessary for hexamer formation, serving as a spacer that allowed the PDZ2 domains to interact with the opposing trimer. Apparently, such a spacer is not necessary for hexamerization of HhoA. In this case, the lack of pillars formed by the LA loops may enable the PDZ domains to interact with the opposing trimer. However, such an HhoA hexamer is expected to have a reduced dimension for the inner chamber. Taken together, these findings suggest that the hexameric HhoA protease complex differs from the complex observed in the crystal structure of E. coli DegP, whose formation was attributed primarily to the protease domain (23).
PDZ domain of HhoA is required for the degradation of sterically challenging substrates.
Here we demonstrated that a Synechocystis sp. strain PCC 6803 HhoA construct lacking the only PDZ domain retained proteolytic activity against β-casein but lost the ability to degrade the sterically more complex resorufin-labeled casein substrate (Fig. 2B). Similarly, deletion constructs of E. coli DegP lacking the PDZ2 domain were found to be impaired for the degradation of resorufin-labeled casein (42) but to be almost as active as the wild type against β-casein (18). DegP deletion constructs lacking the PDZ1 domain or both PDZ domains, however, were proteolytically inactive against both resorufin-labeled casein (38, 42) and β-casein (16). Assuming that the hexameric HhoA complex resembles the structure reported for E. coli DegP, this suggests that hexamerization, which is mediated by the single PDZ domain of Synechocystis sp. strain PCC 6803 HhoA, is necessary for the cleavage of sterically demanding substrates. Similar to the PDZ1 domain in E. coli DegP, the PDZ domain of HhoA may have the additional task of feeding the substrate to the protease domain and/or preventing its premature escape. The constraining cage formed by six protease domains may increase the probability of binding of the sterically hindered substrate to a binding pocket, which would be followed by cleavage of the substrate. In the case of the trimeric HhoAPD the bulky resorufin attached to the side chains of some casein residues would prevent such binding. On the other hand, a sterically less demanding substrate, such as β-casein, may bind to the substrate-binding pocket even in the absence of a constraining cage-like structure and/or without the help of the PDZ domains. The hypothesis that sterically challenging peptides are poor substrates for Deg proteases is additionally supported by the observation that HhoA and HhoAPD could not cleave a range of para-nitroanilide-labeled tetrapeptide substrates commonly used to determine protease substrate specificities (data not shown).
Proteolytic activity of HhoA is temperature dependent.
Our data demonstrated that Synechocystis sp. strain PCC 6803 HhoA showed only residual proteolytic activity at temperatures between 15 and 25°C and that this activity increased almost linearly with increasing temperature up to 50°C (Fig. 4C). It was shown that at lower temperatures, E. coli DegP was proteolytically almost inactive but promoted refolding of chemically denatured MalS (42). This chaperone function was also retained by the catalytically inactive mutant DegPS210A (42). The crystal structure of E. coli DegP, which showed the protein locked in the proteolytically inactive conformation, supported the suggestion that a conformational change is induced by higher temperatures to activate the protease function (23, 42). It is tempting to speculate that Synechocystis sp. strain PCC 6803 HhoA might undergo a similar structural change, but the effect of the temperature-dependent increase in HhoA protease activity may be related to general effects of higher temperatures, such as increased overall reaction rates, increased unfolding of the substrate, and/or increased diffusion of the sterically hindered model substrate into the catalytic chamber.
Conserved general role of HhoA in quality control of protein folding.
Three Deg proteases, designated Deg1, Deg5, and Deg8 (1, 14), which are closely related to Synechocystis sp. strain PCC 6803 HhoA, have been identified in the thylakoid lumen of Arabidopsis thaliana (14, 21). Only one of these three proteases, Deg1, has been characterized in biochemical terms. Comparison of A. thaliana Deg1 with Synechocystis sp. strain PCC 6803 HhoA is particularly interesting in light of the cyanobacterial origin of the chloroplast (31). Like HhoA, A. thaliana Deg1 also possesses only one C-terminally located PDZ domain, forms a hexamer, and degrades unfolded model substrates (6). Deg1 was further suggested to degrade denatured and mistargeted proteins in the thylakoid lumen (6) and to participate in the degradation of photodamaged membrane D1 protein from the reaction center of photosystem II (20). Thus, HhoA and Deg1 appear to perform similar physiological tasks in the quality control of membrane proteins. Comparison of the biochemical properties reported for Deg1 (6) and for Synechocystis sp. strain PCC 6803 HhoA (this study) showed that both enzymes adapted to different environmental conditions. The positive effect of increasing pH on the proteolytic activity of HhoA correlated well with the pH tolerance of Synechocystis sp. strain PCC 6803 cells, which are able to survive under slightly acidic conditions but prefer alkaline conditions for growth, and thus with the pH experienced in the periplasm (24). In contrast, A. thaliana Deg1 was most active at a low pH, pH 6.0, and the activity decreased at a higher pH, which corresponds to the conditions prevalent in the thylakoid lumen (6). HhoA and Deg1 also differ in their responses to stress conditions both on the transcript level and on the protein level. Deg1 was originally discovered as a transiently heat shock-induced protein which showed twofold-higher accumulation at elevated temperatures (15), suggesting that it has a role in the transient heat shock response in higher plants. Northern blot analysis showed no significant increase in hhoA transcript levels (17), and a combined microarray and proteome analysis revealed that 1.8- and 1.28-fold-higher levels of HhoA transcripts and proteins, respectively, accumulated in response to heat shock (43).
We demonstrated that HhoA recognized and cleaved a wide range of substrates, supporting the suggestion that HhoA is a general protease involved in the quality control of extracytoplasmic proteins, including membrane proteins (3). The identification of native substrates of HhoA under different stress conditions and the identification of the substrates of the HtrA and HhoB proteases provide interesting future perspectives for understanding the role of individual enzymes and their combined action in the protein quality control network and their importance for cell survival under stress conditions. Identification of the molecular determinants for the differences observed in the biochemical properties of E. coli DegP, A. thaliana Deg1, and Synechocystis sp. strain PCC 6803 HhoA should provide insights into the adaptation of the protein quality control machinery to changing environmental conditions during evolution.
Acknowledgments
We thank Julia Rottberger for help with the initial stages of the project and Silvia Kuhn for excellent technical assistance. We are grateful to Karsten Schäfer and Alexander Brosig for help with dynamic light scattering data collection and interpretation.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (grant AD92/8-2) and Konstanz University to I.A.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Adam, Z., I. Adamska, K. Nakabayashi, O. Ostersetzer, K. Haußühl, A. Manuell, B. Zheng, O. Vallon, S. R. Rodermel, K. Shinozaki, and A. K. Clarke. 2001. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 125:1912-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma(E) activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 3.Barker, M., R. de Vries, J. Nield, J. Komenda, and P. J. Nixon. 2006. The deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 281:30347-30355. [DOI] [PubMed] [Google Scholar]
- 4.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36-W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavard, D. 1995. Role of DegP protease on levels of various forms of colicin A lysis protein. FEMS Microbiol. Lett. 125:173-178. [DOI] [PubMed] [Google Scholar]
- 6.Chassin, Y., E. Kapri-Pardes, G. Sinvany, T. Arad, and Z. Adam. 2002. Expression and characterization of the thylakoid lumen protease DegP1 from Arabidopsis. Plant Physiol. 130:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 8.Ehrmann, M., and T. Clausen. 2004. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 38:709-724. [DOI] [PubMed] [Google Scholar]
- 9.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 10.Fulda, S., F. Huang, F. Nilsson, M. Hagemann, and B. Norling. 2000. Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 267:5900-5907. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 12.Huang, F., S. Fulda, M. Hagemann, and B. Norling. 2006. Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6:910-920. [DOI] [PubMed] [Google Scholar]
- 13.Huang, F., E. Hedman, C. Funk, T. Kieselbach, W. P. Schroder, and B. Norling. 2004. Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol. Cell Proteomics 3:586-595. [DOI] [PubMed] [Google Scholar]
- 14.Huesgen, P. F., H. Schuhmann, and I. Adamska. 2005. The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 123:413-420. [Google Scholar]
- 15.Itzhaki, H., L. Naveh, M. Lindahl, M. Cook, and Z. Adam. 1998. Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273:7094-7098. [DOI] [PubMed] [Google Scholar]
- 16.Iwanczyk, J., D. Damjanovic, J. Kooistra, V. Leong, A. Jomaa, R. Ghirlando, and J. Ortega. 2007. Role of the PDZ domains in Escherichia coli DegP Protein. J. Bacteriol. 189:3176-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen, T., H. Kidron, H. Taipaleenmaki, T. Salminen, and P. Maenpaa. 2005. Transcriptional profiles and structural models of the Synechocystis sp. PCC 6803 Deg proteases. Photosynth. Res. 84:57-63. [DOI] [PubMed] [Google Scholar]
- 18.Jomaa, A., D. Damjanovic, V. Leong, R. Ghirlando, J. Iwanczyk, and J. Ortega. 2007. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 189:706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 20.Kapri-Pardes, E., L. Naveh, and Z. Adam. 2007. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieselbach, T., and C. Funk. 2003. The family of Deg/HtrA proteases: from Escherichia coli to Arabidopsis. Physiol. Plant. 119:337-346. [Google Scholar]
- 22.Komenda, J., and J. Barber. 1995. Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34:9625-9631. [DOI] [PubMed] [Google Scholar]
- 23.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 24.Kurian, D., K. Phadwal, and P. Maenpaa. 2006. Proteomic characterization of acid stress response in Synechocystis sp. PCC 6803. Proteomics 6:3614-3624. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, W., S. M. Srinivasula, J. Chai, P. Li, J. W. Wu, Z. Zhang, E. S. Alnemri, and Y. Shi. 2002. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9:436-441. [DOI] [PubMed] [Google Scholar]
- 28.Li, X. P., P. Muller-Moule, A. M. Gilmore, and K. K. Niyogi. 2002. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 99:15222-15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtenthaler, H. K. 1987. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148:350-382. [Google Scholar]
- 30.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McFadden, G. I. 2001. Chloroplast origin and integration. Plant Physiol. 125:50-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra, R., M. CastilloKeller, and M. Deng. 2000. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J. Bacteriol. 182:4882-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo, E., S. E. Peters, C. Willers, D. J. Maskell, and I. G. Charles. 2006. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Microb. Pathog. 41:174-182. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 36.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 39.Silva, P., Y. J. Choi, H. A. Hassan, and P. J. Nixon. 2002. Involvement of the HtrA family of proteases in the protection of the cyanobacterium Synechocystis PCC 6803 from light stress and in the repair of photosystem II. Philos. Trans. R. Soc. Lond. B 357:1461-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolenko, A., E. Pojidaeva, V. Zinchenko, V. Panichkin, V. M. Glaser, R. G. Herrmann, and S. V. Shestakov. 2002. The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41:291-310. [DOI] [PubMed] [Google Scholar]
- 41.Spence, E., M. Sarcina, N. Ray, S. G. Moller, C. W. Mullineaux, and C. Robinson. 2003. Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 48:1481-1489. [DOI] [PubMed] [Google Scholar]
- 42.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, I., W. J. Simon, and A. R. Slabas. 2006. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J. Exp. Bot. 57:1573-1578. [DOI] [PubMed] [Google Scholar]
- 44.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 45.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 46.Wilken, C., K. Kitzing, R. Kurzbauer, M. Ehrmann, and T. Clausen. 2004. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483-494. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, R. L., L. L. Brown, D. Kirkwood-Watts, T. K. Warren, S. A. Lund, D. S. King, K. F. Jones, and D. E. Hruby. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]