Abstract
Alcaligenes sp. strain NyZ215 was isolated for its ability to grow on ortho-nitrophenol (ONP) as the sole source of carbon, nitrogen, and energy and was shown to degrade ONP via a catechol ortho-cleavage pathway. A 10,152-bp DNA fragment extending from a conserved region of the catechol 1,2-dioxygenase gene was obtained by genome walking. Of seven complete open reading frames deduced from this fragment, three (onpABC) have been shown to encode the enzymes involved in the initial reactions of ONP catabolism in this strain. OnpA, which shares 26% identity with salicylate 1-monooxygenase of Pseudomonas stutzeri AN10, is an ONP 2-monooxygenase (EC 1.14.13.31) which converts ONP to catechol in the presence of NADPH, with concomitant nitrite release. OnpC is a catechol 1,2-dioxygenase catalyzing the oxidation of catechol to cis,cis-muconic acid. OnpB exhibits 54% identity with the reductase subunit of vanillate O-demethylase in Pseudomonas fluorescens BF13. OnpAB (but not OnpA alone) conferred on the catechol utilizer Pseudomonas putida PaW340 the ability to grow on ONP. This suggests that OnpB may also be involved in ONP degradation in vivo as an o-benzoquinone reductase converting o-benzoquinone to catechol. This is analogous to the reduction of tetrachlorobenzoquinone to tetrachlorohydroquinone by a tetrachlorobenzoquinone reductase (PcpD, 38% identity with OnpB) in the pentachlorophenol degrader Sphingobium chlorophenolicum ATCC 39723.
Nitrophenols, including ortho-nitrophenol (ONP), meta-nitrophenol (MNP), and para-nitrophenol (PNP), have been widely utilized in the pharmaceutical, agricultural, and chemical industries. Due to their stability and high water solubility, these pollutants spread rapidly and then persist in the environment. However, the introduction of large quantities of nitrophenols into the environment has enabled some microorganisms to develop the ability to degrade these compounds. So far, several strains able to degrade nitrophenols have been isolated, including the ONP-degrading strain Pseudomonas putida B2 (42), the MNP-degrading strains P. putida B2 (42) and Ralstonia eutropha JMP 134 (30), and the PNP-degrading organisms Moraxella sp. (34), Pseudomonas sp. strain WBC-3 (20), and Rhodococcus opacus SAO101 (16).
It has been generally accepted that the nitro groups of nitroaromatics are readily removed under aerobic conditions either by partial reduction or via oxidation of the chemical (6, 21). During partial reduction, as illustrated in Pseudomonas pseudoalcaligenes JS45, nitrobenzene is reduced through nitrosobenzene to hydroxylaminobenzene (25). Similar partial reductive metabolism was also observed for the degradation of other nitroaromatics, such as MNP (30), nitrobenzene (28), and chlorinated nitrobenzene (40, 41). In the oxidative pathway, however, the nitro groups are directly removed from the aromatic ring with concomitant nitrite release, catalyzed by nitroarene dioxygenases or monooxygenases. In the case of dioxygenation, two adjacent hydroxyl groups are introduced into an aromatic ring in a single reaction, as reported for the metabolism of 2,4-dinitrotoluene (14, 35), 2-nitrotoluene (10), and nitrobenzene (26). For monooxygenation, a single hydroxyl group is introduced into the aromatic ring, as described for the degradation of ONP (42), PNP (16, 33), and 4-methyl-5-nitrocatechol (8, 14).
Molecular characterization of the initial reactions during the degradation of PNP has recently been reported for R. opacus SAO101, in which genes encoding a two-component PNP/4-nitrocatechol monooxygenase and hydroxyquinol 1,2-dioxygenase were confirmed to be involved in PNP mineralization (16). For the degradation of ONP, a single-component nitrophenol oxygenase was purified from P. putida B2 two decades ago, which converted ONP to catechol with nitrite release (43, 44). However, no amino acid sequence has been determined for this enzyme, and no genetic determinants of any step in the catabolism of ONP have been described. In this study, we report the cloning and sequencing of overlapping fragments covering a continuous 10-kb region from a newly isolated ONP utilizer, Alcaligenes sp. strain NyZ215, and characterization of genes encoding the enzymes involved in the initial steps of ONP catabolism. This fills a gap in our understanding of the genes that encode biodegradation of ONP.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. Alcaligenes sp. strains were grown at 30°C on Luria-Bertani (LB) medium or minimal medium (MM) (19) supplemented with substrates (0.2% yeast extract was added to enhance the biomass when cultures were prepared for enzyme assays), and P. putida PaW340 strains were cultured in LB medium or MM with 1 mg/ml streptomycin (plus 40 μg/ml kanamycin if required) at 30°C. Escherichia coli strains were grown on LB medium at 37°C with 100 μg/ml ampicillin, 40 μg/ml kanamycin, or 100 μg/ml streptomycin as necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Alcaligenes sp. strain NyZ215 | Wild type; ONP+ | This study |
| Alcaligenes sp. strain NyZ216 | Mutant of Alcaligenes sp. strain NyZ215; ONP− | This study |
| P. putida PaW340 | Trp− Strr; plasmid-free derivative of P. putida mt-2, grown on catechol through ortho-cleavage pathway | 13, 39 |
| E. coli DH5α | Host strain | Gibco, BRL |
| E. coli S17-1 λ-pir | Strr; strain for biparental conjugation | 12 |
| Plasmids | ||
| pUC19 | Ampr; cloning and expression vector | Takara |
| pVLT33 | Kmr; RSF1010-lacIq/Ptac hybrid broad-host-range expression vector | 5 |
| pGEM-T | Ampr; vector for cloning Taq polymerase-synthesized PCR fragment | Promega |
| pZWXA | PCR fragment containing onpA cloned into pUC19 | This study |
| pZWXC | PCR fragment containing onpC cloned into pUC19 | This study |
| pZWX33A | PCR fragment containing onpA cloned into pVLT33 | This study |
| pZWX33AB | PCR fragment containing onpAB cloned into pVLT33 | This study |
| pZWXW1 | 5-kb PCR fragment obtained from first cycle of genome walking cloned into pGEM-T | This study |
Cloning of ONP degradation genes and sequence analyses.
Primers C12Of and C12Or (31) based on a conserved region of the catechol 1,2-dioxygenase gene were used to amplify possible genes encoding catechol 1,2-dioxygenase from strain NyZ215. The PCR was run for 30 cycles with an annealing temperature of 48°C. The product was ligated into pGEM-T (Promega) before sequencing. The gene clusters were subsequently isolated by the “genome walking” method (32), starting with the fragments produced by the amplification described above.
DNA sequences were determined by Invitrogen Technologies (Shanghai, China). Analyses of potential open reading frames (ORFs) and comparison of amino acid sequences (or nucleotide sequences) were performed with the ORF finder and BLAST programs on the National Center for Biotechnology Information website. Multiple-sequence alignment and construction of phylogenetic trees (using the neighbor-joining method) were performed with the Mega 3.1 software (17).
RT-PCR.
Total RNA of strain NyZ215 grown on ONP was extracted with an RNeasy mini kit (QIAGEN). The recovered RNA in 10 μl was incubated with 1.5 U of RQ1 RNase-free DNase (Promega) and 20 U of RNase inhibitor (Takara) at 37°C for 30 min to remove any DNA contamination. A reverse transcription-PCR (RT-PCR) was performed by using Moloney murine leukemia virus reverse transcriptase (Promega) and then amplifying the resulting cDNAs for 30 cycles using appropriate primer pairs. The RTA primers (forward, 5′-TTCACAAGCGAGTCAAGGATTT-3′; reverse, 5′-GCCGTCGTGCCAATGTTCTA-3′) amplified an internal 611-bp portion of onpA; the RTB primers (forward, 5′-ATTGTGAGCCTTGAGTTGATGC-3′; reverse, 5′-CGAACGATGCTGTGCGAATAT-3′) amplified an internal 421-bp portion of onpB; the RTC primers (forward, 5′-GTCGCAGCCACCGTTCAATC-3′; reverse, 5′-CACGCTCCTTGACCAACGTG-3′) amplified an internal 374-bp portion of onpC; the RTAB primers (forward, 5′-ACAGCAAGCACGAAAGGATG-3′; reverse, 5′-TTAAAGATGCCACCCGAACG-3′) amplified 438 bp of the onpA-onpB spanning region; and the RTBC primers (forward, 5′-GTCACTCGGGTACTTGAAGGTG-3′; reverse, 5′-GCTTCGTCCATCCGTAGGTC-3′) amplified 529 bp of the onpB-onpC spanning region.
Cloning and expression of onpABC genes in E. coli.
Entire genes, including the Shine-Dalgarno sequence, were amplified from genomic DNA by PCR using the high-fidelity DNA polymerase Pyrobest (Takara). Primer sequences are available upon request. The purified PCR products were treated with restriction enzymes and ligated into similarly treated pUC19 or pVLT33. Subsequently, the resulting plasmids (Table 1) were introduced into E. coli DH5α for expression.
Preparation of cell extracts and enzyme assays.
The method used for preparation of cell extracts described previously (41) was modified slightly so that the cell extracts were resuspended in 20 mM phosphate buffer (pH 7.5). ONP 2-monooxygenase activity was determined spectrophotometrically by measuring the decrease in absorbance at 410 nm due to substrate consumption. The molar extinction coefficients of ONP, 4-methyl-ONP, and 5-fluoro-ONP used were 3.47, 2.80, and 5.07 mM−1 · cm−1, respectively, at a wavelength of 410 nm (43). The standard enzyme assay mixture for ONP 2-monooxygenase contained cell extract, 0.1 mM ONP, 0.4 mM NADPH, and 4 mM Mg2+ in 500 μl (final volume) of phosphate buffer (20 mM, pH 7.5) (43). The extinction coefficient used for calculating NADPH consumption was 6.22 mM−1 · cm−1 at 340 nm. Catechol 1,2-dioxygenase activity was assayed using a previously published method (44), and the extinction coefficient used for the product cis,cis-muconic acid was 16.0 mM−1 · cm−1 at 260 nm (11). One unit of enzyme activity was defined as the amount of enzyme required for the disappearance (or production) of 1 μmol of substrate (or product) per min at 30°C. Specific activities are expressed below in units per gram of protein.
Growth experiment for the derivatives of P. putida PaW340.
Plasmids pVLT33 (5), pZWX33A, and pZWX33AB (Table 1) were used to transform E. coli S17-1 λ-pir (12) for biparental conjugation with P. putida PaW340 as the recipient. The resulting transconjugants were selected on LB medium plates with streptomycin and kanamycin. MM was used for the growth test and was supplemented with tryptophan (20 mg/liter), ONP (0.12 mM), ammonium sulfate (0.5 g/liter), isopropyl-β-d-thiogalactopyranoside (IPTG) (0.2 mM), streptomycin (250 μg/ml), and kanamycin (20 μg/ml). Strain NyZ215, as a positive control, was inoculated into the same medium without IPTG, tryptophan, and antibiotics. Due to the toxicity of ONP to cells, it was supplied by regular addition at a low concentration (0.1 to 0.2 mM) after prior depletion. Cell growth was determined spectrophotometrically by measuring the absorbance at 600 nm at 8- to 12-h intervals over 72 h.
Analytical methods.
After reaction of ONP with cell extracts containing OnpA at 30°C for 25 min, 100 μl of the reaction mixture was mixed with 250 μl methanol and 20 μl ammonium acetate (0.1 M, pH 4.2). The mixture was incubated at 65°C for 5 min and centrifuged at 13,000 × g for 20 min, and the supernatant was analyzed immediately or stored at −20°C. High-performance liquid chromatography (HPLC) analysis was performed at 30°C with a Gilson 715 system equipped with a C18 reversed-phase column (25 cm by 4.6 mm; particle size, 5 μm; Supelco). The mobile phase consisted of methanol (50%) and 0.1 M ammonium acetate (pH 4.2) (50%) at a flow rate of 0.8 ml/min. Both the substrate and the product were monitored at 278 nm using a Gilson 119 UV/VIS detector. Under these conditions, authentic catechol and ONP had retention times of 5.1 and 10.3 min, respectively.
Nitrite was assayed as described elsewhere (18), and protein concentrations were determined by the Bradford method (2) with bovine serum albumin as the standard.
Nucleotide sequence accession numbers.
The sequences of the 1,409-bp 16S rRNA gene, the 10,152-bp ONP degradation gene cluster, and the 7,145-bp chlorocatechol degradation gene cluster from Alcaligenes sp. strain NyZ215 have been deposited in the GenBank database under accession numbers EF540877, EF547253, and EF544605, respectively.
RESULTS
Isolation and characterization of ONP-degrading strain NyZ215.
After several weeks of enrichment, an ONP-degrading bacterial strain, designated NyZ215, was isolated with ONP as the sole source of carbon, nitrogen, and energy from activated sludge in a nitrophenol-manufacturing factory in China. The characteristics of this bacterium included buff, smooth colonies and gram-negative coccal rods. This isolate was identified as an Alcaligenes sp. based on its characteristics and sequence analysis of its 16S rRNA gene.
Accumulation of nitrite in the media during the degradation of ONP indicated that strain NyZ215 had an oxidative pathway for ONP mineralization similar to the pathway in Comamonas sp. strain JS765 for degradation of nitrobenzene (26) and the pathway in P. putida B2 for degradation of ONP (42). The cell extracts of strain NyZ215 grown on ONP or LB medium with 0.5 mM ONP had both ONP 2-monooxygenase and catechol 1,2-dioxygenase activities. However, neither of these activities was detected in cell extracts from the strain grown on LB medium or LB medium with 1 mM catechol. This indicated that the expression of these enzymes was induced by ONP but not by catechol. Strain NyZ215 did not grow on 2-chlorophenol, the isomers MNP and PNP, or the probable catabolic intermediate catechol.
Intriguingly, a spontaneous mutant of strain NyZ215, designated Alcaligenes sp. strain NyZ216, was obtained during repetitive culturing in LB medium. Despite possessing the same morphological characteristics and 16S rRNA gene sequence as strain NyZ215, this mutant was not able to degrade ONP. PCR diagnostic assays, carried out after the identification of ONP-degrading genes, indicated that strain NyZ216 had lost the onpABC genes (data not shown). This implied that the onpABC cluster was involved in the degradation of ONP and located on an independent element of the genome (see below for functions of onpABC).
Cloning and sequence analyses of the ONP degradation gene cluster from strain NyZ215.
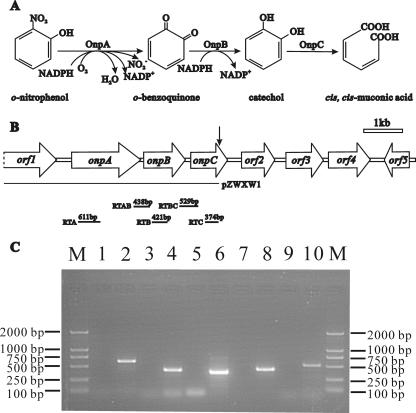
A pair of primers based on a conserved region of the catechol 1,2-dioxygenase gene was initially employed to amplify a PCR product with an anticipated size of 280 bp. To our surprise, two different DNA sequences were found in this product, and they were found to be parts of genes encoding catechol 1,2-dioxygenase and chlorocatechol 1,2-dioxygenase after sequencing. Given that the coexpression of ONP 2-monooxygenase and catechol 1,2-dioxygenase was induced only by ONP in strain NyZ215, it was considered likely that the encoding genes are located in the same operon and physically close. Therefore, the genome walking method for both directions was used to acquire the gene clusters flanking the two different 280-bp sequences. In this way, a 10,152-bp DNA fragment around the catechol 1,2-dioxygenase gene and a 7,145-bp DNA fragment around the chlorocatechol 1,2-dioxygenase gene were obtained in parallel after several cycles of genome walking. From the 10,152-bp fragment, seven complete ORFs were deduced (Fig. 1B). Three of these ORFs, onpA, onpB, and onpC, are transcribed in the same direction. OnpA is similar to salicylate hydroxylases from Pseudomonas stutzeri AN10 (1) (accession no. AAD02157) and P. putida S-1 (38) (accession no. BAA61829), but the level of identity is low (26%). OnpB is similar to the reductase subunit of vanillate O-demethylases from Pseudomonas fluorescens BF13 (3) (accession no. CAB93957) and Pseudomonas sp. strain HR199 (29) (accession no. O05617), with 54 and 48% identity. OnpC is similar to catechol 1,2-dioxygenases from Frateuria sp. strain ANA-18 (24) (accession no. BAA75211) and Acinetobacter lwoffii K24 (15) (accession no. O33950), with 76 and 78% identity, respectively. Notably, the T-vector-based construct pZWXW1 containing a ca. 5-kb fragment (Fig. 1B), obtained after the first cycle of genome walking, was able to convert ONP to a black product which was probably due to oxidation of catechol (22), with concomitant nitrite release. This suggested that this fragment did contain functional genes involved in the transformation of ONP. Subsequent sequencing of this fragment revealed that the onpAB genes were indeed present in this construct.
FIG. 1.
(A) Proposed initial reactions of ONP catabolism catalyzed by the onpABC gene products in vivo. (B) Organization of the ONP gene cluster involved in the initial reactions of ONP degradation in Alcaligenes sp. strain NyZ215. The arrows indicate the sizes and directions of transcription of each ORF or gene. The arrow above onpC indicates the region where the genome walking started in both directions. Plasmid pZWXW1 was constructed by cloning the 5-kb fragment from the first cycle of genome walking into T-vector. E. coli containing this plasmid exhibited ONP 2-monooxygenase activity. The locations of primer sets RTA, RTB, RTC, RTAB, and RTBC and the amplified DNA fragments for RT-PCR are indicated. (C) Analysis of onpABC transcription by RT-PCR. Total RNAs of ONP-grown strain NyZ215 were prepared for RT-PCR, and reactions performed without RT were used as negative controls. Lanes M, molecular size markers; lanes 2, 4, 6, 8, and 10, products amplified using the RTA, RTB, RTC, RTAB and RTBC primer sets, respectively, with products of RT; lanes 1, 3, 5, 7, and 9, corresponding negative controls.
In contrast, the DNA sequence at positions 1145 to 7022 of the 7,145-bp fragment was almost identical (99% identity, with no gaps) to that of a known chlorocatechol degradation gene cluster from P. putida B13 (accession no. AJ617740) (7). This gene cluster codes for a LysR family transcriptional regulator (ClcR), chlorocatechol 1,2-dioxygenase (ClcA), chloromuconate cycloisomerase (ClcB), and dienelactone hydrolase (ClcD), indicating that the 7,145-bp gene cluster is associated with chlorocatechol degradation.
OnpA catalyzes ONP transformation.
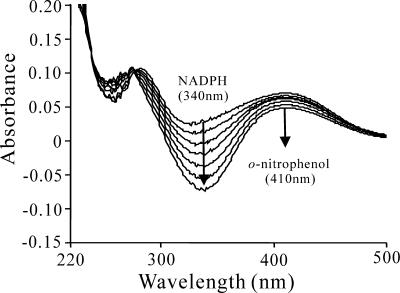
Cell extracts of E. coli DH5α(pZWXA) carrying onpA were found to contain ONP 2-monooxygenase with a specific activity of 44.6 U/g against ONP. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the same cell extracts revealed elevated levels of a ∼60-kDa polypeptide (data not shown), as expected from the deduced amino acid composition. Figure 2 shows the degradation of ONP (λmax, 410 nm) by the cell extracts, together with consumption of NADPH (λmax, 340 nm), as described originally by Zeyer and Kocher (43). ONP 2-monooxygenase activity (57.3 U/g) was also detected in the cell extracts of strain NyZ215 grown on ONP. OnpA exhibited extended substrate specificity for available alkyl and halogenated ONPs (Table 2) but not for the isomers PNP and MNP, salicylate, or vanillate. ONP 2-monooxygenases from E. coli containing onpA and strain NyZ215, as shown in Table 2, had similar relative activities with different substrates or under different conditions, which clearly demonstrated that the onpA gene cloned codes for the native form of ONP 2-monooxygenase in the wild-type strain.
FIG. 2.
Spectral changes associated with the transformation of ONP by OnpA. Sample and reference cuvettes contained 0.1 mM NADPH, 4 mM Mg2+, and cell extracts containing OnpA (about 10 μg/ml) in 500 μl of 20 mM phosphate buffer (pH 7.5). Reactions were started by addition of ONP (at a final concentration of 0.02 mM) to the sample cuvettes, and spectra were recorded every 3 min.
TABLE 2.
Relative activities of the wild-type and recombinant ONP 2-monooxygenases under various conditions or with different substrates
| Conditions or substrate | Relative activity (%)
|
|
|---|---|---|
| Wild type | Recombinant | |
| Conditions for assay with ONPa | ||
| Cell extract + NADPH | 16 | 11 |
| Cell extract + NADPH + Mgb | 100 | 100 |
| Cell extract + NADPH + Mg + FAD | 108 | 108 |
| Cell extract + NADH + Mg | 8 | 8 |
| Cell extract + NADPH + Mg (Tris-Cl buffer) | 73 | 63 |
| Substrates used for assayc | ||
| ONPb | 100 | 100 |
| 4-Methyl-2-nitrophenol | 109 | 107 |
| 5-Fluoro-2-nitrophenol | 17 | 20 |
Most assay mixtures contained ONP (0.1 mM) and cell extract (about 50 μg/ml) in phosphate buffer (20 mM, pH 7.5); the exception was the mixture containing Tris-Cl buffer (50 mM, pH 7.5). The concentrations of other components were as follows: NAD(P)H, 0.4 mM; Mg2+, 4 mM; and flavin adenine dinucleotide (FAD), 0.05 mM.
The activity determined in the enzyme assay (with about 50 μg/ml cell extract, 0.4 mM NADPH, and 4 mM Mg2+ in 500 μl [final volume] of 20 mM phosphate buffer [pH 7.5]) using 0.1 mM ONP was defined as 100%. The specific activities of cell extracts containing wild-type and recombinant ONP 2-monooxygenases were 57.3 and 44.6 U/g, respectively.
All assay mixtures contained 0.1 mM substrate, 4 mM Mg2+, 0.4 mM NADPH, and cell extract (about 50 μg/ml) in phosphate buffer (20 mM, pH 7.5).
Spectrophotometric analysis also revealed that 2.28 mol NADPH was oxidized for each 1 mol of ONP metabolized, which is similar to the oxidation of ONP by nitrophenol monooxygenase from strain B2 (43) or the oxidation of PNP by extracts from Moraxella sp. (33). The product of degradation of ONP was confirmed by HPLC to be catechol since it had the same retention time as authentic catechol. Quantitative analysis indicated that 0.93 mol catechol and 1.04 mol nitrite were produced for each 1 mol of ONP consumed, confirming the stoichiometry of catechol production and nitrite release from ONP degradation. However, there was no evident impact on the conversion of ONP to catechol when cell extracts containing OnpB together with OnpA were added in the assay (data not shown).
OnpC catalyzes the ortho cleavage of catechol and sequential transformation of ONP by OnpAC in vitro.
Catechol 1,2-dioxygenase activity was observed in cell extracts of both strain NyZ215 grown on ONP and E. coli DH5α(pZWXC) containing onpC, with specific activities of 166 and 359 U/g, respectively. No catechol 2,3-dioxygenase activity was detected in either case. During the transformation of ONP by OnpA, an evident isosbestic point appeared in the vicinity of 270 nm, as shown in Fig. 2, indicating formation of a new product which was identified as catechol by HPLC (see above). Cell extracts containing OnpC were subsequently added to the same mixture to allow the sequential reaction to occur. It was found that the product formed from ONP by OnpA could serve as the substrate for OnpC, and the absorption at 260 nm increased, apparently due to the conversion of catechol to cis,cis-muconic acid (11). Based on these observations, it is reasonable to conclude that the product obtained from OnpA was catechol and ONP could be degraded sequentially by ONP 2-monooxygenase and catechol 1,2-dioxygenase in vitro via the same pathway described for strain B2 (42).
Transcription of onpABC in strain NyZ215.
To determine whether onpABC were transcribed in strain NyZ215, RT-PCR was carried out using RNA from NyZ215 grown on ONP as the template. Five amplified DNA products with expected sizes were observed on the agarose gel, as shown in Fig. 1C. However, these products were not evident when RNA from LB medium-grown NyZ215 was used, and they were not always reproducible in three independent experiments (data not shown). This suggested that onpA, onpB, and onpC were transcribed in the presence of ONP and they were located in a single operon.
OnpAB confers on P. putida PaW340 the ability to grow on ONP.
Due to its ability to grow on catechol through the ortho-cleavage pathway (13, 39), P. putida PaW340 was chosen as the recipient for reconstruction of an ONP catabolic pathway by introduction of plasmid pZWX33AB containing onpAB or pZWX33A containing only onpA via conjugation. Both constructs had evident ONP 2-monooxygenase activity in E. coli DH5α or P. putida PaW340. The results showed that the transconjugant containing onpAB was able to grow on ONP within 72 h but the transconjugant with onpA was unable to grow. This implied that onpB also played an important role in the degradation of ONP in vivo.
DISCUSSION
The degradation pathway for ONP and characterization of ONP monooxygenase were reported by Zeyer et al. in the 1980s (42-44). However, studies at the genetic level have not been reported previously. In this study, we cloned and characterized the genes involved in the initial steps of ONP degradation from a newly isolated strain, Alcaligenes sp. strain NyZ215. The single-component ONP 2-monooxygenase encoded by onpA converts ONP to catechol, probably via o-benzoquinone which is spontaneously reduced in the presence NADPH in vitro, whereas o-benzoquinone reductase encoded by onpB, an enzyme that was not described in the previous study of ONP degradation by Zeyer et al. (42-44), may catalyze this reduction in vivo. onpC encodes catechol 1,2-dioxygenase for conversion of catechol to cis,cis-muconic acid. Therefore, the pathway is the same as that previously reported, and the corresponding genes are indicated in Fig. 1A and B.
Salicylate hydroxylase of P. stutzeri AN10 (1) or P. putida S-1 (38) shares the strongest similarity with OnpA, but the levels of identity are quite low for both of them (26%) and OnpA does not exhibit salicylate 1-monooxygenase activity under the conditions used for either the ONP 2-monooxygenase assay or the salicylate 1-monooxygenase assay (1). OnpA (558 amino acids) is also significantly larger than salicylate 1-monooxygenases (around 400 amino acids). Analysis of structural motifs using the software available at http://www.sanger.ac.uk/Software/Pfam suggested that in addition to a flavin adenine dinucleotide-binding domain between positions 1 and 387 common to both enzymes, OnpA has a Cyt-b5 domain between positions 482 and 555, which is not found in salicylate 1-monooxygenase. Of the available sequences in the GenBank database, there is only one homolog with more than 30% identity to OnpA. This is a 563-amino-acid putative monooxygenase (accession no. YP_168356) with an unknown function annotated from the genome sequence of Silicibacter pomeroyi DSS-3 (23), and it exhibits 63% identity to OnpA and also has a putative Cyt-b5 domain. However, the genes adjacent to the gene coding for this putative protein are completely different from the ONP degradation genes in strain NyZ215.
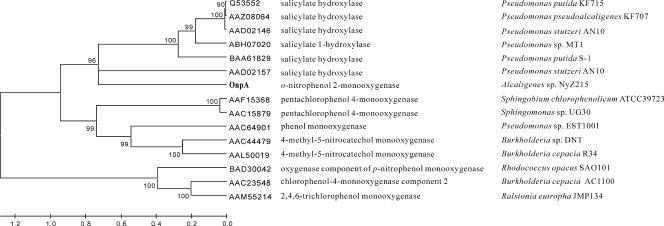
However, it seems that the OnpA of strain NyZ215 and the nitrophenol oxygenase of P. putida B2 share many features, including protein size, substrate range, and metal ions that are stimulators of activity (data not shown). Unfortunately, it is impossible to compare OnpA to the nitrophenol oxygenase from strain B2 at the sequence level as its sequence is still unavailable. To date, there are two other nitroarene monooxygenases that have been genetically characterized: the two-component PNP monooxygenase (NpcAB) of R. opacus SAO101 (16) and the single-component 4-methyl-5-nitrocatechol monooxygenases (DntB) of Burkholderia sp. strain DNT (9) and Burkholderia cepacia R34 (14). However, there is no significant protein sequence similarity among OnpA, NpcA (the oxygenase component), and DntB. A phylogenetic tree of nitroarene monooxygenases also indicates that OnpA, NpcA, and DntB are located in three different branches (Fig. 3). This implies that they evolved from different origins. In contrast, the nitroarene dioxygenases, including nitrotoluene dioxygenase from Pseudomonas sp. strain JS42 (27) and 2,4-dinitrotoluene dioxygenase from B. cepacia R34 (14), all appear to have evolved from naphthalene dioxygenase encoded by the nag operon in Ralstonia sp. strain U2 (46), as suggested previously (14, 45).
FIG. 3.
Phylogenetic relationships of ONP 2-monooxygenase and related proteins having known functions based on the results of a BLASTP search of the GenBank database. Construction of the phylogenetic tree (using the neighbor-joining method) and multiple-sequence alignment were performed with the Mega 3.1 software. Gap and gap length penalties were set at 10. The bootstrap confidence limits are indicated at the nodes, and the scale at the bottom indicates sequence divergence.
OnpB shows about 50% identity to the reductase subunit of the two-component vanillate O-demethylase (VanB) from P. fluorescens BF13 (3) and vanillate O-demethylase oxidoreductase (VanB) of Pseudomonas sp. strain HR199 (29). It also exhibits 38% identity to PcpD, a tetrachlorobenzoquinone reductase which catalyzes the reduction of tetrachlorobenzoquinone to tetrachlorohydroquinone in the pentachlorophenol (PCP) degrader Sphingobium chlorophenolicum ATCC 39723 (4). As with VanBs and PcpD, the software at http://www.sanger.ac.uk/Software/Pfam predicted flavin adenine dinucleotide-binding, NAD-binding, and 2Fe-2S iron-sulfur cluster-binding domains in OnpB. The involvement of OnpB in the degradation of ONP was apparent only when the pathway of ONP degradation had been reconstructed in P. putida PaW340, indicating that OnpB catalyzes a critical step in ONP degradation in vivo, most likely the reduction of o-benzoquinone to form catechol. An analogous example has been reported for PcpD, which may catalyze the reduction of tetrachlorobenzoquinone to tetrachlorohydroquinone, as a mutant lacking functional PcpD was impaired in removal of PCP from the medium and this benzoquinone was found during the degradation of PCP (4). Therefore, OnpB may be a benzoquinone reductase like PcpD rather than the reductase subunit of a two-component monooxygenase like VanBs. However, as the nonenzymatic reduction of o-benzoquinone to catechol occurs very rapidly (within seconds) in the presence of NAD(P)H in vitro (37, 43), the recognition of a reductase activity has been hampered.
The analyses suggest that onpA and onpB are related to salicylate degradation and vanillate degradation, respectively. Thus, this is a further example of an operon encoding xenobiotic biodegradation that is assembled from existing catabolic genes. The presence of putative integrase (ORF1) and transposase (ORF2) genes flanking onpABC implies that a horizontal gene transfer event may have occurred. In addition, onpABC are found in a compact operon (Fig. 1B) with an organization similar to that of the PNP degradation gene cluster in R. opacus SAO101 (16) but different from that of 2,4-dinitrotoluene degradation genes in Burkholderia sp. strain DNT (36) or B. cepacia R34 (14), in which genes encoding the initial reactions are dispersed. Therefore, this compact and functional genetic organization provides the potential for efficient transfer to other hosts, as demonstrated by conversion of strain PaW340 into an ONP utilizer in this study.
Acknowledgments
This work was supported by the Knowledge Innovation Program of the Chinese Academy of Sciences (grant KSCX2-YW-G-009) and by grants 30570021 and 30230010 from National Natural Science Foundation of China.
We are grateful to David Leak at Imperial College London for editing the manuscript and constructive suggestions.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Bosch, R., E. R. Moore, E. Garcia-Valdes, and D. H. Pieper. 1999. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 181:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Civolani, C., P. Barghini, A. R. Roncetti, M. Ruzzi, and A. Schiesser. 2000. Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 66:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai, M., J. B. Rogers, J. R. Warner, and S. D. Copley. 2003. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD). J. Bacteriol. 185:302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 6.Esteve-Núñez, A., A. Caballero, and J. L. Ramos. 2001. Biological degradation of 2,4,6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 65:335-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillard, M., T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Puhler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigler, B. E., S. F. Nishino, and J. C. Spain. 1994. Biodegradation of 4-methyl-5-nitrocatechol by Pseudomonas sp. strain DNT. J. Bacteriol. 176:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigler, B. E., W. C. Suen, and J. C. Spain. 1996. Purification and sequence analysis of 4-methyl-5-nitrocatechol oxygenase from Burkholderia sp. strain DNT. J. Bacteriol. 178:6019-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigler, B. E., W. H. Wallace, and J. C. Spain. 1994. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol. 60:3466-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayaishi, O., M. Katagiri, and S. Rothberg. 1957. Studies on oxygenases. Pyrocatechase. J. Biol. Chem. 229:905-920. [PubMed] [Google Scholar]
- 12.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeenes, D. J., and P. A. Williams. 1982. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J. Bacteriol. 150:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S. I., S. H. Leem, J. S. Choi, Y. H. Chung, S. Kim, Y. M. Park, Y. K. Park, Y. N. Lee, and K. S. Ha. 1997. Cloning and characterization of two catA genes in Acinetobacter lwoffii K24. J. Bacteriol. 179:5226-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa, W., N. Kimura, and Y. Kamagata. 2004. A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 186:4894-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 18.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H., S. J. Wang, and N. Y. Zhou. 2005. A new isolate of Pseudomonas stutzeri that degrades 2-chloronitrobenzene. Biotechnol. Lett. 27:275-278. [DOI] [PubMed] [Google Scholar]
- 20.Liu, H., J. J. Zhang, S. J. Wang, X. E. Zhang, and N. Y. Zhou. 2005. Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. strain WBC-3. Biochem. Biophys. Res. Commun. 334:1107-1114. [DOI] [PubMed] [Google Scholar]
- 21.Marvin-Sikkema, F. D., and J. A. de Bont. 1994. Degradation of nitroaromatic compounds by microorganisms. Appl. Microbiol. Biotechnol. 42:499-507. [DOI] [PubMed] [Google Scholar]
- 22.Mason, H. S. 1949. The chemistry of melanin. VI. Mechanism of the oxidation of catechol by tyrosinase. J. Biol. Chem. 181:803-812. [PubMed] [Google Scholar]
- 23.Moran, M. A., A. Buchan, J. M. Gonzalez, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, S., A. Takashima, J. Takemoto, S. Takenaka, R. Shinke, and K. Aoki. 1999. Cloning and sequence analysis of two catechol-degrading gene clusters from the aniline-assimilating bacterium Frateuria species ANA-18. Gene 226:189-198. [DOI] [PubMed] [Google Scholar]
- 25.Nishino, S. F., and J. C. Spain. 1993. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishino, S. F., and J. C. Spain. 1995. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol. 61:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 28.Park, H. S., S. J. Lim, Y. K. Chang, A. G. Livingston, and H. S. Kim. 1999. Degradation of chloronitrobenzenes by a coculture of Pseudomonas putida and a Rhodococcus sp. Appl. Environ. Microbiol. 65:1083-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priefert, H., J. Rabenhorst, and A. Steinbuchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schenzle, A., H. Lenke, P. Fischer, P. A. Williams, and H. Knackmuss. 1997. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP 134. Appl. Environ. Microbiol. 63:1421-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sei, K., K. Asano, N. Tateishi, K. Mori, M. Ike, and M. Fujita. 1999. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. J. Biosci. Bioeng. 88:542-550. [DOI] [PubMed] [Google Scholar]
- 32.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spain, J. C., and D. T. Gibson. 1991. Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl. Environ. Microbiol. 57:812-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spain, J. C., O. Wyss, and D. T. Gibson. 1979. Enzymatic oxidation of p-nitrophenol. Biochem. Biophys. Res. Commun. 88:634-641. [DOI] [PubMed] [Google Scholar]
- 35.Spanggord, R. J., J. C. Spain, S. F. Nishino, and K. E. Mortelmans. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suen, W. C., and J. C. Spain. 1993. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J. Bacteriol. 175:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, K., T. Gomi, and E. Itagaki. 1991. Intermediate and mechanism of hydroxylation of o-iodophenol by salicylate hydroxylase. J. Biochem. (Tokyo) 109:791-797. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki, K., M. Mizuguchi, K. Ohnishi, and E. Itagaki. 1996. Structure of chromosomal DNA coding for Pseudomonas putida S-1 salicylate hydroxylase. Biochim. Biophys. Acta 1275:154-156. [DOI] [PubMed] [Google Scholar]
- 39.Williams, P. A., and K. Murray. 1974. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120:416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, J. F., C. Y. Jiang, B. J. Wang, Y. F. Ma, Z. P. Liu, and S. J. Liu. 2006. Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl. Environ. Microbiol. 72:1759-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao, Y., J. F. Wu, H. Liu, S. J. Wang, S. J. Liu, and N. Y. Zhou. 2006. Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73. Appl. Microbiol. Biotechnol. 73:166-171. [DOI] [PubMed] [Google Scholar]
- 42.Zeyer, J., and P. C. Kearney. 1984. Degradation of o-nitrophenol and m-nitrophenol by a Pseudomonas putida. J. Agric. Food Chem. 32:238-242. [Google Scholar]
- 43.Zeyer, J., and H. P. Kocher. 1988. Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho-nitrophenol to catechol and nitrite. J. Bacteriol. 170:1789-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeyer, J., H. P. Kocher, and K. N. Timmis. 1986. Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2. Appl. Environ. Microbiol. 52:334-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, N. Y., J. Al Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, N. Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]