Abstract
Pseudomonas putida KT2440(pWW0) can use toluene via the TOL plasmid-encoded catabolic pathways and can use glucose via a series of three peripheral chromosome-encoded routes that convert glucose into 6-phosphogluconate (6PG), namely, the glucokinase pathway, in which glucose is transformed to 6PG through the action of glucokinase and glucose-6-phosphate dehydrogenase. Alternatively, glucose can be oxidized to gluconate, which can be phosphorylated by gluconokinase to 6PG or oxidized to 2-ketogluconate, which, in turn, is converted into 6PG. Our results show that KT2440 metabolizes glucose and toluene simultaneously, as revealed by net flux analysis of [13C]glucose. Determination of glucokinase and gluconokinase activities in glucose metabolism, gene expression assays using a fusion of the promoter of the Pu TOL upper pathway to ′lacZ, and global transcriptomic assays revealed simultaneous catabolite repression in the use of these two carbon sources. The effect of toluene on glucose metabolism was directed to the glucokinase branch and did not affect gluconate metabolism. Catabolite repression of the glucokinase pathway and the TOL pathway was triggered by two different catabolite repression systems. Expression from Pu was repressed mainly via PtsN in response to high levels of 2-dehydro-3-deoxygluconate-6-phosphate, whereas repression of the glucokinase pathway was channeled through Crc.
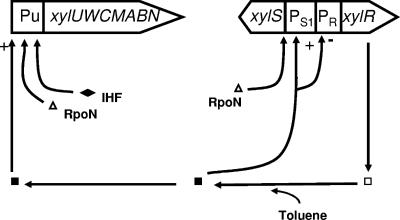
The Pseudomonas putida pWW0 TOL plasmid encodes the catabolic pathways for the mineralization of toluene and xylenes (4). Figure 1 shows the genetic organization of the catabolic operons. The chief regulator involved in the transcriptional control of TOL plasmid catabolic pathways in cells growing on aromatic hydrocarbons is XylR. This regulator drives transcription from the Pu promoter in front of the xylUWCMABN genes, which constitute the upper operon, for conversion of toluene and p- and m-xylenes into the corresponding benzoates, as well as from the Ps1 promoter to increase expression of the xylS gene (Fig. 1). This, in turn, results in the induction of the meta operon for the oxidation of benzoates into Krebs cycle intermediates. Transcription from Pu and Ps1 is mediated by RNA polymerase with RpoN, also known as sigma-54 (37, 39).
FIG. 1.
TOL plasmid promoters under control of the XylR protein. The xylR gene, expressed from two overlapping PR promoters, yielded an inactive XylR protein (□) which, in the presence of toluene, became active (▪) and stimulated transcription (+) from Pu and Ps1 while repressing (−) its own synthesis. The alternative RpoN sigma factor participating in transcription of Pu and Ps1 is indicated. IHF has a positive role in the transcription of Pu (37).
Expression from the Pu promoter is repressed by the presence of alternative C sources. The first indication of TOL pathway susceptibility to catabolite repression was the seminal observation by Worsey and Williams (49) that cells grown in batch cultures on a mixture of acetate and m-xylene contained twofold-lower levels of the upper pathway enzymes than cells grown with m-xylene as the sole source of carbon and energy. Definitive proof of catabolite repression was provided by Duetz et al. (12), who showed that o-xylene did not induce expression of the TOL catabolic pathways in continuous cultures growing either at a high rate under nonlimiting conditions (with an excess of all nutrients) or at a low rate in cultures limited in N, P, or S (all such conditions resulted in excess carbon in the medium).
In Enterobacteriaceae cyclic AMP (cAMP) acts as a signal molecule in catabolite repression (36). However, catabolite repression in Pseudomonadaceae does not involve cAMP; in fact, in P. putida and Pseudomonas aeruginosa, cAMP levels are relatively constant regardless of the growth conditions (34, 38, 41). In Pseudomonadaceae catabolite repression seems to integrate different signals instead, a feature that increases the complexity of the system. Up to five different potential regulators have been related to catabolite repression in P. putida, namely, Crc (20, 29, 38), Crp (called Vfr in P. aeruginosa) (43, 48), CyoB (10, 33), RelA (24, 44), and the Pts system (3, 5). The level of expression from the Pu promoter in wild-type cells growing on glucose or gluconate and toluene was one-third the level in cells growing with only toluene (2, 5, 21), and suppression of upper pathway induction in the presence of these carbon sources was proposed to operate through the ptsN gene product. Indeed, in a PtsN-deficient mutant background Pu expression in the presence of glucose was derepressed (5), and Aranda-Olmedo et al. (2) proposed that addition of the repressing carbon source resulted in PtsN preventing XylR binding to its target upstream activator sequences.
We recently found that in P. putida KT2440 6-phosphogluconate (6PG), the key metabolite of the Entner-Doudoroff pathway, was synthesized by three converging pathways; the glucose dehydrogenase (Gcd) route and the glucokinase (Glk)/glucose-6-phosphate dehydrogenase pathway accounted for almost 90% of the 6PG synthesized by the cells, whereas the remaining 10% was synthesized through direct phosphorylation of gluconate in a reaction mediated by gluconokinase (9). Velázquez et al. (46) suggested that 6PG or the products derived from its metabolism acted as signals for glucose repression of Pu. The present study was undertaken to obtain further insight into the phenomenon of glucose repression of the TOL Pu upper pathway operon promoter from the physiological and molecular points of view. We show here that in the wild-type P. putida KT2440 strain and isogenic mutants deficient in the operation of the Gcd and Glk pathways, toluene and glucose affected each other's metabolism in a process that counterbalanced the total amount of carbon assimilated by the cells. Our study showed that repression of toluene degradation by glucose is signaled by 2-dehydro-3-deoxygluconate-6-phosphate and that effective repression requires a functional PtsN protein, whereas toluene repression of glucose metabolism affects the Glk pathway but not the gluconate pathways. The effect of toluene on the Glk route is channeled via the Crc protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
We used P. putida KT2440, a derivative of P. putida mt-2 (1, 31) (Table 1). Isogenic glk (strain PSC278) and gcd (strain M438) mutants of this strain have been described previously (Table 1 and references therein). These strains were grown at 30°C at 200 rpm in 250-ml conical flasks with 20 ml M9 minimal medium supplemented with Fe citrate, MgSO4 and trace metals (1, 13). Glucose (16 mM), toluene (6 mM), or glucose (16 mM) and toluene (6 mM) were used as carbon sources. For 13C-labeling experiments we used a mixture of 20% (wt/wt) [U-13C]glucose (>99% pure; Martek Biosciences Corporation, Columbia, MD) and 80% (wt/wt) natural glucose.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Reference(s) |
|---|---|---|
| P. putida strains | ||
| KT2440 | Wild type, prototroph; Cmr Rifr | 1, 32 |
| M1044b | edd:mini-Tn5-Km Kmr Rifr | 14 |
| M1128b | eda:mini-Tn5-Km Kmr Rifr | 14 |
| M438b | gcd:mini-Tn5-Km Kmr Rifr | 14 |
| PSC278b | glk::pCHESIΩ-Km Rifr | 9 |
| KT2440 ptsN | Kmr; P. putida KT2440 with a kanamycin resistance cassette interrupting the ptsN gene | 3 |
| KT2440 crc | Gmr; P. putida KT2440 with a gentamicin resistance cassette interrupting the crc gene | 3 |
| KT2440 crp | Kmr; P. putida KT2440 with a kanamycin resistance cassette interrupting the crp gene | 3 |
| KT2440 cyoB | Tcr; P. putida KT2440 with a tetracycline resistance cassette interrupting the cyoB gene | 38 |
| KT2440 relA | Kmr; P. putida KT2440 with a kanamycin resistance cassette interrupting the relA gene | 44 |
| KT2440(pWW0r PmΩ Sm) | Smr; P. putida KT2440 with a kanamycin resistance cassette interrupting the Pm gene | 26 |
| KT2440(pWW0::μ 21) | Kmr; P. putida KT2440 with a kanamycin resistance cassette interrupting the xylE gene | 26 |
| Plasmids | ||
| pRK600 | Helper plasmid; tra+ mob+ Cmr | 5, 19 |
| pS10 | IncP1 SmrxylR; transcriptional Pu::lacZ::tet fusion | 3 |
| pWW0 | IncP9 mob+ tra+ 3MB+ | 49 |
Apr, Cmr, GmR, and Kmr, resistance to ampicillin, chloramphenicol, gentamicin, and kanamycin, respectively.
Obtained from the collection of KT2440 mutants available at our institute.
Analytical procedures, physiological parameters, sample preparation, and gas chromatography-mass spectrometry analysis.
The glucose concentration was determined enzymatically with a commercially available kit (Roche Diagnostics). The cell dry weight, the maximum specific growth rate, the biomass yield, and the specific carbon source consumption rate were determined as described previously (9, 17).
To analyze proteinogenic amino acids, cell aliquots were harvested during the mid-exponential growth phase by centrifugation of 5 to 7 ml of culture broth at 4°C, and the pellets were hydrolyzed in 6 M HCl for 24 h at 110°C in sealed 2-ml Eppendorf tubes. Samples were desiccated, treated, and analyzed as described previously (8, 15, 17, 32).
Analysis of metabolic fluxes.
For METAFoR analysis, mass spectra of four derived amino acids (glycine, serine, proline, and glutamate) were considered. These amino acids are synthesized from single metabolic intermediates, and the mass isotopomer distribution vector of these metabolites was derived from the mass isotopomer distribution vector of the amino acids. The values were used to calculate the fractional contribution of the corresponding reaction to the target metabolite pool with a set of algebraic equations implemented in the MATLAB-based program Fiat Flux, version 1.04, as described by Fischer et al. (16).
Preparation of RNA.
The P. putida KT2440 strain was grown overnight in M9 minimal medium with glucose. Cells were then diluted until the turbidity at 660 nm (OD660) was 0.05 in fresh M9 minimal medium without a carbon source, and three aliquots were removed and supplemented with glucose or glucose plus toluene. Samples were then incubated until the culture reached a turbidity at 660 nm of 0.7. Then 15-ml portions of the cultures were harvested by centrifugation at 7,000 × g for 5 min, and total bacterial RNA was isolated exactly as described by Marqués et al. (27). Extracts were treated with RNase-free DNase I (10 U/μl) in the presence of an RNase inhibitor cocktail (40 U/μl RNaseOUT).
P. putida microarrays.
The genome-wide DNA chip used in this work (printed by Progenika Biopharma) was described previously (50). It consists of an array of 5,539 oligonucleotides (50-mers) spotted in duplicate onto γ-aminosilane-treated slides and covalently linked with UV light and heat. The oligonucleotides represent 5,350 of the 5,421 predicted open reading frames annotated in the P. putida KT2440 genome. The chips also contain homogeneity controls consisting of oligonucleotides for the rpoD and rpoN genes spotted at 20 different positions, as well as duplicate negative controls at 203 predefined positions. Preparation and labeling of RNA for hybridization and data analysis were done as described previously (11, 14, 50).
β-Galactosidase assays.
The pS10 plasmid was transformed into P. putida KT2440(pWW0) and isogenic mutants of this strain. Transformants were selected on M9 minimal medium with 5 mM 3-methylbenzoate as the sole carbon source and 10 μg/ml tetracycline. P. putida(pWW0, pS10) was grown on M9 minimal medium with glucose or citrate (16 mM), and when the cultures reached an OD660 of 0.1, they were supplemented or not supplemented with toluene in the gas phase and incubation was continued at 30°C with shaking until the OD660 of the cultures were 0.6 ± 0.1. The β-galactosidase activity was assayed in permeabilized whole cells using Miller's method (28). Assays were done in triplicate and were repeated at least three times.
Preparation of cell extracts and enzyme assays.
Cell batches in 50 ml of minimal medium with glucose, toluene, or glucose plus toluene as the carbon source were harvested by centrifugation at 7,000 × g for 7 min, washed twice, and frozen at −20°C. Cells were disrupted in a French press at 120 MPa. Whole cells and debris were removed by centrifugation at 11,180 × g (45 min, 4°C). The clear supernatant was used as a cell extract. The protein concentrations in cell extracts were determined by the Bradford method, using bovine serum albumin as the standard. Glucokinase and gluconokinase assays were performed at 30°C at 340 nm with a Shimadzu UV-160A spectrophotometer as described previously (22, 45). Specific activities were calculated based on an NAD(P)H extinction coefficient of 6.3 mM−1 cm−1.
RESULTS
Simultaneous utilization of glucose and toluene by P. putida KT2440 and isogenic mutants of this strain.
P. putida KT2440 (Table 1) growing on M9 minimal medium with glucose, toluene, or glucose plus toluene exhibited high growth rates (Table 2). As reported previously, mutants deficient in the synthesis of Glk or Gcd were still able to grow on glucose, but the growth rates were lower (around 0.4 h−1) than those of the parental strain (0.73 ± 0.03 h−1) (Table 2).
TABLE 2.
Growth rates and physiological parameters of P. putida KT2440(pWW0) and isogenic mutants of this strain growing with different carbon sources
| Strain | Growth rate (h−1) with:
|
qGlu (μmol/mg [cell dry wt] h−1)a
|
qtol (μmol/mg [cell dry wt] h−1)b
|
||||
|---|---|---|---|---|---|---|---|
| Glucose | Toluene | Glucose + toluene | Without toluene | With toluene | Without glucose | With glucose | |
| KT2440(pWW0) | 0.73 ± 0.03c | 0.72 ± 0.02 | 0.74 ± 0.07 | 13.1 ± 0.8 | 5.7 ± 0.5 | 11.9 ± 0.5 | 6.4 ± 0.2 |
| KT2440 gcd(pWW0) | 0.42 ± 0.01 | 0.73 ± 0.04 | 0.45 ± 0.01 | 5.0 ± 0.5 | 2.4 ± 0.1 | 12.4 ± 0.1 | 11.4 ± 0.4 |
| KT2440 glk(pWW0) | 0.38 ± 0.06 | 0.72 ± 0.01 | 0.66 ± 0.01 | 5.1 ± 0.5 | 5.9 ± 0.7 | 11.3 ± 0.2 | 5.5 ± 0.3 |
| KT2440 ptsN(pWW0) | 0.67 ± 0.01 | 0.70 ± 0.01 | 0.71 ± 0.18 | NDd | ND | ND | ND |
| KT2440 crc(pWW0) | 0.51 ± 0.1 | 0.65 ± 0.1 | 0.65 ± 0.04 | ND | ND | ND | ND |
Glucose consumption was determined in cells growing in the absence and in the presence of toluene.
Toluene consumption was determined in cells growing in the absence and in the presence of glucose.
The data are averages ± standard deviations of three to six independent assays, each done in duplicate.
ND, not determined.
The growth rates with toluene of mutants deficient in Gcd or Glk were similar to those of the parental strain (around 0.72 ± 0.03 h−1) (Table 2). When the M438 (gcd mutant) and PSC278 (glk mutant) strains were grown on glucose plus toluene, we found that the growth rate of the gcd mutant was similar to that measured with glucose alone, whereas the glk mutant grew at a higher rate with glucose and toluene (0.66 ± 0.01 h−1) than with glucose alone (Table 2). These results suggest that toluene influenced glucose metabolism, particularly in the gcd mutant, in which glucose assimilation operates via the Glk route.
When we examined the rate of glucose consumption (qglu) by the parental strain and isogenic mutants of it growing in the absence and in the presence of toluene, we found that the qglu for wild-type cells growing in the presence of toluene was about one-half the rate with glucose alone (Table 2). We also found that the rate of toluene assimilation (qtol) in the wild-type strain in the presence of glucose dropped by about 47% compared with the rate of hydrocarbon utilization in the absence of the sugar (Table 2). This indicates that the wild-type cells were able to assimilate glucose and toluene simultaneously.
We also examined qglu and qtol in M438 and PSC278 mutant cells. In these mutants the level of uptake of glucose was lower than that in the parental strain, as expected from their reduced growth rates with this sugar. However, although the glucose uptake rate decreased when toluene was present in the culture medium of the gcd mutant (strain M438), no decrease was observed in glk mutant cells (strain PSC278) (Table 2). When qtol in the glk- and gcd-deficient mutants was examined, we found the opposite effect, namely, that in the presence of glucose the rate of toluene consumption was not affected in the Δgcd mutant, whereas it decreased by almost 50% in the glk mutant. Therefore, KT2440 and isogenic mutants of this strain can use glucose and toluene simultaneously in the early steps of glucose assimilation.
Results of net flux analyses support the simultaneous use of glucose and toluene by P. putida.
To learn more about the simultaneous metabolism of glucose and toluene, we decided to carry out assays using 13C-labeled glucose, as described in Materials and Methods. To this end, we grew wild-type and mutant cells in the absence and in the presence of nonlabeled toluene. We focused our attention on four amino acids: serine and glycine, made from 3-phosphoglycerate (Fig. 2), and glutamate and proline, made from the 2-ketoglutarate Krebs cycle intermediate (Fig. 2). In the absence of toluene, as expected, the percentage of labeled amino acids was about 20%, consistent with the amount of 13C supplied to the cultures (Fig. 3). In the wild type in presence of toluene, the ratio of 13C in glycine and serine was almost 20%, in agreement with the fact that 3-phosphoglycerate is synthesized mainly from [13C]glucose. However, the percentage of 13C in glutamate and proline was slightly less than 10%. This is in agreement with the dilution of 13C in the tricarboxylic acid cycle because of feeding with both glucose and toluene (Fig. 3). The proportion of 13C in glutamate and proline suggested that there were similar contributions by glucose and toluene to the Krebs cycle in wild-type P. putida KT2440 cells. These results are in agreement with the rates of glucose and toluene consumption shown in Table 2. It should be noted that in P. putida KT2440 without the TOL plasmid, the percentage of labeled amino acids was around 20% of the total regardless of the presence of toluene, as expected.
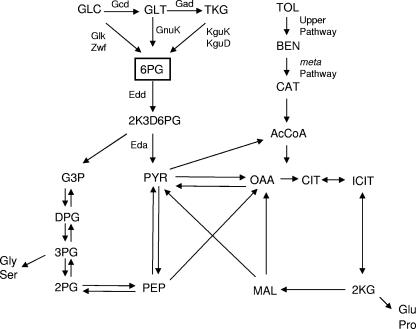
FIG. 2.
Integration of glucose and toluene metabolism into central metabolism in P. putida KT2440. The pathways are based on experimental data reported by Worsey and Williams (49), Velázquez et al. (49), and del Castillo et al. (9). Abbreviations: GLC, glucose; GLT, gluconate; TKG, 2-ketogluconate; 6PG, 6-phosphogluconate; 2K3D6PG, 2-dehydro-3-deoxy-6-phosphogluconate; PYR, pyruvate; PEP, phosphoenolpyruvate; G3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; TOL toluene; BEN, benzoate; CAT, catechol; AcCoA, acetyl coenzyme A; CIT, citrate; ICIT, isocitrate; 2KG, 2-ketoglutarate; MAL, malate; OAA oxaloacetate.
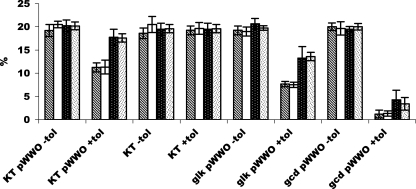
FIG. 3.
13C-labeling patterns in different amino acids derived from [13C]glucose. Bacterial cultures were fed with 20% (wt/wt) [13C]glucose, and 13C levels in serine (hatched bars), glycine (open bars), proline (stippled bars, white dots on black background), and glutamate (stippled bars, black dots on white background) were determined. The strains used are indicated as follows: KT, parental strain; glk pWWO, mutant deficient in glk; and gcd pWWO, mutant deficient in gcd. −tol indicates that toluene was absent, and +tol indicates that toluene was present. The y axis indicates the percentages of 13C/12C in the amino acids.
In the KT2440 gcd and KT2440 glk mutants growing on labeled glucose in the absence of toluene, we obtained about 20% 13C-labeled molecules for the four amino acids tested. This was expected since glucose was the sole C source. However, when toluene was also present, the situation was different than that described above for the wild-type strain. In this case the percentages of labeling in glycine and serine and in glutamate and proline were equivalent, as expected from their common biosynthetic origins. In the PSC278 (glk-deficient) mutant strain the amount of 13C labeling in glycine and serine was 14%, whereas for glutamate and proline it was 8% because of dilution with unlabeled acetyl coenzyme A. In the M438 mutant strain (with the gcd gene knocked out) [13C]glutamate and [13C]proline accounted for only 1% of the total amount, whereas [13C]serine and [13C]glycine accounted for about 4% (Fig. 3). This was interpreted as evidence that toluene provides not only larger amounts of carbon to the Krebs cycle but also an extra flux of carbon necessary to synthesize 3-phosphoglycerate from pyruvate (Fig. 2).
The results described above support the conclusions that glucose and toluene can be assimilated simultaneously and that toluene has a marked negative effect on the Glk pathway, as suggested by our finding that the flux of carbon from glucose in a gcd mutant was significantly limited. This effect was less substantial in the Gcd pathway. Therefore, it seems that toluene has a negative effect on the Glk pathway and a less noticeable effect on the gluconate pathways.
Global transcriptomic response of P. putida KT2440 cells growing with glucose and toluene.
The physiological data reported above indicated that glucose affects the breakdown of toluene and that toluene affects glucose degradation too. To shed light on these effects at the transcriptional level, we decided to carry out a series of assays to compare the transcriptomes of cells growing in the exponential phase with glucose plus toluene and cells growing with glucose alone. A summary of the most relevant results for upregulated genes is shown in Table 3. Tables S1 and S2 in the supplemental material show downregulated genes and all genes identified as genes involved in glucose metabolism, respectively. The most remarkable result was that the sets of genes that formed the upper operon (xylUWCMABN) and the lower operon (xylXYZLTEFGHKT) were induced in response to toluene. The changes varied between 2.93-fold for xylC and 13.43-fold for xylU in the upper operon and between 2-fold for xylF and 4.68-fold for xylZ in the meta operon (Table 3). This indicates that cells induced the set of genes necessary for toluene catabolism. In addition to the TOL plasmid genes for toluene catabolism, we found that another 28 genes were induced more than twofold (Table 3). In agreement with Domínguez-Cuevas et al. (11), among the induced genes were the genes that encoded a number of stress proteins, including the IbpA chaperone (PP1982), two efflux pumps that removed excess solvent (PP3735 and PP3961), proteins involved in oxidative stress (PP3998), a number of proteins for general metabolism, and a number of hypothetical proteins. We also found two genes that encoded regulators, a sensor kinase (PP3413), and a luxR-like regulator (PP3717), all of which were induced.
TABLE 3.
P. putida KT2440 upregulated genes in cells growing on glucose plus toluene compared to cells growing on glucose alone
| Open reading frame and/or gene | Family | Fold changea |
|---|---|---|
| xylU | Probable toluene porin | 13.43 |
| xylW | Probable toluene porin | 6.31 |
| xylA | Toluene monooxygenase | 6.05 |
| xylB | Benzyl alcohol dehydrogenase | 3.05 |
| xylC | Benzaldehyde dehydrogenase | 2.93 |
| xylX | Toluate 1,2-dioxygenase | 2.86 |
| xylZ | Toluate 1,2-dioxygenase | 4.68 |
| xylL | 1,2-Dihydroxy-3,5-cyclohexadiene-1-1-carboxylate | 4.11 |
| xylE | Catechol 2,3-dioxygenase | 2.70 |
| xylF | Hydrolase semialdehyde 2-hydroxymuconic | 2.02 |
| xylG | Dehydrogenase semialdehyde 2-hydroxymuconic | 3.44 |
| xylH | 4-Oxalocrotonate tautomerase | 3.74 |
| xylI | 4-Oxalocrotonate decarboxylase | 2.78 |
| xylK | 4-Hydroxy-2-oxovalerate hydrolase | 3.42 |
| xylM | Toluene monooxygenase | 3.84 |
| xylN | Unknown function | 2.61 |
| xylQ | Acetaldehyde dehydrogenase | 2.48 |
| xylT | Ferredoxine | 2.43 |
| PP0210 | Putative phycobiliprotein | 2.86 |
| PP1074 (glpR) | Glycerol-3-phosphate regulon repressor | 2.79 |
| PP1897 | DNA internalization-related competence protein | 4.36 |
| PP2268 | Phage endodeoxyribonuclease | 2.19 |
| PP2589 | Aldehyde dehydrogenase family protein | 2.52 |
| PP2805 | Monooxygenase flavin-binding family | 2.27 |
| PP3243 | Acetyltransferase GNAT family | 7.38 |
| PP3413 | Sensor histidine kinase/response regulator | 2.38 |
| PP3717 | Transcriptional regulator LuxR family | 2.94 |
| PP3998 | Glutathione S-transferase domain protein | 4.05 |
| PP4538 | Putative acyl carrier protein phosphodiesterase | 3.87 |
| PP4983 | Flavin-containing monamine oxidase family protein | 2.23 |
| PP5340 | Acetylpolyamine aminohydrolase | 2.09 |
| pcaC | 4-Carboxymuconolactone decarboxylase | 2.13 |
| pcaJ | 3-Oxoadipate coenzyme A-transferase, subunit B | 2.15 |
| PP3726 (ech) | Enoyl coenzyme A hydratase/isomerase family protein | 4.91 |
| PP5248 | Hydrolase isochorismatase family | 2.46 |
| PP5255 | Hydrolase isochorismatase family | 2.49 |
| PP1982 (ibpA) | IbpA heat shock protein IbpA | 2.00 |
| PP3735 | ABC transporter ATP-binding protein | 2.00 |
| PP3961 | Putative transporter | 2.00 |
| PP1687 | Hypothetical protein | 2.94 |
| PP2644 | Hypothetical protein | 2.97 |
| PP3353 | Conserved hypothetical protein | 7.04 |
| PP4561 | Conserved hypothetical protein | 2.73 |
| PP4901 | Conserved hypothetical protein | 2.72 |
| PP4981 | Conserved hypothetical protein | 2.02 |
| PP4982 | Conserved hypothetical protein | 2.64 |
| Hypothetical protein pWW0 c57031-56282 | 2.70 | |
| Hypothetical protein pWW0 c65777-65391 | 2.08 | |
| Hypothetical protein pWW0 c94962-94570 | 3.65 | |
| Hypothetical protein pWW0 c66769-66416 | 3.97 |
The changes are averages of at least two assays. The P values were ≤0.05.
The number of downregulated genes was small (11 genes). Four of these genes encoded proteins having unknown functions, five other genes encoded membrane proteins, including an efflux pump (PP3582) (see Table S1 in the supplemental material), and two genes encoded putative general metabolism enzymes.
We specifically analyzed the arrays in detail for glucose metabolism genes (see Table S2 in the supplemental material). Glucose metabolism involves entry of the sugar into the periplasmic space, a process that takes place through specific porins in the outer membrane (OprB porin). Glucose can then be internalized into the cytoplasm via an ABC system for subsequent conversion to glucose-6-phosphate (catalyzed by the Glk enzyme) and 6PG (catalyzed by glucose-6-phosphate dehydrogenase), or alternatively, glucose in the periplasmic space can be oxidized to gluconate and 2-ketogluconate, which are transported to the cytoplasm for conversion into 6PG (Fig. 2). We analyzed the effect of toluene on the different sets of gene clusters involved in glucose metabolism. The set of glucose metabolism genes more affected by the presence of toluene comprised the genes involved in glucose uptake via the ABC glucose transport system, made up of PP1015 through PP1018. This set of genes was repressed between 2- and 2.49-fold. The glk and edd genes, which are part of the same operon, were also repressed almost 2.0-fold. In contrast, the set of genes for the gluconate loops was affected little, if at all (see Table S2 in the supplemental material). Therefore transcriptomic analysis revealed that the catabolism of glucose through the glucokinase pathway is influenced by toluene catabolism, whereas the gluconate loop pathways are not sensitive to the presence of hydrocarbons.
Toluene affects the level of glucokinase activity in cells growing on glucose and toluene.
The physiological and transcriptomic results reported above led us to measure glucokinase activity and gluconokinase activity in wild-type cells growing in the absence and in the presence of toluene. We found that in cells growing on toluene, gluconokinase and glucokinase activities were low, whereas in cells growing on glucose both enzymes were induced. This is in agreement with our previous results (9). The level of glucokinase in wild-type cells growing on glucose and toluene was almost 60% of the level in cells growing on glucose alone (Table 4). No significant effect on gluconokinase activity was observed (not shown). Therefore, the biochemical data support the pattern deduced from our transcriptomic analysis.
TABLE 4.
Glucokinase activities in wild-type and mutant cells
| Strain | Substrate(s) | Glucokinase activity (nmol/min/mg protein)a |
|---|---|---|
| KT2440(pWW0) | Glucose | 27.3 ± 1.9 |
| KT2440(pWW0) | Glucose + toluene | 16.4 ± 1.9 |
| KT2440(pWW0) | Toluene | 1.3 ± 0.3 |
| KT2440 pstN(pWW0) | Glucose | 30.2 ± 1.4 |
| KT2440 pstN(pWW0) | Glucose + toluene | 16.2 ± 2.7 |
| KT2440 pstN(pWW0) | Toluene | 0.9 ± 0.1 |
| KT2440 crc(pWW0) | Glucose | 27.3 ± 5.4 |
| KT2440 crc(pWW0) | Glucose + toluene | 27.6 ± 1.7 |
| KT2440 crc(pWW0) | Toluene | 1.4 ± 0.2 |
| KT2440(pWW0 Δ Pm) | Glucose | 26.3 ± 0.3 |
| KT2440(pWW0 Δ Pm) | Glucose + toluene | 26.7 ± 0.4 |
| KT2440(pWW0 Δ Pm) | Toluene | 1.5 ± 0.2 |
The data are the averages ± standard deviations of at least three independent determinations done in triplicate.
Identification of the glucose metabolite(s) involved in toluene repression.
Wild-type P. putida cells, as well as cells of the gcd and glk mutants bearing the TOL plasmid, were transformed with pS10, a low-copy-number Tcr Smr plasmid bearing a Pu:lacZ fusion and xylR (2). Cells were grown on M9 minimal medium with glucose, toluene, and toluene plus glucose as described in Materials and Methods. In wild-type cells, basal levels of β-galactosidase activity were found when cells were grown on glucose or citrate, and expression was about 20-fold higher in cells growing on toluene (Table 5). The β-galactosidase levels were only 30% of the maximal induced levels when they were measured in wild-type cells growing on glucose plus toluene. In the glk and gcd mutants, the β-galactosidase activity in cells growing on glucose plus toluene was lower than that in cells growing on toluene alone (Table 5). Therefore, this set of results supports previous studies that showed that glucose has a marked effect on toluene metabolism in P. putida.
TABLE 5.
Expression of the Pu promoter fused to lacZ in the wild type and glucose mutants growing under different conditionsa
| Strain | β-Galactosidase activity (Miller units) with:
|
||
|---|---|---|---|
| Glucose | Toluene | Glucose + toluene | |
| KT2440(pWW0) | 310 ± 20 | 6,620 ± 150 | 2,060 ± 40 |
| KT2440 gcd(pWW0) | 185 ± 15 | 6,035 ± 125 | 4,710 ± 700 |
| KT2440 glk(pWW0) | 400 ± 30 | 6,775 ± 100 | 3,870 ± 140 |
| KT2440 ptsN(pWW0) | 450 ± 20 | 8,100 ± 200 | 8,550 ± 190 |
| KT2440 crc(pWW0) | 380 ± 40 | 7,700 ± 500 | 5,680 ± 300 |
The strains used were transformed with pS10, which carries a Pu:′lacZ fusion and the xylR gene. Assays were done in duplicate, and the data are the averages ± standard deviations of at least three independent assays.
Several research groups (9, 46, 47) have shown that P. putida mutants with mutations in the edd or eda genes cannot grow on glucose because the breakdown of 6PG into central metabolites is blocked (Fig. 2). To determine whether glucose or one of its metabolites was responsible for the catabolite repression described above, we decided to grow wild-type and edd and eda mutant cells on citrate and then transfer cells to minimal medium with toluene, glucose, or glucose plus toluene. After incubation for 30 min we measured β-galactosidase activity and found that it had increased from negligible levels to almost 3,000 Miller units (Fig. 4) in the parental strain and 5,000 to 7,000 Miller units in the edd and eda mutants exposed to only toluene. We also found that with glucose plus toluene the level of β-galactosidase activity in the parental strain or in the eda mutant was about 10 to 20% of the maximal activity measured with toluene alone, whereas in the edd mutant the level was close to 90% (Fig. 4). This suggests that the 2-dehydro-3-deoxygluconate-6-phosphate (Fig. 2) that accumulates in the eda mutant cells, not glucose or 6PG, is the true catabolite repressor signal.
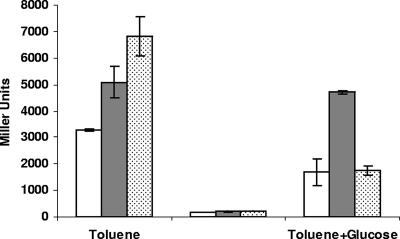
FIG. 4.
Induction of Pu in parental strain P. putida KT2440 and edd and eda mutants of this strain. Bacterial cells were grown on citrate as the sole C source, and when cultures reached the mid-exponential phase (OD660, 0.7 ± 0.1), cells were harvested, washed in M9 minimal medium without a C source, and divided into three aliquots that were supplemented with glucose, toluene, or glucose plus toluene. β-Galactosidase levels were determined 30 min later. The data are the averages and standard deviations of three independent assays. Open bars, P. putida KT2440(pS10); gray bars, edd mutant containing pS10; dotted bars, eda mutant.
To find out whether toluene or its metabolites were responsible for the effect observed on glucose assimilation, we carried out a series of assays with TOL mutants that convert toluene into benzoate (pWW0 ΔPm) or that are blocked at the level of 2,3-dioxygenase (TOL μ21) (Table 1). We found that with these TOL plasmid mutants, in which toluene does not serve as a carbon source, the glucokinase activity levels in cultures with toluene and glucose did not change compared with the levels in cells growing on glucose alone (see Table 4 for data for pWW0 ΔPm). Therefore, we concluded that utilization of the aromatic hydrocarbon as a carbon source is required for toluene to affect glucose metabolism.
Identification of master regulators involved in crossed repression of the toluene and glucose degradation genes.
It was previously shown that in cells growing on glucose, the decrease in Pu expression was mediated by the PtsN protein (3, 5) and that a minor role could be ascribed to Crc (46). However, the potential regulator involved in catabolite repression of glucokinase was unknown. We decided to measure growth rates with glucose and toluene (Table 2) and to determine glucokinase activity and expression from Pu (Pu::′lacZ) in a series of isogenic mutants deficient in one of the global regulators involved in catabolite repression (Crc, Crp, PtsN, RelA, and CyoB). Isogenic mutant cells were transformed with pS10 (Pu::lacZ xylR), and β-galactosidase activity was determined in cells growing on glucose in the absence and in the presence of toluene. In agreement with previous studies, we found that glucose had a marked effect on Pu expression in the parental strain (Table 5). However, in the PstN-deficient background β-galactosidase levels were slightly higher in cells growing on glucose plus toluene than in cells growing on glucose alone (Table 5). The repressing effect was alleviated but not entirely eliminated in a crc mutant background (Table 5), and no major effects on other mutations were found (not shown).
We measured glucokinase and gluconokinase activities in all of these mutant backgrounds. As expected, we found that the activity of gluconokinase, one of the enzymes of the gluconate loop, was equally high regardless of the presence of toluene (not shown). Hence, this confirms that gluconate-metabolizing enzymes are not under catabolite repression by toluene in KT2440. Glucokinase activity was equally high in cells of the Crc mutant growing on glucose and in cells growing on glucose plus toluene (Table 4), whereas in the rest of the mutants this activity was decreased in cells growing on glucose plus toluene (see data for PtsN in Table 4). This suggests that the effect of toluene on glucokinase levels is mediated by the Crc protein.
DISCUSSION
Catabolite repression control refers to the ability of an organism to preferentially metabolize one carbon source over another when both carbon sources are present in the growth medium (3, 6, 42). Most studies of catabolite repression of the TOL plasmid catabolic pathways by carbon sources have been carried out with the nonmetabolizable toluene analogue o-xylene and have concentrated on the effects of sugars, organic acids, and alcohol on the transcriptional activity of the Pu promoter. The rationale behind such assays was to avoid the superimposed carbon load effect that toluene might have on its own metabolism. However, the drawback inherent in the design of such experiments was that they overlooked the potential effects of hydrocarbon metabolism on the assimilation of other C sources. Our study involved a different experimental setup for catabolite repression of the TOL plasmid Pu promoter: two assimilable carbon sources, glucose and toluene, were used simultaneously. Our results for the growth rates support the conclusion that glucose and toluene are good carbon sources and that when cells are exposed to both these carbon sources, cells counterbalance the amounts of total carbon taken up from glucose and toluene, so that cells grow at a rate similar to that observed with glucose or toluene alone. This set of results supports the earlier proposal that expression of the TOL plasmid operons is integrated into overall metabolic control in P. putida (37).
The observation that the total amount of carbon used by a microorganism is drawn from several carbon sources simultaneously has been reported before. Klebsiella oxytoca uses glycerol and sucrose simultaneously (35), and Escherichia coli exposed to limiting amounts of up to seven sugars can use all of them simultaneously (25). We can therefore speak of crossed catabolite repression of toluene and glucose metabolism in P. putida. This contrasts with the stricter catabolite repression described for other pathways. Among cases of strict catabolite repression is the preferential use of glucose over lactose in E. coli (23, 40, 42). This phenomenon has also been described for the metabolism of aromatic compounds, including the control of protocatechuate dioxygenase in Pseudomonas cepacia (52) and the control of the enzymes for the degradation of aniline in Pseudomonas multivorans ANl (18).
We recently showed that the early metabolism of glucose involves three convergent pathways (Fig. 2). Our microarray and enzymatic results support the conclusion that toluene catabolite repression of glucose metabolism was exerted on only one of the pathways, the glucokinase branch, rather than on the gluconate/2-ketogluconate loops. Array data indicated that OprB porin (PP1019), the ABC glucose transport system (PP1015 to PP1018), and the glk/edd operon were repressed in response to the presence of toluene, whereas the catabolic enzymes of the gluconate loops were not repressed. In agreement with these transcriptional data is the finding that the glucokinase levels in cells growing on glucose plus toluene were lower than the levels in cells grown with glucose alone (Table 4). The repressing effect of toluene on glucose metabolism was obvious in the gcd mutant, in which the only pathway for the assimilation of glucose is the Glk pathway. In agreement with repression of glk is the finding that the gcd mutant assimilated very small amounts of glucose in the presence of toluene (Table 2) and the finding that glucokinase activity was repressed (Table 4), which was reflected in the pattern of 13C labeling of glycine, serine, glutamate, and proline (Fig. 3).
Velázquez et al. (46) proposed that either 6PG or 2-dehydro-3-deoxygluconate-6-phosphate could be the metabolic signal switching the Pu promoter pathway on or off. We ruled out the possibility that 6PG is the signal since the effect was not observed in the edd mutant. In contrast, we found evidence supporting the conclusion that the metabolite responsible for the effect was 2-dehydro-3-deoxygluconate-6-phosphate, since in an eda mutant catabolite repression of Pu by glucose was exacerbated (Fig. 4). Since the loss of ptsN rendered Pu unresponsive to glucose (3, 5), as monitored by ′lacZ reporter technology with a P. putida strain carrying the low-copy-number plasmid pS10, we suggest that PtsN/2-dehydro-3-deoxygluconate-6-phosphate acts as the switcher in Pu inhibition. How the 2-dehydro-3-deoxygluconate-6-phosphate signal is transferred to PtsN is unknown, but Cases et al. (5) showed that PtsN inhibition required the phosphorylation of the protein at its phospho-acceptor His68 residue. Therefore, an excess of 2-dehydro-3-deoxygluconate-6-phosphate could result in a permanent PtsN-P state that could, in turn, prevent XylR from binding to its upstream activator sequences (3).
The Crc protein seems to act as the switch for the glucokinase pathway, since in a crc-deficient background glucokinase was not repressed by toluene. This is in agreement with previous results for P. aeruginosa and P. putida (PpG2) which suggested that Crc controlled the expression of glucose-6-phosphate dehydrogenase, the second enzyme in the pathway that converts glucose-6-phosphate into 6PG (6, 7). Our results show that control of the glucokinase pathway in the presence of aromatic hydrocarbons also requires the active metabolism of toluene, although none of the key metabolites between toluene and catechol seemed to act as the specific signal between toluene and catechol. At present, the nature of the chemical signal is unknown, but it may be related to the energy state of the cell since Crc was proposed to launch appropriate responses based on the energy status of the cell (38).
In studies with P. putida, Rojo and colleagues showed that Crc may exert its effect at the posttranscriptional level, at least in the modulation of the alk system (30, 51). Our microarray results support a transcriptional role for Crc, but whether the effect is exerted directly by Crc or indirectly through modulation of transcription of the other regulator(s) remains unknown.
A feature that we have not overlooked is the effect of glucose on the bacterial response to toluene. Domínguez-Cuevas et al. (11) reported that exposure of P. putida cells to toluene resulted in the upregulation of 180 genes and the downregulation of 127 genes. In contrast, when glucose was present, only 50 genes were upregulated in response to toluene and only 11 genes were downregulated (Tables 3 and 4). In agreement with the results of Domínguez-Cuevas et al. (11) was our finding that all toluene assimilation genes were induced, but major differences were found with regard to the induction of stress genes. For instance, in cells growing on glucose plus toluene the IbpA protein was the only chaperone induced, which contrasted with the induction of 23 shock genes with toluene alone. This indicates that glucose metabolism alleviates the toxic effect of toluene.
In short, our results show that KT2440 metabolizes glucose and toluene simultaneously, as revealed by net flux analysis of [13C]glucose. Simultaneous catabolite repression of the glucokinase and TOL pathways was triggered by two different catabolite repression systems; Pu was repressed mainly via PtsN in response to high levels of 2-dehydro-3-deoxygluconate-6-phosphate, whereas repression of the glucokinase pathway was channeled through Crc.
Supplementary Material
Acknowledgments
This study was financed by grant BIO2006-05668 from the Spanish Ministry of Science and Education and by grant GEN2006-27750-C5-J-E from the EU SySMO Programme.
We thank Uwe Sauer and Tobias Führer for assistance with the 13C assays done at the ETH in Zürich. We thank M. Mar Fandila and Carmen Lorente for secretarial assistance and K. Shashok for improving the use of English in the manuscript.
Footnotes
Published ahead of print on 6 July 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abril, M. A., C. Michán, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda-Olmedo, I., P. Marín, J. L. Ramos, and S. Marqués. 2006. Role of the pstN gene product in catabolite repression of Pseudomonas putida TOL toluene degradation pathway in chemostat cultures. Appl. Environ. Microbiol. 72:7418-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranda-Olmedo, I., J. L. Ramos, and S. Marqués. 2005. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl. Environ. Microbiol. 71:4191-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv. Microb. Physiol. 31:1-69. [DOI] [PubMed] [Google Scholar]
- 5.Cases, I., J. Pérez-Martín, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the σ54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 274:15562-15568. [DOI] [PubMed] [Google Scholar]
- 6.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in the pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 7.Collier, D. N., C. Spencer, M. J. Cox, and P. V. Phibbs. 2001. Isolation and phenotypic characterization of Pseudomonas aeruginosa pseudorevertants containing suppressors of the catabolite repression control defective crc 10 allele. FEMS Microbiol. Lett. 196:87-92. [DOI] [PubMed] [Google Scholar]
- 8.Dauner, M., and U. Sauer. 2000. GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol. Prog. 16:642-649. [DOI] [PubMed] [Google Scholar]
- 9.del Castillo, T., J. L. Ramos, J. J. Rodríguez-Herva, T. Führer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinamarca, M. A., A. Ruíz-Manzano, and F. Rojo. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domínguez-Cuevas, P., J. E. González-Pastor, S. Marqués, J. L. Ramos, and V. de Lorenzo. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 281:11981-11991. [DOI] [PubMed] [Google Scholar]
- 12.Duetz, W. A., S. Marqués, B. Wind, J. L. Ramos, and J. G. van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 62:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duque, E., A. Haïdour, F. Godoy, and J. L. Ramos. 1993. Construction of a Pseudomonas hybrid strain that mineralizes 2,4,6-trinitrotoluene. J. Bacteriol. 175:2278-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duque, E., A. J. Molina-Henares, J. de la Torre, M. A. Molina-Henares, T. del Castillo, J. Lam, and J. L. Ramos. 2007. Towards a genome-wide mutant library of Pseudomonas putida strains KT2440. In J. L. Ramos (ed.), Pseudomonas, vol. V, in press Kluwer. London, United Kingdom.
- 15.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880-891. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325:308-316. [DOI] [PubMed] [Google Scholar]
- 17.Führer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helm, V., and H. Reber. 1979. Investigation on the regulation of aniline utilization in Pseudomonas multivorans strain AN1. Eur. J. Appl. Microbiol. Biotechnol. 7:191-199. [Google Scholar]
- 19.Herrero, M., and V. de Lorenzo. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtel, A., S. Marqués, I. Mohler, U. Jakubzik, and K. N. Timmis. 1994. Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J. Bacteriol. 176:1773-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, J. C., and P. V. Phibbs. 1983. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J. Bacteriol. 154:793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inada, T., K. Kimata, and H. Aiba. 1996. Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells 1:293-301. [DOI] [PubMed] [Google Scholar]
- 24.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lendenmann, U., M. Snozzi, and T. Egli. 1996. Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Appl. Environ. Microbiol. 62:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marqués, S., A. Holtel, K. N. Timmis, and J. L. Ramos. 1994. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J. Bacteriol. 176:2517-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marqués, S., J. L. Ramos, and K. N. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta-fission pathway operon around the transcriptional initiation point, the xylTE and the xylFJ regions. Biochim. Biophys. Acta 1216:227-236. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno, R., A. Ruíz-Manzano, L. Yuste, and F. Rojo. 2007. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol. Microbiol. 64:665-676. [DOI] [PubMed] [Google Scholar]
- 31.Nakazawa, T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782-786. [DOI] [PubMed] [Google Scholar]
- 32.Nanchen, A., T. Führer, and U. Sauer. 2007. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principle. Methods Mol. Biol. 358:177-198. [DOI] [PubMed] [Google Scholar]
- 33.Petruschka, L., G. Burchhardt, C. Muller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199-206. [DOI] [PubMed] [Google Scholar]
- 34.Phillips, A. T., and L. M. Mulfinger. 1981. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J. Bacteriol. 145:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñar, G., K. Kovárová, T. Egli, and J. L. Ramos. 1998. Influence of carbon source on nitrate removal by nitrate-tolerant Klebsiella oxytoca CECT 4460 in batch and chemostat cultures. Appl. Environ. Microbiol. 64:2970-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 38.Rojo, F., and A. Dinamarca. 2004. Catabolite repression and physiological control, p. 365-387. In J. L. Ramos (ed.), Pseudomonas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 39.Ruíz, R., M. I. Aranda-Olmedo, P. Domínguez-Cuevas, M. I. Ramos-González, and S. Marqués. 2004. Transcriptional regulation of the toluene catabolic pathways, p. 509-537. In J. L. Ramos (ed.), Pseudomonas, vol. II, Kluwer Academic Publishers, New York, NY. [Google Scholar]
- 40.Santillán, M., and M. C. MacKey. 2004. Influence of catabolite repression and inducer exclusion on the bistable behavior of the lac operon. Biophys. J. 86:1282-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel, L. S., P. B. Hylemon, and P. V. Phibbs, Jr. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J. Bacteriol. 129:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 43.Suh, S. J., L. J. Runyen-Janecky, T. C. Maleniak, P. Hager, C. H. MacGregor, N. A. Zielinski-Mozny, P. V. Phibbs, Jr., and S. E. West. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561-1569. [DOI] [PubMed] [Google Scholar]
- 44.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 45.Temple, L., A. Sage, G. E. Christie, and P. Phibbs, Jr. 1994. Two genes for carbohydrate catabolism are divergently transcribed from a region of DNA containing the hexC locus in Pseudomonas aeruginosa PAO1. J. Bacteriol. 176:4700-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velázquez, F., I. di Bartolo, and V. de Lorenzo. 2004. Genetic evidence that catabolites of the Entner-Doudoroff pathway signal C source repression of the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 186:8267-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicente, M., and J. L. Cánovas. 1973. Glucolysis in Pseudomonas putida: physiological role of alternative routes from the analysis of defective mutants. J. Bacteriol. 116:908-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.West, S. E., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuste, L., A. B. Hervás, I. Canosa, R. Tobes, J. I. Jiménez, J. Nogales, M. M. Pérez-Pérez, E. Santero, E. Díaz, J.-L. Ramos, V. de Lorenzo, and F. Rojo. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8:165-177. [DOI] [PubMed] [Google Scholar]
- 51.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183:6197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zylstra, G. J., R. Olsen, and D. P. Ballou. 1989. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase. J. Bacteriol. 171:5907-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.