Abstract
Growing bacterial L forms are reputed to lack peptidoglycan, although cell division is normally inseparable from septal peptidoglycan synthesis. To explore which cell division functions L forms use, we established a protocol for quantitatively converting a culture of a wild-type Escherichia coli K-12 strain overnight to a growing L-form-like state by use of the β-lactam cefsulodin, a specific inhibitor of penicillin-binding proteins (PBPs) 1A and 1B. In rich hypertonic medium containing cefsulodin, all cells are spherical and osmosensitive, like classical L forms. Surprisingly, however, mutant studies showed that colony formation requires d-glutamate, diaminopimelate, and MurA activity, all of which are specific to peptidoglycan synthesis. High-performance liquid chromatography analysis confirmed that these L-form-like cells contain peptidoglycan, with 7% of the normal amount. Moreover, the β-lactam piperacillin, a specific inhibitor of the cell division protein PBP 3, rapidly blocks the cell division of these L-form-like cells. Similarly, penicillin-induced L-form-like cells, which grow only within the agar layers of rich hypertonic plates, also require d-glutamate, diaminopimelate, and MurA activity. These results strongly suggest that cefsulodin- and penicillin-induced L-form-like cells of E. coli—and possibly all L forms—have residual peptidoglycan synthesis which is essential for their growth, probably being required for cell division.
Bacterial L forms are spherical, osmosensitive variants isolated from many different species and reported to have no peptidoglycan cell wall, although they grow and divide indefinitely. In contrast, cell division in normal bacteria is indissociable from cell wall synthesis; a septum, consisting of a double layer of peptidoglycan, is laid down at midcell and then split to form the new poles of the two daughter cells. The division of L forms in the apparent absence of peptidoglycan is thus paradoxical. We wished to explore which cell division functions L forms require, using Escherichia coli K-12 as the model.
Septal synthesis is carried out by an assembly of cell division proteins organized in a ring-like membrane-associated complex surrounding the midcell (19, 52). The structural basis of this ring is the FtsZ protein, a bacterial tubulin homologue. In E. coli, some 15 cell division proteins are known to be recruited into the FtsZ ring.
The study of L forms goes back to 1935, when Emmy Klieneberger succeeded in establishing a pure culture of the curious mycoplasma-like organism that appeared systematically in cultures of Streptobacillus moniliformis (26, 27). It consisted of spherical, osmosensitive cells that grew on plates of hypertonic complex medium containing serum. Klieneberger called the culture “L1” in honor of the Lister Institute in London, where she had emigrated in 1933 when the German authorities fired her as a Jew. Similar morphological variants, also called L forms, were subsequently isolated from other bacterial species (10, 11).
In 1942, L forms were found to be resistant to penicillin (40). This provided a convenient tool for isolating L forms from essentially any cultivable bacterial species. The protocol that emerged was to spread a heavy inoculum of bacteria on a plate of hypertonic complex medium containing serum, broth, and penicillin. After a growth period, usually of several weeks, an agar block is cut from the plate, inverted, and spread on a plate of the same medium for a new growth period. Such “passages” are repeated serially, often for years, until finally a stable L form appears, able to grow indefinitely and no longer able to revert to normal morphology when cultivated in the absence of penicillin. Two such L forms of E. coli K-12 are extant, one isolated in 1987 after heavy mutagenesis (38) and the other in 1969 without mutagenesis (43).
There is little information on the genetics of L forms, but it is clear that a number of mutations accumulate during the many passages; the two extant stable L forms of E. coli K-12 both carry numerous uncharacterized mutations (see Discussion). To study the cell division functions required for the propagation of E. coli in the absence of peptidoglycan, we needed a genetically defined lineage. We therefore sought a procedure to convert all cells of a given culture (of known genotype) to the L form on command, as it were. With appropriate genetic constructs, the resulting L-form cells could then be tested for their ability to divide after depletion of one or another cell division protein. Using the β-lactam cefsulodin, we developed a protocol that converts an entire population of wild-type E. coli overnight to viable, growing cells that are osmosensitive and spherical. In the present report, we characterize these L-form-like cells. To our surprise, physiological, genetic, and biochemical evidence, presented here, showed that they still synthesize 7% of the normal amount of peptidoglycan and that this residual synthesis is essential for their propagation, most likely for cell division.
Wild-type E. coli can grow in the presence of penicillin only if embedded within the agar layer of a rich, hypertonic plate, where it produces minicolonies of spherical L-form cells (31). However, we present genetic and physiological evidence that this growth, too, absolutely requires residual peptidoglycan synthesis.
We conclude that cefsulodin- and penicillin-induced L-form-like cells of E. coli retain the ability to synthesize small amounts of peptidoglycan and that this residual synthesis is essential.
MATERIALS AND METHODS
Bacterial strains and plasmids.
With one exception, all experiments presented here were carried out with MG1655 (2) and derivatives obtained by P1 transduction (36) and transformation (42). The exception is the experiment with VIP205, genotype MC1061 [araD139 Δ(araA-leu) Δ(codB-lacI) galK16 galE1 mcrA mcrB1 hsdR2 relA1 spoT1 rpsL150] (Ω-Kmr-lacIq-Ptac)::ftsZ (16). The chromosomal alleles introduced into MG1655 (and the references and sources for their descriptions) are as follows: ΔdapA::Eryr (8) and dapB17::Mu (6); dapE::Cmr (Coli Genetic Stock Center, http://cgsc.biology.yale.edu); gltS murI::Kmr (12); murA::Kmr (5); ΔmrcA::Kmr (34) and ΔmrcB::Kmr (9); ΔrcsA::Cmr, ΔrcsB::Cmr, ΔrcsC::Cmr, ΔrcsD::Cmr, and ΔrcsF::Cmr (13); and cpsE3::Tn10 (49).
The plasmids used are as follows: pJFK1183H(Kmr) and pUM1Ba(mrcB+ Kmr) (35) and pBAD(murA+ Cmr). The latter was constructed by isolating a 1.4-kb KpnI-XbaI fragment carrying the murA+ gene from pBAD30-Z(Ampr) (5) and recloning it in pBAD33(Cmr) (22) digested by the same enzymes.
The altered gltS allele permitting efficient d-glutamate uptake was transduced into strain MG1655 pyrE zib-563::Tn10, selecting Ura+ and screening for cotransductants that were Tcs and able to grow on glutamate as the sole carbon source; one such strain was then transduced to murI::Kmr. The dapB17::Mu allele was introduced into this gltS murI::Kmr strain by cotransduction with carA::Tn10; the latter allele was then removed by transduction to Arg+ Pyr+. The murI::Kmr allele was removed by transduction to argE::Tn10 and screening for d-glutamate prototrophy; one such strain was then transduced to Arg+ Tcs to produce a dapB17::Mu gltS strain.
For strains MG1655 lacY::Cmr ΔmrcB/pJFK1183H (Kmr) and MG1655 lacY::Cmr ΔmrcB/pUM1Ba (mrcB+ Kmr), the Kmr cassette in the mcrB gene, flanked by two res sites, was removed using the nonreplicative plasmid pJMSB8 carrying the resolvase (29); this plasmid is propagated in strain S17-1(λpir).
Media and growth conditions.
L-form-like cells were grown in M medium, a rich hypertonic medium specially designed for this work. It contains beef extract (Difco), 3 g/liter; Bacto peptone (Difco), 10 g/liter; yeast extract (Difco), 5 g/liter; NaCl, 5 g/liter; MgSO4, 0.01 M; sucrose, 0.23 M. M plates contained in addition 1.2% Bacto agar (Difco). For routine growth, LB broth (36) was used; for some experiments, LB broth was prepared without added NaCl. Diaminopimelate (DAP) and dl-glutamate, when needed, were used at 30 and 100 μg/ml, respectively. Antibiotic concentrations were as follows: for chloramphenicol, 20 μg/ml; for ampicillin, 100 μg/ml; for tetracycline, 10 μg/ml; for erythromycin, 200 μg/ml; for piperacillin, 3 or 5 μg/ml; for aztreonam, 0.1 μg/ml; for A22 [S-(3,4-dichlorobenzyl)isothiourea; Calbiochem], 8 μg/ml; and for cefsulodin (Sigma), 30 μg/ml.
Unless indicated otherwise, all bacterial growth was at 30°C. Liquid cultures were agitated vigorously.
Muropeptide analysis.
Total peptidoglycan and the degree and type of cross-linking were measured as described previously (18, 50). In brief, 250-ml overnight cultures of strain MG1655 grown with aeration in M medium with or without cefsulodin were centrifuged, resuspended in ice-cold water, and dropped into a boiling 6% sodium dodecyl sulfate (SDS) solution. After boiling for 15 h, the crude sacculi were collected by ultracentrifugation, washed free of SDS with water, and digested with α-amylase for 2 h at 37°C and with pronase for 90 min at 60°C. After the addition of 1% SDS, the sacculi were boiled for 30 min; SDS was then removed by repeated centrifugation-resuspension. The murein samples were digested with the amidase cellosyl. Muropeptides were analyzed after reduction with sodium borohydride by separation with reversed-phase high-performance liquid chromatography (HPLC) and quantification of the UV absorption of the muropeptides. The total amount of peptidoglycan was calculated as the sum of the absorptions of all muropeptides.
Protein assay.
To compare the amounts of peptidoglycan in bacteria growing in M medium with and without cefsulodin, we normalized the amount of muropeptides to the amount of protein, evaluated with a DC protein assay (Bio-Rad). To validate this, we measured the protein concentration relative to the optical density at 600 nm (OD600). Bacterial cultures were prepared exactly as for the peptidoglycan assay. After boiling with SDS, the protein concentration was measured. The two cultures had the same protein concentrations for a given OD600 (220 μg/ml for an OD600 of 1).
Microscopy.
Bacterial suspensions were placed at 40°C and diluted twofold with a 2% solution of low-melting-point agarose (GIBCO) kept at the same temperature; 8 μl of the mixture was placed on a prewarmed slide, 2 μl of a 200-μg/ml solution of FM 4-64 (Molecular Probes) was added, and a coverslip was placed on the droplet. Bacteria were examined in a Leica DMRE2 videomicroscope by use of a wide-field 100× objective fitted with a mercury arc lamp, a red filter (SC4), and a high-resolution CoolSnap HQ camera (Photometrics). Each cell was photographed in 35 to 54 focal planes by use of a piezoelectric motor PI. The images were analyzed with Image J 1.36b software (NIH). This program was used to obtain the pictures shown in Fig. 1 and the cell diameter distribution in Fig. 2.
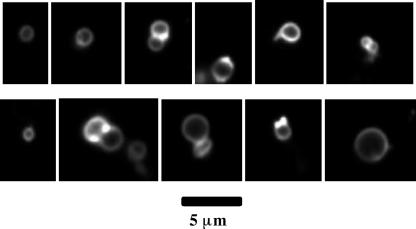
FIG. 1.
L-form-like cells of E. coli MG1655. Cells were stained with FM 4-64, a fluorescent membrane dye (see Materials and Methods). Each image was taken in the focal plane giving the maximum diameter.
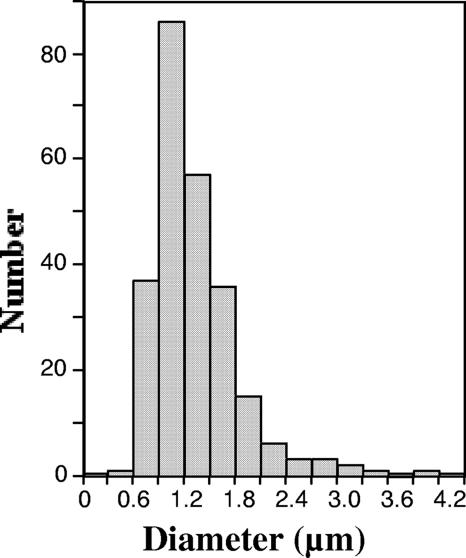
FIG. 2.
Diameter distribution of L-form-like E. coli MG1655. In all, 248 cells were measured.
RESULTS
L-form-like growth in the presence of cefsulodin.
In 1958, Lederberg and St. Clair reported that the E. coli K-12 strain Y10 made L-form colonies with 10 to 50% efficiency when plated within the agar layer of a hypertonic complex medium containing 1,000 U/ml penicillin; no L-form growth occurred on the surfaces of the plates or in liquid medium of the same composition (31). We reproduced these results with the wild-type E. coli K-12 strain MG1655 and our rich hypertonic M medium (see Materials and Methods). Growth was observed at an agar concentration of 1.2% but not at 0.6%, and there was no growth on the surfaces of these plates or in liquid M penicillin medium.
To carry out analyses on L-form cells, it is convenient to propagate them in liquid culture or on the surfaces of plates to avoid the difficult problem of separating the fragile cells from the agar. We therefore sought an alternative protocol that could produce L-form growth on the surfaces of M medium plates or in liquid culture. We focused in particular on penicillin-binding proteins (PBPs) 1A and 1B. These enzymes catalyze the polymerization of glycan chains, with lipid-linked disaccharide pentapeptide units as the substrate (23). PBPs 1A and 1B are bifunctional enzymes, possessing both transglycosylase (chain lengthening) and transpeptidase (cross-linking) activity. Genetic studies in 1978 established that E. coli grows normally in the absence of PBP 1A or PBP 1B but lyses in normal (isotonic) media when both are genetically inactivated (47). The β-lactam cefsulodin specifically inhibits the transpeptidase activity of PBPs 1A and 1B, causing lysis (37). We investigated the possibility of inducing L-form growth on the plate surface by use of cefsulodin instead of penicillin.
The effect of different concentrations of cefsulodin on the growth of MG1655 on M plates is shown in Table 1. At 30 or 100 μg/ml, mucoid colonies appear overnight and contain only spherical cells. If the osmolarity of the medium is further increased, growth is slowed down considerably. If it is lowered, ultimately the cells no longer form colonies in the presence of cefsulodin; on plates of LB medium to which no NaCl is added; for example, the presence of 30 μg/ml cefsulodin reduces the plating efficiency 200-fold.
TABLE 1.
Growth, cell morphology, and colony aspect on cefsulodin plates
| Cefsulodin concn (μg/ml) | EOPa | Cells | Colonies |
|---|---|---|---|
| 0 | ≡1.0 | Rods | Nonmucoid |
| 10 | 0.92 | Rods | Nonmucoid |
| 30 | 0.62 | Spheres | Mucoid |
| 100 | 0.10 | Spheres | Mucoid |
| 200 | 0.001 | Spheres | Mucoid |
EOP compared to the titer on an M plate without cefsulodin. Overnight cultures in liquid M medium were diluted and spread on the surfaces of M plates containing cefsulodin at the indicated concentrations. Plates were incubated for 24 h at 30°C.
We next studied the effect of temperature on the plating efficiency in the presence of cefsulodin. On M plates containing 30 μg/ml cefsulodin, the efficiency of plating (EOP), 62% at 30°C (Table 1), dropped to 10% at 37°C and 5% at 42°C. In the following work, cefsulodin, when used, was added at 30 μg/ml, and all cultures and plates were incubated at 30°C.
We tested the ability of the spherical, L-form-like cells appearing on M cefsulodin plates to continue growing under the same conditions. A colony was resuspended in M medium and assayed on M plates with or without cefsulodin. The EOP on the former was 25 to 50% compared to the titer in the absence of cefsulodin. The colonies on the M cefsulodin plate were mucoid and contained only spherical cells, whereas those on the M plate without cefsulodin were nonmucoid, and the cells in them had reverted to rod-shaped morphology.
A colony from an M cefsulodin plate inoculated into M cefsulodin liquid medium exhibited exponential growth after a lag period, with a doubling time of 60 min, compared to 30 min in M medium without cefsulodin. Growth during this phase was balanced, as judged by a constant ratio of viable cell concentration to optical density (OD600); at saturation, both cultures reached an OD600 of between 1.0 and 2.0 (data not shown). It is also possible to establish L-form-like growth directly in M cefsulodin liquid medium, using as the inoculum cells grown in M medium without antibiotic. Cells growing in M cefsulodin liquid cultures are uniformly spherical (see below).
L-form-like growth in mutants lacking PBP 1A or 1B.
PBPs 1A and 1B are specified by the mrcA and mrcB genes, respectively. The synthetic lethality of mrcA and mrcB mutations (47) suggests that these proteins are to some degree functionally redundant. To study the effect of the genetic inactivation of mrcA or mrcB on cefsulodin-induced L-form-like cells, we constructed ΔmrcA and ΔmrcB derivatives of MG1655. As expected, both grew normally in M medium, with rod-shaped morphology. On M cefsulodin plates, the ΔmrcA strain, totally lacking PBP 1A, behaved like the wild type: overnight, it formed mucoid colonies (EOP of about 25%) containing only spherical cells. The ΔmrcB mutant, in contrast, grew extremely poorly on M cefsulodin plates. It formed visible colonies only after at least 4 days of incubation, with an EOP of about 0.1; the cells in the colonies were spherical.
β-Lactams inactivate the transpeptidase activity of PBPs but not the transglycosylase (23). The above observations suggest that the transglycosylase activity of PBP 1B is more important than that of PBP 1A for L-form-like growth in the presence of cefsulodin. A more important role for PBP 1B was also observed when PBP 2 or PBP 3 was specifically inhibited; under these conditions, cells lysed rapidly in the absence of PBP 1B but continued growing for several generations in the absence of PBP 1A (14).
Transformation of the ΔmrcB strain with a plasmid carrying the mrcB+ gene under Plac control restored rapid growth on M cefsulodin plates containing the lac operon inducer IPTG (isopropyl-β-d-thiogalactopyranoside) (3 × 10−5M), with an EOP of 0.6 in 24 h. Under these conditions of PBP 1B overexpression, however, the cells in the colonies were rod shaped or filamentous rather than spherical.
Osmosensitivity and envelope disorganization in L-form-like cells.
Classical L forms are generally osmosensitive. We tested the osmosensitivity of cefsulodin-induced L-form-like cells by subjecting them to an osmotic downshift. A culture of spherical MG1655 cells grown in M cefsulodin medium was plated on twofold-diluted LB medium to which no NaCl had been added. It had an EOP of 7 × 10−4, whereas rod-shaped MG1655 cells grown in M medium without cefsulodin had an EOP of 1.0 in this medium.
When an M cefsulodin culture was centrifuged, washed, and resuspended in distilled water, there was massive lysis. This was caused by the osmotic downshift, not by the centrifugation, since washing and resuspending in M cefsulodin medium gave 100% recovery. Rod-shaped cells grown without cefsulodin were not affected by washing and resuspension in distilled water.
These results show clearly that cefsulodin-induced L-form-like cells are osmosensitive.
Osmosensitive cells are fragile. To evaluate the degree of spontaneous lysis in our hypertonic M medium, we grew a culture in the presence of the lac operon inducer IPTG (5 × 10−4 M) with and without cefsulodin to see how much β-galactosidase was liberated in the medium. Enzyme levels were low (580 and 750 Miller units, respectively). For osmosensitive spherical cells (culture with cefsulodin), about 20% of the total enzyme activity remained in the supernatant (after a 15-min centrifugation at 15,000 rpm) or in the filtrate (after passage through a 0.2-μm membrane filter); for rod-shaped cells, about 11% remained.
In the course of the above-described experiments, we made a striking observation. The usual β-galactosidase assay includes a permeabilization step (treatment with chloroform and SDS), since the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) can diffuse across the outer membrane but not across the cytoplasmic membrane. Although ONPG can be taken up by lactose permease, transport is considerably slower than hydrolysis by β-galactosidase and thus limits the rate of ONPG hydrolysis. In rod-shaped cells growing in M IPTG medium, the omission of the permeabilization step gave an apparent specific activity only 12% that of permeabilized cells; this was primarily enzyme released by spontaneous lysis. With spherical cells, in sharp contrast, fully 50 to 60% of the enzyme activity was detected in unpermeabilized cells, well above the level of lysis. This strongly suggests that the cefsulodin-induced L-form-like cells are permeable to ONPG. This in turn indicates that their cytoplasmic membrane, although intact, is somewhat disorganized.
In gram-negative bacteria, the outer membrane, tightly associated with the peptidoglycan layer, is the first barrier protecting the cells against toxic products. In L forms of gram-negative species, the outer membrane has been variously reported to be absent or present in altered form, called, respectively, protoplast- and spheroplast-type L forms (21). Indeed, advantage has been taken of this to establish a protein display, in which proteins anchored in the outer leaflet of the cytoplasmic membrane are exposed at the surface of protoplast-type L-form cells (20).
We tested the sensitivities of our cefsulodin-induced L-form-like cells to three toxic agents which, at low concentrations, are excluded from E. coli by the outer membrane: SDS, rifampin, and novobiocin (39). MG1655 grew normally, with an EOP of near 1, on M plates containing 3.5% SDS, 5 μg/ml rifampin, or 150 μg/ml novobiocin. Growth was abolished, however, if the plates also contained cefsulodin (EOP < 2 × 10−4 in all cases). L-form-like cells are thus hypersensitive to these three compounds.
Some L forms of E. coli have been found to be resistant to various bacteriophages (48). We tested our L-form-like cells for sensitivity to phage λ by spotting λvir on a lawn of cells on an M cefsulodin plate. Lysis was observed. On a lawn of the MG1655 lamB strain, lacking the λ receptor, there was no visible lysis. Phage λh80cI, which uses the TonB-activated FhuA outer membrane protein as the receptor, lysed both strains. These observations indicate that the outer membrane proteins LamB and FhuA are present in L-form-like cells and recognizable by the phage.
We conclude that our L-form-like cells have an outer membrane which is in a somewhat disorganized state.
Cell shape and size distribution.
We studied the morphology of our L-form-like cells. To see cell contours clearly, the bacteria were stained with FM 4-64, a dye which enters the membrane and there becomes fluorescent. Taking serial pictures in successive focal planes allowed us to get precise measurements of individual cells (see Materials and Methods).
MG1655 cells from an M cefsulodin culture were uniformly spherical (Fig. 1). Cell size was heterogeneous, as has been observed with classical L forms. The average diameter was 1.33 μm, standard deviation 0.50 μm, with 89% of the cells having diameters between 0.6 and 1.8 μm (Fig. 2), corresponding to a 27-fold range in volume (0.11 to 3.0 μm3). Dividing cells revealed that cell division is often asymmetrical, producing daughter cells of unequal sizes (Fig. 1).
These observations are in sharp contrast to the situation with rod-shaped cells growing in the absence of cefsulodin: cell size was more uniform, with less than a threefold range in volume; the average volume was larger (3.2 μm3); and cell division took place precisely at midcell.
MreB independence and capsule dependence of L-form-like growth.
The rod shape of wild-type E. coli is ensured in part by the actin-like protein MreB (4). This is an essential protein (30), although its precise role has not been clearly established. A specific inhibitor of MreB, the chemical A22, has been described (17, 25).
On M plates, A22 inhibits the growth of MG1655. On M cefsulodin plates, in striking contrast, the L-form-like cells are completely resistant to A22 (Table 2).
TABLE 2.
MreB independence of L-form-like growth
| Presence of cefsulodina | EOP when A22 was:
|
|
|---|---|---|
| Absent | Present | |
| Absent | ≡1.0 | <10−4 |
| Present | 0.6 | 0.5 |
A culture of MG1655 in M medium was assayed on four types of M plates: with or without cefsulodin (30 μg/ml) and with or without A22 (8 μg/ml). Plates were incubated for 24 h.
This strongly suggests that the normally essential actin-like protein MreB, involved in maintaining rod shape, is not required for the growth of spherical L-form-like cells.
Cells growing on M cefsulodin plates form mucoid colonies, indicating overproduction of capsular polysaccharide, which in E. coli K-12 consists of colanic acid. The enzymes that carry out this biosynthesis are specified by a group of genes regulated by the RcsBCD stress system (24, 32). We inactivated the synthesis of colanic acid by introducing into MG1655 either a regulatory mutation (ΔrcsB, ΔrcsC, ΔrcsD, ΔrcsA, or ΔrcsF) or a cpsE::Tn10 allele inactivating the structural gene of an enzyme in the colanic acid biosynthetic pathway (49). The resulting strains were all unable to grow overnight on M cefsulodin plates (EOP < 5 × 10−4).
These results show that L-form-like growth on the surface of an M cefsulodin plate requires a protective colanic acid capsule.
Growth within the agar layer of an M plate can also protect the cells. We looked to see whether these conditions obviate the requirement for capsule. The MG1655 cpsE::Tn10 mutant was unable to grow within the agar layer of an M cefsulodin plate (EOP < 5 × 10−5). Thus, agar cannot replace the capsule requirement for L-form-like growth.
L-form-like growth requires ongoing peptidoglycan synthesis.
Since L forms are thought to have no cell wall, we attempted to establish L-form growth by blocking a specific step in peptidoglycan synthesis. We first studied an auxotroph for d-glutamate, the second amino acid of the pentapeptide side chain of muramic acid. A d-glutamate auxotroph of MG1655 grew efficiently as rods on M d-glutamate plates (and as spheres on M cefsulodin d-glutamate plates), but it was unable to grow in the absence of d-glutamate (EOP < 10−4, with or without cefsulodin).
We next examined MG1655 derivatives that require DAP, the third amino acid of the muramic acid side chain. We constructed dapB::Mu and dapE::Cmr derivatives of MG1655 (see Materials and Methods). Both grew efficiently as rods on M DAP plates (and as spheres on M cefsulodin DAP plates), but again neither could grow in the absence of DAP (EOP < 2 × 10−6 for both strains, with or without cefsulodin).
In a final attempt to obtain L-form growth by means of a genetic block to cell wall synthesis, we constructed a strain in which the murA gene product can be depleted. This cytoplasmic enzyme catalyzes the first reaction in the synthesis of the muramic acid side chain. We constructed an MG1655 ΔmurA derivative carrying a plasmid with the murA gene under control of the araBAD promoter, expressed only in the presence of exogenous l-arabinose. The resulting strain grew efficiently as rods on M plates containing 5 × 10−4 M l-arabinose (and as spheres on M cefsulodin l-arabinose plates), but it was unable to form colonies on M plates lacking l-arabinose (EOP < 2 × 10−5, with or without cefsulodin).
Although we were unable to induce L-form growth on the surfaces of M plates when cell wall synthesis was genetically blocked, it was conceivable that the difficulty lay in the initial establishment of L-form-like growth but not in the subsequent propagation of preestablished L-form-like cells. We therefore tested these conditions again, using for the inoculum spherical L-form-like cells from cultures pregrown in the presence of cefsulodin.
We prepared liquid cultures of murI::Kmr and dapB::Mu mutant and murI::Kmr dapB::Mu double mutant strains in M medium containing cefsulodin, DAP, and d-glutamate and of the ΔmurA/pBADmurA+ strain in M medium containing cefsulodin and l-arabinose. The cultures, which contained only spherical L-form-like cells, were all assayed on M plates with or without cefsulodin and with or without the supplements (either DAP plus d-glutamate or l-arabinose). None of the mutants could grow without its supplement, with or without cefsulodin (all EOPs < 10−3).
We conclude that a tight genetic block in peptidoglycan synthesis completely prevents the propagation of L-form-like cells of MG1655 on the surfaces of M cefsulodin plates. This in turn suggests that these cells have residual peptidoglycan synthesis which is essential for their propagation.
Peptidoglycan in L-form-like cells.
To test this hypothesis directly, we assayed the cells for peptidoglycan. We grew MG1655 for about 20 generations in liquid M cefsulodin medium and, in parallel, in liquid M medium without cefsulodin. The cells were harvested and subjected to the procedure for peptidoglycan purification and hydrolysis (see Materials and Methods). HPLC analysis of the muropeptides revealed clearly that the L-form-like cells did indeed contain a low amount of peptidoglycan, about 7% as much as the rod-shaped cells. This estimate may neglect very short glycan chains not cross-linked to higher-molecular-weight peptidoglycan and thus not pelleted in the ultracentrifugation step (see Materials and Methods).The glycan chains were on the average one-third shorter in the L-form-like cells, as estimated by the fraction of anhydrosugars (Table 3). The cross-linking patterns were also somewhat different in the two samples. The muropeptides from the L-form-like cells had more DAP-DAP cross-links and fewer DAP-d-Ala cross-links. The latter, which are d-d cross-links, can be formed by PBPs 1A, 1B, 2, and 3; this presumably accounts for their relative underrepresentation in the presence of cefsulodin, which inhibits the transpeptidase activity of PBPs 1A and 1B. The former, which are l-d cross-links, are not formed by PBPs; l-d-cross-linking enzymes have not been identified in E. coli (23).
TABLE 3.
Analysis of muropeptides in L-form-like cells
| Cell typea | Muropeptides/proteinb | % DAP-DAP cross-linkage | % Total dimers | DAP-DAP dimers/total dimers | % Anhydromuropeptides | Average chain length | % Total cross-linking |
|---|---|---|---|---|---|---|---|
| Rods | 370 × 103 | 5.7 | 32 | 0.18 | 9.5 | 10.6 | 37 |
| Spheres | 25 × 103 | 13 | 29 | 0.38 | >14.5 | 6.9 | 35 |
MG1655 cells were grown for 20 generations in liquid M medium without cefsulodin (rods) or with 30 μg/ml cefsulodin (spheres). Cells were harvested and peptidoglycan was extracted and analyzed (see Materials and Methods).
Total muropeptide was calculated by integrating all peaks after HPLC separation; total protein was measured as described in Materials and Methods.
Thus, our L-form-like cells must synthesize peptidoglycan, as indicated by the genetic results. The quantitative differences observed in cross-linking and average chain length are probably attributable to the inactivation of the transpeptidase activity of PBPs 1A and 1B; in fact, the residual peptidoglycan synthesized in the presence of cefsulodin is relatively normal in structure.
Cell division requirements of L-form-like cells.
Penicillin G inhibits all PBPs and prevents the surface growth of L-form-like cells, whereas cefsulodin, which inactivates only PBPs 1A and 1B, does not. It would thus seem that one or more of the other PBPs are required for surface growth. Since PBP 3 is required for cell division (45) and PBP 2 has been implicated in the division of spherical cells (51), we investigated the ability of L-form-like cells to grow in the presence of specific inhibitors of PBP 2 or PBP 3.
Amdinocillin specifically inhibits PBP 2 (45, 46). Spherical MG1655 cells grown overnight in liquid M cefsulodin medium were plated on M cefsulodin plates with or without 3 μg/ml amdinocillin. No growth was observed in the presence of amdinocillin (EOP < 1 × 10−3), indicating that PBP 2 activity is required for L-form-like growth.
Piperacillin and aztreonam are specific inhibitors of PBP 3 (FtsI), which is required for septation (45). Spherical MG1655 cells grown overnight in liquid M cefsulodin medium were plated on M cefsulodin plates with or without 3 μg/ml piperacillin or 0.1 μg/ml aztreonam. Both antibiotics prevented the growth of L-form-like cells (EOP < 5 × 10−4), indicating that growth requires PBP 3. Furthermore, aztreonam has been reported not to prevent the recruitment of the final division protein, FtsN, into the septal ring (M. Wissel and D. Weiss, cited in reference 1), suggesting that L-form-like growth specifically requires PBP 3 transpeptidase activity.
For both spherical and rod-shaped cells, the antibiotic concentrations used were the lowest that gave a plating efficiency less than 10−3.
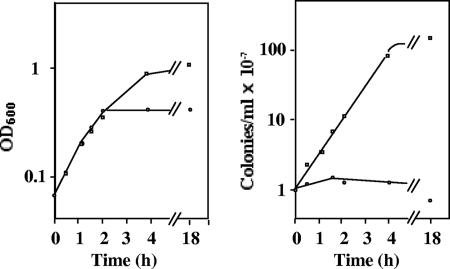
To see whether PBP 3 is required for the cell division of L-form-like cells, we looked at the effect of piperacillin in liquid M medium. To an exponentially growing culture of MG1655 in liquid M cefsulodin medium we added 5 μg/ml piperacillin. The OD600 of the culture continued to increase exponentially at the same rate as in the control for 2.5 generations, and then the increase stopped. The concentration of viable cells, in contrast, rapidly stopped increasing, with a total increment of less than 50% (Fig. 3). Microscope examination of the cells 4 h after piperacillin addition revealed large spherical cells and frequent clusters of small spherical granules. After overnight incubation, the OD600 had not changed and the viable cell count had dropped a mere twofold.
FIG. 3.
Growth of L-form-like cells in the presence of piperacillin. An exponentially growing culture of strain MG1655 in M cefsulodin medium was separated into two parts. At time zero, piperacillin (5 μg/ml) was added to one of them. At the indicated times, the OD600 of each culture was measured (left), and the cultures were diluted in M medium and assayed on M cefsulodin plates (right). Plates were counted after 24 h incubation. Symbols: squares, culture without piperacillin; circles, culture with piperacillin.
The same experiment was carried out on rod-shaped MG1655 cells growing exponentially in M medium without cefsulodin. In the presence of piperacillin, the OD600 increased for three generations, although at a rate lower than that for the untreated control. The concentration of viable cells increased by about 50%, remained constant for 2 hours, and then rapidly dropped 100-fold. The cells initially formed filaments; during overnight incubation, they lysed (data not shown).
These observations confirm that cefsulodin-induced L-form-like cells require PBP 2 and PBP 3 activity and strongly suggest that the latter is required, as for rod-shaped cells, for cell division.
The FtsZ protein is found in all bacteria, including cell wall-less mycoplasmas. In bacteria with a cell wall, FtsZ forms the midcell ring which, when completed with the other cell division proteins, carries out septum synthesis. To test whether FtsZ is required for the division of cefsulodin-induced L-form-like cells, we used strain VIP205, in which the chromosomal ftsZ gene has been put under exclusive Ptac control; the strain grows only in the presence of an inducer of the lac operon (16). On M plates containing 3 × 10−5 M IPTG, strain VIP205 formed normal rod-shaped cells; on M cefsulodin IPTG plates, it formed spherical cells (EOP, 0.6). In the absence of IPTG, no growth was observed in the presence or absence of cefsulodin (EOP < 2 × 10−4). Thus, the cell division protein FtsZ is required for colony formation on M cefsulodin plates.
Penicillin-induced L-form-like cells.
E. coli MG1655 is unable to grow on the surfaces of M plates containing 1,000 U/ml penicillin G (EOP < 2 × 10−7). L-form-like cells of wild-type MG1655 pregrown in M cefsulodin medium are also unable to grow on the surfaces of M penicillin plates (EOP < 2 ×10−4). The EOP within the agar, however, is high, about 0.5, and the cells in the colonies are spherical; such cells were considered unstable L forms by Lederberg and St. Clair (31). We were unable to separate the cells from the agar to test for the presence of peptidoglycan and to evaluate osmosensitivity, etc. We could, however, examine the genetic requirements for this L-form-like growth in the presence of penicillin.
Cultures of murI::Kmr, dapA::Eryr, and ΔmurA/pBADmurA+ derivatives of MG1655 were grown in liquid M medium containing d-glutamate, DAP, and l-arabinose, respectively. They were then plated within the agar layers of M plates containing 1,000 U/ml penicillin G, with or without the appropriate growth requirement. The mutants all grew in M penicillin agar when the growth supplement was present (EOP ≥ 0.1), but none could grow in the absence of its requirement (EOP < 5 × 10−5 in all cases). Penicillin-resistant growth within the agar layer also required a colanic acid capsule, as evidenced by the inability of the cpsE::Tn10 mutant to grow under these conditions (EOP < 2 × 10−5).
The above results indicate that growth within the agar layer of M penicillin plates requires d-glutamate, DAP, and MurA activity. This strongly suggests that L-form-like growth in the presence of penicillin (within the agar layer), like that in the presence of cefsulodin (on the plate surface), requires residual peptidoglycan synthesis.
DISCUSSION
Normal bacterial cell division is indissociable from septal peptidoglycan synthesis. In apparent contradiction with this, L-form derivatives of bacteria have been reported to have no peptidoglycan, yet they can grow and divide indefinitely. In the present work, we present a protocol for quantitatively converting all cells in a culture of a genetically well-defined E. coli strain to growing L-form-like cells. This can be done by adding the β-lactam cefsulodin, a specific inhibitor of PBPs 1A and 1B, to a culture of an E. coli K-12 strain like MG1655 growing in a rich hypertonic medium such as our M medium. The procedure works both on plates and in liquid culture. Like classical L forms described in the literature, the cells are spherical, osmosensitive, smaller on the average than rod-shaped cells, and more heterogeneous in size. Using appropriate mutants, we found to our surprise that the propagation of these L-form-like cells requires d-glutamate, DAP, and MurA activity, all specific to peptidoglycan synthesis. Direct measurement revealed that these cells do in fact contain peptidoglycan, about 7% of the amount in rod-shaped cells. Coupled with the genetic results, we conclude that this residual peptidoglycan synthesis is essential.
Like many workers before us, we were unable to find conditions under which wild-type E. coli is able to establish L-form growth on the surfaces of hypertonic plates containing penicillin. However, MG1655 can grow within the agar layer of M penicillin plates, producing spherical cells. Again to our surprise, we found that the propagation of penicillin-induced L-form-like cells within the agar layer requires d-glutamate, DAP, and MurA activity. This strongly suggests that L-form-like growth within M penicillin agar also requires residual peptidoglycan synthesis. We were unable to assay the peptidoglycan content of these cells. We nevertheless conclude that the propagation of both cefsulodin- and penicillin-induced L-form-like cells requires residual peptidoglycan synthesis.
What is the source of this residual peptidoglycan? In the presence of cefsulodin, rapid L-form-like growth clearly requires PBP 1B transglycosylase activity and PBP 2 and PBP 3 transpeptidase activity. These enzymes probably account for the residual peptidoglycan synthesis in the presence of cefsulodin. During growth within the agar layer in the presence of 1,000 U/ml penicillin G, the cells are likely to express various stress responses. We cannot say at present whether these protect one or more PBPs from total inactivation or permit the expression of alternative (unknown) peptidoglycan-synthesizing enzymes.
The L-form-like growth described here has an absolute requirement for a colanic acid capsule, the synthesis of which is governed by the RcsBCD system together with the additional regulators RcsA and RcsF (24, 32). The Rcs stress response is induced when the cell envelope is perturbed (24). It is also induced when PBPs 1A and 1B are specifically inactivated (41), consistent with our observations that cefsulodin treatment causes a general disorganization of both the cytoplasmic and the outer membrane.
What is the evidence that classical L forms have no peptidoglycan? The initial speculation that L forms lack a cell wall came from their mycoplasma-like morphology; indeed, they were initially thought to be mycoplasmas. Electron microscopy showed in many cases that there is no visible cell wall in L forms of different bacterial species, including E. coli (21). Biochemical analyses of cell wall constituents in L forms have given variable results, with numerous reports in which muramic acid, DAP, d-glutamate, or glucosamine was or was not detected in extracts of L forms of various bacteria, usually with little quantification. We are unaware of any published data that eliminate the possibility of 7% residual peptidoglycan in an established L form. We therefore speculate that a low level of residual peptidoglycan synthesis may be a requirement for the propagation of all L forms.
Little is known of what mutations L forms can tolerate. It was recently reported that an established E. coli L form isolated nearly 40 years ago has acquired mutations in several genes required for peptidoglycan synthesis and cell division (44). From the sequence of 36 kb of L-form DNA, the authors deduced that the FtsA, FtsW, and MurG proteins have one or two amino acid changes each; the ftsQ gene has an amber triplet at codon 132 (of 276 codons), and the mraY gene has a frame shift in codon 294 that should produce a protein of 298 amino acids (instead of 360). The functional consequences in rod-shaped cells of the missense mutations and of the truncation of MraY are unknown. The truncated FtsQ protein would almost certainly be nonfunctional for cell division in rod-shaped cells, although a low level of amber suppression could provide the 22 molecules of intact FtsQ estimated to be required for division (7). Further characterization of this classical L form should establish clearly whether or not the cells carry out residual peptidoglycan synthesis and, if they do, whether it is essential for their propagation.
What should be called an L form has been discussed since 1939, when it was shown that many bacterial species gave rise to forms similar to the L1 culture that Klieneberger had isolated from S. moniliformis. Since 1942, the methodology for establishing L forms has routinely involved numerous passages on complex hypertonic penicillin plates over an extended period of time. The first growing cells obtained, unstable L forms, are spherical and osmosensitive, and they revert to normal morphology in the absence of penicillin. After further passages, often extending over several years, stable (nonreverting) derivatives are obtained, and some authors have suggested that only these should be called L forms (28). Others, however, have presented convincing evidence that stabilization is a secondary event which simply prevents the reconstitution of a normal cell wall in the absence of penicillin but does not affect L-form growth (31).
What then is an L form? In the absence of a recognized authority empowered to establish such definitions, the wisest course is to describe clearly the origin and cultivation of the organisms used, whatever name they go by. This, unfortunately, is not always the case in the L-form literature. In the present work, to avoid confusion, we have called our spherical, osmosensitive cells “L-form-like.”
The L-form-like growth of E. coli described here, whether induced by cefsulodin or penicillin, requires residual peptidoglycan synthesis, amounting in the former case to 7% of that of wild-type cells. This raises the question of the function of this peptidoglycan. The situation is in some ways reminiscent of the paradox of chlamydial species, which are reputed to have no cell wall yet seem to require peptidoglycan synthesis, probably for cell division (33). The amount of peptidoglycan in L-form-like E. coli cells is far too little to form a sacculus covering the entire cell (53). Although techniques for locating this peptidoglycan within the cell are not presently available, the following arguments suggest that it may be at the division site of the spherical cells. PBP 3, which is specific to the septation process, is required for the propagation of cefsulodin-induced L-form-like cells, and when it is inhibited by piperacillin, there is a rapid block of the viable cell count (Fig. 3). The transglycosylase activity of PBP 1B is required for rapid growth of the L-form-like cells, and this protein has been shown to interact directly with PBP 3 (3); PBP 1B has also been implicated in cell division under certain conditions (15). The central cell division protein FtsZ is required for the propagation of L-form-like cells. PBP 2, although not normally a cell division protein, has been implicated in the division of spherical cells (51), and it is required for L-form-like growth. The simplest hypothesis to account for these observations is that cell division in cefsulodin-induced L-form-like cells, and possibly in all L forms, takes place, as in rods, by means of peptidoglycan synthesized in the division plane and indispensable for cytokinesis.
Acknowledgments
We thank Tanneke den Blaauwen for an extremely constructive dialogue and Eliora Ron, Miguel Angel de Pedro, and Conrad Woldringh for helpful comments on preliminary versions of our manuscript. M. Kohiyama kindly provided the FM 4-64. Christophe Chamot carried out the microscopy at the service “Imaging of Dynamic Processes in Cell and Developmental Biology” (Institut Jacques Monod). For strains and plasmids received we are grateful to Mary Berlyn, David Clarke, Didier Mazel, Dominique Mengin-Lecreulx, Miguel Vicente, Waldemar Vollmer, and Kevin Young.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Arends, S. J. R., and D. S. Weiss. 2004. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 186:880-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 3.Bertsche, U., T. Kast, B. Wolf, C. Fraipont, M. E. G. Aarsman, K. Kannenberg, M. von Rechenberg, M. Nguyen-Distèche, T. den Blaauwen, J.-V. Höltje, and W. Vollmer. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61:675-690. [DOI] [PubMed] [Google Scholar]
- 4.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. D., E. I. Vivas, C. T. Walsh, and R. Kolter. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukhari, A. I., and A. L. Taylor. 1971. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J. Bacteriol. 105:844-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson, M. J., J. Barondess, and J. Beckwith. 1991. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol. 173:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demarre, G., A.-M. Guérout, C. Matsumoto-Mashimo, D. A. Rowe-Magnus, P. Marlière, and D. Mazel. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245-255. [DOI] [PubMed] [Google Scholar]
- 9.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dienes, L. 1939. Organisms of Klieneberger and Streptobacillus moniliformis. J. Infect. Dis. 65:24-42. [Google Scholar]
- 11.Dienes, L. 1942. The significance of the large bodies and the development of L type of colonies in bacterial cultures. J. Bacteriol. 44:37-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublet, P., J. van Heijenoort, J.-P. Bohin, and D. Mengin-Lecreulx. 1993. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J. Bacteriol. 175:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrières, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 14.García del Portillo, F., and M. A. De Pedro. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol. 172:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García del Portillo, F., M. A. De Pedro, D. Joseleau-Petit, and R. D'Ari. 1989. Lytic response of Escherichia coli cells to inhibitors of penicillin-binding proteins 1a and 1b as a timed event related to cell division. J. Bacteriol. 171:4217-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrido, T., M. Sánchez, P. Palacios, M. Aldea, and M. Vicente. 1993. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 12:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitai, Z., M. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 18.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 19.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 20.Gumpert, J., and C. Hoischen. 1998. Use of cell wall-less bacteria (L-forms) for efficient expression and secretion of heterologous gene products. Curr. Opin. Biotech. 9:506-509. [DOI] [PubMed] [Google Scholar]
- 21.Gumpert, J., and U. Taubeneck. 1983. Characteristic properties and biological significance of stable protoplast type L-forms. Experientia Suppl. 46:227-241. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y.-H., L. Ferrières, and D. J. Clarke. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157:206-212. [DOI] [PubMed] [Google Scholar]
- 25.Iwai, N., K. Nagai, and M. Wachi. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 66:2658-2662. [DOI] [PubMed] [Google Scholar]
- 26.Klieneberger, E. 1936. Further studies on Streptobacillus moniliformis and its symbiont. J. Pathol. Bacteriol. 42:587-598. [Google Scholar]
- 27.Klieneberger, E. 1935. The natural occurrence of pleuropneumonia-like organisms in apparent symbiosis with Streptobacillus moniliformis and other bacteria. J. Pathol. Bacteriol. 40:93-105. [Google Scholar]
- 28.Klieneberger-Nobel, E. 1960. L-forms of bacteria, p. 361-386. In I. C. Gunsalus and R. Y. Stanier (ed.), The Bacteria, vol. 1. Academic Press, New York, NY. [Google Scholar]
- 29.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Molin, and V. De Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 177:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 31.Lederberg, J., and J. St. Clair. 1958. Protoplasts and L-type growth of Escherichia coli. J. Bacteriol. 75:143-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 33.McCoy, A. J., and A. T. Maurelli. 2006. Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14:70-77. [DOI] [PubMed] [Google Scholar]
- 34.Meberg, B. M., F. C. Sailer, D. E. Nelson, and K. D. Young. 2001. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J. Bacteriol. 183:6148-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisel, U., J. V. Höltje, and W. Vollmer. 2003. Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli. J. Bacteriol. 185:5342-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Noguchi, H., M. Matsuhashi, and S. Mitsuhashi. 1979. Comparative studies of penicillin-binding proteins in Pseudomonas aeruginosa and Escherichia coli. Eur. J. Biochem. 100:41-49. [DOI] [PubMed] [Google Scholar]
- 38.Onoda, T., A. Oshima, S. Nakano, and A. Matsuno. 1987. Morphology, growth and reversion in a stable L-form of Escherichia coli K12. J. Gen. Microbiol. 133:527-534. [DOI] [PubMed] [Google Scholar]
- 39.Onufryk, C., M.-L. Crouch, F. C. Fang, and C. A. Gross. 2005. Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 187:4552-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce, C. H. 1942. Streptobacillus moniliformis, its associated L1 form, and other pleuropneumonia-like organisms. J. Bacteriol. 43:780. [Google Scholar]
- 41.Sailer, F. C., B. M. Meberg, and K. D. Young. 2003. β-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol. Lett. 226:245-249. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Schuhmann, E., and U. Taubeneck. 1969. Stabile L-Formen verschiedener Escherichia coli-Stämme. Z. Allg. Mikrobiol. 9:297-313. [DOI] [PubMed] [Google Scholar]
- 44.Siddiqui, R. A., C. Hoischen, O. Holst, I. Heinze, B. Schlott, J. Gumpert, S. Diekmann, F. Grosse, and M. Platzer. 2006. The analysis of cell division and cell wall synthesis genes reveals mutationally inactivated ftsQ and mraY in a protoplast-type L-form of Escherichia coli. FEMS Microbiol. Lett. 258:305-311. [DOI] [PubMed] [Google Scholar]
- 45.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spratt, B. G. 1977. The mechanism of action of mecillinam. J. Antimicrob. Chemother. 3(Suppl. B):13-19. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, H., Y. Nishimura, and Y. Hirota. 1978. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 75:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taubeneck, U., and E. Schuhmann. 1966. Stabile, penicillininduzierte L-Formen von E. coli B. Z. Allg. Mikrobiol. 6:341-343. [Google Scholar]
- 49.Trisler, P., and S. Gottesman. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ursinus, A., F. van den Ent, S. Brechtel, M. A. de Pedro, J.-V. Höltje, and W. Vollmer. 2004. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 186:6728-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vinella, D., D. Joseleau-Petit, D. Thévenet, P. Bouloc, and R. D'Ari. 1993. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, relieved by FtsZ overexpression. J. Bacteriol. 175:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 53.Wientjes, F. B., C. L. Woldringh, and N. Nanninga. 1991. Amount of peptidoglycan in cell walls of gram-negative bacteria. J. Bacteriol. 173:7684-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]