Abstract
A metabolic isotope-labeling strategy was used in conjunction with nano-liquid chromatography-electrospray ionization mass spectrometry peptide sequencing to assess quantitative alterations in the expression patterns of subunits within cellulosomes of the cellulolytic bacterium Clostridium thermocellum, grown on either cellulose or cellobiose. In total, 41 cellulosomal proteins were detected, including 36 type I dockerin-containing proteins, which count among them all but three of the known docking components and 16 new subunits. All differential expression data were normalized to the scaffoldin CipA such that protein per cellulosome was compared for growth between the two substrates. Proteins that exhibited higher expression in cellulosomes from cellulose-grown cells than in cellobiose-grown cells were the cell surface anchor protein OlpB, exoglucanases CelS and CelK, and the glycoside hydrolase family 9 (GH9) endoglucanase CelJ. Conversely, lower expression in cellulosomes from cells grown on cellulose than on cellobiose was observed for the GH8 endoglucanase CelA; GH5 endoglucanases CelB, CelE, CelG; and hemicellulases XynA, XynC, XynZ, and XghA. GH9 cellulases were the most abundant group of enzymes per CipA when cells were grown on cellulose, while hemicellulases were the most abundant group on cellobiose. The results support the existing theory that expression of scaffoldin-related proteins is coordinately regulated by a catabolite repression type of mechanism, as well as the prior observation that xylanase expression is subject to a growth rate-independent type of regulation. However, concerning transcriptional control of cellulases, which had also been previously shown to be subject to catabolite repression, a novel distinction was observed with respect to endoglucanases.
Clostridium thermocellum, a thermophilic, strictly anaerobic gram-positive bacterium, has the highest rate of cellulose utilization of any bacterium, and for this reason it is deemed of great significance to the pursuit of biofuel production from the cellulosic materials in plant biomass (3, 6, 20, 32). The organism achieves hydrolysis of crystalline cellulose by virtue of a large cell surface-bound protein complex known as the cellulosome, the structure of which consists of a central noncatalytic scaffoldin protein (CipA) bearing up to nine catalytic subunits (44). The attachment of a given subunit is mediated by the interaction of its type I dockerin (Doc1) domain with one of the nine cohesin type I domains of CipA (26). CipA is, in turn, bound to the cell surface by virtue of the interaction of its type II dockerin domain with the type II cohesin domain of one of three S-layer anchor proteins, SdbA, Orf2p, or OlpB (6). CipA also contains a type III cellulose-binding module for attachment of the complex to cellulose (13).
Previous studies have shown that cellulolytic activity in C. thermocellum is regulated by either carbon source or growth rate (or both) and that changes with respect to one or the other are reflected in overall cellulase production (47) and in the cellulosomal subunit profile (4, 11, 28, 35). Catabolite repression by nonlimiting concentrations of readily metabolized carbon sources has been the standing hypothesis for cellulase regulation in C. thermocellum for more than 20 years (12). The immediate availability of energy results in an increased growth rate and leads to the repression of genes required to mine energy from crystalline cellulose. Lower growth rates and cellulose as a substrate seem to promote cellulase production, as has been demonstrated for the processive glycoside hydrolase family 48 (GH48) exoglucanase CelS, both at the protein (4) and the mRNA level (7, 38), as well as for the transcription of the GH5 endoglucanases celB and celG and the GH9 endoglucanase celD (9). Transcription of the scaffoldin gene cipA and cell surface anchoring genes olpB and orf2p is likewise controlled by growth rate and/or carbon source, which is not the case for another cell surface gene, sdbA (8, 38).
Sequencing and annotation of the C. thermocellum ATCC 27405 genome led to the discovery of more than 60 open reading frames coding for products with putative Doc1 domains (50), that is, proteins that can potentially bind to CipA and contribute to cellulosomal activities. Among these are genes for endoglucanases, exoglucanases, xylanases, and other hemicellulases. The predicted catalytic activity or function of about one-quarter of these genes is unknown. Considering the number of “dockable” candidate open reading frames, relatively few, or about one-third, of the products of these genes have been identified from the cellulosome complex itself. The participation in the cellulosome of the remaining putative gene products remains moot.
Low expression levels and overlapping and/or novel biochemical activity not detected by frequently used activity assays can account for the difference between the number of cellulosomal proteins predicted and the number of those that have been biochemically characterized. Mass spectrometry (MS) has become an increasingly popular tool in the study of proteins due to its high sensitivity and mass accuracy, and its quantitative applications are being progressively refined (36). The most wide-ranging C. thermocellum cellulosome study until now coupled a two-dimensional gel electrophoresis system with protein mass fingerprinting by matrix-assisted laser desorption ionization MS, giving rise to the simultaneous identification of 13 docking components from a cellulose-grown culture (50).
In the present study, we report quantitative differences between the subunit profiles of cellulosomes from cells grown in liquid batch cultures on Avicel (crystalline cellulose) versus cellobiose as the carbon source. In comparing the cellulosomes from cells grown on these two substrates, we expected to detect several novel gene products and also to uncover differences in protein expression that can shed more light on our understanding of the regulation of cellulosomal cellulases and hemicellulases. A metabolic isotope-labeling strategy was used in conjunction with nano-liquid chromatography-electrospray ionization MS (nano-LC-ESI-MS) peptide sequencing to assess alterations in the expression patterns within cellulosomes grown under different conditions. Moreover, a peptide-counting technique was applied to approximate the relative abundance of each cellulosome component per sample.
MATERIALS AND METHODS
Metabolic labeling and cellulosome purification.
C. thermocellum strain ATCC 27405 was grown anaerobically at 58°C in 100-ml batch cultures with ATCC medium 1191, prepared without sodium sulfide and containing either Avicel PH101 (Fluka-Biochemika) or cellobiose (Sigma-Aldrich) at 0.2% (wt/vol). An Avicel-grown reference culture was prepared similarly, substituting 99% 15N-enriched NH4Cl (Cambridge Isotope Laboratories, Andover, MA) for the nitrogen source in the medium and pyridoxal-HCl for pyridoxine-HCl. A 5% inoculum of unlabeled Avicel-grown cells was passed three times into 15NH4Cl-containing medium before inoculation of the final reference batch, which was consequently enriched with 15N to an estimated 98.9%. All cultures were harvested for protein isolation in late stationary phase (70 h). Each test culture was mixed 1:1 (vol/vol) with the reference culture. Supernatants were collected by centrifuging culture mixtures at 10,000 × g for 10 min. To 900 ml of each mixture was added 14 mg of phosphoric acid-swollen cellulose, and cellulosomes were prepared by the affinity digestion method adapted by Zhang et al. (45), using Pierce Slide-A-Lyzer cassettes (molecular weight cutoff of 10,000). After a 5-h digestion and dialysis period at 58°C, the contents of the cassettes were removed and precipitated with 4 volumes of cold acetone. The precipitates were collected by centrifugation, dried down in a SpeedVac, and suspended in 50 mM Tris-HCl, pH 7.4, each to a final concentration of approximately 10 mg · ml−1, as verified by Bradford assay.
Analysis of purified cellulosomes by nano-LC-ESI-MS.
The resulting purified cellulosomes were separated by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. Sample lanes from the gel were excised and divided into 15 gel bands, with each band containing on average roughly 11 μg of protein. The protein in each gel band was subsequently reduced, alkylated, and digested with trypsin TPCK (N-tosyl-l-phenylalanine chloromethyl ketone; Sigma-Aldrich), as described previously (24). The resulting peptide mixtures were removed from the gel pieces using excess extraction buffer, dried, and then made up in equal volumes of 8% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid. Peptide samples were injected quantitatively for separation on a PicoFrit BioBasic C18 nanocolumn (New Objective; 10-cm length by 75-μm inner diameter; 5-μm particle size; 300-Å pore size) with a 60-min solvent gradient, ranging from 3% to 50% acetonitrile in 0.1% formic acid, at a flow rate of 1 μl · min−1. Before flowing to the column, the sample was cleaned of impurities using a C18 peptide trap. Under these conditions, most peptides eluted in about 30 s or 500 nl. Detection and sequencing of peptide ions was accomplished by an LTQ ion trap MS (Thermo Electron, San Jose, CA), equipped with an ESI nanosource and operating in positive mode with a voltage of 1.4 kV applied at a liquid junction just upstream of the column. An initial full MS survey scan (∼10 ms) was performed for the m/z range of 400 to 2,000, followed by several data-dependent scans (∼33 ms each). The seven most abundant ions from the survey scan were subjected to tandem MS (MS/MS) for sequencing using pulsed-Q dissociation for ion fragmentation. A triggering threshold of three times the noise level (signal-to-noise ratio [S/N]) was applied for MS/MS events. Peptide ions that triggered an MS/MS more than once within a 30-s window were placed on an exclusion list for 3 min to improve the possibility of detecting less abundant ions.
Database screening and success criteria.
Using SEQUEST from BioWorks 3.3 (Thermo Electron), the peptide sequence results were searched against the 16 February 2007 release of the C. thermocellum genome available at NCBI courtesy of the Department of Energy, Joint Genome Institute (http://www.ncbi.nlm.nih.gov; Refseq accession number NC_009012). The database was digested in silico with trypsin and indexed for carboxymethylation of cysteine residues to include masses within the range of 400 to 3,500 Da. A peptide tolerance of ±2 atomic mass units was implemented. Charge state analysis was performed during DTA file filtering, and a series of high-stringency filters was applied to the search results. Singly, doubly, and triply charged peptide ions required SEQUEST cross-correlation (XC) scores of at least 1.8, 2.5, and 3.5, respectively. Peptide and protein hits also needed probability scores, as calculated by BioWorks, of less than 10−3. Moreover, only proteins identified on the basis of two or more unique peptides were considered in the final analysis. The SignalIP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) was used to verify that proteins contained an N-terminal peptide signaling secretion from the cell (10).
RelEx analysis.
DTA files were filtered separately using DTASelect (39), which assembles the peptides into proteins using the same XC score stringency factors as above. The filtered DTA files were then analyzed by RelEx (33), which generates extracted ion chromatograms of peptide isotope pairs and uses the areas under each curve to calculate a peptide signal ratio of sample to isotope-labeled reference. An extracted ion chromatogram pair was rejected if the S/N ratio was below 3 or if the correlation factor, the measure of the overlap of the curves, was below 0.9. Protein ratios were calculated as averages of the ratios of the peptides matched to them. The ratio of each unlabeled Avicel-grown protein to 15N-labeled Avicel-grown protein was divided by the ratio of the corresponding unlabeled cellobiose-grown protein to 15N-labeled Avicel-grown protein. The quotient of the ratios is the ratio of unlabeled Avicel-grown protein to cellobiose-grown protein. In such a way, this strategy corrects for any systematic errors introduced during sample preparation (33). All ratios were normalized to that obtained for the comparison of CipA.
emPAI analysis.
The exponentially modified protein abundance index (emPAI), which was shown to bear a linear relationship to protein concentration, is defined as 10PAI minus 1, where PAI is the ratio of the number of MS-observed peptides for a given protein over its theoretically observable peptides (19). The unique peptide parent ions matched for a given protein were counted as its observed peptides. For theoretical peptides, the relative hydrophobicity of a protein's in silico tryptic digest products (no missed cleavages) was calculated using the Sequence Specific Retention Calculator available at http://hs2.proteome.ca/SSRCalc/SSRCalc.html (25). Peptide retention times were predicted based on relative hydrophobicity and coefficients derived from our data set. Theoretical peptides were accepted within a retention time window of 12 to 68 min and a mass window of 400 to 3,500 Da. All emPAI values were normalized to that obtained for CipA, assuming that one CipA protein exists per cellulosome.
RESULTS
Detection and relative abundance of cellulosomal proteins induced by Avicel or cellobiose.
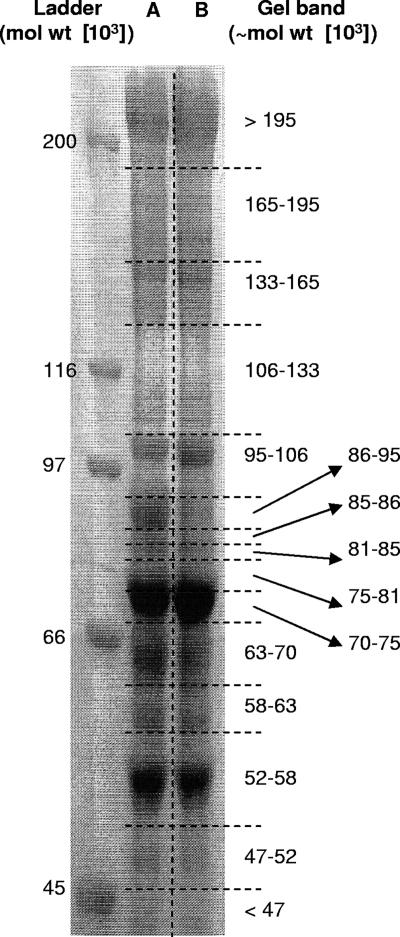
For investigation of substrate-induced changes to the cellulosomal subunit profile of C. thermocellum, cellulosome complexes were isolated from the extracellular materials of batch cultures grown to late stationary phase on either Avicel or cellobiose. Prior to cellulosome isolation, each culture was mixed with an equal volume of a 15N-labeled Avicel-grown culture for quantitation at a later step. Purified cellulosomes were denatured, and the components were separated by SDS-PAGE. Proteins in the gel bands (Fig. 1) were trypsin digested and extracted for analysis.
FIG. 1.
C. thermocellum cellulosomal protein separated by SDS-PAGE (6%), stained with Coomassie blue. Lane A, 1:1 (vol/vol) mixture of unlabeled cellobiose-grown and 15N-labeled Avicel-grown cellulosomes from late stationary phase (170 μg of total protein); lane B, 1:1 (vol/vol) mixture of unlabeled Avicel-grown and 15N-labeled Avicel-grown cellulosomes from late stationary phase (170 μg of total protein). Molecular weight (mol wt) markers are shown at left. At right, the approximate molecular weight ranges for the division of the gel bands for trypsin digestion are shown.
In total, 41 cellulosomal proteins in the C. thermocellum database were detected between the two samples, 35 on Avicel (Table 1) and 34 on cellobiose (Table 2), with 29 common to both samples. Thus, a similar number of subunits were detected under the two growth conditions. A total of 36 docking components were identified, including 16 subunits that have never been observed experimentally as components of the cellulosome. The specificity of the methodology is such that the matching of only two unique peptides to one protein out of the 3,238 proteins in the C. thermocellum database resulted in a probability of at worst 10−5 that another protein could have been matched. The molecular weights of the proteins identified generally corresponded to the gel bands in which they were detected; deviations from this trend suggested possible proteolysis or glycosylation. The 17 new proteins identified in this study are indicated in Tables 1, 2, and 3 by boldface. The reference protein from Avicel-grown cells did not interfere with the identification of cellulosomal proteins from cellobiose-grown cells in the mixed sample as SEQUEST analysis could not identify 15N-labeled peptides given the LC conditions and MS parameters applied. This was tested in an earlier experiment (data not shown), where 15N-labeled cellulosomes were isolated independently and analyzed by nano-LC-ESI-MS. No proteins were identified using SEQUEST and the same criteria as described above.
TABLE 1.
C. thermocellum Avicel-grown cellulosomal components identified by nano-LC-ESI-MS, ranked by emPAIa
| GenInfo identifier | Protein | (Putative) function or activityc | No. of peptide ionsd | emPAI | emPAI/CipAe | Doc1/CipA (mol%)f | PProg | XC score | Coverage (%)h | Mol wt (103) | Gel band mol wt (103) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 125975556 | CipA | Scaffoldin | 42 | 5.92 | 1.00 | 2.2E-12 | 378 | 34 | 196.7 | >195 | 44 | |
| 125972933 | CelK | Exoglucanase (GH9) | 39 | 4.12 | 0.70 | 11.0 | 2.0E-12 | 350 | 35 | 100.6 | 95-106 | 23 |
| 125974579 | CelS | Exoglucanase (GH48) | 29 | 3.56 | 0.60 | 9.4 | 6.9E-10 | 240 | 32 | 83.5 | 75-81 | 44 |
| 125973097 | CelR | Endoglucanase (GH9) | 28 | 3.19 | 0.54 | 8.5 | 9.3E-11 | 250 | 31 | 82.1 | 81-86 | 50 |
| 125975557 | OlpB | Cell-surface anchor | 27 | 2.75 | 0.47 | 3.4E-13 | 210 | 26 | 248.0 | 165-195 | 31 | |
| 125973339 | GH5 | 15 | 2.59 | 0.44 | 6.9 | 1.3E-09 | 140 | 25 | 63.0 | 59-63 | ||
| 125972791 | CelA | Endoglucanase (GH8) | 14 | 2.46 | 0.41 | 6.4 | 1.1E-08 | 120 | 22 | 52.6 | 52-59 | 16 |
| 125973315 | CelE | Endoglucanase (GH5), CE2 | 24 | 2.24 | 0.38 | 6.0 | 9.1E-13 | 200 | 32 | 90.2 | 86-95 | 18 |
| 125973142 | CelJ | Endoglucanase (GH9), GH44 | 42 | 2.16 | 0.37 | 5.8 | 2.4E-13 | 390 | 24 | 178.0 | 165-195 | 16 |
| 125974342 | XynC | Xylanase (GH10) | 18 | 1.57 | 0.26 | 4.1 | 2.0E-10 | 160 | 24 | 69.5 | 81-95 | 16 |
| 125974464 | XynZ | Xylanase (GH10), CE1 | 18 | 1.57 | 0.26 | 4.1 | 2.0E-09 | 150 | 20 | 92.2 | 63-70 | 50 |
| 125972934 | CbhA | Exoglucanase (GH9) | 30 | 1.51 | 0.26 | 4.1 | 3.2E-11 | 280 | 22 | 137.0 | 133-165 | 54 |
| 125975294 | CelT | Endoglucanase (GH9) | 14 | 1.29 | 0.22 | 3.4 | 6.0E-11 | 120 | 21 | 68.5 | 59-70 | 27 |
| 125975353 | CelG | Endoglucanase (GH5) | 9 | 1.15 | 0.20 | 3.1 | 1.1E-07 | 90 | 16 | 63.2 | 81-85 | 30 |
| 125975452 | XynA | Xylanase (GH11), CE4 | 12 | 1.15 | 0.20 | 3.1 | 1.6E-08 | 100 | 14 | 74.4 | 47-52 | 17 |
| 125973912 | XghA | Xyloglucanase (GH74) | 21 | 1.13 | 0.19 | 3.0 | 3.4E-10 | 180 | 22 | 92.3 | 86-95 | 50 |
| 125973263 | GH9 | 15 | 0.94 | 0.16 | 2.5 | 7.4E-08 | 140 | 17 | 82.1 | 85-95 | ||
| 125973062 | CelF | Endoglucanase (GH9) | 12 | 0.90 | 0.15 | 2.4 | 3.3E-08 | 120 | 19 | 82.0 | 81-85 | 34 |
| 125973254 | -b | Cell-surface anchor | 21 | 0.82 | 0.14 | 5.6E-09 | 178 | 19 | 140.5 | 133-165 | ||
| 125974678 | GH5 | 11 | 0.71 | 0.12 | 1.9 | 4.0E-10 | 100 | 9.3 | 103.1 | 106-133 | ||
| 125972735 | LicBb | Lichenase (GH16) | 4 | 0.62 | 0.11 | 1.7 | 4.1E-07 | 28.2 | 8.4 | 37.9 | <47 | 49 |
| 125975293 | ManAb | Mannanase (GH26) | 7 | 0.61 | 0.10 | 1.6 | 7.5E-10 | 70.2 | 19 | 67.0 | 63-70 | 15 |
| 125973143 | CelQb | Endoglucanase (GH9) | 9 | 0.58 | 0.10 | 1.6 | 1.0E-13 | 70.2 | 14 | 79.8 | 70-81 | 2 |
| 125972556 | GH26 | 5 | 0.53 | 0.09 | 1.4 | 1.2E-07 | 40.2 | 5.8 | 67.3 | 63-70 | ||
| 125973055 | CelB | Endoglucanase (GH5) | 5 | 0.53 | 0.09 | 1.4 | 6.1E-08 | 50.2 | 7.6 | 63.9 | 63-70 | 7 |
| 125975558 | Orf2p | Cell-surface anchor | 7 | 0.50 | 0.08 | 5.9E-08 | 70.2 | 15 | 74.9 | 86-95 | 8 | |
| 125972796 | -b | GH9 | 5 | 0.49 | 0.08 | 1.3 | 3.7E-06 | 50.2 | 11 | 62.6 | 59-63 | |
| 125975243 | GH9 | 8 | 0.44 | 0.07 | 1.1 | 6.7E-06 | 80.2 | 9.3 | 80.2 | 85-86 | ||
| 125972926 | GH5 | 3 | 0.33 | 0.06 | 0.9 | 1.7E-06 | 30.1 | 6.3 | 59.9 | 59-63 | ||
| 125972567 | CelNb | Endoglucanase (GH9) | 4 | 0.27 | 0.05 | 0.8 | 3.1E-06 | 40.2 | 5.7 | 82.1 | 85-86 | 50 |
| 125973343 | CelD | Endoglucanase (GH9) | 4 | 0.23 | 0.04 | 0.6 | 6.2E-06 | 30.2 | 5.2 | 72.4 | 59-70 | 21 |
| 125972954 | GH9 | 4 | 0.21 | 0.04 | 0.6 | 7.3E-06 | 40.2 | 2.9 | 89.4 | 95-106 | ||
| 125972792 | ChiA | Chitinase (GH18) | 2 | 0.20 | 0.03 | 0.5 | 8.9E-08 | 20.2 | 4.8 | 55.4 | 47-52 | 48 |
| 125973914 | GH53 | 2 | 0.17 | 0.03 | 0.5 | 5.5E-07 | 20.1 | 6.0 | 47.0 | <47 | ||
| 125973158 | -b | Endopygalactorunase | 2 | 0.16 | 0.03 | 0.5 | 7.0E-07 | 20.2 | 5.2 | 64.5 | 59-63 |
Proteins in boldface are those that have never been observed experimentally in purified cellulosomes previous to this study.
Found only in the Avicel sample.
CE, carbohydrate esterase family.
Number of unique parent peptide ions matched (including different charge states, modifications).
emPAI normalized to the value obtained for CipA.
Molar percentage per CipA for Doc1-containing subunits, calculated as 100 × (emPAI/CipA)/[Σ(emPAI/CipA)docking subunits].
Probability of finding a match as good or better than the observed match by chance.
Percentage of amino acid coverage to the matched protein.
TABLE 2.
C. thermocellum cellobiose-grown cellulosomal components identified by nano-LC-ESI-MS, ranked by emPAIa
| GenInfo identifier | Protein | (Putative) function or activityc | No. of peptide ionsd | emPAI | emPAI/CipAe | Doc1/CipA (mol%)f | PProg | XC score | Coverage (%)h | Mol wt (103) | Gel band mol wt (103) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 125974464 | XynZ | Xylanase (GH10), CE1 | 25 | 2.70 | 1.25 | 14.0 | 4.0E-13 | 200 | 31 | 92.2 | 86-95 | 50 |
| 125975556 | CipA | Scaffoldin | 25 | 2.16 | 1.00 | 1.5E-10 | 230 | 30 | 196.7 | >195 | 44 | |
| 125975452 | XynA | Xylanase (GH11), CE4 | 17 | 1.97 | 0.91 | 10.2 | 1.9E-11 | 170 | 31 | 74.4 | 59-85 | 17 |
| 125974342 | XynC | Xylanase (GH10) | 16 | 1.31 | 0.61 | 6.8 | 8.1E-11 | 160 | 20 | 69.5 | 63-70 | 16 |
| 125972791 | CelA | Endoglucanase (GH8) | 9 | 1.22 | 0.56 | 6.3 | 7.2E-10 | 90.2 | 22 | 52.6 | 52-59 | 16 |
| 125973912 | XghA | Xyloglucanase (GH74) | 22 | 1.21 | 0.56 | 6.3 | 1.6E-10 | 210 | 27 | 92.3 | 86-95 | 50 |
| 125973339 | GH5 | 9 | 1.15 | 0.53 | 5.9 | 3.2E-05 | 80.2 | 13 | 63.0 | 59-63 | ||
| 125972933 | CelK | Exoglucanase (GH9) | 18 | 1.12 | 0.52 | 5.8 | 6.7E-09 | 180 | 21 | 100.6 | 95-106 | 23 |
| 125973315 | CelE | Endoglucanase (GH5), CE2 | 14 | 0.99 | 0.46 | 5.1 | 1.1E-12 | 120 | 20 | 90.2 | 86-95 | 18 |
| 125972556 | GH26 | 8 | 0.98 | 0.45 | 5.0 | 1.4E-08 | 60.2 | 9.5 | 67.3 | 63-70 | ||
| 125973055 | CelB | Endoglucanase (GH5) | 7 | 0.82 | 0.38 | 4.2 | 6.4E-07 | 70.2 | 15 | 63.9 | 63-70 | 7 |
| 125975557 | OlpB | Cell-surface anchor | 11 | 0.71 | 0.33 | 1.2E-10 | 90.2 | 14 | 248.0 | 165-195 | 31 | |
| 125974678 | GH5 | 10 | 0.63 | 0.29 | 3.2 | 7.6E-07 | 88.2 | 11 | 103.1 | 106-133 | ||
| 125975073 | XynDb | Xylanase (GH10) | 7 | 0.58 | 0.27 | 3.0 | 4.7E-07 | 70.2 | 15 | 71.6 | 70-75 | 50 |
| 125975353 | CelG | Endoglucanase (GH5) | 5 | 0.53 | 0.25 | 2.8 | 3.9E-08 | 50.1 | 9.2 | 63.2 | 63-70 | 30 |
| 125973097 | CelR | Endoglucanase (GH9) | 8 | 0.51 | 0.23 | 2.6 | 9.6E-08 | 90.2 | 8.6 | 82.1 | 81-85 | 50 |
| 125973062 | CelF | Endoglucanase (GH9) | 7 | 0.45 | 0.21 | 2.3 | 1.3E-05 | 70.2 | 6.4 | 82.0 | 81-85 | 34 |
| 125972934 | CbhA | Exoglucanase (GH9) | 11 | 0.40 | 0.19 | 2.1 | 5.5E-07 | 108 | 8.6 | 137.0 | 133-165 | 54 |
| 125972926 | GH5 | 3 | 0.33 | 0.15 | 1.7 | 1.6E-07 | 30.1 | 6.5 | 59.9 | 59-63 | ||
| 125973786 | -b | GH43 | 5 | 0.32 | 0.15 | 1.7 | 2.9E-06 | 50.1 | 7.8 | 74.5 | 63-75 | |
| 125975243 | GH9 | 6 | 0.31 | 0.14 | 1.6 | 1.7E-06 | 60.2 | 6.5 | 80.2 | 85-86 | ||
| 125975294 | CelT | Endoglucanase (GH9) | 4 | 0.27 | 0.12 | 1.3 | 3.6E-05 | 40.2 | 6.5 | 68.5 | 59-63 | 27 |
| 125973263 | GH9 | 5 | 0.25 | 0.11 | 1.2 | 8.9E-08 | 50.2 | 6.6 | 82.1 | 85-86 | ||
| 125974579 | CelS | Exoglucanase (GH48) | 4 | 0.23 | 0.11 | 1.2 | 9.8E-08 | 40.1 | 5.4 | 83.5 | 75-81 | 44 |
| 125972792 | ChiA | Chitinase (GH18) | 2 | 0.20 | 0.09 | 1.0 | 7.3E-08 | 20.1 | 2.5 | 55.4 | 47-52 | 48 |
| 125973142 | CelJ | Endoglucanase (GH9), GH44 | 6 | 0.18 | 0.08 | 0.9 | 1.3E-07 | 54.2 | 3.4 | 178.0 | 165-195 | 16 |
| 125973914 | GH53 | 2 | 0.17 | 0.08 | 0.9 | 5.0E-05 | 20.1 | 6.0 | 47.0 | <47 | ||
| 125972954 | GH9 | 3 | 0.15 | 0.07 | 0.8 | 6.3E-05 | 28.2 | 3.3 | 89.4 | 95-106 | ||
| 125972540 | -b | GH43, α-l-arabinofuranosidase B | 3 | 0.15 | 0.07 | 0.8 | 1.0E-05 | 30.2 | 4.1 | 79.0 | 63-75 | |
| 125975558 | Orf2p | Cell-surface anchor | 2 | 0.12 | 0.06 | 5.5E-05 | 20.2 | 1.9 | 74.9 | 85-95 | 8 | |
| 125973343 | CelD | Endoglucanase (GH9) | 2 | 0.11 | 0.05 | 0.6 | 2.8E-06 | 20.2 | 1.8 | 72.4 | 59-63 | 21 |
| 125973822 | SdbAb | Cell-surface anchor | 2 | 0.10 | 0.05 | 3.9E-05 | 20.2 | 4.6 | 68.6 | 63-70 | 29 | |
| 125974626 | -b | GH30, α-l-arabinofuranosidase B | 2 | 0.09 | 0.04 | 0.4 | 1.4E-06 | 20.2 | 2.7 | 110.6 | 106-133 | |
| 125973429 | XynYb | Xylanase (GH10), CE1 | 2 | 0.07 | 0.03 | 0.3 | 9.5E-06 | 20.2 | 1.2 | 119.6 | 106-133 | 42 |
Proteins in boldface are those that have never been observed experimentally in purified cellulosomes previous to this study.
Found only in the cellobiose sample.
CE, carbohydrate esterase family.
Number of unique parent peptide ions matched (including different charge states, modifications).
emPAI normalized to the value obtained for CipA.
Molar percentage per CipA for Doc1-containing subunits, calculated as 100 × (emPAI/CipA)/[Σ(emPAI/CipA)docking subunits].
Probability of finding a match as good or better than the observed match by chance.
Percentage of amino acid coverage to the matched protein.
TABLE 3.
Fractional differences in expression of C. thermocellum Avicel-grown cellulosomal components relative to cellobiose-grown components by RelEx, ranked by P value, and normalized to CipAa
| GeneInfo identifier | Protein name or type | Fractional difference in expression
|
Overall change on Avicel | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
14N Avicel/15N Avicel
|
14N cellobiose/15N Avicel
|
14N Avicel/14N cellobiose
|
||||||||||
| Ratio 1 | SD | No. of peptides | Ratio 2 | SD | No. of peptides | Ratio 1/ratio 2 | Pb | CipAc | SEd | |||
| 125975557 | OlpB | 1.404 | 0.299 | 80 | 0.134 | 0.016 | 7 | 10.453 | <0.0001 | 2.063 | 0.672 | Increase |
| 125975556 | CipA | 1.574 | 0.244 | 179 | 0.311 | 0.047 | 108 | 5.067 | <0.0001 | 1.000 | 0.305 | Increase |
| 125973339 | GH5 | 1.358 | 0.138 | 14 | 0.314 | 0.031 | 10 | 4.320 | <0.0001 | 0.853 | 0.220 | None |
| 125974678 | GH5 | 1.171 | 0.131 | 7 | 0.751 | 0.074 | 6 | 1.559 | <0.0001 | 0.308 | 0.081 | Decrease |
| 125972791 | CelA | 0.939 | 0.112 | 21 | 0.659 | 0.067 | 9 | 1.426 | <0.0001 | 0.281 | 0.075 | Decrease |
| 125973142 | CelJ | 1.959 | 0.220 | 48 | 0.121 | 0.019 | 3 | 16.191 | <0.0001 | 3.195 | 0.926 | Increase |
| 125972933 | CelK | 1.776 | 0.353 | 76 | 0.158 | 0.015 | 14 | 11.214 | <0.0001 | 2.213 | 0.680 | Increase |
| 125975294 | CelT | 1.338 | 0.244 | 14 | 0.197 | 0.011 | 3 | 6.791 | <0.0001 | 1.340 | 0.386 | None |
| 125973062 | CelF | 1.041 | 0.250 | 18 | 0.161 | 0.007 | 5 | 6.452 | <0.0001 | 1.273 | 0.415 | None |
| 125972934 | CbhA | 1.030 | 0.180 | 39 | 0.166 | 0.019 | 6 | 6.211 | <0.0001 | 1.226 | 0.369 | None |
| 125973097 | CelR | 1.441 | 0.325 | 27 | 0.252 | 0.032 | 7 | 5.719 | <0.0001 | 1.129 | 0.380 | None |
| 125974342 | XynC | 1.105 | 0.102 | 21 | 0.528 | 0.030 | 13 | 2.094 | <0.0001 | 0.413 | 0.100 | Decrease |
| 125974464 | XynZ | 1.043 | 0.264 | 32 | 4.323 | 1.244 | 48 | 0.241 | <0.0001 | 0.048 | 0.021 | Decrease |
| 125975452 | XynA | 1.095 | 0.25 | 27 | 2.144 | 0.684 | 32 | 0.511 | <0.0001 | 0.101 | 0.045 | Decrease |
| 125974579 | CelS | 0.932 | 0.138 | 81 | 0.028 | 0.006 | 4 | 33.274 | <0.0001 | 6.567 | 2.171 | Increase |
| 125973912 | XghA | 1.035 | 0.151 | 24 | 1.662 | 0.332 | 33 | 0.623 | <0.0001 | 0.123 | 0.040 | Decrease |
| 125975353 | CelG | 0.947 | 0.245 | 10 | 0.333 | 0.035 | 4 | 2.842 | 0.0004 | 0.561 | 0.198 | Decrease |
| 125972556 | GH26 | 0.871 | 0.090 | 3 | 1.748 | 0.197 | 5 | 0.499 | 0.0004 | 0.098 | 0.026 | Decrease |
| 125973315 | CelE | 1.032 | 0.187 | 23 | 0.736 | 0.205 | 9 | 1.401 | 0.0005 | 0.277 | 0.110 | Decrease |
| 125973263 | GH9 | 1.438 | 0.408 | 12 | 0.546 | 0.061 | 4 | 2.633 | 0.0008 | 0.520 | 0.194 | Decrease |
| 125973055 | CelB | 0.996 | 0.080 | 4 | 1.320 | 0.231 | 6 | 0.754 | 0.0291 | 0.149 | 0.043 | Decrease |
| 125972954 | GH9 | 1.565 | 0.245 | 2 | 0.938 | 0.073 | 2 | 1.669 | 0.0742 | 0.329 | 0.091 | None |
| 125975243 | GH9 | 0.978 | 0.185 | 8 | 1.110 | 0.102 | 6 | 0.881 | 0.1426 | 0.174 | 0.052 | None |
| 125972792 | ChiA | 1.199 | 0.054 | 2 | 0.774 | 0.253 | 2 | 1.548 | 0.1463 | 0.306 | 0.121 | None |
| 125973914 | GH53 | 1.372 | 0.257 | 2 | 1.007 | 0.198 | 2 | 1.362 | 0.2528 | 0.269 | 0.093 | None |
Proteins in boldface are those that have never been observed experimentally previous to this study.
P value, probability that the null hypothesis is true, based on a two-tailed Student t test of ratio1 versus ratio 2.
Normalized to the value obtained for CipA.
Standard error was calculated using the simple quotient rule of error propagation, where a protein's ratio on Avicel, its ratio on cellobiose, CipA's ratio on Avicel, and CipA's ratio on cellobiose were considered random and independent.
The emPAI method (19) was used to relate the number of unique peptides matched to a protein to the relative abundance of that protein in each sample. While attempts to standardize the emPAI method on our system revealed a divergence from linearity at higher concentrations such that higher-abundance proteins would be underestimated, the method nevertheless supplies a basis for informed analysis as to the abundance of particular proteins per cellulosome preparation. Since the affinity digestion method used to isolate cellulosomes pulls the complex down “by the CipA,” all relative abundance values (emPAI and RelEx below) were normalized to that obtained for CipA. This provided a protein-per-CipA basis for comparison between samples.
There are significant differences in the relative abundances of docking subunits per CipA between the two data sets as per molar percentage calculated from emPAI values. Exoglucanases accounted for a total molar percentage of 24.4% of the total moles per CipA of all docking subunits detected when cells were grown on Avicel but only 9.2% when cells were grown on cellobiose. The molar percentage of CelS dropped from 9.4% on Avicel to 1.2% on cellobiose, while values for the GH9 exoglucanases CelK and CbhA changed from 11.0 to 5.8% and 4.1 to 2.1%, respectively. Components with known endoglucanase activity accounted for a total molar percentage of 40.0% when cells were grown on Avicel, but this decreased to 26.1% on cellobiose. In total, GH9 cellulases decreased from 43.6% on Avicel to 19.2% on cellobiose, whereas enzymes containing a GH5 domain increased slightly from 20.2% on Avicel to 23.0% on cellobiose. The GH5 fold is predominantly associated with cellulases, but it has also been linked to hemicellulolytic activity (37). A new GH5 enzyme (gi 125973339) was detected among the most abundant catalytic subunits in both samples (6.9% on Avicel and 5.9% on cellobiose). It has a predicted mass of 63.0 kDa and exhibits SDS-PAGE migration properties similar to those of CelB and CelG, with masses of 63.9 and 63.2 kDa, respectively. Its overlap with these proteins might explain why it was not identified previously. Overall, the molar percentage of hemicellulases increased from 19.9% on Avicel to 50.3% on cellobiose. Docking subunits with xylanase activity accounted for a total of 11.3% of all docking subunits detected when cells were grown on Avicel, but their contribution increased to 34.3% when cells were grown on cellobiose. Other hemicellulases accounted for a total molar percentage of 8.6% on Avicel and 15.1% on cellobiose. GH9 cellulases were the most abundant group of enzymes per CipA when cells were grown on Avicel, while hemicellulases were the most abundant group on cellobiose.
Other notable differences between the two samples concern the 13 components detected exclusively in one sample but not the other. Detected only in Avicel-grown cellulosomes were GH9 endoglucanases CelN and CelQ, the GH16 lichenase LicB, the GH26 mannanase ManA, a new GH9 cellulase, a new subunit with putative endopygalactorunase activity, and a new cell-surface anchor protein predicted to have the same number of type II cohesin domains as OlpB but no SLH (S-layer homology) domain. XynD and XynY, both with GH10 xylanase activity, were detected exclusively in cellobiose-grown cellulosomes, along with the cell-surface anchor protein SdbA, a new bifunctional GH30/α-l-arabinofuranosidase B hemicellulase, a new GH43 glycosidase, and a new bifunctional GH43/α-l-arabinofuranosidase B glycosidase.
Relative differences in abundance of cellulosomal components induced by Avicel or cellobiose.
Simultaneous quantitative differences in the expression of all but four cellulosomal components common to both Avicel and cellobiose were measured by means of metabolically 15N-labeled peptides as internal standards. While emPAI supplied a means of determining the relative abundance of proteins in a given sample, RelEx provided a highly reliable way to compare the amount of a particular protein present in two samples. Sample-to-reference ratios were determined separately for Avicel- and cellobiose-grown cellulosomes, and the ratio of ratios represented the fractional difference between proteins grown on either substrate. Normalization of ratio values to the value obtained for the scaffoldin protein CipA allowed for comparison of changes in protein expression per cellulosome complex. That the average ratio of unlabeled Avicel-grown protein to 15N-labeled protein was 1.23 with a standard deviation of 0.29 (Table 3) suggests that our methodology was accurate (and precise) at determining ratios between cellulosomal proteins from two separate samples.
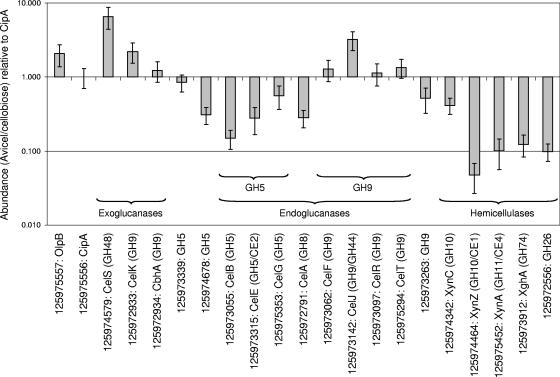
From the total of 29 proteins found in both samples, RelEx was able to determine a ratio of sample-to-reference for 25 protein pairs, given the S/N and correlation filters adopted (Table 3). The null hypothesis was rejected for all but four of these, for which it was determined that P was ≥0.05. There was no significant change in expression for these four proteins: two new GH9 cellulases and two hemicellulases, ChiA and a new GH53 subunit, whether obtained from Avicel- or cellobiose-grown cells. Proteins for which significant differences were observed are represented visually over a logarithmic scale in Fig. 2.
FIG. 2.
Fractional differences in expression of C. thermocellum Avicel-grown cellulosomal components relative to cellobiose-grown components by RelEx, normalized to CipA, over a logarithmic scale. Docking components are grouped by function and activity. CE, carbohydrate esterase family. Only proteins passing the null value with a P of <0.05 are shown. Columns rising above 1 represent proteins determined to have greater expression in the Avicel-grown sample. Columns falling below 1 represent proteins with higher expression in the cellobiose-grown sample. Error bars traversing 1 signify no change in expression between the two samples.
The grouping of proteins by structural function or enzymatic activity revealed several trends. Cell-surface anchor protein OlpB demonstrated higher expression during growth on Avicel than on cellobiose (Table 3), suggesting an increased anchoring requirement for a greater number of cellulosomes. Expression of exoglucanases was either higher in Avicel-grown cellulosomes or showed no change compared to growth on cellobiose. As expected, based on the results of a previous study, cellobiohydrolase CelS showed the greatest difference in favor of growth on Avicel of any docking enzyme. GH9 endoglucanases either demonstrated higher expression on Avicel (CelJ) than on cellobiose or exhibited no significant change between the two substrates (CelT, CelF, and CelR). On the other hand, GH8 endoglucanase CelA and GH5 endoglucanases (CelB, CelE, and CelG) showed lower expression on Avicel than on cellobiose. One new enzyme from each of GH9 and GH5 demonstrated higher expression in cells grown on cellobiose. All hemicellulases compared displayed higher expression per cellulosome when cells were grown on cellobiose.
Noncellulosomal proteins detected.
Four noncellulosomal proteins with signal peptides for secretion were detected (not shown in Tables 1 or 2). The GH9 endoglucanase CelI (gi 125972564) was detected in the cellobiose cellulosome sample (53). It was identified by two unique peptides. From the Avicel-grown sample only, three unique peptides were matched to a predicted 34-kDa protein (gi 125972914) with similarity (E value of 3E-32) to RbsB (COG1879), a ribose-binding protein in Escherichia coli. This protein also has a lipid attachment site to anchor it to the membrane. In both Avicel- and cellobiose-grown cellulosome preparations, 17 and 10 unique peptides, respectively, matched to a predicted 50-kDa protein (gi 125973535) with similarity (E value of 1E-42) to UgpB (COG1653), a periplasmic glycerol-3-phosphate-binding protein in E. coli. Finally, seven unique peptides from both samples were matched to a predicted 113-kDa protein (gi 125974833) with a possible (E value of = 0.006) SLH domain (pfam00395) for anchoring it to the cell wall and also an immunoglobulin-like fold, which may behave like a carbohydrate binding domain. This protein had been recently observed in the cell membrane fraction (42). All three of the latter proteins were observed in considerable abundance (at least 25% amino acid coverage) in the total extracellular protein fraction from cells grown on cellobiose (data not shown). Their high abundance and, more particularly, the presence in each of them of a possible carbohydrate binding domain point to the possibility that these proteins are contaminants of the cellulosome preparations, consistently copurifying with cellulosome-cellulose complexes. This possibility does not, however, preclude the alternative: that they may in fact be specifically associated with these complexes and play roles in secondary cellulosomal product-related function, perhaps in the uptake of cellodextrins in the manner of RbsB from Bacillus subtilis (43) or MalX from Streptococcus pneumoniae (14), both lipoproteins involved in sugar transport in gram-positive bacteria.
DISCUSSION
This article presents the most comprehensive proteomic study of the C. thermocellum cellulosome to date. Until the recent use of two-dimensional gels and MS-based methods to improve the compositional detail of the C. thermocellum cellulosome (42, 50), most of the work concerning the identification of cellulosomal components had so far been done by means of enzymatic assay (44) or Western blot analysis (2, 15-17, 22, 27, 29-31, 48, 49, 53, 54). The detection of 16 new Doc1-containing proteins represents a 70% increase in the number of docking subunits observed in cellulosomes. However, it should be noted that in general the proteins detected in highest abundance were known, which attests to the fact that the more abundant proteins are the more discoverable. Yet one new GH5 enzyme (gi 125973339) containing a predicted galactose-binding domain was found in considerable abundance under both growth conditions and may prove to be a subunit of some importance upon further investigation.
The three known docking subunits to escape detection were the noncatalytic docking component CseP (53), the serine protease inhibitor PinA (22), and the bifunctional component CelH (42); however, all three of these were observed by us in earlier trials (data not shown) in which either no reference protein was mixed in or the reference had not been 15N-enriched to 99%. CseP and PinA were detected on both substrates, whereas CelH, which has both a GH5 and a GH26 domain, was detected only on cellobiose. CelO, the only known GH5 exoglucanase in C. thermocellum (52), is the only previously cloned docking gene product never to be detected by us.
XynD was detected exclusively on cellobiose even though it had been discovered on cellulose by MS (50), and ManA and LicB were detected exclusively on Avicel, whereas they had previously been observed on cellobiose by Western blot analysis (15, 49). These discrepancies could be explained by the differences between the protein identification methods used in the previous studies and the method used in the present work.
Growth on the different substrates revealed a similar mix of cellulosomal components that were present in significantly different relative amounts. Differences in the relative expression levels of individual components grown on either carbon source demonstrated GH family-specific regulatory patterns, providing evidence in support of existing hypotheses for cellulosomal component regulation as well as contributing a novel distinction with respect to endoglucanase synthesis.
The exoglucanase CelS exhibited the greatest increase of any docking component during growth on Avicel compared to cellobiose. The increase of CelS on Avicel versus cellobiose had already been observed at the protein level by SDS-PAGE (4) and Western blot analysis (7). This result also agrees with changes in celS transcript levels per cell between growth on cellulose and cellobiose (7). Exoglucanases are the key enzymes in cellulase mixtures effective on crystalline cellulose (40), so it was not surprising that exoglucanase CelK also increased on Avicel, even while the expression of CbhA did not change significantly.
Docking proteins with known endoglucanase activity demonstrated varied expression patterns. The GH5 endoglucanases CelB, CelE, and CelG demonstrated higher expression when cells were grown on cellobiose than on Avicel. The same was true for CelA from GH8. In contrast, CelJ from GH9 showed increased expression on Avicel, while the expression of other GH9 endoglucanases, CelF, CelR and CelT, did not change significantly. The detection of CelN and CelQ on Avicel and not cellobiose may be taken as another indication of increased GH9 endoglucanase production on Avicel. The differential expression of GH9 versus GH5 endoglucanases poses an apparent discrepancy with the recent transcript analysis of Dror et al. (9), who observed increased transcript levels per cell of each of the endoglucanase genes celB and celG from GH5 and celD from GH9 when cells were grown at a low versus a high growth rate and also on cellulose versus cellobiose. Thus, while our results with respect to GH9 endoglucanases agree with these previous findings at the transcript level, the increase of GH5 endoglucanases and of CelA on cellobiose was a somewhat unanticipated result. One possible explanation for the difference between the trends observed at the mRNA and protein levels is that GH9 endoglucanase genes may be more responsive to catabolite repression than celA or GH5 endoglucanase genes, such that the former would be more repressed on cellobiose than either of the latter.
The data suggest that the organism has a “cellulolytic preference” for GH9 endoglucanases when degradation of crystalline cellulose is required. In total, cellulosomal GH9 cellulases contained in the C. thermocellum genome outnumber GH5 enzymes by 14 to 8. This preference could be due to what distinguishes them from CelA and GH5 endoglucanases: the presence, in many instances, of a type IIIc carbohydrate binding module, which has been shown to participate in the catalytic activity of the enzyme (1, 2) and to be responsible for processivity (5, 41). What is more, GH9 endoglucanases carry out different modes of attack on cellulose, resulting in cellodextrins of different lengths (1). CelR, which was the most abundant endoglucanase in cellulosomes from Avicel-grown cells, is one such enzyme, a processive GH9 endoglucanase that produces cellotetraose as its primary hydrolysis product (51), which is more energetically favorable for the cell than production of cellobiose (46).
Finally, with respect to hemicellulases, all subunits with xylanase or xyloglucanase activity decreased on Avicel, as per RelEx and emPAI analysis. XynC production has previously been shown to increase on cellobiose (4, 9), and xynC transcript levels have been found to increase on cellobiose in a growth rate-independent fashion (9). In this study, XynZ, XynA, XynC, and XghA were among the five most abundant docking components in cellobiose-grown cellulosomes, along with CelA. XynD and XynY were not detected in the Avicel sample, possibly because their signals were overwhelmed by those of more abundant subunits. On the other hand, their exclusive detection on cellobiose might be taken as another indication of increased xylanase production on cellobiose. Other new subunits with glycosidase and arabinofuranosidase activities were detected exclusively on cellobiose. The trend of increased hemicellulase production on cellobiose could also explain the increase in the bifunctional subunit CelE, which has a family 2 carbohydrate esterase domain in addition to a GH5. As for other hemicellulases, no change was noted for ChiA, and the appearance of LicB and ManA on Avicel but not cellobiose suggests that transcription of these genes was repressed on cellobiose. In the case of manA, Stevenson et al. (38) reported a 10-fold reduction in its transcript level on cellobiose compared to cellulose. Thus, while xylanase transcription is growth rate independent and increases on cellobiose, chitinase, lichenase, and mannanase appear to be under a different type of regulation mechanism. C. thermocellum is unable to utilize the pentose sugars produced by the action of xylanases and other hemicellulases (6, 12); therefore, the apparent role of hemicellulases is to expose cellulose to the action of cellulases. When the organism is not mining energy from cellulose, as when it is grown on cellobiose, in general it appears to prepare itself to mine cellulose from plant wall materials, hemicellulose and lignin, as it would in its natural ecosystem.
In conclusion, this work provides a global view of the C. thermocellum cellulosome. During growth on two substrates, the organism produced a wide variety of dockable hydrolytic enzymes, accounting for two-thirds of the genes containing Doc1 sequences. Of the remaining unobserved putative dockable gene products, there are six various hemicellulases, one GH9 cellulase, and about 16 proteins of unknown function, which may be inducible using more complex substrates. An understanding of the mechanisms by which bacteria regulate the expression of the various cellulases and hemicellulases at their disposal will be important to the eventual production of optimal enzyme cocktails or designer cellulosomes used in the breakdown of cellulosic materials for the transition from an oil-based to a carbohydrate-based economy.
Acknowledgments
We thank Emma Master and Reginald Storms for their help in reviewing the manuscript.
This work was supported by research grants from the Natural Sciences and Engineering Research Council of Canada (grant numbers 312357-06 and 330781-06) and the Canada Foundation for Innovation (grant number 202359) as well as a Petro-Canada Young Innovator Award to V.J.J.M.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Arai, T., A. Kosugi, H. Chan, R. Koukiekolo, H. Yukawa, M. Inui, and R. Doi. 2006. Properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl. Microbiol. Biotechnol. 71:654-660. [DOI] [PubMed] [Google Scholar]
- 2.Arai, T., H. Ohara, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Sequence of celQ and properties of CelQ, a component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 57:660-666. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., J.-P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 6.Demain, A. L., M. Newcomb, and J. H. D. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J. Bacteriol. 185:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dror, T. W., A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2005. Regulation of major cellulosomal endoglucanases of Clostridium thermocellum differs from that of a prominent cellulosomal xylanase. J. Bacteriol. 187:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953-971. [DOI] [PubMed] [Google Scholar]
- 11.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Martinez, D. V., A. Shinmyo, A. Madia, and A. L. Demain. 1980. Studies on cellulase production by Clostridium thermocellum. Eur. J. Appl. Microbiol. Biotechnol. 9:189-197. [Google Scholar]
- 13.Gerngross, U. T., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 14.Gilson, E., G. Alloing, T. Schmidt, R. Claverys, R. Dudler, and M. Hofnung. 1988. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 7:3971-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halstead, J. R., P. E. Vercoe, H. J. Gilbert, K. Davidson, and G. P. Hazlewood. 1999. A family 26 mannanase produced by Clostridium thermocellum as a component of the cellulosome contains a domain which is conserved in mannanases from anaerobic fungi. Microbiology 145:3101-3108. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, H., K. I. Takagi, M. Fukumura, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1997. Sequence of xynC and properties of XynC, a major component of the Clostridium thermocellum cellulosome. J. Bacteriol. 179:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, H., M. Takehara, T. Hattori, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1999. Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum. Appl. Microbiol. Biotechnol. 51:348-357. [DOI] [PubMed] [Google Scholar]
- 18.Hazlewood, G. P., K. Davidson, J. H. Clarke, A. J. Durrant, J. Hall, and H. J. Gilbert. 1990. Endoglucanase E, produced at high level in Escherichia coli as a lacZ′ fusion protein, is part of the Clostridium thermocellum cellulosome. Enzyme Microb. Technol. 12:656-662. [DOI] [PubMed] [Google Scholar]
- 19.Ishihama, Y., Y. Oda, T. Tabata, T. Sato, T. Nagasu, J. Rappsilber, and M. Mann. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4:1265-1272. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, E. A., M. Sakajoh, G. Halliwell, A. Madia, and A. L. Demain. 1982. Saccharification of complex cellulosic substrates by the cellulase system from Clostridium thermocellum. Appl. Environ. Microbiol. 43:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joliff, G., P. Beguin, and J.-P. Aubert. 1986. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 14:8605-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, S., Y. Barak, R. Lamed, E. A. Bayer, and M. Morrison. 2006. The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors. Mol. Microbiol. 60:1344-1354. [DOI] [PubMed] [Google Scholar]
- 23.Kataeva, I., X.-L. Li, H. Chen, S.-K. Choi, and L. G. Ljungdahl. 1999. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J. Bacteriol. 181:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinter, M., and N. E. Sherman. 2000. Protein sequencing and identification using tandem mass spectrometry. John Wiley and Sons, Inc., New York, NY.
- 25.Krokhin, O. V., S. Ying, J. P. Cortens, D. Ghosh, V. Spicer, W. Ens, K. G. Standing, R. C. Beavis, and J. A. Wilkins. 2006. Use of peptide retention time prediction for protein identification by off-line reversed-phase HPLC-MALDI MS/MS. Anal. Chem. 78:6265-6269. [DOI] [PubMed] [Google Scholar]
- 26.Kruus, K., A. C. Lua, A. L. Demain, and J. H. D. Wu. 1995. The anchorage function of CipA (CelL), a scaffolding protein of the Clostridium thermocellum cellulosome. Proc. Natl. Acad. Sci. USA 92:9254-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurokawa, J., E. Hemjinda, T. Arai, T. Kimura, K. Sakka, and K. Ohmiya. 2002. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 59:455-461. [DOI] [PubMed] [Google Scholar]
- 28.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Techol. 7:37-41. [Google Scholar]
- 29.Leibovitz, E., H. Ohayon, P. Gounon, and P. Beguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaire, M., and P. Beguin. 1993. Nucleotide sequence of the celG gene of Clostridium thermocellum and characterization of its product, endoglucanase CelG. J. Bacteriol. 175:3353-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Beguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacCoss, M. J., C. C. Wu, H. Liu, R. Sadygov, and J. R. Yates. 2003. A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem. 75:6912-6921. [DOI] [PubMed] [Google Scholar]
- 34.Mishra, S., P. Beguin, and J. P. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morag, E., E. A. Bayer, and R. Lamed. 1990. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J. Bacteriol. 172:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong, S.-E., and M. Mann. 2005. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1:252-262. [DOI] [PubMed] [Google Scholar]
- 37.Shallom, D., and Y. Shoham. 2003. Microbial hemicellulases. Curr. Opin. Microbiol. 6:219-228. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson, D. M., and P. J. Weimer. 2005. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl. Environ. Microbiol. 71:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabb, D. L., W. H. McDonald, and J. R. Yates. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teeri, T. T. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15:160-167. [Google Scholar]
- 41.Tormo, J., R. Lamed, A. J. Chirino, E. Morag, E. A. Bayer, Y. Shoham, and T. A. Steitz. 1996. Crystal structure of a bacterial family III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15:5739-5751. [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, T. I., J. C. Combs, B. C. Lynn, and H. J. Strobel. 2007. Proteomic profile changes in membranes of ethanol-tolerant Clostridium thermocellum. Appl. Microbiol. Biotechnol. V 74:422-432. [DOI] [PubMed] [Google Scholar]
- 43.Woodson, K., and K. M. Devine. 1994. Analysis of a ribose transport operon from Bacillus subtilis. Microbiology 140:1829-1838. [DOI] [PubMed] [Google Scholar]
- 44.Wu, J. H. D., W. H. Orme-Johnson, and A. L. Demain. 1988. Two components of an extracellular protein aggregate of Clostridium thermocellum together degrade crystalline cellulose. Biochemistry 27:1703-1709. [Google Scholar]
- 45.Zhang, Y., and L. R. Lynd. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an enzyme-linked immunosorbent assay-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 75:219-227. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y.-H. P., and L. R. Lynd. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. USA. 102:7321-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y.-H. P., and L. R. Lynd. 2005. Regulation of cellulase synthesis in batch and continuous cultures of Clostridium thermocellum. J. Bacteriol. 187:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zverlov, V. V., K. P. Fuchs, and W. H. Schwarz. 2002. Chi18A, the endochitinase in the cellulosome of the thermophilic, cellulolytic bacterium Clostridium thermocellum. Appl. Environ. Microbiol. 68:3176-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zverlov, V. V., K. P. Fuchs, W. H. Schwarz, and G. A. Velikodvorskaya. 1994. Purification and cellulosomal localization of Clostridium thermocellum mixed linkage β-glucanase LicB (1,3-1,4-β-d-glucanase). Biotechnol. Lett. 16:29-34. [Google Scholar]
- 50.Zverlov, V. V., J. Kellermann, and W. H. Schwarz. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646-3653. [DOI] [PubMed] [Google Scholar]
- 51.Zverlov, V. V., N. Schantz, and W. H. Schwarz. 2005. A major new component in the cellulosome of Clostridium thermocellum is a processive endo-β-1,4-glucanase producing cellotetraose. FEMS Microbiol. Lett. 249:353-358. [DOI] [PubMed] [Google Scholar]
- 52.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2002. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology 148:247-255. [DOI] [PubMed] [Google Scholar]
- 53.Zverlov, V. V., G. A. Velikodvorskaya, and W. H. Schwarz. 2003. Two new cellulosome components encoded downstream of celI in the genome of Clostridium thermocellum: the nonprocessive endoglucanase CelN and the possibly structural protein CseP. Microbiology 149:515-524. [DOI] [PubMed] [Google Scholar]
- 54.Zverlov, V. V., G. V. Velikodvorskaya, W. H. Schwarz, K. Bronnenmeier, J. Kellermann, and W. L. Staudenbauer. 1998. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J. Bacteriol. 180:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]