Abstract
Most digestive tracts contain a complex consortium of beneficial microorganisms, making it challenging to tease apart the molecular interactions between symbiont and host. The digestive tract of Hirudo verbana, the medicinal leech, is an ideal model system because it harbors a simple microbial community in the crop, comprising the genetically amenable Aeromonas veronii and a Rikenella-like bacterium. Signature-tagged mutagenesis (STM) was used to identify genes required for digestive tract colonization. Of 3,850 transposon (Tn) mutants screened, 46 were identified as colonization mutants. Previously we determined that the complement system of the ingested blood remained active inside the crop and prevented serum-sensitive mutants from colonizing. The identification of 26 serum-sensitive mutants indicated a successful screen. The remaining 20 serum-resistant mutants are described in this study and revealed new insights into symbiont-host interactions. An in vivo competition assay compared the colonization levels of the mutants to that of a wild-type competitor. Attenuated colonization mutants were grouped into five classes: surface modification, regulatory, nutritional, host interaction, and unknown function. One STM mutant, JG736, with a Tn insertion in lpp, encoding Braun's lipoprotein, was characterized in detail. This mutant had a >25,000-fold colonization defect relative to colonization by the wild-type strain at 72 h and, in vitro, an increased sensitivity to sodium dodecyl sulfate, suggesting the presence of an additional antimicrobial property in the crop. The classes of genes identified in this study are consistent with findings from previous STM studies involving pathogenic bacteria, suggesting parallel molecular requirements for beneficial and pathogenic host colonization.
Most animals house a diverse microbial community within their digestive tracts. For example, the human colon contains about 400 different microbial species (3), while some insects house simpler gut communities containing tens of species (8). The microbial community carries out many important functions for the host, including preventing pathogenic bacteria from colonizing (64), providing essential vitamins (34, 51), stimulating the immune system (18), and aiding digestion (23). Despite these important roles, little is known about the molecular interactions underlying digestive tract associations, in part due to the complexity of the consortia and the obligate nature of some symbioses (32).
Naturally occurring, simple model systems can overcome these challenges and provide new information that can be tested subsequently in more complex systems (17, 48). The digestive tract symbiosis of the medicinal leech, Hirudo verbana, has unique features that aid in its use as a model to identify and characterize these molecular interactions (17). The microbial community residing in the crop of the leech is unusually simple. It is dominated by Aeromonas veronii and a Rikenella-like bacterium (16, 66). The medicinal leech feeds exclusively on vertebrate blood, consuming as much as five times its body weight (52). The ingested blood meal, now called the intraluminal fluid, is stored in the crop, where water and salts are absorbed. Similar functions occur in the mammalian large intestine. The ability of the leech to consume blood and the concurrent secretion of anticoagulants and vasodilators are the underlying basis of the benefit of leech therapy and led to the approval by the FDA of the leech as a medical device in treating venous congestion after reconstructive surgery (44). A clinical trial demonstrated the benefit of leech therapy for patients suffering from osteoarthritis (35). When patients with local or general immune suppression receive leech therapy, wound infections by the gut symbionts can occur. Presently these infections can be prevented by preemptive antibiotic treatment (30). One of the symbionts, A. veronii, is genetically amenable (46), while the Rikenella-like bacterium has not been cultured outside the leech (66). Recently, epifluorescent microscopic observations revealed that A. veronii and the Rikenella-like bacterium form mixed-species, polysaccharide-embedded microcolonies while colonizing the leech crop (27). In addition to being a symbiont, A. veronii can also cause wound infections, septicemia, and diarrhea in humans (26). The unusual simplicity of the microbiota and the availability of genetic tools, an accessible host animal, and a defined food source allow one to gain fundamental insights into microbe-host and microbe-microbe interactions by investigating this model system (17).
The intriguing simplicity of the microbiota in the crop is likely due to multiple factors (17). Previously we have shown that the complement system of the ingested vertebrate blood remains active inside the crop and kills sensitive bacteria (25). Bacterial surface structures can provide resistance to complement-mediated killing. For example, mutants containing a defect in lipopolysaccharide (LPS) have been shown to be serum sensitive; this was the first class of colonization mutants discovered in this symbiosis (6). The growth of Pseudomonas aeruginosa and Staphylococcus aureus strains inside the leech was inhibited in a complement-independent manner, suggesting the presence of other barriers that contribute to the specificity of this symbiosis (25). One recently discovered barrier consists of leech hemocytes, macrophage-like cells that patrol the gut and remove sensitive bacteria (55). A type III secretion system is required by A. veronii for protection against phagocytosis (55). The presence of multiple layers enforcing specificity requires a back-and-forth interaction between host and symbiont, as has been shown in well-established model systems such as the symbioses of Sinorhizobium meliloti with leguminous plants and of Vibrio fischeri with the Hawaiian bobtail squid, Euprymna scolopes (9, 22, 39, 62).
In this study, we utilized signature-tagged mutagenesis (STM) to uncover the molecular requirements of the beneficial digestive tract association between A. veronii and the medicinal leech. STM is an improvement on transposon (Tn) mutagenesis in which each Tn carries a unique sequence, or “signature tag” (19). The power of STM is that it allows for the screening of multiple Tn mutants in one animal and for the recovery of mutants with a colonization defect by utilizing a signature-tagged Tn to track each strain present in a pool of mutants. Essentially, each mutant acts as a probe that reveals insights about the microenvironment and identifies genes required for successful colonization of the host. STM has been used primarily to identify virulence factors in mammalian pathogens such as Yersinia enterocolitica, Salmonella enterica serovar Typhimurium, and Vibrio cholerae (13, 19, 33). STM has also been applied to the mutualistic association between Xenorhabdus nematophila and the nematode Steinernema carpocapsae, leading to the identification of genes required for successful beneficial colonization (20).
MATERIALS AND METHODS
Bacterial strains.
The A. veronii STM mutants were derived from HM21R, and HM21RS was used as the competitor strain in competition assays (Table 1) (16, 46). Escherichia coli strain S17-1 harboring pUTminiTn5Km2STM (19) was used to generate A. veronii STM mutants. A. veronii was cultured at 30°C and E. coli at 37°C in LB or on LB agar plates (50). Ringer's solution for leeches (115.0 mM NaCl, 1.8 mM CaCl2, 4.0 mM KCl, 10.0 mM Tris maleate [pH 7.4]) was supplemented with 10 g/liter tryptone and 5 g/liter yeast extract to simulate the osmotic conditions inside the crop (52). The growth medium was supplemented with the appropriate antibiotics at the following concentrations: ampicillin, 100 μg/ml; kanamycin (Km), 100 μg/ml; rifampin, 100 μg/ml for selection and 10 μg/ml for maintenance; streptomycin, 100 μg/ml; chloramphenicol, 1 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| A. veronii | ||
| HM21R | Spontaneous Rfr mutant of wild-type HM21 | 16 |
| HM21RS | Spontaneous Smr mutant of HM21R | 46 |
| JG736 | HM21R lpp-1 (lpp::mTn5Km2STM); Rfr Kmr | This study |
| E. coli | ||
| TOP10 | Cloning strain; Smr | Invitrogen |
| S17-1λpir | Strain used for conjugation with A. veronii | 14 |
| DH5αλpir | Cloning strain capable of maintaining suicide vectors | 5 |
| CC118λpir | Carrying helper plasmid pEVS104 | 57 |
| Plasmids | ||
| pUTminiTn5Km2 | Contains Tn used for mutant generation; Kmr | 19 |
| pEVS104 | Helper plasmid; tra trb Kmr | 57 |
| pCR2.1 | Bacterial cloning vector; Kmr Apr | Invitrogen |
| pAS10 | Derivative of pCR2.1 containing lpp; Kmr Apr | This study |
| pMMB207 | Low-copy-number broad-host-range vector; Cmr | 36 |
| pAS11 | Derivative of pMMB207 containing lpp; Cmr | This study |
Rfr, rifampin resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Smr, streptomycin resistance.
Selection of Tn's.
Previous studies reported occasional weak hybridization signals and cross-hybridization of different signature tags carried on miniTn5Km2STM (19, 20). We screened tags for signal strength and lack of cross-hybridization, obtaining 55 signature tags. One signature tag that hybridized to several of the selected tags was spotted onto the membranes as a positive control.
Generation of mutants.
Fifty-five E. coli S17-1 strains, each harboring a pUTminiTn5Km2STM plasmid carrying a unique tag, were used as donor strains in conjugations with HM21R (46). Transconjugants were recovered on LB plates with Km and rifampin and were picked into microtiter plate wells (Corning Incorporated, Corning, NY). Frozen stocks were prepared from overnight cultures by addition of 50 μl of 80% glycerol to each well and were stored at −80°C.
STM screen.
The STM screen was performed in a manner similar to that described by Hensel et al. (19) with the following modifications. Cells were grown to mid-log phase and pooled. From a portion of this “input” pool, the DNA was extracted (46). Another portion was used to inoculate 5 ml of fresh sheep blood (containing heparin [25 U/ml; Sigma Chemical Co., St. Louis, MO]) with 200 CFU/ml of each strain (16). The blood was collected from sheep at the University of Connecticut's School of Agriculture (Storrs, CT) in accordance with an approved IACUC protocol and was warmed to 37°C prior to inoculation. Animals were fed as described previously (16). After 42 h, animals were dissected, and intraluminal fluid was collected, serially diluted, and plated as described previously (16).
From approximately 20,000 CFU of the “output,” DNA was extracted (46). The tags from the input and output pools were PCR amplified with primers P2 (5′-TACCTACAACCTCAAGCT) and P4 (5′-TACCCATTCTAACCAAGC) (19). Each reaction mixture contained 1 to 2 μg of DNA, 1× thermal polymerase buffer (New England Biolabs, Ipswich, MA), 8 mM MgSO4, 400 μM each deoxynucleoside triphosphate (dNTP), 1.4 μM each primer, and 1 U of Vent DNA polymerase (New England Biolabs) in a final volume of 50 μl. The amplification conditions were as follows: (i) 5 min at 95°C; (ii) 25 cycles of 30 s at 95°C, 45 s at 50°C, and 10 s at 75°C. Two microliters of the PCR mixture was separated on a 3% NuSieve (Cambrex Bio Science Rockland, Inc., Rockland, ME) gel, and the 80-bp product was gel extracted using β-agarase (New England Biolabs). The tags were labeled with digoxigenin (DIG; Roche Applied Science, Indianapolis, IN) by PCR with primers P2 and P4. Each reaction mixture contained 4 μl of the digested gel slice; 1× thermal polymerase buffer; 180 μM dTTP; 5 μM dUTP; 200 μM (each) dATP, dGTP, and dCTP; and 1 μM each primer. Vent Exo− polymerase (0.5 U; New England Biolabs) was added once the reaction temperature reached 95°C, in a final volume of 20 μl. The amplification conditions were those described above. The primer sequences were removed from the tag by digesting 2 μl of the PCR product with HindIII. Fifty microliters of double-distilled H2O was added to the digest, and the mixture was heated for 5 min at 95°C and then used to probe a membrane.
Membranes were prepared by PCR amplification of the 56 tags using primer pair P2-P4. Each reaction mixture contained 10 to 20 ng of plasmid DNA extracted with a QIAGEN miniprep kit (QIAGEN Sciences, Germantown, MD), 1× buffer, 5 mM MgCl2, 400 μM each dNTP, 1 μM each primer, and 1.25 U of AmpliTaq polymerase (Applied Biosystems, Foster City, CA) in a final volume of 50 μl. The amplification was carried out as described above except that the extension was performed at 72°C. Five microliters from each PCR mixture was spotted onto a positively charged nylon membrane (Roche Applied Science). The membrane was placed in a denaturing solution (1.5 M NaCl, 0.5 M NaOH) for 3 min, in a neutralizing solution (1.5 M NaCl, 0.5 M Tris-HCl [pH 7.4]) for 5 min, in a fresh neutralizing solution for 1 min, and in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 min. DNA was cross-linked to the membrane using a UV cross-linker.
The membrane was prehybridized with 5 ml of DIG hybridization solution (Roche Applied Science) for 30 min at 42°C. The DIG-labeled probe was added to 2 ml of fresh DIG hybridization solution, and the membrane was incubated overnight at 42°C. Washing, probing, and detection were performed as follows: (i) two washes with 20 ml 2× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min, (ii) two washes with 20 ml 0.1× SSC-0.1% SDS at 65°C for 5 min each, (iii) one wash with 20 ml of washing buffer (0.1 M maleic acid, 0.15 M NaCl [pH 7.5], 0.3% [wt/vol] Tween 20) at room temperature for 5 min, (iv) blocking with 40 ml of 1× blocking solution for 30 min at room temperature, (v) one wash with 20 ml of antibody solution (alkaline phosphatase-conjugated anti-digoxigenin in 1× blocking solution) for 30 min at room temperature, (vi) two washes with 20 ml of washing buffer for 5 min each, (vii) equilibration with 20 ml of detection buffer (0.1 M Tris-HCl, 0.1 M NaCl [pH 9.5]) for 3.5 min, and (viii) detection by applying 2 ml CDP-Star ready-to-use solution (Roche Applied Science) and incubating the membrane for 5 min at room temperature. Signals were detected using an AI Fluorochem 8800 detection system.
Competition assay.
The competition assay used in this study compares the colonization ability of a test strain against that of a competitor strain, HM21RS, by inoculating a blood meal with 250 CFU/ml of each strain (46). The conditions were identical to those for the assay described previously (46) except that heat-inactivated blood (Quad 5, Ryegate, MT) was used. At least three animals were examined at each time point: 6, 15, 24, 42, and 72 h postfeeding. The limit of detection was 10 CFU/ml.
Inverse PCR.
Inverse PCR was used to amplify the DNA flanking the Tn insertion (40). The procedure was performed as described previously (55) except that the following restriction enzymes were used: PstI, SalI, SphI, ApaI, or NcoI in a 20-μl reaction mixture.
Fosmid construction.
The fosmid for JG736 was constructed as described previously (55).
DNA sequencing and analysis.
DNA was sequenced and analyzed as described previously (55).
Southern blot analysis.
Five micrograms of genomic DNA was digested with either XhoI or XbaI, and equal amounts of each digest were separated on a 0.6% SeaKem LE agarose gel (Cambrex Bio Science). Genomic DNA was transferred to a positively charged nylon membrane (Roche Applied Science) using a Vacu Gene XL vacuum apparatus (Pharmacia, Uppsala, Sweden) under neutral transfer conditions. The membrane was treated as described under “STM screen” above except that step i was performed at 42°C; the wash solution in step ii contained 0.5× SSC-0.1% SDS, and each wash lasted 15 min; the washing in step iii, the blocking, and step v were performed at 42°C; and in step vi, each wash lasted 15 min.
Serum sensitivity assay.
Serum was obtained from fresh sheep blood by centrifugation at 2,000 × g for 10 min at 4°C. The serum was collected and centrifuged again under the same conditions. After overnight growth, HM21R and all STM mutants were patched with cotton swabs onto LB plates. After drying, 25 μl of serum was pipetted onto the plate. The plates were incubated at 30°C overnight and observed for a zone of no growth.
Phenotypic tests.
The following phenotypic tests were performed on HM21R and each STM mutant: growth in blood, beta-hemolysis, prototrophy, autoinducer production, siderophore production, and motility. Growth in blood was assessed by inoculating heat-inactivated blood with 250 CFU/ml of mutant and competitor strains. The inoculated blood was incubated at room temperature (23°C). Aliquots were removed at 0, 18, 24, and 42 h after inoculation and were plated as described for the competition assay. For the 42-h time point, four to eight replicates were performed. Beta-hemolysis was determined on blood agar plates (Difco Columbia blood agar base; Becton Dickinson and Company, Sparks, MD) after overnight growth at 30°C. Prototrophy was assessed on M9 minimal medium plates containing glucose (50). Siderophore production was evaluated on chrome azurol S plates, which change color as iron is removed from chrome azurol S (43). Autoinducer production was determined by cross-streaking the strains onto LB plates against Chromobacterium violaceum, which turns purple in the presence of acylated homoserine lactone-type autoinducers (31). Motility was assessed using 0.45% LB agar plates and light microscopy at a magnification of ×1,000.
Complementation of the lpp mutant.
Primer pair lpp-F (5′-CTGTGGTGGGGACTATGCAAAG)-lpp-R (5′-GGGGAATAAGGTGCTGTGGG) was used to amplify lpp. The reaction mixture contained 100 ng of DNA, 1× PCR buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.2 μM each primer, and 1 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) in a final volume of 50 μl. The amplification conditions were as follows: (i) 2 min at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. A TA cloning kit was used to clone the PCR product into pCR2.1 according to the manufacturer's instructions (Invitrogen). The resulting plasmid, pAS10, was isolated using a QIAGEN plasmid purification kit (QIAGEN Sciences). The cloned 421-bp fragment was ligated into the broad-host-range vector pMMB207 from pAS10 by utilizing the EcoRI restriction sites (36). The resulting plasmid, pAS11, was transformed into chemically competent DH5αλpir cells (50). pAS11 and pMMB207 were introduced into HM21R and JG736 by conjugation with the E. coli helper strain CC118λpir carrying pEVS104 (57). The resulting strains were competed against HM21RS.
SDS sensitivity assay.
Cells were grown to mid-log phase and diluted 50-fold in microtiter plates (Corning Incorporated) that contained LB either without SDS or with 0.3125%, 0.625%, 1.25%, 2.5%, or 5% SDS. The cultures were incubated at 30°C overnight with shaking (220 rpm), and the optical density at 595 nm (OD595) was measured in a model 550 microplate reader (Bio-Rad, Hercules, CA) to determine the growth yield. Duplicate experiments were performed, each with four replicates.
Statistical analysis.
The data were analyzed using GraphPad Prism, version 2.01. A two-tailed, one-sided t test was used to test whether the competitive index (CI) differed significantly (P ≤ 0.05) from 1. The CI values obtained in the animal were compared to the CI values obtained in blood by using a two-tailed t test. Complementation in the animal was assessed with the nonparametric Mann-Whitney test.
Nucleotide sequence accession number.
The NCBI nucleotide accession number of the lpp locus is EF579800.
RESULTS AND DISCUSSION
Identification of colonization mutants.
The power of STM is the ability to screen multiple mutants in each animal and to recover mutants with an attenuated colonization phenotype (19). While earlier studies used pools of mutants generated with Tn's carrying random signature tags, more-recent studies selected tags that produced a strong hybridization signal and did not cross-hybridize (20, 29). We used 55 tags that met these criteria to generate STM mutants. A total of 3,850 A. veronii miniTn5Km2STM mutants were screened for colonization defects by inoculating fresh sheep blood with pools containing 55 mutants and feeding it to leeches. The animals were dissected 42 h after feeding. By that time, the symbiont population had reached its maximum level and modifications of the ingested blood meal had taken effect. These changes include the osmolarity of the intraluminal fluid, the activity of the mammalian complement system, and the infiltration of hemocytes (16, 52, 55). The content of the crop, the intraluminal fluid, was diluted and plated onto a selective medium. From the recovered bacteria, the output pool, DNA was isolated. The PCR-amplified tags were labeled with DIG and used to probe membranes containing each tag. The hybridization signal from the output pool was compared to the signal from an input pool in order to identify mutants with no hybridization signal or reduced hybridization signals. These mutants were assembled into new pools and retested in duplicate animals. Mutants with no signal or a consistently reduced signal were considered presumptive colonization mutants and verified as described below. This secondary screen reduced the number of false positives by 68%.
In previous studies, we demonstrated that the complement system of the ingested vertebrate blood remains active inside the leech. As a consequence, serum resistance is essential in order for bacteria to successfully colonize the leech gut (6, 25). The goal of this investigation was to discover new classes of mutants; hence, the 83 presumptive colonization mutants were first tested for serum sensitivity. Twenty-six serum-sensitive mutants were identified and will be characterized in a separate study. The isolation of these serum-sensitive mutants validated our screening technique.
In the STM screen, the mutants competed with wild-type bacteria; accordingly, the colonization phenotype of the 57 serum-resistant mutants was verified in a competition assay using heat-inactivated blood. The heat treatment inactivated the mammalian complement system and ensured that the colonization defect of the mutant was due to an intrinsic property of the leech or a heat-resistant property of the ingested blood (25). The competitor strain in the competition assay, HM21RS, served as an internal standard, improving reproducibility. Twenty serum-resistant mutants (0.5%) had a statistically significantly reduced ability to colonize the leech crop 42 h after feeding (Table 2). Including the serum-sensitive mutants, 1.2% of the mutants screened had a colonization phenotype. The CI values exhibit a large range in the severity of attenuation, demonstrating the sensitivity and dynamic range of the STM screen. Twelve mutants possessed slight but significant colonization defects (1.5- to 10-fold), while one mutant had a >10,000-fold defect.
TABLE 2.
Characterization of mutants
| Predicted function and strain(s) | Disrupted gene or gene product | GenBank accession no. | Identity, similarity,a e value | Symbiosisb | Growth in bloodc |
|---|---|---|---|---|---|
| Surface modification | |||||
| JG535 | Glycosyltransferase | YP_727333 | 49, 68, e−86 | 0.0099***/*** | 1.27 |
| JG736 | Outer membrane lipoprotein | YP_926710 | 56, 72, e−11 | 0.0031***/** | 0.60 |
| JG738 | Methyltransferase type 11 | YP_944741 | 24, 49, e−9 | 0.018***/*** | 1.26 |
| Regulatory | |||||
| JG574 | rRNA operon | X74684 | 99, 99, e0 | 0.15**/* | 0.61 |
| JG697 | GTP-binding protein | YP_857656 | 97, 100, e−70 | 0.092***/— | 0.06 |
| JG730 | Response regulator | YP_855452 | 77, 85, e−81 | 0.00020***/** | 2.44 |
| JG741 | Exoribonuclease II | YP_855087 | 95, 98, e−76 | 0.28*/* | 0.76 |
| Nutritional | |||||
| JG537 | Phosphate ABC transporter | YP_270310 | 92, 96, e−10 | 0.033***/* | 0.80 |
| JG698 | GufA protein | YP_856244 | 95, 97, e−38 | 0.23*/— | 0.01 |
| JG750 | Threonine/serine transporter | NP_755741 | 85, 94, e−168 | 0.10*/** | 1.34 |
| Host interaction | |||||
| JG752 | ascU | CAD56767 | 100, 100, e−40 | 0.13***/*** | 0.73 |
| Unknown | |||||
| JG521 | Hypothetical protein | YP_737079 | 33, 54, e−19 | 0.10**/* | 1.19 |
| JG523 | No similarity detected | 0.32*/* | 1.07 | ||
| JG532 | Conserved hypothetical protein | ZP_01669898 | 53, 69, e−38 | 0.63**/* | 0.36 |
| JG533 | KAP family P-loop domain protein | YP_856709 | 71, 80, e−80 | 0.33**/— | 0.60 |
| JG538 | Peptidase S15 | ZP_01542412 | 66, 78, e−59 | 0.43*/** | 1.87 |
| JG573 | Hypothetical protein | NP_930352 | 52, 70, e−70 | 0.0090***/* | 2.07 |
| JG735 | Hypothetical protein | XP_001153743 | 27, 34, e−5 | 0.000076***/** | 0.95 |
| JG751 | Hypothetical protein | YP_856595 | 84, 88, e−95 | 0.33*/* | 0.92 |
| JG753 | No similarity detected | 0.19**/*** | 1.46 | ||
| Wild type | |||||
| HM21R | 1.3 | 1.07 |
Percent identity and percent similarity by a BLASTX or BLASTN search.
Mean in vivo CI values. CI values were calculated by dividing the mutant/competitor strain output ratio by the input ratio of the mutant to the competitor. The first set of asterisks after each value represents the results of a two-tailed, one-sided t test that was used to determine whether the CI differed from 1, while the second set of asterisks represents the results of an unpaired two-tailed, two-sided t test that was used to determine whether the in vivo CI differed from the in vitro CI. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; —, not significantly different.
Mean in vitro CI values.
Phenotypic characterization of colonization mutants.
One possible cause of the colonization defect could be a decreased ability to grow in blood. We determined the CI values for the 20 mutants in heat-inactivated blood at 42 h and compared the CI values obtained inside the leech to the CI values in blood for the 42-h time point. Seventeen mutants had significantly lower CI values inside the leech gut, indicating that the observed defect was not due simply to a growth defect in blood (Table 2). Based on previous studies on the leech digestive tract and symbioses in general (16, 17, 27), we hypothesized that genes affecting beta-hemolysis, prototrophy, siderophore production, autoinducer production, and motility could play a role in this association. Hemolysis would be important to obtain nutrients contained within the erythrocytes. Motility is often required by beneficial and pathogenic bacteria for host colonization (41), and in a recent study, Kikuchi and Graf discovered the presence of Aeromonas and Rikenella-like bacteria together in polysaccharide-embedded microcolonies that are reminiscent of a floating granular biofilm or a biofilm associated with erythrocytes (27). If attachment to the Rikenella-like bacteria is required for enhanced colonization, then directional motility could be important. In the leech crop, free iron is likely to be bound by transferrin, suggesting that siderophore production by A. veronii could be required for iron acquisition. Quorum sensing has been shown to be important for many microbe-host interactions, and Aeromonas species have been shown to produce an acylated homoserine lactone-type autoinducer (59). Autoinducer production could play an important role in any number of steps in the colonization process, including biofilm formation (42, 59).
All of the mutants had wild-type phenotypes except for two mutants, JG532 and JG736, which were motile when observed under the microscope but produced smaller halos in soft agar. Halo sizes for JG532 and JG736 were significantly smaller (9.8 mm [P < 0.0001] and 16 mm [P ≤ 0.0139], respectively) than that for the parent, HM21R (17.9 mm), 24 h after inoculation. (An unpaired two-tailed, two-sided t test was used to determine if the halo sizes differed.)
While the size of our mutant bank (3,850) was not large enough to saturate the genome, the identification of many mutants without detectable phenotypes indicates that the STM screen yielded new information about the colonization requirements.
Molecular characterization of the mutants.
Southern blot analysis was performed using the DIG-labeled Km resistance gene of the Tn as the probe and revealed that all colonization mutants had Tn insertions at a single locus (data not shown). One of the mutants, JG730, appeared to have the donor plasmid incorporated at the site of the Tn insertion. The DNA flanking the Tn insertion was identified by inverse PCR and DNA sequencing. In all cases, this analysis identified the end of the Tn, which confirmed that the inverse PCR product was not the result of a random amplification but was the DNA flanking the Tn. The analysis of the flanking DNA provided important information about the possible function of the inactivated gene (Table 2). The 20 colonization mutants were organized into five different categories based on their predicted functions.
(i) Surface modification mutants.
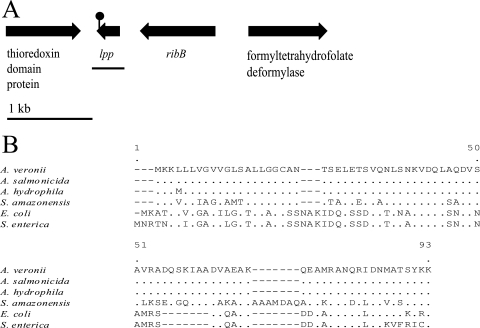
Three mutants have Tn insertions in genes predicted to encode proteins involved in cell surface modification or stability. These three mutants were among those with the most attenuated colonization phenotypes, and the CI values in the animals were significantly lower than those for cells grown in heat-treated blood (Table 2). One of these mutants, JG535, has a Tn insertion in a gene encoding a predicted glycosyltransferase. Another cell surface modification mutant, JG738, has a Tn insertion in a predicted methyltransferase type 11 gene. According to the Pfam and COG databases, this protein has conserved domains that are present in glycosyltransferases (56, 61). Glycosyltransferases are typically responsible for the biosynthesis of LPS, the capsule, or an extracellular matrix (68, 71); however, some glycosyltransferases have been shown to posttranslationally modify proteins in bacteria (4). For A. veronii, intact LPS is required for resistance to the complement system (6), and based on the serum resistance phenotype, we predict that the LPS is not dramatically altered. Smaller modifications of LPS, glycosylated surface proteins, the capsule, or exopolysaccharide formation may be required for successful colonization. The study by Kikuchi and Graf revealed that A. veronii and a Rikenella-like bacterium form mixed-species polysaccharide-embedded microcolonies (27). The third cell surface modification mutant, JG736, has a Tn insertion in lpp, encoding Braun's major outer membrane lipoprotein, which has been shown previously to be important in membrane stability as well as bacterial virulence (15, 67, 69). This mutant is characterized in more detail below.
The discovery of genes encoding proteins involved in cell surface modification and membrane integrity indicates that not only pathogens but also beneficial symbionts encounter membrane stress during the colonization process. While some of this similarity may be due to the analysis of simple digestive tract symbioses where powerful antimicrobial processes have to be overcome, the complex mammalian gut also releases a large number of antimicrobial compounds into the gut lumen, including bile and secreted antibodies.
(ii) Regulatory mutants.
Bacteria colonizing host environments, both beneficial symbionts and pathogens, need to sense the host environment and modulate gene expression and protein levels accordingly (63). We identified four mutants with Tn insertions in genes likely associated with regulating gene expression, enzymatic activity, or protein synthesis rates. One of these mutants, JG730, has a Tn insertion in a homolog of pvrR, a gene encoding a response regulator of a two-component regulatory system. Two-component regulatory systems are often utilized by bacteria to sense environmental conditions and transmit the information via a phosphorelay system to a regulator that mediates changes in gene expression, interacts with other proteins, or affects enzyme activity (2). PvrR is a CheY-type response regulator. While it does not possess a DNA-binding domain, it has a predicted EAL domain that could be involved in cyclic di-GMP signaling (49). An intriguing aspect of EAL domain-containing proteins is that many of them are involved in the transition from motile to sessile lifestyles.
JG574 has a Tn insertion in the regulatory region of a ribosomal operon. While the colonization defect in the animal was moderate (a sixfold decrease), the CI was significantly lower than that in blood. Analysis of the genome sequence of Aeromonas hydrophila identified 10 copies of the ribosomal operon, and other aeromonads also have many copies, which could potentially compensate for this defect (38, 53). In Sinorhizobium meliloti, which has three copies of the ribosomal operons, certain copies are expressed only during specific stages of the growth curve (47). In E. coli, the deletion of one rRNA operon resulted in a decreased growth rate under fast-growth conditions but not in media supporting slower growth (58). If the symbionts grow faster inside the leech than in blood, this phenomenon could explain why the mutant had a significantly lower CI inside the leech.
Another regulatory mutant with a threefold lower CI, JG741, has a Tn insertion in a putative exoribonuclease II, which could be involved in mRNA degradation and tRNA precursor end processing. The final regulatory mutant, JG697, has reduced CI values in both the leech and blood, indicating a more general defect.
Previous STM studies of digestive tract pathogens or beneficial symbionts (13, 20, 33) identified several genes encoding regulatory proteins, supporting the notion that the recognition of and adaptation to the host environment is critical in these associations independently of the effect on the host. Inhabiting a niche outside the host, overcoming host defenses, coordinating gene expression with host responses, and transmission between generations necessitate precise regulatory mechanisms (62). In contrast, obligately intracellular symbionts have lost most regulatory genes. For example, Buchnera spp., the obligate symbionts of aphids, contain less than 2.5% of the regulatory genes found in E. coli, which can be explained by the constant environment in which these symbiotic bacteria reside (37).
(iii) Nutritional mutants.
Other STM studies revealed a large number of nutritional genes to be important for colonization of the host by pathogens or symbionts (20, 54). We identified three colonization mutants with Tn insertions in genes required for transporting nutrients, and two of them had significantly lower CI values in the leech than in blood. One of these two mutants, JG750, has a Tn insertion in a gene encoding a threonine/serine transporter, suggesting that there is greater competition for these amino acids inside the leech. The increased demand for amino acids in the crop is likely to be due to the presence of the Rikenella-like symbionts, because the leech is thought to absorb nutrients in the intestinum and not in the crop.
The other mutant with a reduced CI, JG537, has a Tn insertion in a homolog of pstC, a gene predicted to encode a component of a high-affinity phosphate ABC transporter. In E. coli, the Pst (phosphate-specific transport) protein is expressed under low phosphate concentrations. The Pst operon is composed of five genes, pstS, pstC, pstA, pstB, and phoU, and belongs to the Pho regulon. The gene order was conserved in the A. hydrophila genome except that pstS was not detected (53). In E. coli, the pst operon is responsible for the transport of phosphate into the cell in environments with low phosphate concentrations and repression of the Pho regulon (1, 45). This suggests that there are low phosphate levels in the lumen of the crop. Interestingly, besides being involved in phosphate acquisition from the environment, an intact Pst system has been shown to be required for full pathogenicity in two animal virulence models for E. coli. Deletion of the Pst system in E. coli O78 has been shown to cause sensitivity to rabbit serum (the mutant remained resistant to avian serum) and increased susceptibility to polymyxin and acid shock, as well as severely attenuated virulence in a chicken experimental infection model (28). Another study showed that a mutation in pstC from E. coli O115 leads to sensitivity to rabbit serum and reduced virulence in a pig septicemia model (12). The link between the pst operon and membrane stability remains to be determined. Whether phosphate limitation, membrane stability, or both are responsible for the observed colonization defect remains to be demonstrated.
While STM screens involving gram-negative pathogens have identified a large number of mutants defective in amino acid synthesis (54), our screen only revealed three nutritional mutants with slight colonization defects, none of which had Tn insertions in genes involved with amino acid biosynthesis. This result is not surprising, considering that with the exception of certain group B vitamins, blood is a rich source of nutrients, containing fatty acids, glucose, and amino acids.
(iv) Host interaction mutant.
Another mutant, JG752, has a Tn insertion in a homolog of ascU, a gene encoding a cytoplasmic membrane component of the type III secretion system. This mutant was characterized in another study (55).
(v) Hypothetical proteins.
The remaining nine mutants were classified as having insertions in genes with unknown functions. Two of these mutants, JG573 and JG735, had CI values dramatically lower (100-fold and 10,000-fold, respectively) than those for the wild type. Sequencing efforts have dramatically increased the number of hypothetical proteins, which often account for 26% of the genes identified (24). The characterization of hypothetical proteins has been challenging because of the lack of phenotypes, but in this case the presence of a strong colonization phenotype provides us with the ability to characterize hypothetical proteins by creating specific mutations and testing the abilities of the mutated genes to complement the colonization defect. An animal phenotype can be used to assess complementation and the importance of individual protein domains. Alternatively, physiological stresses determined to be important for host colonization by the characterization of other mutants could hint at potential roles for the hypothetical proteins.
Characterization of the lpp mutant.
For further characterization, we chose to characterize a mutant, JG736, with a dramatic colonization defect and in vitro phenotypes in order to assess complementation. JG736 has a >300-fold-reduced ability to colonize the crop of the leech at 42 h and has the Tn inserted into a gene with homology to lpp. We cloned and sequenced the DNA flanking the Tn insertion of JG736. Sequence analysis verified that the Tn insertion is located in a gene with the highest similarity to Aeromonas salmonicida lpp (100% deduced amino acid identity) (Fig. 1B). The closest homolog that has been characterized biochemically is E. coli Lpp. Based on the 42% amino acid identity and conserved features described below, we term the inactivated gene lpp (Fig. 1B). The N terminus of Lpp contains a leader sequence for exporting the protein, and its C-terminal end is anchored to the peptidoglycan by a conserved lysine residue, thus tethering the outer membrane to the cell wall (7). The Lpp proteins in E. coli and Salmonella serovar Typhimurium (15, 21) have been shown to be important for membrane structure and function; strains carrying mutations in lpp are more susceptible to toxic compounds such as the surfactants SDS and Triton X-100 as well as to certain antibiotics (21). It has been observed that E. coli lpp mutants undergo membrane blebbing and leak periplasmic contents (21). A Salmonella serovar Typhimurium lpp mutant has been shown to be less virulent in mice than the wild type, inducing a dramatically weaker inflammatory response (15).
FIG. 1.
(A) lpp locus. The region used for complementation (black line) encompasses lpp as well as 92 bp downstream and 89 bp upstream of lpp. The transposon insertion site is indicated by the lollipop. (B) Amino acid alignment of Lpp proteins from A. veronii, A. salmonicida, A. hydrophila, Shewanella amazonensis, E. coli, and S. enterica.
Querying the recently published genome sequence from A. hydrophila (53) revealed the presence of an lpp-like sequence that was not annotated, possibly due to its small size, with 99% identity to the deduced amino acid sequence of A. veronii Lpp. The organization of the locus is conserved between the two species. A gene encoding a thioredoxin domain protein is located downstream of lpp, while ribB and a gene encoding a formyltetrahydrofolate deformylase are upstream of lpp (Fig. 1A). Based on the analysis of the open reading frames flanking lpp, the Tn insertion is unlikely to have a downstream effect (Fig. 1A). The gene encoding Lpp is 240 bp long, and the Tn is inserted after the 230th bp. While initially it seemed surprising that a Tn insertion that close to the end of an open reading frame can result in a phenotype, previous studies have shown that the final amino acid, a conserved lysine (Fig. 1B), is essential for anchoring the protein to the peptidoglycan (11, 70).
Complementation of JG736 with lpp.
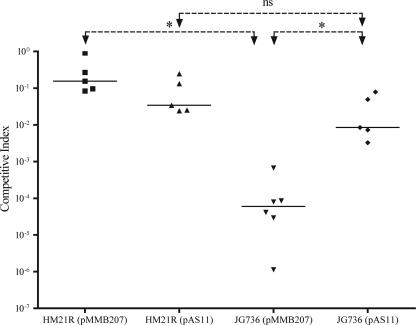
The observed colonization defect was linked to the disruption of lpp by complementing JG736 with lpp and its presumptive promoter region on pAS11, which was derived from the broad-host-range vector pMMB207 (Fig. 1A). The complementation was assessed 72 h after feeding by competing either HM21R (the parent strain) or JG736, each carrying either pAS11 or pMMB207, against the Smr competitor strain HM21RS in the leech. The complemented mutant colonized the leech crop at significantly higher levels than the mutant carrying the empty control plasmid (Fig. 2). Colonization by JG736(pAS11) did not reach wild-type levels, which could be due to poor maintenance of the plasmid or a copy number effect. These results demonstrate that the colonization defect in the leech is due to the lack of a functional lpp.
FIG. 2.
Complementation of JG736. The observed phenotype was linked to the disruption of lpp by complementing the JG736 colonization defect. The parent strain, HM21R, and JG736, each carrying either the broad-host-range vector pMMB207 or pAS11 (pMMB207 containing the complementing region) were tested against the competitor strain, HM21RS, and assessed at 72 h. The CI [calculated as (mutantoutput/competitoroutput)/(mutantinput/competitorinput)] for each animal is shown. Horizontal lines represent median CI values. Asterisks indicate a statistically significant difference between data sets (P < 0.05 by the Mann-Whitney test); ns, not significantly different.
SDS sensitivity assay.
Previously it has been shown that lpp mutants can be more susceptible to toxic compounds due to the decreased stability of the mutant's cell membrane (21). We examined the sensitivities of HM21R and JG736 carrying either pMMB207 or pAS11 to SDS. As expected, JG736(pMMB207) was significantly more sensitive to SDS than the parent strain, HM21R(pMMB207) (Table 3). The complemented mutant, JG736(pAS11), reached a significantly higher OD than JG736(pMMB207) at all SDS concentrations except 0.625%, indicating that a functional Lpp was required for the increased resistance to SDS. The complemented mutant also grew to levels similar to those of the parent strain carrying either vector (Table 3). These results are consistent with previous studies demonstrating the altered integrity of the cell membrane for lpp mutants.
TABLE 3.
SDS sensitivity assay
| Strain (plasmid) | OD595 (mean ± SD) at the following SDS concn (%):
|
|||||
|---|---|---|---|---|---|---|
| 0 | 0.3125 | 0.625 | 1.25 | 2.5 | 5.0 | |
| HM21R(pMMB207) | 0.95 ± 0.06 | 0.26 ± 0.04 | 0.13 ± 0.06 | 0.12 ± 0.06 | 0.09 ± 0.03 | 0.06 ± 0.03 |
| HM21R(pAS11) | 0.88 ± 0.01 | 0.22 ± 0.03 | 0.10 ± 0.05 | 0.17 ± 0.05 | 0.15 ± 0.09 | 0.13 ± 0.06 |
| JG736(pMMB207) | 0.87 ± 0.03 | 0.09 ± 0.04 | 0.11 ± 0.05 | 0.06 ± 0.06 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| JG736(pAS11) | 0.94 ± 0.02 | 0.19 ± 0.01 | 0.11 ± 0.03 | 0.16 ± 0.11 | 0.20 ± 0.18 | 0.10 ± 0.09 |
Motility assay.
The motility assay performed on the STM mutants revealed that JG736 was slightly less motile than the parent strain. We then examined the motilities of the parent strain, HM21R, carrying either pMMB207 or pAS11 and JG736 carrying either pMMB207 or pAS11 at various temperatures. JG736(pMMB207) was significantly less motile than the parent strain, HM21R(pMMB207) (Table 4). The motility phenotype of the complemented mutant, JG736(pAS11), was similar to that of the wild type, indicating that the mutation in lpp was responsible for the decreased motility (Table 4). The motility defect exhibited by JG736 is possibly due to an indirect effect on the proton motive force affecting flagellar rotation. However, the dramatic colonization defect exhibited by JG736 is not likely to be due to the slight motility defect we observed in soft agar. Another motility mutant, JG532, was less motile than JG736 yet possessed only a 1.58-fold colonization defect at 42 h, compared to a >300-fold defect for JG736. This suggests that the colonization defect of JG736 is most likely caused by increased sensitivity to antimicrobial peptides rather than by decreased motility.
TABLE 4.
Motility assay
| Strain (plasmid) | Diam (mm)a of colony at:
|
||
|---|---|---|---|
| 23°C | 30°C | 37°C | |
| HM21R(pMMB207) | 8.58 ± 1.16 | 14.00 ± 1.46 | 17.20 ± 1.68 |
| HM21R(pAS11) | 8.08 ± 0.92 | 12.80 ± 1.35 | 17.50 ± 0.71 |
| JG736(pMMB207) | 3.58 ± 0.66 | 9.40 ± 2.04 | 11.50 ± 2.45 |
| JG736(pAS11) | 6.92 ± 0.80 | 11.10 ± 1.43 | 16.40 ± 1.78 |
Values are means ± standard deviations, measured 24 h after inoculation.
Osmolarity.
While leeches feed, the ingested blood meal is being modified to concentrate the blood and achieve an osmolarity inside the crop that is isosmotic to the leech hemolymph (52). Water and salts are absorbed from the crop, and within 6 h after feeding, the weight of the leech is reduced by 25% due to the removal and loss of water. One hypothesis is that the change in osmolarity and the decreased stability of the bacterial membrane led to the decreased colonization ability of JG736. However, no increased sensitivity to osmolarity was observed. There was no difference in plating efficiency between JG736 and wild-type cells when they were grown in a medium simulating the osmotic conditions of leech hemolymph and then plated onto regular LB agar (data not shown). These results suggest that changes in osmolarity are not responsible for the observed colonization defect of JG736.
Temporal competition phenotype.
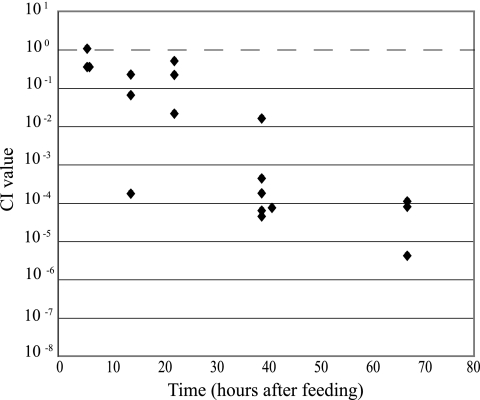
The ability of the lpp mutant JG736 to colonize the digestive tract of the leech was tested in a competition assay over time (Fig. 3). JG736 did not exhibit a colonization defect at 6 h postfeeding; however, from 15 h onward, JG736 had a statistically significantly reduced ability to colonize the crop. The inability of JG736 to successfully colonize the leech crop was most evident at 72 h, when the mutant showed a >25,000-fold colonization decrease compared to the competitor strain. The temporal changes in colonization dynamics suggest that the digestive tract physiology changes beyond 6 h after feeding in a way that is detrimental to JG736.
FIG. 3.
Decreased ability of the lpp mutant, JG736, to colonize the leech. JG736 was coinoculated with the competitor strain in a 1:1 ratio. The CI [(mutantoutput/competitoroutput)/(mutantinput/competitorinput)] was calculated for each animal and plotted over time. A CI of 1 (dashed line) indicates that the mutant and competitor strain colonize to equal levels. A CI below 1 indicates that the mutant is outcompeted and has a colonization defect. The decrease in the CI is statistically significant from 15 h onward (P < 0.05 by a single-sided, two-tailed t test).
We have shown that a functional lpp is required for successful colonization of the leech digestive tract. A likely explanation for the observed colonization defect is that the intraluminal fluid inside the leech is modified in a manner that increases membrane stress. One intriguing hypothesis is that antimicrobial peptides are secreted by the leech or the microbial symbionts. Antimicrobial peptides have been reported for many invertebrates, including the leech (60, 65), but none from the leech crop have yet been identified. Further analysis of the lpp mutant may lead to the identification of another factor that contributes to the simplicity of this digestive tract symbiosis.
Comparison of pathogenic and mutualistic associations.
Our findings have greatly increased the understanding of the molecular requirements for the benign colonization of digestive tracts. Interestingly, the classes of genes involved in host colonization are similar to those reported in previous studies utilizing STM with pathogenic bacteria and one symbiotic bacterium (13, 20, 33, 54). The proportion of genes identified as being important for the colonization of the leech (1.2%) was more than two times higher than that found to be required for the symbiotic colonization of the nematode host by X. nematophila (20). However, STM studies with gram-negative mammalian pathogens such as Yersinia enterocolitica, Salmonella serovar Typhimurium, and Vibrio cholerae have shown that slightly more than 2% of their STM mutants had attenuated virulence (10, 13, 33, 54). The percentage is affected by the sensitivity and reproducibility of the colonization assay as well as by the complexity of the interactions.
In this study, we identified 20 mutants that had reduced abilities to compete in the crop of the medicinal leech. Only three of these mutants had a similar defect in blood. The remaining mutants provided new insights into the interaction between A. veronii and H. verbana. The importance of regulatory genes for colonization demonstrates the ability of A. veronii to recognize the host environment and regulate genes accordingly. Future studies of the response regulator with a dramatic colonization defect may provide information about how the symbiont recognizes the host environment and regulates gene expression or enzymatic activity.
We have previously shown the importance of intact LPS for serum resistance (6) and the importance of a type III secretion system to prevent phagocytosis (55). The characterization of the lpp mutant suggests that there are additional antimicrobial properties that prevent the colonization of sensitive bacteria. Interestingly, only 53% of the genes identified in our STM screen possessed homologs in the closely related organism A. hydrophila. These genes could account for the ability of A. veronii to colonize the leech digestive tract. Future functional characterization of these genes will not only enhance the understanding of the A. veronii-leech symbiosis but also pave the way for the discovery of conserved mechanisms for digestive tract colonization.
Acknowledgments
We thank D. Holden and H. Goodrich-Blair for sending strains and R. Rio and Y. Kikuchi for helpful comments. We also thank D. Gage for the use of the microplate reader.
This research was supported by U.S. NSF career award MCB 0448052 to J.G. S.K. was supported by Swiss National Science Foundation grant 31-63775 to J.G.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Aguena, M., E. Yagil, and B. Spira. 2002. Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268:518-524. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1996. Understanding signal transduction during bacterial infection. Trends Microbiol. 4:141-146. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 5.Boccazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri fusaro. J. Bacteriol. 182:2611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braschler, T. R., S. Merino, J. M. Tomas, and J. Graf. 2003. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl. Environ. Microbiol. 69:4268-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415:335-377. [DOI] [PubMed] [Google Scholar]
- 8.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 11.Choi, D. S., H. Yamada, T. Mizuno, and S. Mizushima. 1987. Molecular assembly of the lipoprotein trimer on the peptidoglycan layer of Escherichia coli. J. Biochem. (Tokyo) 102:975-983. [DOI] [PubMed] [Google Scholar]
- 12.Daigle, F., J. M. Fairbrother, and J. Harel. 1995. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect. Immun. 63:4924-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 15.Fadl, A. A., J. Sha, G. R. Klimpel, J. P. Olano, D. W. Niesel, and A. K. Chopra. 2005. Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar Typhimurium systemic infection. Infect. Immun. 73:1081-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, J. 1999. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect. Immun. 67:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf, J., Y. Kikuchi, and R. V. Rio. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 18.Hamann, L., V. El-Samalouti, A. J. Ulmer, H. D. Flad, and E. T. Rietschel. 1998. Components of gut bacteria as immunomodulators. Int. J. Food Microbiol. 41:141-154. [DOI] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 20.Heungens, K., C. E. Cowles, and H. Goodrich-Blair. 2002. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 45:1337-1353. [DOI] [PubMed] [Google Scholar]
- 21.Hirota, Y., H. Suzuki, Y. Nishimura, and S. Yasuda. 1977. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc. Natl. Acad. Sci. USA 74:1417-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch, A. M., and M. J. McFall-Ngai. 2000. Fundamental concepts in symbiotic interactions: light and dark, day and night, squid and legume. J. Plant Growth Regul. 19:113-130. [DOI] [PubMed] [Google Scholar]
- 23.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 24.Iliopoulos, I., S. Tsoka, M. A. Andrade, P. Janssen, B. Audit, A. Tramontano, A. Valencia, C. Leroy, C. Sander, and C. A. Ouzounis. 2001. Genome sequences and great expectations. Genome Biol. 2:INTERACTIONS0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indergand, S., and J. Graf. 2000. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 66:4735-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi, Y., and J. Graf. 2007. Spatial and temporal population dynamics of a naturally occurring, two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamarche, M. G., C. M. Dozois, F. Daigle, M. Caza, R. Curtiss III, J. D. Dubreuil, and J. Harel. 2005. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 73:4138-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 30.Mackay, D. R., E. K. Manders, G. C. Saggers, D. R. Banducci, J. Prinsloo, and K. Klugman. 1999. Aeromonas species isolated from medicinal leeches. Ann. Plast. Surg. 42:275-279. [DOI] [PubMed] [Google Scholar]
- 31.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 32.McFall-Ngai, M. J. 2000. Negotiations between animals and bacteria: the ‘diplomacy’ of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A 126:471-480. [DOI] [PubMed] [Google Scholar]
- 33.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 34.Metges, C. C. 2000. Contribution of microbial amino acids to amino acid homeostasis of the host. J. Nutr. 130:1857S-1864S. [DOI] [PubMed] [Google Scholar]
- 35.Michalsen, A., S. Klotz, R. Ludtke, S. Moebus, G. Spahn, and G. J. Dobos. 2003. Effectiveness of leech therapy in osteoarthritis of the knee: a randomized, controlled trial. Ann. Intern. Med. 139:724-730. [DOI] [PubMed] [Google Scholar]
- 36.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 37.Moran, N. A., and P. H. Degnan. 2006. Functional genomics of Buchnera and the ecology of aphid hosts. Mol. Ecol. 15:1251-1261. [DOI] [PubMed] [Google Scholar]
- 38.Morandi, A., O. Zhaxybayeva, J. P. Gogarten, and J. Graf. 2005. Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J. Bacteriol. 187:6561-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2:632-642. [DOI] [PubMed] [Google Scholar]
- 40.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 42.Parsek, M. R., and E. P. Greenberg. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13:27-33. [DOI] [PubMed] [Google Scholar]
- 43.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 44.Rados, C. 2004. Beyond bloodletting: FDA gives leeches a medical makeover. FDA Consum. 38:9. [PubMed] [Google Scholar]
- 45.Rao, N. N., and A. Torriani. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4:1083-1090. [DOI] [PubMed] [Google Scholar]
- 46.Rio, R. V., M. Anderegg, and J. Graf. 2007. Characterization of a catalase gene from Aeromonas veronii, the digestive-tract symbiont of the medicinal leech. Microbiology 153:1897-1906. [DOI] [PubMed] [Google Scholar]
- 47.Rosado, M., and D. J. Gage. 2003. Transcriptional control of a rRNA promoter of the nodulating symbiont Sinorhizobium meliloti. FEMS Microbiol. Lett. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Ruby, E. G., and M. J. McFall-Ngai. 1992. A squid that glows in the night: development of an animal-bacterial mutualism. J. Bacteriol. 174:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188:8327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 51.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 52.Sawyer, R. T. 1986. Leech biology and behavior. Clarendon Press, Oxford, United Kingdom.
- 53.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shea, J. E., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 55.Silver, A. C., Y. Kikuchi, A. A. Fadl, J. Sha, A. K. Chopra, and J. Graf. 2007. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. USA 104:9481-9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 57.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 58.Stevenson, B. S., and T. M. Schmidt. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 70:6670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tasiemski, A., F. Vandenbulcke, G. Mitta, J. Lemoine, C. Lefebvre, P. E. Sautiere, and M. Salzet. 2004. Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J. Biol. Chem. 279:30973-30982. [DOI] [PubMed] [Google Scholar]
- 61.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vollaard, E. J., H. A. Clasener, H. K. van Saene, and N. F. Muller. 1990. Effect on colonization resistance: an important criterion in selecting antibiotics. DICP 24:60-66. [DOI] [PubMed] [Google Scholar]
- 65.Wang, X., X. Wang, Y. Zhang, X. Qu, and S. Yang. 2003. An antimicrobial peptide of the earthworm Pheretima tschiliensis: cDNA cloning, expression and immunolocalization. Biotechnol. Lett. 25:1317-1323. [DOI] [PubMed] [Google Scholar]
- 66.Worthen, P. L., C. J. Gode, and J. Graf. 2006. Culture-independent characterization of the digestive tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yem, D. W., and H. C. Wu. 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 133:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, H., J. W. Peterson, D. W. Niesel, and G. R. Klimpel. 1997. Bacterial lipoprotein and lipopolysaccharide act synergistically to induce lethal shock and proinflammatory cytokine production. J. Immunol. 159:4868-4878. [PubMed] [Google Scholar]
- 70.Zhang, W. Y., and H. C. Wu. 1992. Alterations of the carboxyl-terminal amino acid residues of Escherichia coli lipoprotein affect the formation of murein-bound lipoprotein. J. Biol. Chem. 267:19560-19564. [PubMed] [Google Scholar]
- 71.Zhang, Y. L., E. Arakawa, and K. Y. Leung. 2002. Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 70:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]