Abstract
MetR (formerly Smu.1225), a regulator of the LysR family, controls key genes for methionine supply in Streptococcus mutans. An S. mutans metR mutant is unable to transport l-methionine and to grow in the absence of this amino acid. Accordingly, MetR activates transcription by binding to the promoter regions of two gene clusters and smu.1487, whose products are involved in methionine biosynthesis (MetEF and Smu.1487) and uptake (AtmBDE). Transcriptional activation by MetR requires the presence of a 17-bp palindromic sequence, the Met box. Base substitutions in the Met box hinder the formation of a MetR-DNA complex and abolish MetR-dependent activation, showing that Met boxes correspond to MetR recognition sites. Activation by MetR occurs in methionine-depleted medium and is rapidly triggered under nonactivating conditions by the addition of homocysteine. This intermediate of methionine biosynthesis increases the affinity of MetR for DNA in vitro and appears to be the MetR coeffector in vivo. Homocysteine plays a crucial role in methionine metabolic gene regulation by controlling MetR activity. A similar mechanism of homocysteine- and MetR-dependent control of methionine biosynthetic genes operates in S. thermophilus. These data suggest a common mechanism for the regulation of the methionine supply in streptococci. However, some streptococcal species are unable to synthesize the homocysteine coeffector. This intriguing feature is discussed in the light of comparative genomics and streptococcal ecology.
The sulfur-containing amino acid methionine is of major importance in the cellular metabolism of all living organisms. Methionine is the universal initiator of protein synthesis, and its derivative S-adenosylmethionine (SAM) and autoinducer-2 (AI-2) (see Fig. 1) are involved in a variety of cellular processes. SAM serves as the major biological methyl donor in methylation reactions, which are essential for the biosynthesis of phospholipids, proteins, DNA, and RNA (11), and the signaling molecule AI-2 is involved in interspecies communication and gene regulation in both gram-negative and gram-positive bacteria (7, 21, 46). Methionine availability is limiting for the growth of several lactic acid bacteria (4) and group B streptococci (39), and methionine biosynthetic genes are essential for the virulence of Brucella melitensis (28), Haemophilus parasuis (22), and Salmonella enterica (9).
FIG. 1.
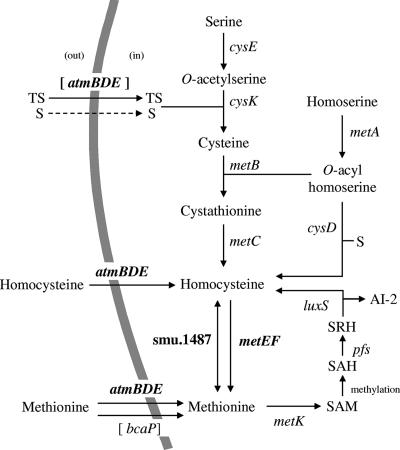
Sulfur amino acid biosynthesis and uptake pathways in S. mutans. The pathways were deduced from (i) the analysis of S. mutans genome sequence data (http://www.ncbi.nlm.nih.gov/genomes), (ii) knowledge of sulfur amino acid biosynthetic pathways in other bacteria, such as L. lactis (41), and (iii) the experimental results of this work. Genes of S. mutans proposed to encode proteins involved in the corresponding reaction are shown; genes belonging to the MetR and homocysteine regulon are in bold, and genes for transporters potentially involved in the uptake of thiosulfate and methionine are in brackets. S, sulfide; TS, thiosulfate; SAH, S-adenosylhomocysteine; SRH, S-ribosylhomocysteine; dashed-line arrow, not-yet-identified transporters; AtmBDE, ABC transporter belonging to the MUT family and associated with d-methionine, l-methionine (low concentrations), selenomethionine, and homocysteine in S. mutans; BcaP (formerly Smu.16), transporter potentially involved in methionine uptake; CysE, serine O-acetyltransferase; CysK, O-acetylserine sulfhydrylase; MetB, cystathionine γ-synthase; MetC, cystathionine β-lyase; MetA, homoserine O-acyltransferase; CysD, O-acylhomoserine sulfhydrylase; Smu.1487, putative homocysteine S-methyltransferase; MetE, methionine synthase; MetF, putative methylenetetrahydrofolate reductase; MetK, methionine adenosyltransferase; Pfs, S-adenosylhomocysteine nucleosidase; LuxS, S-ribosylhomocysteinase.
Methionine can be either supplied from the environment by the cell or synthesized de novo (see Fig. 1). Two methionine transport systems in microorganisms have been described previously: an ABC transporter that belongs to the methionine uptake transporter (MUT) family (23, 33, 48) and BcaP of the permease transporter family (8). Methionine can be produced directly by the methylation of homocysteine by a methionine synthase (MetE) in conjunction with a methylenetetrahydrofolate reductase (MetF) (see Fig. 1) (16, 27). Homocysteine can be synthesized from (i) cysteine by the transsulfuration pathway (MetA, MetB, and MetC), (ii) sulfide by the sulfhydrylation pathway (MetA and CysD), and (iii) methionine by the SAM recycling pathway (MetK, Pfs, and LuxS). Both the transsulfuration and sulfhydrylation pathways start with O-acylhomoserine, synthesized by the acylation of homoserine by homoserine acyltransferase (MetA). In the transsulfuration pathway, homocysteine is formed from O-acylhomoserine and cysteine in two steps, with the intermediary formation of cystathionine. This process requires the sequential action of a cystathionine γ-synthase (MetB) and a cystathionine β-lyase (MetC). In the sulfhydrylation pathway, homocysteine is directly synthesized from O-acylhomoserine and sulfide by O-acylhomoserine sulfhydrylase (CysD). Finally, in the SAM recycling pathway, methionine serves as a substrate for the synthesis of homocysteine, after its activation by ATP to form SAM. The utilization of SAM as a methyl donor results in the formation of S-adenosylhomocysteine, which is degraded into AI-2 and homocysteine by the successive action of an S-adenosylhomocysteine nucleosidase (Pfs) and an S-ribosylhomocysteinase (LuxS) (see Fig. 1).
While methionine biosynthetic enzymes and metabolic pathways are well conserved among bacteria, the regulation of methionine metabolism often involves very different mechanisms. Two transcriptional regulators, MetJ and MetR, regulate the expression of methionine biosynthetic genes in Escherichia coli and Salmonella enterica serovar Typhimurium (16, 45). The E. coli met genes (except for metH) are negatively regulated by the MetJ repressor, with SAM serving as a corepressor (37). Several of these genes are also under the positive control of MetR, a LysR-type transcriptional regulator (LTTR), with homocysteine as a coeffector (3, 5, 30, 43, 44). In Bacillus subtilis, the met genes are controlled by a mechanism of premature termination of transcription at the conserved S-box motif (1, 17). Here, SAM binds directly to the 5′ untranslated part of the mRNA at the S-box motif to modulate the expression of the downstream genes (31, 32). In the food bacterium Lactococcus lactis, a unique regulator, FhuR (CmbR), controls most of the genes of the cysteine and methionine biosynthesis pathways, including those involved in the interconversion and transport of these amino acids (41). The LTTR FhuR directly activates the expression of these genes with the first intermediate of cysteine synthesis, O-acetylserine (OAS), as a coeffector (10, 41). Very little is known about the regulation of methionine metabolism in other streptococci. In group B streptococci, the LTTR MtaR regulates the synthesis of an uncharacterized methionine transporter (39). However, neither the signal nor the MtaR-binding site has been characterized. It is noteworthy that MtaR shows only weak similarity to FhuR (31% identity) and that no FhuR-binding sites are found in the streptococcal genomes (15, 41). The control of methionine metabolism in other streptococci may be different from the characterized control system in L. lactis.
These considerations prompted us to study the possible role of MetR (formerly Smu.1225), an MtaR-like protein (81% identical to MtaR), in the control of methionine metabolism in Streptococcus mutans, the primary etiological agent of human dental caries. We showed that MetR activated the expression of several genes whose products are involved in methionine synthesis (MetEF and Smu.1487) and uptake (AtmBDE) by binding directly to their promoter regions. Both MetR binding and MetR-dependent transcriptional activation required the presence of a 17-bp palindromic sequence, the Met box. We also showed that homocysteine, an intermediate of methionine biosynthesis, triggered the MetR-dependent expression of smu.1487 and the metEF and atmBCDE clusters in vivo and increased the affinity of the regulator for its target sequences in vitro. Finally, we demonstrated that the characterized MetR-dependent regulation of methionine metabolism in S. mutans was operative in S. thermophilus and, therefore, may widely apply to streptococci.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli TG1 was used for plasmid propagation. E. coli was grown in Luria-Bertani medium at 37°C (29). S. mutans strains were grown at 37°C in M17 glucose medium or in chemically defined medium (42) containing glucose (20 g/liter) as the carbon source and all amino acids except methionine and cysteine (CDM). The CDM was supplemented with 1 mM sulfide (Na2S; Sigma), 1 mM sulfite (Na2SO3; Merck), 0.5 mM disulfite (Na2S2O5; Merck), 1 mM sulfate (Na2SO4; Merck), 0.5 mM thiosulfate (Na2S2O3; Merck), 0.67 mM d-methionine (Sigma), or 0.7 mM seleno-l-methionine (Sigma) when indicated. The CDM was supplemented with 0.67 mM l-methionine (Merck; CDM+M), 1.65 mM l-cysteine (Sigma; CDM+C), or l-cysteine and l-methionine (CDM+CM) as noted. The effect of OAS (Sigma), thiosulfate, N-acetylserine (Sigma), sulfide, or dl-homocysteine (Sigma) on transcription was tested by the addition of 4 mM concentrations of these compounds to exponentially growing cells (optical density at 600 nm [OD600] of 0.2) in CDM+CM. When required, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (0.04 g/liter), isopropyl-1-β-d-thiogalactopyranoside (IPTG; 0.04 g/liter), ampicillin (100 μg/ml for E. coli), erythromycin (8 μg/ml for S. mutans and 100 μg/ml for E. coli), and tetracycline (3 μg/ml for S. mutans) were added to the culture medium.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE Δthi(lac-proAB) hsdD5 (F′ traD36 proAB lacIqZΔM15) | 14 |
| ER2566 | F− λ−fhuA2 [lon] ompT (lacZ::T7 gene 1) gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::mini-Tn10-Tets)2 R(zgb-210::Tn10-Tets) endA1 [dcm] | New England Biolabs |
| JIM9042 | ER2566 containing pJIM5793 for MetR-His8 purification; Apr | This work |
| JIM8859 | TG1 containing pGEM-T easy; Apr | This work |
| S. thermophilus | ||
| CNRZ1066 | Wild-type strain isolated from yogurt manufactured in France | INRA |
| JIM8315 | CNRZ1066 ΔmetR Emr | S. Petrya |
| S. mutans | ||
| UA159 | S. mutans wild-type strain (ATCC 700610) | ATCC |
| JIM8860 | UA159 ΔmetR Emr | This work |
| JIM8861 | UA159 ΔmetR Tetr | This work |
| JIM8862 | UA159 ΔatmB Emr | This work |
| JIM8863 | UA159 ΔatmBCDE Emr | This work |
| JIM8864 | UA159 ΔcysD Emr | This work |
| JIM8865 | UA159 ΔmetE Emr | This work |
| JIM8866 | UA159 Δsmu.1487 Emr | This work |
| JIM8881 | UA159 ΔmetC Emr | This work |
| JIM8885 | UA159 ΔmetC ΔcysD Emr Tetr | This work |
| JIM8867 | UA159 containing pJIM5778 integrated at the PmetE locus; Emr | This work |
| JIM8868 | UA159 containing pJIM5779 integrated at the PmetE locus; Emr | This work |
| JIM8869 | UA159 containing pJIM5780 integrated at the PmetE locus; Emr | This work |
| JIM8870 | UA159 containing pJIM5781 integrated at the PmetE locus; Emr | This work |
| JIM8871 | UA159 containing pJIM5782 integrated at the PmetE locus; Emr | This work |
| JIM8872 | UA159 containing pJIM5783 integrated at the PmetE locus; Emr | This work |
| JIM8873 | JIM8861 containing pJIM5778 integrated at the PmetE locus; Emr Tetr | This work |
| Plasmids | ||
| pGEM-T easy | Apr M13ori pBR322ori; linear T overhangs vector | Promega |
| PTYB1 | Apr; expression vector used in IMPACT-CN protein purification system | New England Biolabs |
| pJIM4900 | Emr ori+ ΔrepA; integrative promoter probe vector containing luxAB genes | S. Petrya |
| pJIM5793 | 937-bp fragment carrying fusion of metR and the His8 coding sequence cloned into NdeI-NsiI sites of pTYB1 | This work |
| pJIM5778 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE on a 908-bp fragment | This work |
| pJIM5779 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE with the box 1 deletion on a 908-bp fragment | This work |
| pJIM5780 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE with the box 2 deletion on a 908-bp fragment | This work |
| pJIM5781 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE with the box 1 and 2 deletions on a 908-bp fragment | This work |
| pJIM5782 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE with the box 1 and 2 and P2 deletions on a 710-bp fragment | This work |
| pJIM5783 | PmetE-lux-SpeI site fusion of pJIM4900 and pGEM-T easy containing PmetE with the P2 deletion on a 797-bp fragment | This work |
Unpublished data.
DNA manipulation procedures.
DNA manipulations and E. coli transformation were performed as described previously (41). All enzymes for DNA technology were used according to the manufacturers' specifications. The transformation of S. mutans by natural competence was performed as described by Murchison et al. (34). Primers used in this work were synthesized by Sigma-Genosys (Table 2). DNA sequencing of both strands was performed by using a fluorescence sequencing procedure.
TABLE 2.
Primers used in this study
| Primer function or corresponding gene | Sequencea (5′-3′) |
|---|---|
| Primers for gene deletion | |
| atmB up | CAGTTATACTAAAGAGCGTGCC |
| CCAACCAACCAACCAACCGCTAAAGCCAATCCTAAAAGACC | |
| atmB down | CTGTTGAAGCAGATTCTTCAGCTCC |
| CCTTCCTTCCTTCCTTCCCTGCCAAAGGCGTTGATGAACCTG | |
| atmBCDE up | CAGTTATACTAAAGAGCGTGCC |
| CCAACCAACCAACCAACCGCTAAAGCCAATCCTAAAAGACC | |
| atmBCDE down | CCTGTCCGACACGATTAACAGC |
| CCTTCCTTCCTTCCTTCCCGCTAACTCGAAAAGTAAGTCATCG | |
| cysD up | GGAATCACGCGAGCGGAACAAATGC |
| CCAACCAACCAACCAACCCTCCATTCTACTCCATTATTCGGT | |
| cysD down | GCGATGGCTCTAACAGGAGAACCTG |
| CCTTCCTTCCTTCCTTCCCGAAGATTTGCGCCTAGCACTTGA | |
| metE up | GTCAACATTCCAATGGCAGCTATC |
| CCAACCAACCAACCAACCGTCATTTCGTCCTCCTCTTAGCC | |
| metE down | CATTACCAAGTTACTAAAAGCTTCC |
| CCTTCCTTCCTTCCTTCCGCTGCAACTAAGGAAGTTCGTC | |
| metR up | CTTCCAAAATCACCGCATCTTTGTTGGC |
| CCAACCAACCAACCAACCACGCATGCCTTTATTTTAGCACA | |
| metR down | GCTTAATGTTGATTTTCTCTTTCAGAAC |
| CCTTCCTTCCTTCCTTCCTAGTATGCAAAAAGGCGACAGC | |
| smu.1487 up | CAACCGTCAGGTCAAACAAGCG |
| CCAACCAACCAACCAACCGTGTCATATACATGCCTCTTTCTAG | |
| smu.1487 down | GTCAACTGCGGTAGATGAATCAG |
| CCTTCCTTCCTTCCTTCCCAAACAAGGCCAAGAAATCGCTAAG | |
| Primers for gene amplification | |
| eryR | GGTTGGTTGGTTGGTTGGCAGCAAAGAATGGCGGAAACG |
| GGAAGGAAGGAAGGAAGGCTACATTCCCTTTAGTAACGTG | |
| metR-His8 | TTGTGCTAAAATAAACATATGCGTATTCAACAG |
| AATAAAAAATGCATCTACTAATGGTGATGGTGATGGTGATGGT GTTTATCAAATTTAACTTCT | |
| Primers for QRT-PCR analysis of S. mutans | |
| atmB | GCTCGTAATTTAGCATCCGTTG |
| CCATTGTTTAGAGCCTTCGTTC | |
| cysD | GCGTGCAGTACCAATCTATCAG |
| CGGCTGTAGTCGGATTAGTGAT | |
| metA | GAATATGACCGAGAAACCTTGG |
| CGAGATTCCAACGCATAATAGG | |
| metE | TGGGTCCAATCTTATGGTTCTC |
| CAGGACGATCAGTCAGACTTTG | |
| metR | TGACCAGGGTATCCATAACTCC |
| ATAGCCGTCAAGACCAATCATC | |
| luxS | AGTTTGGACCTAAAGGCGATCT |
| TGACGAATCAGCTTAGCCAGTA | |
| urk | AGCGCGGTCGTAGTTTAGATAG |
| AACGAGATTGCTGACACCTTCT | |
| smu.1387c | ACTCTGCCAATACAGCCATGAT |
| AGCAATGCTAGTATCACGCTCA | |
| smu.1486c | GATGAAGTGGCAGATATGGACA |
| ATCAACCGTCAGGTCAAACAAG | |
| smu.1487 | GTTGGATCTCTTCTTCGTCCAG |
| TCTGCTGTTTCTGTTTCACGAT | |
| smu.1936c | GTTTGGCAGGTGTTATGATTGA |
| TAGTTGTTTCTGCCGTCTGATG | |
| tuf | CAGCGTCGATTGAAGCGTAGTC |
| TTACGGAACATTTCAACACCAG | |
| Primers for QRT-PCR analysis of S. thermophilus | |
| atmB | GAATTGGCTAAAGACAAGGGTG |
| GGAAAGCATTAACGTCGATTTC | |
| metE | AGATTACACACGTTTACCGGCT |
| CTTAGTTTGAGGGAATGAACCG | |
| tuf | CATTGGTACAATCGGACACGTTG |
| CAGCGTCGATTGAAGCGTAGTC | |
| Primers for promoter amplification | |
| metE | GTCAACATTCCAATGGCAGCTATC |
| GTCATTTCGTCCTCCTCTTAGCC | |
| metE with box 1 deletion | GTCAACATTCCAATGGCAGCTATC |
| AGTATTTGTTACAGTTTTCATCTCTAA | |
| GTCATTTCGTCCTCCTCTTAGCC | |
| TTAGAGATGAAAACTGTAACAAATACT | |
| metE with box 2 deletion | GTCAACATTCCAATGGCAGCTATC |
| GTATATGTACAGTTTTAAGCTCTATATG | |
| GTCATTTCGTCCTCCTCTTAGCC | |
| CATATAGAGCTTAAAACTGTACATATAC | |
| metE with box 1 and 2 deletions | GTCAACATTCCAATGGCAGCTATC |
| GTACAGTTTTAAGCTCTATATGTAGTAT | |
| TTGTTACAGTTTTCATCTCTAA | |
| GTCATTTCGTCCTCCTCTTAGCC | |
| TTAGAGATGAAAACTGTAACAAATACTA | |
| CATATAGAGCTTAAAACTGTAC | |
| metE with box 1 and 2 and P2 deletions | GAACCTGAAATGTTTTTGAATACAAG |
| GTCAACATTCCAATGGCAGCTATC | |
| metE with P2 deletion | GTTTCAAGGGTTTTTGTGTTTTTATG |
| GTCAACATTCCAATGGCAGCTATC | |
| Primers for transcription start site mapping | |
| atmB | GCTTTATTCGGCTGTGAATAATCTG |
| CGGACTGATATATGTCTCAGCTAC | |
| cysD | CTGCTGCCATACCAGAAGCTGTTG |
| GATGAGCAGTATCGCCAATTTGCC | |
| metA | CAAGATACTCATGGGCCGAATATCC |
| GAATGATGACTCGTCATGTAGAG | |
| metE | GACCATTCGGGAACAATATAGTG |
| GTCTAAACGCAGTTCCTTAGCACCAGC | |
| urk | CACTGCGTGTATGCTGTGTGTAGTC |
| CGAATGTCATCATCGGTATCAAC | |
| smu.1387 | GGCAGCTTGCCTGCCAGTAAATCCTTC |
| GGATAGGAATTCCTTAATAAGAAGGAG | |
| smu.1486c | GCTAGGTTGTGGTCGATCATCTTGTG |
| CAACCGTCAGGTCAAACAAGCG | |
| smu.1487 | GTGTAGCCGTGGTCTGCAATGTAACG |
| TCTGCTGTTTCTGTTTCACGAT | |
| smu.1936c | CCTGTCTGAGCCAACTCCCGGATC |
| CGTTTAGTCGCTACAGATCCAGC | |
| Primers for gel retardation probe | |
| atmB | CCTTGGAGGAAGCTATCTTCCAG |
| GCTAAAGCCAATCCTAAAAGACC | |
| cysD | TAGCGTTCACGGGATTAGAC |
| GCTCTCCTTTTATTATTAATTAGCTCC | |
| cysK | GTTAACACTACTATTTATGTGG |
| CTTCAGGAACAACATTGTTGAG | |
| metA | GGCAACCATTGAAATGATTGAAAGG |
| GAGTAATTGGCATGAAAATATCTCC | |
| metE | GTCACTTGAAATGATAAGAGGAG |
| GTCATTTCGTCCTCCTCTTAGCC | |
| metR | CCTTACCAACTAACTGGCAAGGTTAC |
| GGCAGCCTCATTCATGCTGCCAG | |
| smu.1487 | GTGTGTAAATGGTTATCGCG |
| GTCAAGCAGATCAGCTGGAC | |
| metE with box 1 and 2 deletions | GTCACTTGAAATGATAAGAGGAG |
| GTACAGTTTTAAGCTCTATATGTAGTAT | |
| TTGTTACAGTTTTCATCTCTAA | |
| GTCATTTCGTCCTCCTCTTAGCC | |
| TTAGAGATGAAAACTGTAACAAATACT | |
| ACATATAGAGCTTAAAACTGTAC |
Upper and lower sequences correspond to primer pairs.
Gene deletion in S. mutans.
The chromosomal atmB, atmBCDE, cysD, metC, metE, metR (smu.1225), and smu.1487 genes in the S. mutans UA159 strain were deleted as follows. Two DNA fragments of about 750 bp corresponding to the upstream and the downstream regions of each gene were amplified by PCR using specific primers (Table 2). For each gene, a second PCR amplification was carried out using these two PCR fragments and an antibiotic (erythromycin or tetracycline) resistance cassette as a template to synthesize a fragment carrying the antibiotic resistance cassette flanked by the upstream and downstream chromosomal regions of the gene. This PCR fragment was used to transform S. mutans strain UA159. The antibiotic resistance cassette was integrated at the corresponding locus by a double-crossover event leading to the deletion of the targeted gene. The mutants obtained are listed in Table 1. Regions flanking the deletions were verified by sequencing to ensure the absence of PCR-induced mutations.
l-Methionine uptake and substrate specificity of the AtmBDE transporter.
S. mutans cells were grown at 37°C to mid-exponential growth phase (OD600 of 0.4) in 2 ml of CDM+C for the wild type and 2 ml of CDM+CM for the metR mutant. Cells were harvested, washed twice with CDM containing 100 μg of chloramphenicol/ml, and resuspended in 2 ml of CDM containing chloramphenicol. l-[14C]methionine (68 Ci/mol; MP Biomedicals) was added to a concentration of 1.5 μM, and cells were incubated at 30°C for uptake experiments. Samples (200 μl) were withdrawn at intervals and filtered through 0.45-μm-pore-size Durapore membranes (catalog no. HVLP02500; Millipore). Filters were washed with 3 ml of CDM and dried, and the radioactivity was measured by scintillation counting. The effect of the addition of unlabeled amino acids or sulfur compounds at a concentration of 15 μM to the reaction mixture containing 1.5 μM l-[14C]methionine was determined. A 5-min transport kinetics analysis was performed to ensure the linearity of the curves. The inhibition of l-methionine uptake at 2 min was determined.
Construction of lux transcriptional fusions and determination of Lux activity in S. mutans.
The luxAB reporter genes were placed under the control of the metE promoter region. The metE-lux fusions were integrated at the metE locus of S. mutans as described previously (19). Briefly, chromosomal DNA from S. mutans UA159 was used as a template to amplify DNA fragments containing the wild-type or mutant metE promoter variants. PCR fragments were cloned into the pGEM-T easy vector, sequenced, and introduced into the streptococcal integration vector pJIM4900 upstream of the luxAB genes (Table 1). The resulting plasmids were integrated by a single-crossover event at the metE locus in the S. mutans UA159 chromosome. Strains carrying a single copy of the integrated plasmids were identified by PCR with specific primers. The resulting strains (Table 1) contained a PmetE-lux transcriptional fusion as well as an intact copy of metE. Luciferase assays were carried out with a Lumat LB9501 apparatus (Berthold Technologies, Bad Wildbad, Germany) (19).
RNA isolation, transcriptional start site mapping, and real-time QRT-PCR.
Total RNA was isolated as described by Sperandio et al. (41). Residual chromosomal DNA was removed by treating total RNA preparations with RNase-free DNase I according to the protocol of the manufacturer (Roche). The RNA concentration was determined by absorbance at 260 nm, and the quality of RNA preparations was checked as described previously (20). The transcriptional start sites of the atmB, cysD, metA, metE, smu.1387c, smu.1486c, smu.1487, and urk genes were determined by using a kit for the 5′-3′ rapid amplification of cDNA ends (Roche). Real-time quantitative reverse transcription-PCR (QRT-PCR) was carried out using first-strand cDNA as the template. The primers used are listed in Table 2. The synthesis of cDNA and quantitative PCR were carried out as described previously (41). For each gene, QRT-PCR experiments were performed in triplicate. Results were normalized by using the gene coding for the translational elongation factor Tu (smu.714).
Purification of histidine-tagged MetR protein and gel retardation assay.
The metR gene carrying a histidine (His8) tag coding sequence at its 3′ end was generated from the S. mutans UA159 chromosome by PCR with specific primers (Table 2). The purified PCR product was digested with NdeI and NsiI and ligated into the corresponding sites in vector pTYB1 (New England Biolabs), resulting in pJIM5793. E. coli ER2566 carrying plasmid pJIM5793 was grown in Luria-Bertani medium at 30°C (Table 1). IPTG (0.5 mM) was added when the OD600 of the culture reached 0.3 to induce the expression of the fusion protein. The culture was further incubated overnight at 16°C. All subsequent steps were carried out at 4°C. Cells were harvested, resuspended in buffer A (25 mM Tris HCl [pH 7.5], 0.5 M NaCl, and 20 mM imidiazole), lysed by sonication, and centrifuged at 20,000 × g for 1 h. The cleared bacterial lysate was loaded onto a 2-ml resin column (nickel-nitrilotriacetic acid agarose; QIAGEN) and extensively washed with buffer A. Purified MetR-His8 was eluted using 200 mM imidiazole and dialysed against 1 liter of buffer (50 mM Tris·HCl [pH 8], 0.2 M NaCl, 1 mM dithiothreitol, and 50% glycerol). The purified MetR-His8 sample was analyzed by Coomassie blue staining of a sodium dodecyl sulfate-polyacrylamide gel, and its concentration was determined by the method of Bradford (Bio-Rad).
DNA probes of approximately 300 bp corresponding to the promoter regions of atmB, cysD, cysK, metA, metE, metR, and smu.1487 were generated by PCR using specific primers (Table 2) and labeled at their 5′ ends with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs). Unincorporated nucleotides were removed with a PCR purification kit (QIAGEN). Protein-DNA complexes were formed by incubating the 32P-labeled DNA substrates (0.1 nM) with different amounts of MetR-His8 in 10 μl of binding buffer (50 mM Tris [pH 8], 100 mM NaCl, 0.5 mM EDTA, 2 mM MgSO4, 1 mM dithiothreitol, 20% glycerol) in the presence of 0.1-g/liter concentrations of poly(dI-dC)-poly(dI-dC) (GE Healthcare) and bovine serum albumin. Reaction mixtures were incubated at room temperature for 10 min, and samples were run at 4°C in Tris-borate-EDTA (1×) on a 6% polyacrylamide gel. Radioactive bands were detected and quantified using storage PhosphorImager screens and ImageQuant software (Molecular Dynamics).
RESULTS
Methionine metabolism in S. mutans.
S. mutans methionine metabolic genes were identified by the analysis of the potential sulfur amino acid biosynthetic pathways in this bacterium based on the genome sequence of strain UA159 (Fig. 1). Cysteine appears to be synthesized from serine with sulfide or thiosulfate by the sequential action of CysE and CysK. Homocysteine, which may be produced from cysteine (by MetA, MetB, and MetC), sulfide (by MetA and CysD), or methionine (by MetK, Pfs, and LuxS), may be converted into methionine by MetEF and possibly by a putative homocysteine S-methyltransferase (Smu.1487). In addition to synthesizing methionine, S. mutans may take up methionine from the environment, as the atmBCDE cluster encodes components of an ABC transporter belonging to the MUT family (48) and the Smu.16 permease is homologous to BcaP (34% identity), which is involved in methionine uptake in L. lactis (8) (Fig. 1).
To test this in silico analysis, the ability of strain UA159 to grow in the presence of various sulfur sources was examined (Table 3). Growth was observed on CDM plates containing thiosulfate or sulfide, showing that these compounds can be used to synthesize cysteine and methionine, as well as on plates containing cysteine, which can thus be converted into methionine, in agreement with the results of the in silico analysis (Fig. 1). The growth of unusual, translucent pinpoint colonies on agar plates with CDM containing methionine as the sole sulfur source was observed, although S. mutans UA159 does not encode the enzymes known to be involved in the synthesis of cysteine from methionine, suggesting the existence of a yet-unknown conversion pathway.
TABLE 3.
Growth of S. mutans UA159 and its derivatives on different sulfur sourcesa
| Supplement(s) in CDM | Morphology of colonies of S. mutans UA159 | Morphology of colonies of strain expressing:
|
||||
|---|---|---|---|---|---|---|
| ΔmetR | ΔmetC or ΔmetC and ΔcysD | ΔmetE | ΔatmB | ΔatmBCDE | ||
| None | − | − | NT | NT | NT | NT |
| Sulfite | − | − | NT | NT | NT | NT |
| Disulfite | − | − | NT | NT | NT | NT |
| Sulfate | − | − | NT | NT | NT | NT |
| Thiosulfate | + | − | ± | − | − | − |
| Sulfide | + | − | ± | − | ± | ± |
| C | + | − | ± | − | + | + |
| C and M | + | + | + | + | + | + |
| M | ± | ± | ± | ± | ± | ± |
| M and thiosulfate | + | + | + | + | + | + |
| M and sulfide | + | + | + | NT | NT | NT |
| M (50 μM) | ± | ± | NT | NT | − | − |
| d-Methionine (670 μM) | ± | ± | NT | NT | − | − |
| d-Methionine (50 μM) | ± | ± | NT | NT | − | − |
| C and seleno-l-methionine | ± | ± | NT | NT | + | + |
| C, M, and seleno-l-methionine | + | + | NT | NT | + | + |
Washed precultures of S. mutans UA159 and its derivatives were streaked onto CDM plates supplemented with different sulfur sources, and the abilities of the strains to grow were evaluated as a function of colony morphology on agar plates. C, l-cysteine; M, l-methionine (used at 670 μM if not otherwise specified); +, normal colonies; ±, pinpoint translucent colonies; −, no colonies; NT, not tested. Pinpoint colonies were approximately 10 times smaller than normal colonies, and growth on CDM+CM or M17 medium restored a normal colony morphology.
A functional study of mutants was performed to confirm the roles of atmB, atmBCDE, metE, cysD, and smu.1487 gene products in S. mutans methionine metabolism. The ΔcysD and Δsmu.1487 strains grew similarly to the wild-type strain under all conditions tested (data not shown). The growth patterns of the ΔmetC and ΔmetC ΔcysD strains were in agreement with the proposed role of MetC as a cystathionine β-lyase and showed the nonfunctionality of the sulfhydrylation pathway (via CysD) (Table 3). The inability of a metE mutant to grow in the absence of methionine, even in the presence of other sulfur compounds, indicates that MetE is the unique functional methionine synthase of S. mutans UA159.
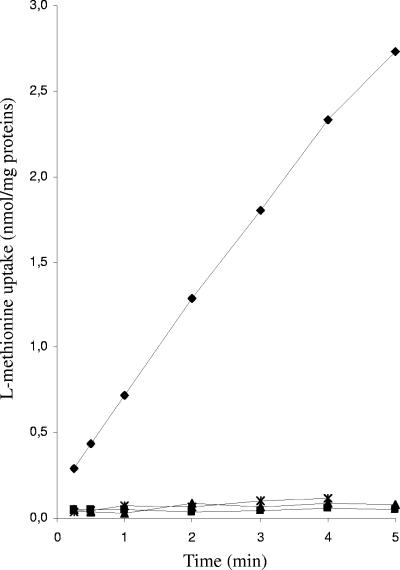
ΔatmB and ΔatmBCDE mutants were unable to grow on CDM containing thiosulfate or d-methionine as the sole sulfur source (Table 3), indicating the involvement of the AtmBDE system in the assimilation of these compounds. The mutants grew similarly to the wild-type strain with a high concentration of l-methionine (670 μM) but not with a low concentration (50 μM), suggesting that the AtmBDE uptake system has a strong affinity for l-methionine. This conclusion is supported by the observation that the growth of the ΔatmB and ΔatmBCDE mutants in CDM+C was not affected by the addition of 700 μM seleno-l-methionine, a toxic analog of l-methionine, whereas the growth of the wild type was greatly reduced. The AtmBDE transport system was further investigated by comparing l-methionine uptake in the wild-type strain to that in the ΔatmB and ΔatmBCDE mutant strains. l-Methionine uptake was detected in the wild-type strain but not in the atm mutants (Fig. 2). Moreover, a decrease in l-[14C]methionine uptake in the wild-type strain was measured when a 10-fold excess of nonradioactive l-methionine (96% reduction), selenomethionine (96% reduction), d-methionine (89% reduction), or homocysteine (47% reduction) was added, demonstrating the direct role of AtmBDE in the uptake of these compounds. Therefore, AtmBDE plays an essential role in the transport of various sulfur compounds.
FIG. 2.
Methionine uptake in various S. mutans strains. Time course for l-[14C]methionine uptake into S. mutans strains UA159 (wild type; ⧫), JIM8862 (ΔatmB; ▴), JIM8863 (ΔatmBCDE; ▪), and JIM8860 (ΔmetR; ×).
Involvement of MetR (formerly Smu.1225) in S. mutans methionine metabolism.
In order to examine the role of Smu.1225, an LTTR of S. mutans, a Δsmu.1225 mutant was constructed by the chromosomal replacement of the entire smu.1225 gene with an erythromycin resistance cassette. The growth of the mutant in the presence of various sulfur compounds was tested (Table 3). Interestingly, growth in the absence of methionine was impaired even when other sulfur compounds were provided in the medium, as observed for the metE mutant. Furthermore, the Δsmu.1225 mutant was unable to transport l-methionine at a low concentration, similar to mutants with deletions of the atm cluster genes (Fig. 2). These results suggest that Smu.1225 is required for the expression of genes involved in methionine biosynthesis and uptake and that metE and the atm cluster may be its targets. Smu.1225 was therefore renamed MetR for methionine regulator.
Identification of potential MetR target genes in S. mutans.
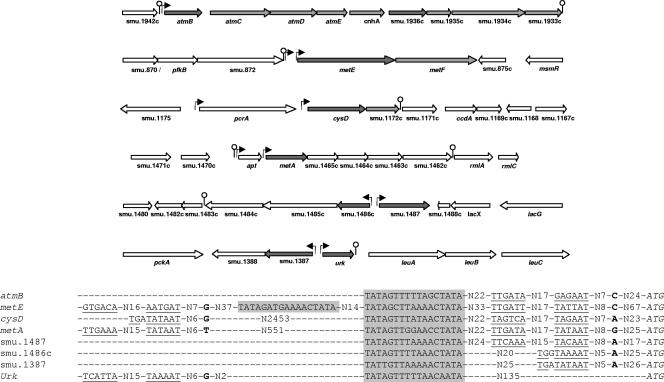
LTTRs generally recognize operators having a T-N11-A consensus sequence, displaying partial dyad symmetry, and located 50 to 80 bp upstream of the transcriptional start sites of regulated genes (38). A search for motifs of this type in the promoter regions of the metEF and atmBCDE clusters was performed. To facilitate this search, the transcriptional start sites of these genes were mapped (Fig. 3). The sequence TAGYTTWWAKCTA was found 58 and 70 bp upstream of the respective transcriptional start sites of these clusters. This sequence is part of a motif (TATAGTTTNAAACTATA) previously identified upstream of methionine metabolic genes of streptococci (36). A search of the entire genome of S. mutans revealed the presence of this motif in the promoter regions of the cysD, metA, smu.1487, smu.1486c, smu.1387, urk, and smu.1936c genes. The transcriptional start sites of these genes were determined in order to check whether the motif was positioned as expected for an LTTR DNA-binding sequence (Fig. 3). No transcriptional start site upstream of smu.1936c could be mapped, suggesting that this gene forms part of the same operon as the atmBCDE cluster. metE, cysD, and metA were transcribed from two promoters, one of which was properly located downstream of the motif. Finally, the motif was found to be distant from the start sites of the smu.1486c, smu.1387, and urk genes (Fig. 3), which have no homology with known methionine metabolic genes, suggesting that these genes are not regulated by MetR. This finding was later confirmed by QRT-PCR (results not shown), and therefore, the study of these genes was discontinued. In conclusion, the conserved 17-nucleotide sequence identified in the promoter regions of atmB, metE, cysD, metA, and smu.1487 appears to be a good candidate for MetR operators and is hereinafter designated the Met box.
FIG. 3.
Genetic organization and sequence alignment of promoter regions with potential Met boxes in S. mutans. In the upper panel, dark gray arrows correspond to genes with Met boxes in their promoter regions and light gray arrows correspond to methionine metabolism genes. Experimentally mapped promoters are indicated by black arrows. No transcription start site located less than 4 kb upstream of smu.1936c was detected, suggesting the coexpression of smu.1936c and the atm cluster. Lollipop symbols indicate predicted transcription terminators. In the lower panel, the ATG start codon of each gene is indicated in italics. The experimentally determined sites of transcription initiation are given in bold letters. The deduced extended −10, −10, and −35 boxes for each promoter are underlined. Met boxes are shaded. The Met boxes of the atmB, metE, cysD, metA, and smu.1487 genes are positioned as expected for an LTTR DNA-binding sequence.
In vitro analysis of MetR binding to promoter regions containing a Met box motif.
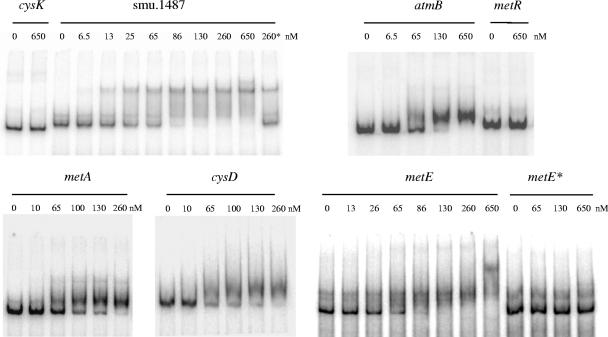
MetR interaction with promoter regions containing a Met box motif was assessed in vitro by gel mobility shift assays. For this purpose, MetR carrying a histidine tag at its C terminus (MetR-His8) was purified. Radiolabeled PCR fragments corresponding to the different promoter regions were incubated with increasing amounts of MetR-His8 (Fig. 4). The bands corresponding to the promoter regions of the cysK and metR genes, devoid of the Met box motif, showed no shift even at the highest concentration of MetR-His8. In contrast, retardation of the bands corresponding to the promoter regions of the smu.1487, metA, cysD, metE, and atmB genes, which contain Met boxes, was observed. Moreover, the complex of MetR-His8 and smu.1487 could be disrupted by adding a 50-fold excess of an unlabeled smu.1487 fragment but not by adding a 50-fold excess of a cysK promoter fragment (Fig. 4 and data not shown). Finally, pairs of base substitutions in the metE MetR boxes (substitutions T73 → G and A85 → G and substitutions T104 → G and A116 → G) affecting the conserved LTTR sequence (T-N11-A) completely abolished the shift of the modified fragment in the presence of MetR-His8 (Fig. 4). These results indicate that the Met box motif is required for the formation of the MetR-DNA complexes. We conclude that MetR specifically interacts with promoter regions containing a Met box and that substitutions within this box in the metE promoter region alter the binding of MetR.
FIG. 4.
Gel mobility shift analyses of MetR binding to different promoter regions. Labeled DNA probes (0.1 nM) corresponding to atmB, cysD, cysK, metA, metE, metE* (a metE fragment with substitutions in the Met boxes), metR, and smu.1487 were incubated with different amounts of MetR-His8 (0 to 650 nM) and analyzed by nondenaturing polyacrylamide gel electrophoresis (see Materials and Methods for details). 260*, a 0.1 nM labeled smu.1487 fragment was incubated with 260 nM MetR-His8 in the presence of a 50-fold excess of an unlabeled smu.1487 fragment.
Transcriptional analysis of metR and Met box-containing genes.
The transcription of metE, smu.1487, atmB, smu.1936c, cysD, metA, and metR in cells grown in CDM+M, CDM+C, or CDM+CM was measured by QRT-PCR (Table 4). Methionine depletion significantly affected metE, smu.1487, atmBCDE, and smu.1936c expression, while cysteine depletion had no effect. metE, with a 39-fold increase in expression, was the gene most strongly induced in the absence of methionine. In contrast, the expression of cysD, metA, or metR was not modulated under the same conditions. These results indicate that metE, smu.1487, atmB, and smu.1936c are regulated at the transcriptional level through a mechanism dependent on methionine availability.
TABLE 4.
Effect of the presence of cysteine, methionine, or homocysteine in the growth medium on the expression of several S. mutans genesa
| Gene | Organizationb | Mean ratio ± SD of gene expression levels in cells grown in:
|
||
|---|---|---|---|---|
| CDM+M-thiosulfate vs CDM+CM-thiosulfate | CDM+C vs CDM+CM | CDM+CM-homocysteine vs CDM+CMc | ||
| metE | metEF | 1.4 ± 0.02 | 39.4 ± 4.1 | 38 ± 7.3 |
| smu.1487 | smu.1487 | 1.7 ± 0.09 | 13.1 ± 2.0 | 15.7 ± 1.8 |
| atmB | atmBCDE-cnhA-smu.1936c-smu.1935c-smu.1934c-smu.1933c | 1.1 ± 0.17 | 3.1 ± 0.6 | 3.9 ± 0.1 |
| smu.1936c | atmBCDE-cnhA-smu.1936c-smu.1935c-smu.1934c-smu.1933c | 1.7 ± 0.15 | 2.5 ± 0.4 | 2.3 ± 0.3 |
| cysD | pcrA-cysD | 1.3 ± 0.17 | 1.3 ± 0.02 | 1.7 ± 0.1 |
| metA | apt-metA | 1.4 ± 0.16 | 1.3 ± 0.1 | 1.0 ± 0.1 |
| metR | metR | 1.4 ± 0.09 | 0.7 ± 0.03 | 1.1 ± 0.1 |
Gene expression in the S. mutans wild-type strain was evaluated by real-time QRT-PCR.
Gene clusters were deduced from a sequence analysis and transcriptional start site mapping (Fig. 3).
The effect of homocysteine on gene expression was evaluated 45 min after the addition of 4 mM homocysteine to exponentially growing cells (OD600 of 0.2).
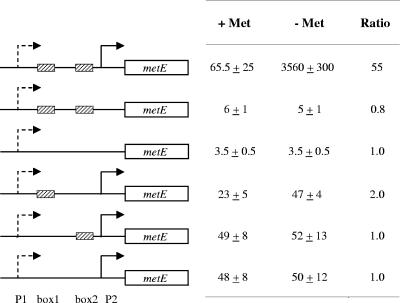
To assess the role of the Met boxes in this regulation, the effects of different modifications in the metE promoter region were examined using metE-lux transcriptional fusions (Fig. 5). Reporter activity from fusions with the complete metE promoter region (P1 and P2) was 55-fold higher in cells grown in CDM+C than in cells grown in CDM+CM, in agreement with the QRT-PCR results. In contrast, when P2 was deleted, a low and constant level of transcription in both CDM+C- and CDM+CM-grown cells was observed. These results show (i) the minor role fulfilled by P1 in metE transcription and (ii) the role of P2 in the methionine-dependent induction of metE. Lastly, no significant modulation of reporter activity was found for promoter regions containing no or only one of the two Met boxes (Fig. 5), indicating that the methionine-dependent regulation of P2 requires the two Met boxes.
FIG. 5.
Effects of substitutions in the Met boxes of PmetE-lux transcriptional fusions. The left panel is a schematic representation of transcriptional fusions between the wild-type or modified metE promoter region and luciferase reporter genes. Striped boxes and arrows represent the Met boxes and the metE promoters, respectively. Consistent with its minor role in the expression of metE, P1 is represented by dashed-line arrows. The right panel shows luciferase activity levels in cells expressing the corresponding fusion and cultivated in CDM+CM (+ Met) or CDM+C (− Met). The values reported are averages of those obtained for at least three independent cultures at an OD600 of 0.4. Ratios were calculated by dividing the values obtained in the absence of methionine by the values obtained in the presence of methionine.
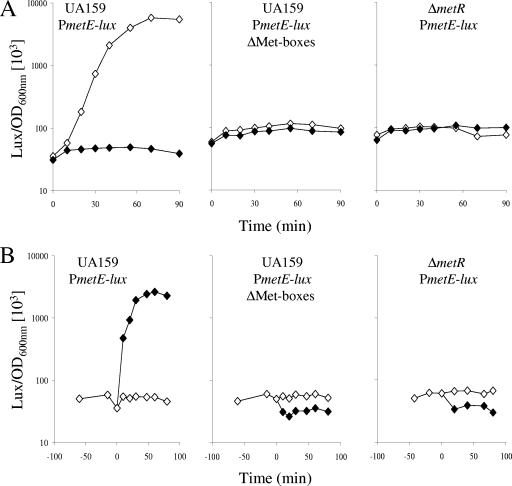
To confirm the role of MetR in this control, the expression of the metE-lux fusion in a metR mutant was monitored. Since this mutant is auxotrophic for methionine, cells were grown in rich medium containing methionine and cysteine to an OD600 of 0.2, washed, and resuspended in CDM+C or CDM+CM, and the luciferase activity was monitored (Fig. 6A). As expected, metE expression in the wild-type cells held in methionine-depleted medium was rapidly triggered (Fig. 6A, left panel), and this induction was abolished in cells carrying the promoter fusion with modified Met boxes (Fig. 6A, central panel). Methionine-dependent transcriptional induction of the metE-lux fusion was also abolished in the metR mutant, demonstrating the activator role of MetR in metE transcription (Fig. 6A, right panel). In the same way, the levels of atmB, smu.1936c, and smu.1487 expression in the metR mutant decreased two-, two-, and sevenfold, respectively, compared to the levels in the wild type (data not shown).
FIG. 6.
Effects of Met box or MetR inactivation on methionine (A)- and homocysteine (B)-dependent regulation of metE transcription. Levels of luciferase activity (amount of Lux per OD600 unit) from the UA159 wild-type (JIM8867 and JIM8870) and ΔmetR mutant (JIM8873) strains expressing the wild-type PmetE-lux fusion and the Met box mutant PmetE-lux fusion (ΔMet-boxes) were measured. (A) Cells were grown in M17 medium to early exponential phase (OD600 of 0.2). Bacterial cultures were split in two, washed, and resuspended in CDM+C (⋄) or CDM+CM (⧫) (time zero). Cells were held in these media for 90 min, and luciferase activity was measured. (B) Cells were grown in CDM+CM to early exponential phase (OD600 of 0.2). Bacterial cultures were split in two, and 4 mM homocysteine was added to one of the aliquots. The level of luciferase activity from cells cultivated in the presence (⧫) or in the absence (⋄) of 4 mM homocysteine was measured. The x axis shows the period of incubation after the addition of homocysteine (time zero). One curve representative of the results from at least three experiments is presented.
Taken together, these results show (i) that the transcription of metEF, smu.1487, and the atmBCDE-cnhA-smu.1936c-smu.1935c-smu.1934c-smu.1933c cluster is controlled by a signal dependent on the methionine supply in the medium and (ii) that the MetR activator and the Met boxes are required for this control in vivo.
Homocysteine induces MetR-dependent transcription and increases the affinity of MetR for DNA.
Transcriptional activation by LTTR proteins requires coeffectors, which are typically intermediates of the regulated pathways (38). To identify a possible MetR coeffector, the effect of the addition of several metabolites to CDM+CM (nonactivating conditions) on the expression of the PmetE-lux fusion was tested. No change in the luciferase activity upon the addition of N-acetylserine, thiosulfate, OAS, or sulfide was observed (data not shown). In contrast, luciferase activity increased rapidly upon the addition of homocysteine (Fig. 6B, left panel). This induction was abolished in strains with either modified Met boxes in the promoter region or a metR mutation (Fig. 6B, central and right panels). Homocysteine also triggered the transcription of smu.1487, atmB, and smu.1936c to levels similar to those observed in the absence of methionine but did not affect the expression of the methionine-independent cysD, metA, and metR genes (Table 4). Homocysteine induction was therefore specific to the MetR- and methionine-dependent genes and required MetR and Met boxes.
In order to further test the role of homocysteine as a coeffector of MetR in vivo, we attempted to modify the intracellular homocysteine pool. Homocysteine appears to be produced from cystathionine by MetC and/or eventually from O-acyl homoserine and sulfide by CysD, while it is converted into methionine by MetE (Fig. 1) (see “Methionine metabolism in S. mutans” above). The intracellular homocysteine concentration is thus expected to increase in a metE mutant and to decrease in a cysD metC double mutant. In agreement with these assumptions, the levels of expression of atmB and smu.1487 were (i) fourfold higher in a metE mutant than in the wild-type strain in the absence of methionine (conditions activating homocysteine synthesis) and (ii) four- to eightfold lower in a cysD metC double mutant in the presence of methionine, as assessed by QRT-PCR. These changes were specific to MetR-controlled genes, since under the same conditions, the level of urk or luxS expression remained unchanged. We conclude that alterations of the homocysteine intracellular pool affect MetR-dependent transcription, suggesting that homocysteine is the MetR coeffector in vivo.
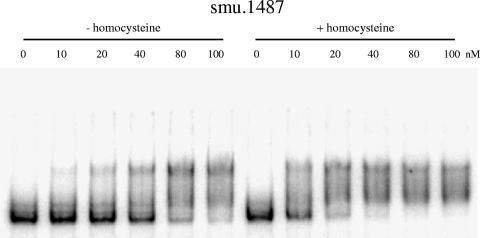
The effect of homocysteine on MetR binding to DNA was assessed by gel shift assays. At low concentrations of MetR-His8, the bands corresponding to the promoter regions of smu.1487, metE, and atmB were retarded in the presence of homocysteine compared to those in the absence of homocysteine (Fig. 7 and data not shown). Typically, the concentration of MetR-His8 required to shift 50% of the smu.1487 promoter probe (as an approximation of the Kd) was 10 nM in the presence of homocysteine, while it was 50 nM in the absence of homocysteine. Therefore, MetR displayed an increased affinity for its target sequences in the presence of homocysteine. The role of homocysteine as a coeffector for S. mutans MetR is thus supported by both in vivo and in vitro data.
FIG. 7.
Effect of homocysteine on MetR-DNA complex formation. This effect was measured by gel mobility shift assays. The promoter region of smu.1487 was generated by PCR and radiolabeled. Labeled DNA probes (0.1 nM) were incubated with the indicated amounts of MetR-His8 (0 to 100 nM) in the presence (+) or absence (−) of 4 mM homocysteine. The samples were analyzed by nondenaturing polyacrylamide gel electrophoresis (see Materials and Methods for details).
MetR-dependent regulation in other streptococci.
A motif similar to the Met box was previously identified upstream of several genes involved in methionine biosynthesis and uptake in other streptococci (36). Interestingly, a protein showing a high degree of similarity to MetR (78 to 90% identity) is present in these species, suggesting a common regulatory pathway for methionine metabolism in streptococci. To test this hypothesis, we inactivated the metR-like gene of S. thermophilus. The promoter regions of its atmB (formerly str0297)- and metE (formerly str0785)-like genes contain Met boxes. The level of induction of these genes in S. thermophilus wild-type and metR mutant strains by homocysteine was measured by QRT-PCR. In the wild-type strain, the addition of homocysteine induced the expression of the metE- and atmB-like genes 9.3- and 2.2-fold, respectively. In contrast, homocysteine had no effect on the expression of these genes in the metR mutant strain. These results indicate that MetR and homocysteine control the expression of two Met box-containing genes in S. thermophilus, confirming that similar regulatory mechanisms operate in S. thermophilus and S. mutans.
DISCUSSION
In this study, we have shown that key genes for methionine synthesis and uptake in S. mutans are under the control of MetR, an LTTR, which requires (i) Met boxes for in vitro DNA binding and in vivo transcriptional activation and (ii) homocysteine as a coeffector. The Met box motif has previously been proposed to contribute to the control of methionine metabolic genes in streptococci on the basis of the findings from a comparative genomic study (36). This regulation is reminiscent of the control of the E. coli methionine synthase gene, which is activated by an LTTR with homocysteine serving as the coeffector (30, 43, 44), as shown for S. mutans (this work). However, the S. mutans and E. coli regulatory binding sites are different (references 16 and 43; also this study), suggesting a functional convergence of these regulatory systems rather than a direct phylogenic relationship. In contrast, to our knowledge, among gram-positive bacteria this control is unique to streptococci. Methionine metabolism genes are controlled by the S-box termination-antitermination system in Bacillales and “Clostridia,” whereas their regulation likely involves a tRNA-dependent T-box system in “Lactobacillales” (17, 18, 36). Neither S-box nor T-box motifs were found in the promoter regions of methionine metabolic genes in S. mutans and other streptococci.
Methionine synthesis and uptake in S. mutans.
The methionine metabolic pathways in S. mutans were studied by genomic and functional analyses. They resemble the previously described pathways in L. lactis (41), with two differences: (i) the use of thiosulfate as a sulfur source and (ii) the absence of known methionine-to-cysteine conversion pathways.
Two pathways may allow the synthesis of the direct precursor of methionine, homocysteine, from cysteine or sulfide: the transsulfuration (MetA, MetB, and MetC) and sulfhydrylation (MetA and CysD) pathways. Here, we showed that only the transsulfuration pathway ensured rapid growth under our experimental conditions. However, the observed residual growth abilities of the metC and metC cysD mutants suggest the existence of an additional pathway for homocysteine biosynthesis. Possibly it involves sulfhydrylation carried out by MetB, as a homolog in S. anginosus was shown to posses O-acetylhomoserine sulfhydrylase activity in addition to cystathionine γ-synthase activity in vitro (47). Concerning the conversion of homocysteine into methionine, we have shown that the methionine synthase MetE was essential for the growth of S. mutans in the absence of methionine. This result indicates that Smu.1487, which was proposed to be a methionine synthase on the basis of results from genomic studies (36) and whose transcription was induced in the absence of methionine (this work), is not able to catalyze this reaction. In L. lactis, the homologous protein (YhcE; 31% identity) is involved in methionine recycling into homocysteine (41), a pathway that appears not to be functional in S. mutans. Further studies are required to elucidate the role of this protein in S. mutans.
In addition to synthesizing methionine de novo, S. mutans can take up methionine from the environment. Here, we show that several transport systems are involved in l-methionine uptake by S. mutans, as observed previously for E. coli, B. subtilis, and L. lactis (8, 23, 24, 25, 41). One of these systems, the AtmBDE system, belongs to the MUT family of ABC transporters (48). This system is also able to take up d-methionine, selenomethionine, and homocysteine. Surprisingly, mutants with deletions in the atm cluster do not grow with thiosulfate as the sole sulfur source, suggesting a role of the Atm proteins in thiosulfate transport, while other members of the MUT family are typically involved in methionine uptake (23, 33, 41, 48) and have not been reported to transport thiosulfate. Interestingly, the S. mutans genome does not contain genes encoding proteins homologous to previously described thiosulfate transporters in other bacteria (26, 40). Whether AtmBDE is directly involved in thiosulfate transport or required for the expression of still-uncharacterized thiosulfate assimilation genes remains to be determined.
Regulatory scheme for the control of methionine synthesis and uptake in S. mutans.
MetR activates in vivo the transcription of two gene clusters and smu.1487, encoding methionine synthase (MetE), a potential homocysteine S-methyltransferase (Smu.1487), and an l-methionine transporter (AtmBDE). An essential step in this activation is the binding of MetR to the promoter regions, which requires a 17-bp palindromic sequence, the Met box. Interestingly, a Met box is also found in the promoter regions of two other methionine metabolic genes (metA and cysD). Although MetR interacts with their promoter regions in vitro, the expression of these genes is not modulated by MetR in vivo. The presence of a second promoter, which is located upstream of the Met box and is likely MetR independent, may explain the absence of MetR control under our experimental conditions.
Most LTTR proteins require a coeffector to activate the transcription of their targets (38). Here, we propose that homocysteine is the MetR coeffector in vivo. First, the addition of homocysteine to a methionine-supplemented medium triggered the expression of MetR-dependent genes. Second, mutations likely affecting the intracellular homocysteine concentration altered the expression of the MetR-controlled genes. Finally, homocysteine enhanced the DNA-binding affinity of MetR for its target promoters in vitro.
Interestingly, although most LTTR proteins regulate their own transcription (38), the levels of expression of metR were found to be similar under all conditions tested, in agreement with the inability of MetR to form a complex with its own promoter, which lacks a Met box. The amount of MetR in the cells, therefore, would be constant, and the activation of MetR-controlled genes would depend mainly on the availability of the coeffector, as proposed for several other LTTRs (6, 35). These results support a model in which homocysteine plays a crucial regulatory role in S. mutans.
A corollary of the model is that, in the absence of methionine, homocysteine is produced in the cell at a level sufficient to activate MetR-dependent regulation. In contrast, in the presence of methionine, the homocysteine concentration should decrease to relieve activation. In the presence of methionine, the MetR regulon was rapidly activated upon the addition of homocysteine, confirming the limitation of the homocysteine concentration under the repressing conditions. Therefore, this model implies (i) sufficient levels of expression of the genes involved in homocysteine biosynthesis (metA, metB, metC, cysD, metK, pfs, and luxS) and (ii) the control of homocysteine synthesis by a signal dependent on methionine availability. Such controls have been described previously. Homoserine O-acyltransferase (MetA) is under allosteric control by methionine and SAM in E. coli and B. subtilis (2, 16). In addition, the synthesis of MetK, the first enzyme of the SAM recycling pathway, is regulated by SAM through a riboswitch mechanism in enterococci and possibly in streptococci (12, 13). Whether such control systems modulate the cellular homocysteine pool in S. mutans remains to be determined.
Control of methionine synthesis and uptake genes in streptococci.
The regulation of methionine metabolism by MetR with homocysteine as the coeffector as found in S. mutans may well be conserved in other streptococci. First, genes for MetR-like regulators are present in all sequenced streptococcal genomes. Second, the expression of genes involved in methionine uptake and biosynthesis is modulated by homocysteine and MetR in at least two streptococci, S. mutans and S. thermophilus. Moreover, MtaR, the MetR-like regulator in S. agalactiae, positively controls the synthesis of a still-uncharacterized methionine transporter (39). The genome of this bacterium encodes a probable methionine ABC transporter (the product of gbsS1688-gbsS11686-gbsS11685 and gbs0957) showing a high degree of similarity to the S. mutans AtmBDE transporter. The promoter regions of the S. agalactiae genes also contain Met boxes, in agreement with the decreased level of methionine transport observed in an mtaR mutant (36, 39). Finally, Met boxes are present upstream of methionine metabolism genes in several streptococci. However, the number of MetR-regulated genes may vary greatly among streptococci (36; E. Guédon, unpublished data), from one gene (metE in L. lactis or atmB in S. pyogenes) to four to five transcriptional units (S. thermophilus and S. pneumoniae). The differences may be related to the absence of the corresponding genes in some species, such as S. pyogenes, which encodes no protein homologous to Smu.1487, CysD, MetA, or MetE. They may also reflect different regulation mechanisms in some species, such as L. lactis, which contains a complete methionine biosynthetic pathway in which a master regulator controls most of the genes involved in cysteine and methionine biosynthesis (41).
Surprisingly, MetR seems to operate in several streptococci lacking the known proteins involved in homocysteine synthesis from cysteine or sulfide (MetA, MetB, MetC, and CysD), such as pathogenic species (S. pyogenes, S. agalactiae, and S. uberis). Therefore, in these species, MetR-dependent regulation may rely on the presence of an external homocysteine source (which may be imported by an AtmBDE-like system, as shown for S. mutans) or the synthesis of homocysteine from methionine via the SAM recycling pathway. Both hypotheses find support in the fact that all sequenced streptococcal genomes encode an Atm-like system and a complete SAM recycling pathway. The natural presence of homocysteine in blood and tissues would support the former hypothesis, at least for the pathogenic streptococci. Alternatively, homocysteine may not be the in vivo MetR coeffector in these streptococci. Further work will be required to confirm the validity of the S. mutans signaling pathway for pathogenic streptococci, in which MetR-dependent genes play an important role in survival in vivo (39).
Acknowledgments
We thank Eliane Milohanic, Alain Chopin, and Maarten van de Guchte for critical reading of the manuscript. We are grateful to Sandrine Petry for the generous gift of the metR mutant of S. thermophilus (JIM8315).
Brice Sperandio had a grant from the Ministère de la Recherche et de l'Education Nationale.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brush, A., and H. Paulus. 1971. The enzymic formation of O-acetylhomoserine in Bacillus subtilis and its regulation by methionine and S-adenosylmethionine. Biochem. Biophys. Res. Commun. 45:735-741. [DOI] [PubMed] [Google Scholar]
- 3.Cai, X. Y., M. E. Maxon, B. Redfield, R. Glass, N. Brot, and H. Weissbach. 1989. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc. Natl. Acad. Sci. USA 86:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-37. [DOI] [PubMed] [Google Scholar]
- 5.Cowan, J. M., M. L. Urbanowski, M. Talmi, and G. V. Stauffer. 1993. Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175:5862-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dangel, A. W., J. L. Gibson, A. P. Janssen, and F. R. Tabita. 2005. Residues that influence in vivo and in vitro CbbR function in Rhodobacter sphaeroides and identification of a specific region critical for co-inducer recognition. Mol. Microbiol. 57:1397-1414. [DOI] [PubMed] [Google Scholar]
- 7.De Keersmaecker, S. C., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14:114-119. [DOI] [PubMed] [Google Scholar]
- 8.den Hengst, C. D., M. Groeneveld, O. P. Kuipers, and J. Kok. 2006. Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J. Bacteriol. 188:3280-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejim, L. J., V. M. D'Costa, N. H. Elowe, J. C. Loredo-Osti, D. Malo, and G. D. Wright. 2004. Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72:3310-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontecave, M., M. Atta, and E. Mulliez. 2004. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29:243-249. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, R. T., F. J. Grundy, and T. M. Henkin. 2006. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 13:226-233. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs, R. T., F. J. Grundy, and T. M. Henkin. 2007. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc. Natl. Acad. Sci. USA 104:4876-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilson, T. J. 1984. Ph.D. thesis. University of Cambridge, Cambridge, England.
- 15.Golic, N., M. Schliekelmann, M. Fernandez, M. Kleerebezem, and R. van Kranenburg. 2005. Molecular characterization of the CmbR activator-binding site in the metC-cysK promoter region in Lactococcus lactis. Microbiology 151:439-446. [DOI] [PubMed] [Google Scholar]
- 16.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 17.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 18.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 8:d20-31. [DOI] [PubMed] [Google Scholar]
- 19.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedon, E., B. Sperandio, N. Pons, S. D. Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895-3909. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, C. E., D. S. Metcalf, and J. I. MacInnes. 2003. A search for virulence genes of Haemophilus parasuis using differential display RT-PCR. Vet. Microbiol. 96:189-202. [DOI] [PubMed] [Google Scholar]
- 23.Hullo, M. F., S. Auger, E. Dassa, A. Danchin, and I. Martin-Verstraete. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res. Microbiol. 155:80-86. [DOI] [PubMed] [Google Scholar]
- 24.Kadner, R. J. 1974. Transport systems for l-methionine in Escherichia coli. J. Bacteriol. 117:232-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadner, R. J., and W. J. Watson. 1974. Methionine transport in Escherichia coli: physiological and genetic evidence for two uptake systems. J. Bacteriol. 119:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kertesz, M. A. 2001. Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 152:279-290. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, D., and J. Gomes. 2005. Methionine production by fermentation. Biotechnol. Adv. 23:41-61. [DOI] [PubMed] [Google Scholar]
- 28.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Mares, R., M. L. Urbanowski, and G. V. Stauffer. 1992. Regulation of the Salmonella typhimurium metA gene by the MetR protein and homocysteine. J. Bacteriol. 174:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel, B. A., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel, B. A., F. J. Grundy, V. P. Kurlekar, J. Tomsic, and T. M. Henkin. 2006. Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression. J. Bacteriol. 188:3674-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merlin, C., G. Gardiner, S. Durand, and M. Masters. 2002. The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184:5513-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murchison, H. H., J. F. Barrett, G. A. Cardineau, and R. Curtiss III. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32:3340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saint-Girons, I., C. Parsot, M. M. Zakin, O. Barzu, and G. N. Cohen. 1988. Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. Crit. Rev. Biochem. 23(Suppl. 1):S1-S42. [DOI] [PubMed] [Google Scholar]
- 38.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 39.Shelver, D., L. Rajagopal, T. O. Harris, and C. E. Rubens. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J. Bacteriol. 185:6592-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sirko, A., M. Zatyka, E. Sadowy, and D. Hulanicka. 1995. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 177:4134-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guedon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanowski, M. L., and G. V. Stauffer. 1989. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J. Bacteriol. 171:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbanowski, M. L., and G. V. Stauffer. 1989. Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]
- 46.Xavier, K. B., and B. L. Bassler. 2005. Interference with AI-2-mediated bacterial cell-cell communication. Nature 437:750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, Y., M. Negishi, and Y. Nakano. 2003. Homocysteine biosynthesis pathways of Streptococcus anginosus. FEMS Microbiol. Lett. 221:277-284. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Z., J. N. Feige, A. B. Chang, I. J. Anderson, V. M. Brodianski, A. G. Vitreschak, M. S. Gelfand, and M. H. Saier, Jr. 2003. A transporter of Escherichia coli specific for l- and d-methionine is the prototype for a new family within the ABC superfamily. Arch. Microbiol. 180:88-100. [DOI] [PubMed] [Google Scholar]