Abstract
Cell-to-cell heterogeneity in gene expression and growth parameters was assessed in the facultative methylotroph Methylobacterium extorquens AM1. A transcriptional fusion between a well-characterized methylotrophy promoter (PmxaF) and gfpuv (encoding a variant of green fluorescent protein [GFPuv]) was used to assess single-cell gene expression. Using a flowthrough culture system and laser scanning microscopy, data on fluorescence and cell size were obtained over time through several growth cycles for cells grown on succinate or methanol. Cells were grown continuously with no discernible lag between divisions, and high cell-to-cell variability was observed for cell size at division (2.5-fold range), division time, and growth rate. When individual cells were followed over multiple division cycles, no direct correlation was observed between the growth rate before a division and the subsequent growth rate or between the cell size at division and the subsequent growth rate. The cell-to-cell variability for GFPuv fluorescence from the PmxaF promoter was less, with a range on the order of 1.5-fold. Fluorescence and growth rate were also followed during a carbon shift experiment, in which cells growing on succinate were shifted to methanol. Variability of the response was observed, and the growth rate at the time of the shift from succinate to methanol was a predictor of the response. Higher growth rates at the time of the substrate shift resulted in greater decreases in growth rates immediately after the shift, but full induction of PmxaF-gfpuv was achieved faster. These results demonstrate that in M. extorquens, physiological heterogeneity at the single-cell level plays an important role in determining the population response to the metabolic shift examined.
A growing body of evidence shows that isogenic populations of exponentially growing microorganisms have substantial cell-to-cell heterogeneity at both the gene expression and growth rate levels (6, 9, 10, 12-16, 19, 20, 29-31, 36, 39). Cell-to-cell heterogeneity in gene expression has been shown to arise from fluctuations in the global gene expression machinery of the cell, which has been termed “extrinsic noise,” “global noise,” or “gene expression capacity” (9, 12, 27, 29, 30). In some cases the source of the variation has been shown to be cell-to-cell differences in transcription, mRNA stability, and/or translation (5, 27). Two recent studies have also measured mRNA in individual cells (13, 19). The results of these studies suggest that cell-to-cell variation in gene expression is not due to fluctuations in low-copy mRNA numbers but rather appears to be due to variations at the bulk mRNA stability and/or translational level.
Significant cell-to-cell variations have also been reported for generation times and growth rates. It was shown as early as 1932 that bacteria and yeast cells exhibit two- to threefold variation in individual cell division time and that the previous division time does not influence the subsequent division time; in other words, the variation appears to be stochastic (15). More recent studies with Escherichia coli found a similar range of division times (36), and studies of growth rates in yeast also found a broad range (9).
Although cell-to-cell variation in gene expression and growth rate have been suggested to generate phenotypic diversification (1, 3, 7, 17, 18, 34), very little is known concerning the connection between stochasticity in gene expression and resultant phenotypic diversity. One prediction is that gene expression might correlate with growth rate, and in the study of yeast cited above (9), a positive correlation was obtained between the output of the mating pheromone response pathway in yeast and the growth rate (increase in cell volume). However, similar studies have not yet been reported for bacteria.
Further information suggests that even minor subpopulations of bacteria in a physiological state significantly different from that of the population average (for instance, bacteria growing very slowly) can play a major role in the population response, especially under stressful conditions (1, 3, 7, 17, 18, 34). It has been suggested that cell-to-cell phenotypic heterogeneity generates physiologically distinct subpopulations that are resistant to stress (7).
We are interested in how various parts of metabolism are integrated at the transcript, protein, and flux levels in the facultative methylotroph Methylobacterium extorquens AM1. This bacterium has two strongly contrasting modes of metabolism, growth on multicarbon compounds, which is energy limited, and growth on one-carbon compounds, which is limited by reducing power (37). In addition, growth on one-carbon compounds involves high flux through the toxic intermediate formaldehyde, raising the possibility of fluctuating stress conditions (8, 24). Our studies involve perturbing the metabolic state, either by stress or by changing the growth substrate, and following the metabolic response at the global level. Therefore, in this system it is important to understand cellular heterogeneity within the populations and ultimately to link the physiological state to the phenotypic response. In this study, we began to characterize heterogeneity at the individual cell level in M. extorquens AM1 by simultaneously monitoring expression of a variant of green fluorescent protein (GFPuv) from a well-characterized methylotrophy promoter (PmxaF) (21, 40) and measuring the change in cell size over time in a flowthrough system coupled to a laser scanning microscope. We monitored these parameters both in stable growth conditions and during the response to a change in substrate from a multicarbon compound (succinate) to a one-carbon compound (methanol).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. extorquens AM1 strains were grown in batch culture at 28°C in minimal salts media supplemented with either 0.3% (vol/vol) methanol or 0.4% succinate as a growth substrate (2, 38). The strains and plasmids used for this study are listed in Table 1. When appropriate, antibiotics were added (50 μg/ml rifamycin and 10 μg/ml tetracycline).
TABLE 1.
M. extorquens AM1 strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| AM1 | Rifr derivative | 28 |
| CM174 | katA::(loxP-trrnB-PmxaF-gfpuv-tT7) | 23 |
| XG4 | ΔmotA-fliN-fliM-fliG | This study |
| TS66 | ΔmotA-fliN-fliM-fliG ΔmotC-motB | This study |
| TS70 | ΔmotA-fliN-fliM-fliG ΔpomA-motB | This study |
| TSX | Nonmotile mutant (ΔmotA-fliN-fliM-fliG ΔmotC-motB ΔpomA-motB) | This study |
| TSXCM174 | Nonmotile mutant with katA::(loxP-trrnB-PmxaF-gfpuv-t17) | This study |
| Plasmids | ||
| pGFPuv | Commercial vector | BD Biosciences (CloneTech) |
| pCR2.1 | PCR cloning vector | Invitrogen |
| pCM157 | Broad-host-range cre expression vector (Tetr) | 22 |
| pCM158 | Broad-host-range cre expression vector (Kanr) | 22 |
| pCM174 | Insertional expression vector (trrnB-MCS-tT7) with PmxaF-gfpuv | 23 |
| pCM184 | Broad-host-range allelic exchange vector | 22 |
| pRK2073 | Helper plasmid expressing IncP tra functions | Lab collection |
| pXG1 | pCR2.1 with motA upstream flank | This study |
| pXG2 | pCR2.1 with fliG downstream flank | This study |
| pXG3 | pCM184 with motA upstream flank | This study |
| pXG4 | pXG3 with fliG downstream flank | This study |
| pTS63 | pCR2.1 with motC upstream flank | This study |
| pTS64 | pCR2.1 with motB downstream flank | This study |
| pTS65 | pCM184 with motC upstream flank | This study |
| pTS66 | pTS65 with motB downstream flank | This study |
| pTS67 | pCR2.1 with pomA upstream flank | This study |
| pTS68 | pCR2.1 with motB downstream flank | This study |
| pTS69 | pCM184 with pomA upstream flank | This study |
| pTS70 | pCR2.1 with motB downstream flank | This study |
Construction of vectors for deletion of flagellar gene clusters.
Allelic exchange vectors were constructed from pCM184 (22). Fragments (∼600 bp) flanking the ΔmotA-fliN-fliM-fliG, ΔmotC-motB, and ΔpomA-motB flagellar gene clusters were PCR amplified and inserted into pCR2.1 (Invitrogen, Carlsbad, CA) to make pXG1, pXG2, pTS63, pTS64, pTS67, and pTS68. The ΔmotA-fliN-fliM-fliG mutant insertion vector was generated by inserting the ∼560-bp BglII-NdeI upstream fragment from pXG1 into pCM184 to make pXG3. The ∼600-bp SacI-SacII downstream fragment from pXG2 was inserted into pXG3 to make pXG4. The ΔmotC-motB mutant insertion vector was constructed by inserting the ∼630-bp BglII-NotI upstream fragment from pTS63 into pCM184 to make pTS65 and then inserting the ∼630-bp ApaI/SacI downstream fragment from pTS64 into pTS65 to make pTS66. The ΔpomA-motB mutant insertion vector was made by inserting the ∼740-bp upstream fragment from pTS67 into pCM184 to make pTS69 and then inserting the ∼670-bp downstream fragment from pTS68 into pTS69 to make pTS70.
Construction of the nonmotile mutant TSX.
Using the insertion vectors described above (pXG4, pTS66, and pTS70), flagellar gene cluster mutations were sequentially introduced by electroporation (35) into M. extorquens AM1 and screening for kanamycin resistance (33 μg/ml) and tetracycline sensitivity (22). All mutations were confirmed by PCR analysis. Antibiotic markers in mutants were removed using the cre-lox system with pCM157 as described previously (22, 23). The motility of M. extorquens AM1 strain TSX was assessed by visual observation with a Zeiss Axioplan microscope using a 100× 1.3 N.A. objective (Thornwood, NY). The first two constructions were motile (TS66 and TS70), while the final construction (TSX) showed no detectable motility.
Fluorimetry analysis.
Fluorescence measurements were carried out with a Shimadzu RF-5301PC fluorimeter (Columbia, MD). GFPuv excitation was performed at 405 nm, and emission was monitored at 509 nm with excitation-emission slit widths of 5:5.
Flow system setup for microscopy.
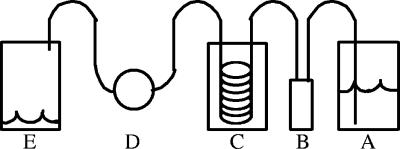
A continuous-flow system was assembled to facilitate microscope experiments (Fig. 1).
FIG. 1.
Flow system used to conduct single-cell experiments. A, medium reservoir; B, Global FIA Milligat pump; C, air equilibration system; D, FCS2 cell culture chamber; E, waste.
(i) Design.
The tubing and adapters used were all obtained from Upchurch Scientific (Oak Harbor, WA). All tubing used was PEEK tubing (outside diameter, 1/16 in.; inside diameter, 0.75 mm). One-liter Pyrex bottles were used for a medium reservoir and waste. The bottle caps for the reservoir and waste were modified to house a 0.2-μm syringe filter and to accommodate the intake or output lines. The intake line was weighted with an inline solvent filter housing to ensure that the intake stayed submerged. Fluid control was conducted with a Global FIA Milligat pump (Fox Island, WA) controlled using LabView (National Instruments, Austin, TX). A gas equilibration system was constructed to equilibrate the medium with air before it entered the culture chamber. This system consisted of a 250-ml Pyrex bottle with a modified cap that had four holes drilled into it. Two holes housed the inlet and outlet lines, and the other two holes allowed the interior of the bottle to stay equilibrated with atmospheric gases. The inlet and outlet lines were connected to 50 ft of silastic tubing (inside diameter, 0.51 mm; outside diameter, 0.94 mm; VWR, West Chester, PA) within the Pyrex bottle. A luer inline check valve was used for injection of cell samples into the culture chamber. A Bioptechs FCS2 closed system chamber (Butler, PA) was used for cell culturing, with a 0.5-mm gasket generating a volume of 350 μl. For carbon shift experiments, a 0.5-cm channel was cut into a solid 0.5-mm gasket to reduce mixing of medium containing the two substrates. For all experiments, the coverslip for the FCS2 chamber was treated with a 0.01% (wt/vol) poly-l-lysine solution (Sigma Aldrich, St. Louis, MO) so that cells could be anchored in place by their flagella (4). The slides were immersed in the solution for 10 min and allowed to cure in air at 37°C for 1 h. The temperature of the medium prior to entry into the culture chamber was controlled by immersion of the medium reservoir and gas equilibration system in a water bath. The temperature of the FCS2 chamber was maintained with an objective heater and a chamber heater (Bioptechs, Butler, PA). For carbon shift experiments, two sets of a medium reservoir, a pump, and a gas equilibration system were connected to the rest of the flow system using a four-port switch valve.
(ii) System preparation and maintenance.
Prior to experiments, the flow system was primed by flowing medium at a rate of 10 μl/s for 30 min to eliminate any air bubbles. All experiments were conducted at a flow rate of 1 μl/s. For cleaning and sterilization, the system was flushed with 10% hypochlorite for 30 min, followed by distilled H2O for at least 2 h, all at a flow rate of 10 μl/s. The entire flow system was then autoclaved.
Microscopy.
Microscopy experiments were conducted with a Zeiss LSM 510 META using a 100× 1.45 N.A. objective (Thornwood, NY). All experiments were conducted using minimal medium supplemented with rifamycin and a carbon source. Cells were injected into the FCS2 chamber and allowed to settle onto the poly-l-lysine-treated glass slide for 10 min. Once the cells were attached, 10 to 15 locations were marked using the microscope's software and monitored for the duration of the experiment. GFPuv excitation was obtained with a 488-nm argon laser at 1% power, and emissions were detected through a 505-nm longpass filter in channel 3. For analysis of growth, images were acquired every 10 min. However, for observation of the amount of fluorescence per cell, images were taken every 30 min at 1% laser power to minimize photobleaching effects. Microscope experiments were conducted for up to 96 h. The Zeiss LSM 510 META imaging software (version 3.2, SP2) was used for image analysis, and data were imported into Excel.
RESULTS
Flowthrough system to observe individual tethered cells.
To microscopically observe single cells of M. extorquens AM1 growing on methanol or succinate, a flowthrough culture system for the Zeiss LSM 510 META microscope was designed and set up as described in Materials and Methods. To facilitate long-term observations of large numbers of individual cells in this flowthrough system, cells were attached to a poly-l-lysine-coated glass slide by their polar flagella. A nonmotile mutant was necessary to facilitate long-term observations. This M. extorquens AM1 mutant (TSXCM174) was constructed by deleting three clusters of genes required for the flagellum motor function. This strain could not be distinguished from the corresponding motile strain in bulk culture based on the growth rate, dynamics of the growth curve, and gene expression from the mxaF promoter-gfpuv fusion (23) used in this study (data not shown). In the flowthrough system, attached cells were observed over several division times, and after division, the daughter cells were swept away by the flow before they could attach. Using this system, it was possible to monitor the length of time between cell divisions, the size of the cells over time (growth rate), and the GFPuv fluorescence intensity at each time point for actively growing individual attached cells. Under these growth conditions with methanol or succinate as a carbon source, cells grew well and showed no discernible lag between divisions.
Distribution of cell doubling times.
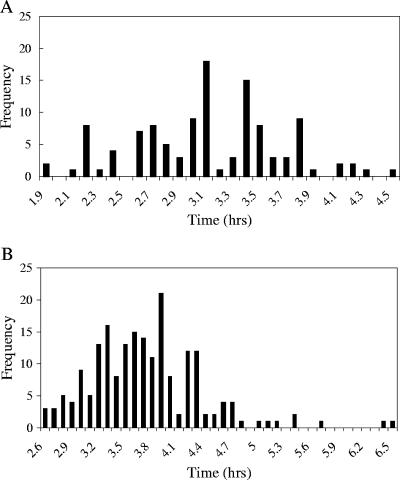
To determine the range and distribution of cell doubling times, cells were observed in the flowthrough growth system during growth with either succinate or methanol. In many cases, cells were observed for multiple divisions. The time between divisions was determined, and the data are expressed in Fig. 2 as doubling times. The variability was high, ranging from 2.5- to 2.6-fold, respectively, for the two growth conditions. The doubling times ranged between 1.9 and 4.6 h on succinate (mean doubling time, 3.12 ± 0.55 h) and between 2.6 and 6.6 h on methanol (mean doubling time, 3.73 ± 0.63 h). These means are significantly lower than those obtained for batch cultures (∼4 and ∼5.5 h, respectively). From the data available, the distribution of division rates did not appear to follow either a Gaussian or lognormal distribution; rather, a more random pattern was observed (Fig. 2). The cells with the longest and shortest generation times made up a few percent of the total population.
FIG. 2.
(A) Distribution of doubling times for individual cells grown on succinate (n = 115). (B) Distribution of doubling times for individual cells grown on methanol (n = 195).
Distribution of cell size and growth rates.
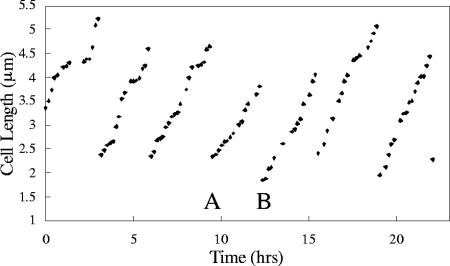
Growth dynamics was monitored for a group of individual tethered cells for a length of time sufficient for multiple consecutive divisions, and cell size was monitored as a function of time. For individual cells followed over multiple divisions, significant variability was observed for cell length before and after each division, for the growth rate (increase in length over time) between divisions, and for the duration of time between the divisions. There was no obvious correlation between the size of a cell at the time of division and the subsequent growth rate or between previous and subsequent growth rates (data not shown). Figure 3 shows an example of one such set of results. A cell was followed over several growth periods and divisions, and significant variability was observed from one division to the next in terms of cell size at the time of division and growth rate. For example, the growth rate in growth period B was 33% higher than the growth rate in the previous growth period, growth period A. In addition, the cell length at the time of division preceding growth period A was 20% greater than the cell length at the end of growth period A.
FIG. 3.
Plot of length versus time for an individual TSXCM174 cell with several successive divisions during growth on methanol. For growth period A, the growth rate was 0.55 μm/h; for growth period B, the growth rate was 0.73 μm/h.
Distribution of fluorescence from promoter-GFPuv fusion.
In order to analyze gene expression from a promoter of interest, a transcriptional fusion between a well-characterized methylotrophy promoter, PmxaF (the promoter for the methanol dehydrogenase operon), and gfpuv (23) was inserted into the chromosome of the nonmotile strain in a standard insertion site for this organism (see Materials and Methods). The intensity of GFPuv-based fluorescent emissions (in relative fluorescence units [RFU]) from single cells containing this PmxaF-gfpuv fusion was measured in both succinate- and methanol-grown cells, normalized to cell size, and expressed in RFU/μm2 (average RFU/pixel of a whole cell). The fluorescence intensity per μm2 of individual cells was variable for cells from both growth conditions (1.6- and 1.7-fold, respectively), but the variability was not as great as that of the growth rate. Calculated using approximately 1,000 cells, the mean relative fluorescence for a cell grown on succinate was 1,993 ± 468 RFU/μm2, with a range of 1,202 to 2,084 RFU/μm2, and the mean relative fluorescence for a cell grown on methanol was 3,075 ± 243 RFU/μm2, with a range of 1,649 to 2,610 RFU/μm2. These means are similar to previous results showing that the difference in expression of the promoter in succinate- and methanol-grown bulk cultures is about 1.5- to 2-fold (21, 40).
Relationship of fluorescence to growth rate and cell size.
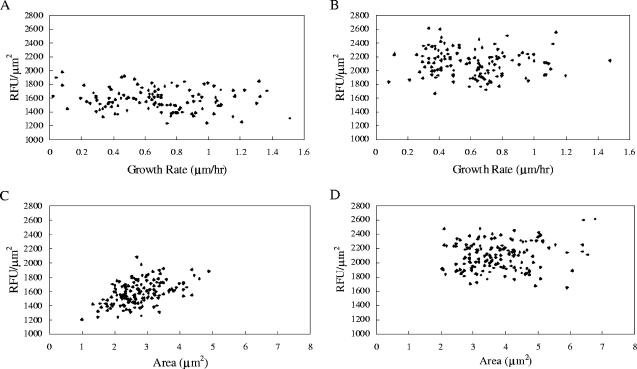
In order to determine whether fluorescence resulting from expression from the mxaF promoter correlated with growth rate or cell size, these parameters were measured for individual cells with a single data set from experiments in which cells were grown in the flowthrough system either on methanol or on succinate. The data collected from 25 individual cells at different time points are shown in Fig. 4. When RFU/μm2 was plotted against these two parameters, no correlation was observed for either parameter for methanol-grown cells. For succinate-grown cells, no correlation was observed for growth rate, but a small positive correlation (R2 = 0.26) was found for cell size.
FIG. 4.
Single-cell fluorescence intensity normalized to cell size (RFU/μm2) as a function of growth rate (μm/h) and cell area (μm2) during growth on succinate (A and C) and on methanol (B and D). The R2 value for panel C is 0.26.
Response during carbon shift.
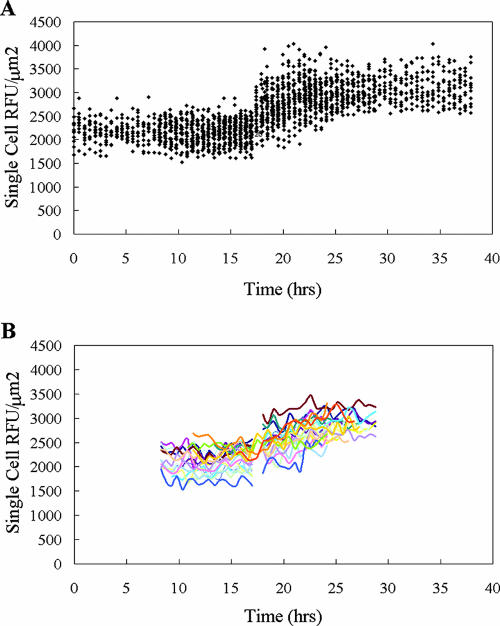
In order to assess whether variability in gene expression and/or growth rate influenced the response of individual cells during the transition from succinate growth to methanol growth, these parameters were measured for individual cells during a shift from succinate to methanol as the growth substrate. Relative fluorescence as a function of time was observed for approximately 1,000 random cells in each experiment for up to 20 h before the shift and for up to 20 h after the shift, and 25 individual cells were followed throughout the transition. The data from one of the succinate-to-methanol shift experiments are shown in Fig. 5. The range for the amount of fluorescence per μm2 in individual cells was about 1.6-fold both before and after the shift (Fig. 5A). However, when the dynamics of individual cells were tracked throughout the experiment, the patterns of these cells also showed significant variability during the experiment, both before and after the shift (Fig. 5B). Variability was observed not only in the patterns of fluorescence with time but also in the total increase in fluorescence of the mxaF promoter and in the time required to achieve full induction. While fluorescence emissions from individual cells were variable, the difference in intensity between succinate and methanol growth could be easily discerned and was in keeping with the earlier results from batch cultures (21, 40).
FIG. 5.
(A) Plot of fluorescence versus time for approximately 1,000 cells (dots) during a carbon shift from succinate to methanol. The shift was made at approximately 17 h. (B) Fluorescence data for 25 individual single cells tracked from 8 to 30 h in the experiment.
The growth rate was also measured for the same 25 cells described above immediately before the shift, immediately after the shift, and throughout the remainder of the postshift adaptation period. Again, significant variability was observed for individual cells, and the range was on the order of twofold for this data set. Immediately after the shift, all cells showed a change in growth rate, and the magnitude of the change was also variable.
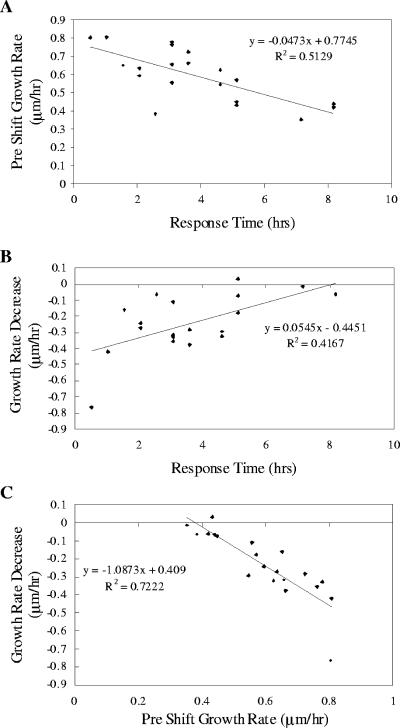
The data set for these 25 cells undergoing the transition from succinate to methanol was analyzed for correlations with regard to the following parameters: growth rates and RFU/μm2 immediately before and after the shift, change in growth rate immediately after the shift, total increase in fluorescence after the shift, and the time until the cells exhibited full induction of PmxaF-gfpuv. Each of these variables was compared to each of the other variables to assess correlations and trends. Only two trends were observed (Fig. 6). Cells growing faster before the switch to methanol achieved full induction of the mxaF promoter faster than more slowly growing cells (Fig. 6A), and the cells that showed faster induction were also the cells that exhibited a greater decline in growth rate immediately after the carbon shift (Fig. 6B). These trends suggested that the cells growing faster before the switch should exhibit a greater decline in growth rate immediately after the carbon shift, and this correlation was confirmed (Fig. 6C).
FIG. 6.
(A) Growth rate on succinate prior to carbon shift as a function of time for full mxaF response after the transition to methanol. (B) Decrease in the growth rate after the carbon shift from succinate to methanol as a function of time for full mxaF response after the transition to methanol. (C) Decrease in the growth rate after the carbon shift from succinate to methanol as a function of the growth rate before the shift.
DISCUSSION
In this study, cell-to-cell heterogeneity was assessed in M. extorquens AM1 by comparing expression from a promoter-gfpuv fusion to growth parameters. A flowthrough system was designed to grow cells in a culture chamber placed under a microscope to allow acquisition of data for individual cells over multiple divisions. Most single-cell analyses to date have involved cells imbedded in soft agar (3, 15, 25, 32), which involves microcolony formation and the possible development of chemical and physical gradients. Such environmental heterogeneity might alter intrinsic biological heterogeneity. The system used in this study involving a commercial flowthrough chamber, attachment by flagella, and a nonmotile mutant allows observation of individual cells maintained in a constant environment. In this system, cells grew significantly faster than cells in batch cultures, with mean growth rates that were on the order of 25 to 35% greater, especially during growth on methanol. Although the reason for this is not known, the flowthrough system does not allow buildup of wastes or cell signaling molecules, which could contribute to this difference. Other parameters measured in this system correlated well with population-based culture data when population means were calculated from the single-cell data, including fluorescence per cell from the PmxaF-gfpuv fusion, magnitude of induction of the promoter as measured by fluorescence, and time for induction after the shift from succinate to methanol. These results suggest that this single-cell analysis system generates response results comparable to those of population-based culture systems.
It has previously been reported for E. coli that both growth rate and gene expression are highly variable between cells in isogenic populations (10, 13, 15, 16, 19, 20, 29, 36, 39). We have confirmed a similar level of cell-to-cell heterogeneity for both growth characteristics and expression from the PmxaF-gfpuv fusion for a more slowly growing bacterium, M. extorquens AM1, using the flowthrough system noted above. As described previously for division times in bacteria (15), no correlation was obtained between a specific division time and the next division time. In addition, we found no correlation between cell size at the time of division and the immediately previous or subsequent division time or growth rate or between growth rate and the immediately previous or subsequent division time or growth rate. Such results are consistent with suggestions from studies cited above that the variability results from stochastic processes in the cell.
This experimental system allowed assessment of correlations between expression of the mxaF promoter, as judged by the GFPuv reporter, and growth parameters. Several studies have suggested that cell-to-cell variations in gene expression are the result of extrinsic noise, that is, variations in gene expression capacity between cells (11, 27, 29, 30). However, the sources of cell-to-cell variations in growth rate are not known. If these variations were due to cell-to-cell variations in gene expression capacity, then a correlation would be expected between growth rate and gene expression, as measured by the gfpuv reporter construct. For instance, a cell with higher-than-average gene expression might be expected to also exhibit a higher-than-average growth rate, as noted in a previous study with yeast (9). However, no such correlation was obtained in our study, suggesting that in M. extorquens AM1, the mechanisms generating heterogeneity in expression of the gfpuv reporter fusion and growth rate are different. If growth rate is not controlled by overall gene expression capacity, other possibilities are functions such as energy metabolism rate or cell pool parameters, such as the NADH/NAD+ ratio, the energy charge, amino acid pool levels, etc.
The existence of physiological heterogeneity predicts that individual cells in a population will respond differently to the same environmental perturbation, depending on their physiological state at the time of the perturbation. In such a case, the population average would not capture the true response at the cellular level. In order to test this hypothesis directly in M. extorquens AM1, we used the flowthrough system to monitor individual cells during the metabolic transition from succinate growth to methanol growth. In M. extorquens AM1, a shift from multicarbon compounds to one-carbon compounds results in a major change in metabolism; there is a shift from energy limitation to reducing power limitation, with over 100 genes involved in methylotrophy (8, 37). Therefore, at the individual cell level this transition might be expected to depend on parameters such as growth rate and gene expression. Under these conditions, we found that the faster-growing cells had an advantage. A trend was observed in which the faster-growing cells showed the greatest drop in growth rate immediately after the switch, suggesting that they were the most stressed. The same cells also recovered fastest and induced the mxaF promoter the maximum amount in the shortest time.
These results are in contrast to the example of antibiotic persistence in E. coli, in which the mostly slowly growing cells have an advantage over the faster-growing cells. However, with antibiotic persistence, most of the cells in the population die, and the slowly growing cells continue to grow slowly for multiple generations (3). In the perturbation described here, viability is maintained and growth is affected only transiently. Under these conditions it is likely that more rapid growth poises the cells to respond more quickly. During the transition to growth on methanol, cultures not only experience transient starvation but also accumulate formaldehyde due to an initial imbalance between the formaldehyde production and consumption fluxes (8, 24). Thus, it would be expected that faster-growing cells would metabolize more rapidly and would experience a greater initial drop in the growth rate from more rapid starvation or formaldehyde accumulation or both. However, these cells would also be poised to recover most quickly, by rapidly inducing methylotrophic pathways and thus allowing formaldehyde detoxification as well as energy extraction and biosynthesis from methanol. As technology to study both gene expression and physiological parameters in large numbers of single cells expands, it should be possible to test this hypothesis to determine the underlying causes for the behavior of individual cells during this transition. However, these results demonstrate that the preexisting physiological state of individual M. extorquens cells does dictate a differential response to a shift from multicarbon to methylotrophic growth.
Simulations predict that physiological heterogeneity is an important selective factor for populations subjected to intermittent environmental change (17, 18). Methylobacterium strains live on the surface of leaves, utilizing methanol emitted by stomata (33), and can also be found in freshwater and soil environments. These environments are characterized by highly fluctuating methanol concentrations (26), and it is possible that the heterogeneity of response demonstrated in this study contributes to the success of natural populations of Methylobacterium under these conditions.
Acknowledgments
We thank Joseph Chao for assistance with microscopy and Deirdre Meldrum for use of facilities.
This work was supported by grant P50 HG02360 from NHGRI for a Center of Excellence in Genomic Sciences.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Aertsen, A., and C. W. Michiels. 2005. Diversify or die: generation of diversity in response to stress. Crit. Rev. Microbiol. 31:69-78. [DOI] [PubMed] [Google Scholar]
- 2.Attwood, M. M., and W. Harder. 1972. A rapid and specific enrichment procedure for Hyphomicrobium spp. Antonie Leeuwenhoek 38:369-377. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, W. W., and P. W. Bankston. 1988. Measurement of live bacteria by Nomarski interference microscopy and stereologic methods as tested with macroscopic rod-shaped models. Appl. Environ. Microbiol. 54:105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, B., S. Balasubramanian, G. Ananthakrishna, T. V. Ramakrishnan, and G. V. Shivashankar. 2004. Tracking operator state fluctuations in gene expression in single cells. Biophys. J. 86:3052-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becskei, A., B. B. Kaufmann, and A. van Oudenaarden. 2005. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 37:937-944. [DOI] [PubMed] [Google Scholar]
- 7.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova, L. V., S. W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colman-Lerner, A., A. Gordon, E. Serra, T. Chin, O. Resnekov, D. Endy, C. G. Pesce, and R. Brent. 2005. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437:699-706. [DOI] [PubMed] [Google Scholar]
- 10.Elfwing, A., Y. LeMarc, J. Baranyi, and A. Ballagi. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl. Environ. Microbiol. 70:675-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science 297:1183-1186. [DOI] [PubMed] [Google Scholar]
- 12.Fraser, H. B., A. E. Hirsh, G. Giaever, J. Kumm, and M. B. Eisen. 2004. Noise minimization in eukaryotic gene expression. PLoS Biol. 2:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golding, I., J. Paulsson, S. M. Zawilski, and E. C. Cox. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123:1025-1036. [DOI] [PubMed] [Google Scholar]
- 14.Kaern, M., T. C. Elston, W. J. Blake, and J. J. Collins. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6:451-464. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, C. D., and O. Rahn. 1932. The growth rate of individual bacterial cells. J. Bacteriol. 23:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang, Y., I. Biran, and D. R. Walt. 2004. Simultaneously monitoring gene expression kinetics and genetic noise in single cells by optical well arrays. Anal. Chem. 76:6282-6286. [DOI] [PubMed] [Google Scholar]
- 17.Kussell, E., R. Kishony, N. Q. Balaban, and S. Leibler. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kussell, E., and S. Leibler. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075-2078. [DOI] [PubMed] [Google Scholar]
- 19.Le, T. T., S. Harlepp, C. C. Guet, K. Dittmar, T. Emonet, T. Pan, and P. Cluzel. 2005. Real-time RNA profiling within a single bacterium. Proc. Natl. Acad. Sci. USA 102:9160-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maloney, P. C., and B. Rotman. 1973. Distribution of suboptimally induced β-d-galactosidase in Escherichia coli. The enzyme content of individual cells. J. Mol. Biol. 73:77-91. [DOI] [PubMed] [Google Scholar]
- 21.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 22.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 23.Marx, C. J., and M. E. Lidstrom. 2004. Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology 150:9-19. [DOI] [PubMed] [Google Scholar]
- 24.Marx, C. J., S. Van Dien, and M. E. Lidstrom. 2005. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 3:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metris, A., Y. Le Marc, A. Elfwing, A. Ballagi, and J. Baranyi. 2005. Modeling the variability of lag times and the first generation times of single cells of E. coli. Int. J. Food Microbiol. 100:13-19. [DOI] [PubMed] [Google Scholar]
- 26.Nemecek-Marshall, M., R. C. MacDonald, J. J. Franzen, C. L. Wojciechowski, and R. Fall. 1995. Methanol emission from leaves (enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development). Plant Physiol. 108:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman, J. R., S. Ghaemmaghami, J. Ihmels, D. K. Breslow, M. Noble, J. L. DeRisi, and J. S. Weissman. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441:840-846. [DOI] [PubMed] [Google Scholar]
- 28.Nunn, D. N., and M. E. Lidstrom. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J. Bacteriol. 166:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedraza, J. M., and A. van Oudenaarden. 2005. Noise propagation in gene networks. Science 307:1965-1969. [DOI] [PubMed] [Google Scholar]
- 30.Rosenfeld, N., J. W. Young, U. Alon, P. S. Swain, and M. B. Elowitz. 2005. Gene regulation at the single-cell level. Science 307:1962-1965. [DOI] [PubMed] [Google Scholar]
- 31.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, E. J., R. Madden, G. Paul, and F. Taddei. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PloS. Biol. 3:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sy, A., A. C. Timmers, C. Knief, and J. A. Vorholt. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ Microbiol. 71:7245-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thattai, M., and A. van Oudenaarden. 2004. Stochastic gene expression in fluctuating environments. Genetics 167:523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyama, H., C. Anthony, and M. E. Lidstrom. 1998. Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol. Lett. 166:1-7. [DOI] [PubMed] [Google Scholar]
- 36.Umehara, S., Y. Wakamoto, I. Inoue, and K. Yasuda. 2003. On-chip single-cell microcultivation assay for monitoring environmental effects on isolated cells. Biochem. Biophys. Res. Commun. 305:534-540. [DOI] [PubMed] [Google Scholar]
- 37.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C3 and C4 metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 38.Van Dien, S. J., Y. Okubo, M. T. Hough, N. Korotkova, T. Taitano, and M. E. Lidstrom. 2003. Reconstruction of C(3) and C(4) metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 149:601-609. [DOI] [PubMed] [Google Scholar]
- 39.Wakamoto, Y., J. Ramsden, and K. Yasuda. 2005. Single-cell growth and division dynamics showing epigenetic correlations. Analyst 130:311-317. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, M., and M. E. Lidstrom. 2003. Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology 149:1033-1040. [DOI] [PubMed] [Google Scholar]