Abstract
Bacillus subtilis OhrR is the prototype for the one-Cys family of organic peroxide-sensing regulatory proteins. Mutational analyses indicate that the high sensitivity of the active site cysteine (C15) to peroxidation requires three Tyr residues. Y29 and Y40 from the opposing subunit of the functional dimer hydrogen bond with the reactive Cys thiolate, and substitutions at these positions reduce or eliminate the ability of OhrR to respond to organic peroxides. Y19 is also critical for peroxide sensing, and the Ala substitution mutant (OhrR Y19A) is less susceptible to oxidation at the active site C15 in vivo. The Y19A protein also displays decreased sensitivity to peroxide-mediated oxidation in vitro. Y19 is in van der Waals contact with two residues critical for protein function, F16 and R23. The latter residue makes critical contact with the DNA backbone in the OhrR-operator complex. These results indicate that the high sensitivity of the OhrR C15 residue to oxidation requires interactions with the opposed Tyr residues. Oxidative modification of C15 likely disrupts the C15-Y29′-Y40′ hydrogen bond network and thereby initiates conformational changes that reduce the ability of OhrR to bind to its operator site.
The cellular response to oxidative stress in bacteria is largely regulated at the transcriptional level (12, 14). In several cases, the corresponding regulator also functions as a direct sensor of oxidative stress, an example of the “one-component” regulatory strategy (22). Regulatory proteins that sense oxidative stress employ a variety of mechanisms ranging from the oxidation of metal centers, as in SoxR and PerR, to the oxidation of cysteine, as in OxyR and OhrR (14, 21). In most cases, these oxidation events reversibly alter protein conformation to effect changes in regulator activity.
OhrR, a member of the MarR family of transcription factors, controls the organic peroxide-inducible expression of the organic hydroperoxide resistance gene, ohr, which encodes a thiol-dependent peroxidase (peroxiredoxin) (5, 11). The ohr gene was first identified in Xanthomonas campestris pv. phaseoli by virtue of its ability to restore organic peroxide resistance to an Escherichia coli ahpC mutant (15), and related proteins are found in a wide variety of bacteria (1). Expression of ohr is strongly and selectively induced by organic peroxides, and this regulation is mediated by OhrR (6, 14, 17, 20). In addition to X. campestris and Bacillus subtilis, ohrR homologs have been identified in Pseudomonas aeruginosa (16), Enterococcus faecalis (18), Actinobacillus pleuropneumoniae (19), and Staphylococcus aureus (3). Genomic analyses suggest that ohr genes are often linked to ohrR (20).
B. subtilis contains two ohr paralogs: ohrA and ohrB (6). Expression of ohrB is controlled by σB, the general stress response σ factor, and is induced by heat shock or entry into stationary phase (23). In contrast, ohrA is repressed by OhrR and selectively induced by organic peroxides (6). OhrR binds to an inverted repeat sequence overlapping the ohrA promoter site and thereby blocks transcription initiation (6, 7).
Insights into the mechanism of peroxide sensing by OhrR have emerged from genetic and biochemical studies (7). B. subtilis OhrR has a single, conserved cysteine residue (C15), which is critical for organic hydroperoxide sensing. The crystal structure of the reduced, dimeric OhrR repressor in complex with DNA has been solved (9). This structure reveals that the reactive C15 is hydrogen bonded to Y29′ and Y40′ from the opposing subunit in the dimer. This reactive Cys residue is ionized at physiological pH (pKa ∼5.2), apparently because of its location at the amino terminus of an alpha-helix (9). In vitro studies demonstrated that exposure of OhrR to the model organic hydroperoxide cumene hydroperoxide (CHP) results in the initial oxidation of C15 to the sulfenic acid (7). Derepression is correlated with the subsequent reaction of the C15 sulfenate with a low-molecular-weight thiol, to generate a mixed disulfide, or with the protein backbone, to generate a sulfenamide derivative (10). Other OhrR homologs function by a distinct mechanism involving the reversible formation of an intersubunit protein disulfide (17). Indeed, most OhrR homologs contain one or more additional Cys residues in the carboxyl-terminal domain of the protein, suggesting that this two-Cys mechanism may be quite common.
Here, we generated mutant OhrR proteins with alterations in residues hypothesized to affect the orientation and ionization of the active site C15 thiol. Our results demonstrate that the two tyrosine residues that bond with the C15 thiolate are critical for genetic derepression. Further, the Y29A mutant displays a greatly reduced sensitivity to oxidants both in vivo and in vitro. A third tyrosine residue in the vicinity of the active site, Y19, is also critical for in vivo regulation and affects the sensitivity of C15 to oxidants. Analysis of these and related results provided insights into the conformational changes that occur upon protein oxidation and ultimately lead to protein dissociation from the ohrA operator site.
MATERIALS AND METHODS
Creation of OhrR variants.
OhrR variants containing single amino acid substitutions were generated by PCR mutagenesis and expressed ectopically using pXT as previously described (7). Using B. subtilis chromosomal DNA as a PCR template, appropriate base changes were introduced to code for the single amino acid substitutions. All substitutions were confirmed by DNA sequencing. The OhrR variants were integrated at the thrC site in an ohrR mutant containing a PohrA-cat-lacZ operon fusion, HB2012 [CU1065 ohrR::kan SPβc2Δ2::Tn917::φ(ohrA-cat-lacZ)] (7). The strains were checked for the integration by PCR, and the correct substitutions were confirmed by DNA sequencing of the PCR product.
Immunoblot analyses.
To measure OhrR protein expression levels (see Fig. 2A), 30 μg of total protein from cell extracts from log-phase cells was resolved by 15% Tris-Tricine sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to an Immune-Blot polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked in Tris-borate-saline buffer supplemented with 5% skim milk and 0.5% Tween 20 for 1 h at room temperature. The membrane was then washed three times for 10 min each time in Tris-borate-saline buffer supplemented with 0.5% Tween 20 and incubated with anti-OhrR rabbit antibody for 2 h at room temperature. The washing procedure was repeated and was followed by incubation with alkaline phosphatase-coupled secondary goat anti-rabbit antibody for 1 h (Bio-Rad). After thorough washing of the membrane, OhrR was visualized by adding 5- bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium substrate (Bio-Rad). Cells were also treated with 100 μM CHP for 1 min and precipitated with 10% trichloroacetic acid (TCA) prior to immunoblotting (see Fig. 3C).
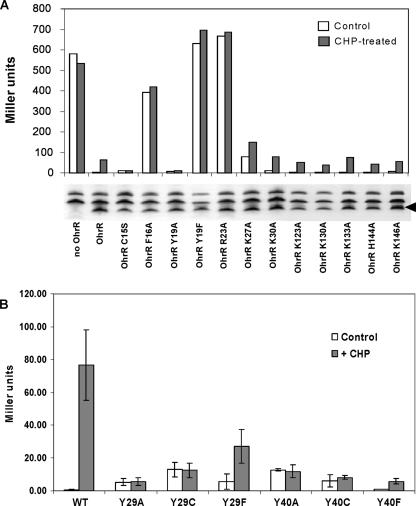
FIG. 2.
Regulatory activity of OhrR mutants as analyzed using a PohrA-cat-lacZ reporter fusion. (A) Effects of substitutions for a positively charged amino acid on OhrR repressor activity and responsiveness to CHP. In addition, substitutions were tested for the F16 and Y19 residues located adjacent to, or one helical turn away from, the active site C15 residue. As a control, the noninducibility of the C15S mutant is also shown. Under the conditions used, exposure of cells containing WT OhrR to CHP leads to ∼50 to 100 Miller units of activity after 15 min of treatment (OhrR bars). An immunoblot showing the protein expression level for each mutant is below the corresponding bars, and the position of the specific OhrR band is indicated by the arrowhead on the right. (B) Mutation of Y29 or Y40 reduces or prevents induction in response to CHP treatment. Cells were either not treated (open bars) or treated with 100 μM CHP (filled bars) for 15 min. The error bars indicate the standard deviations of three independent experiments.
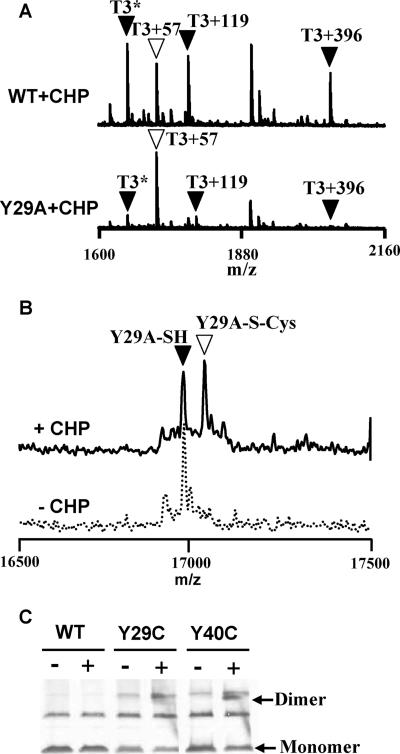
FIG. 3.
Y29A mutant displays reduced oxidation at C15 both in vivo and in vitro. (A) To compare the sensitivity of C15 in the WT and Y29A mutant proteins to oxidation, cells were treated with 100 μM CHP for 1 min, and the epitope (FLAG)-tagged proteins were recovered by immunoprecipitation. MALDI-TOF MS analysis of a tryptic digest of the recovered protein from the WT revealed the expected presence of oxidized C15-containing peptides (filled triangles), including (from left to right) the sulfenamide (T3*), S-cysteinylated peptide (T3+119), and S-thiolated peptide modified with the 398-Da thiol (T3+396), as previously described (10). The open triangle indicates the peak (T3+57) corresponding to reduced OhrR alkylated with IA. In comparison, the Y29A mutant protein was present predominantly in the reduced, and therefore IA-modified, state even after CHP treatment. (B) Y29A mutant protein is only slowly S cysteinylated in vitro. Three hundred nanomolar Y29A OhrR protein was treated with or without 3 μM CHP in the presence of 10 μM cysteine for 10 min. The protein was then TCA precipitated and subjected to ESI-MS. The filled triangle indicates reduced OhrR, whereas the open triangle indicates cysteinylation of OhrR. Note that the Y29A mutant was about half-modified after 10 min of treatment, whereas WT OhrR was similarly modified after only 1 min of treatment and was fully modified by 5 min (see Fig. 4B). (C) Immunoblot analysis of oxidation products of Y29C and Y40C in vivo. Cells were not treated (−) or treated (+) with 100 μM CHP for 1 min before TCA precipitation and analysis by nonreducing SDS-PAGE. A small amount of OhrR dimer was detected for the Y40C mutant (arrow), indicating the presence of a small amount of disulfide cross-linked dimer. Little if any disulfide cross-linked dimer was detected for Y29C. The origin of the upper cross-reactive band, present in the cells containing mutant proteins, was unclear, but this band was not reduced by DTT, indicating that it was unlikely to represent disulfide-linked dimer (data not shown).
β-Galactosidase assay.
Overnight precultures were grown in LB with appropriate antibiotics, transferred at a 1:100 dilution into fresh medium containing 20 mM xylose (to induce OhrR expression), and grown at 37°C with aeration until the optical density at 600 nm was ∼0.4. CHP was added to each culture, which was then incubated for 15 min before the β-galactosidase assay was performed as described previously (13).
Monitoring OhrR oxidation in B. subtilis.
OhrR variants carrying a C-terminal FLAG epitope tag were generated and analyzed as previously described (10). Two hundred milliliters of cells in LB at an optical density at 600 nm of ∼0.4 were not treated or were treated with 100 μM CHP for 1 min. OhrR-FLAG was recovered in the presence of 100 mM iodoacetamide (IA), separated by 15% SDS-PAGE, subjected to in-gel trypsin digestion, and analyzed using an Applied Biosystems 4700 matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometer.
Expression and purification of mutant OhrR proteins.
Genes encoding the Y19A, Y29A, and R23A OhrR mutant proteins were introduced into the expression vector pET16b (Novagen), overexpression plasmids were introduced into strain BL21(DE3)(pLysS) by transformation, and proteins were purified following overexpression as previously described (7). The molecular weight of each protein was checked by electrospray ionization (ESI)-mass spectrometry (MS), and the results showed agreement with the calculated values. OhrR concentrations were determined using the calculated values of ɛ280 (14,440 M−1 cm−1 for the wild type [WT] and corrected values for the mutant proteins), and aliquots were stored at −70°C in buffer A (20 mM Tris [pH 8.0], 100 mM NaCl, 5% [vol/vol] glycerol) containing 1 mM dithiothreitol (DTT) and 1 mM EDTA. For in vitro oxidation experiments, the DTT and EDTA were removed using Bio-Spin 6 chromatography columns immediately prior to use.
FA.
Fluorescence anisotropy (FA) experiments were done as previously described (10), using a 6-carboxyfluorescein-labeled DNA fragment containing the ohrA operator site generated by annealing 5′-6-carboxyfluorescein-TACAATTAAATTGTATACAATTAAATTGTA-3′ (Integrated DNA Technologies) and its unlabeled complementary strand. FA measurements (excitation wavelength, 495 nm [slit width, 15 nm]; emission wavelength, 520 nm; [slit width, 20 nm]; integration time, 1 s) were determined with 50 nM DNA and 300 nM OhrR (monomer) in 3 ml of 20 mM Tris (pH 8.0) containing 150 mM NaCl and 5% (vol/vol) glycerol. FA values were recorded automatically every 10 s using a Perkin-Elmer LS55 luminescence spectrometer. The g-factor for the experiments was 1.07 ± 0.01.
Monitoring OhrR oxidation in vitro.
To monitor protein oxidation using ESI-MS (see Fig. 3B and 5B), purified OhrR proteins (300 nM monomer) were incubated in 1 ml of buffer A and treated with or without 3 μM CHP in the presence of 10 μM cysteine for 10 min. The protein was then TCA precipitated and subjected to ESI-MS analysis. To monitor oxidation using 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) modification, 300 nM Y19A was treated with 3 μM CHP, and the protein was recovered by 10% TCA precipitation. The protein pellet was incubated in a buffer containing 200 mM Tris (pH 8.0), 1% SDS, 5% (vol/vol) glycerol, and 20 mM AMS for 1 h in the dark. The OhrR protein was resolved by 16% Tris-Tricine SDS-PAGE.
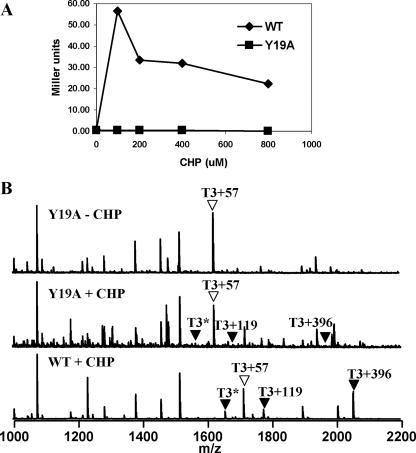
FIG. 5.
OhrR Y19A mutant is insensitive to CHP-mediated oxidation in vivo. (A) Induction of a PohrA-cat-lacZ reporter fusion was monitored in cells containing WT or Y19A OhrR after treatment with 0, 100, 200, 400, or 800 μM CHP for 15 min. (B) Analysis of the in vivo oxidation state of C15 in the Y19A mutant protein. Cells expressing the Y19A-FLAG OhrR protein were treated with 100 μM CHP for 1 min, and then IA was added to modify all reduced thiols. OhrR was recovered by immunoprecipitation, and the tryptic peptides were analyzed by MALDI-TOF MS. The resulting C15-containing peaks are indicated as described in the legend to in Fig. 3.
RESULTS
Effects of basic amino acids on C15 thiolate reactivity.
Induction of the OhrR-repressed ohrA gene by organic peroxides, such as CHP, requires the reactive C15 thiolate (6). In the analogous peroxide sensor OxyR, the high reactivity of C199 has been attributed to proximity to R266, which stabilizes the reactive thiolate anion (4). We investigated the possible role of basic amino acids in the vicinity of C15 by phenotypic characterization of Ala substitution mutants (Fig. 1A). In most cases (K30A, K123A, K130A, K133A, H144A, and K146A), the mutant proteins retained repressor activity, and the ability to be induced by CHP was comparable to that of the WT. In contrast, the K27A and R23A mutants displayed partial loss and complete loss of repressor activity, respectively. Note that in all cases, the amount of the mutant protein detected in vivo was comparable or slightly reduced relative to that of the WT as detected by immunoblotting and there was no correlation between low protein levels and loss of activity. We concluded that these functional defects were not due to a lack of stable protein.
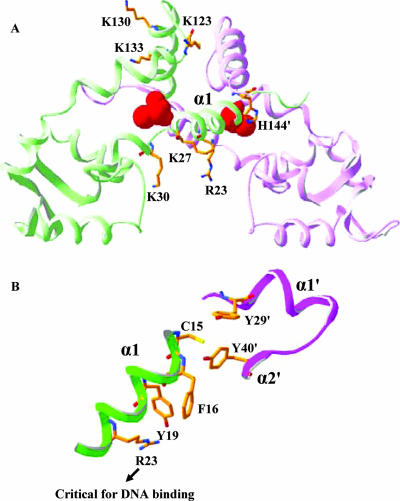
FIG. 1.
Location of OhrR amino acids targeted for site-directed mutagenesis. (A) Ribbon diagram of OhrR showing the two monomers (green and pink), with C15 displayed as red van der Waals spheres. The DNA-binding helices are at the bottom. The side chains for most of the amino acids that have been mutated are shown; the exception is K146, which was not resolved in the crystal structure (9). (B) Close-up of the OhrR active site illustrating the close proximity of C15 to the Y29′ and Y40′ residues from the other subunit. Residues are represented by sticks, and carbon, nitrogen, oxygen, and sulfur atoms are gold, blue, red, and yellow, respectively. The protein diagrams were created using Swiss-Pdb Viewer (8).
The determination of the high-resolution structure of OhrR (9), both free and in complex with operator DNA, led to an alternate hypothesis in which it was proposed that the low pKa of C15 (pKa ∼5.2) is due to the α1 helix macrodipole (Fig. 1A). It was also noted that C15 (or S15 in the higher-resolution structure of the C15S mutant protein) is within hydrogen-bonding distance of Y29′ and Y40′ (9). Thus, the high reactivity of C15 may not require stabilization of the thiolate anion by proximity to a positively charged side chain, consistent with the results of our mutagenesis studies (Fig. 2A). Finally, this structure can explain the reduced function of the K27A mutant and loss of function of the R23A mutant: both side chains are engaged in contacts with the DNA backbone (9). The loss of DNA-binding activity for the R23A mutant was also confirmed in vitro with purified protein using an FA-based DNA-binding assay (data not shown).
Effects of Y29 and Y40 on Cys15 thiolate reactivity.
To investigate the contributions of Y29 and Y40 to the OhrR active site (Fig. 1B), each residue was replaced with alanine, cysteine, or phenylalanine. All six mutant proteins were functional repressors, although the Ala and Cys substitution mutants completely lost peroxide responsiveness (Fig. 2B). Even replacement of the Tyr residues with Phe greatly reduced the extent of derepression, highlighting the importance of the Tyr hydroxyl groups.
The loss of responsiveness in the Y29 and Y40 mutants could have resulted from either an inability of CHP to oxidize C15 or a failure of the oxidized protein to undergo the necessary conformational changes required for derepression. To directly monitor the products of oxidation in vivo, we investigated the fate of the Y29A mutant protein by MALDI-TOF MS analysis. As shown previously (10), WT OhrR is oxidized in cells to generate both mixed disulfides and the cyclic sulfenamide (Fig. 3A). These products can be monitored by recovery of OhrR by immunoprecipitation from cells, in situ tryptic digestion, and mass analysis of the resulting tryptic peptides. In untreated cells, reduced OhrR can be quantitatively modified by IA to generate the carboxyamido derivative (T3+57) (10). Upon exposure to CHP, OhrR forms mixed disulfide products with Cys (T3+119) and with a structurally undefined 398-Da thiol (T3+396). In addition, there is a nonalkylatable peptide (T3*) corresponding to the cyclic sulfenamide (Fig. 3A). In contrast to WT OhrR, cells containing the Y29A mutant protein yielded only very small peaks corresponding to the sulfenamide and S-thiolated products after treatment with 100 μM CHP (Fig. 3A). These results are consistent with the idea that a Y29A substitution greatly reduces the reactivity of OhrR to oxidants. Although it is possible that the Y29A mutant protein is oxidized to the sulfenate but defective in subsequent S thiolation, this seems unlikely since the yield of sulfenamide is also greatly reduced. The reaction between the C15 sulfenate and the adjacent backbone amide to form the cyclic sulfenamide should not be altered by the Y29A substitution.
The susceptibility of C15 to oxidation in the Y29A mutant protein was also monitored in a purified in vitro system. After treatment with a 10-fold molar excess of CHP for 10 min in the presence of 10 μM cysteine, approximately one-half of the Y29A protein was S cysteinylated as judged by ESI-MS analysis (Fig. 3B). In contrast, under the same conditions WT OhrR was quantitatively S cysteinylated (10) (Fig. 4B). The sulfenic acid form of Y29A was not detected, consistent with the observation that the initially formed sulfenate is rapidly trapped as the mixed disulfide in reaction mixtures containing free cysteine. Together, these in vivo (MALDI-TOF MS) and in vitro (ESI-MS) results support the hypothesis that Y29 (and likely Y40) functions to stabilize and possibly orient the reactive C15 thiolate within the OhrR active site.
FIG. 4.
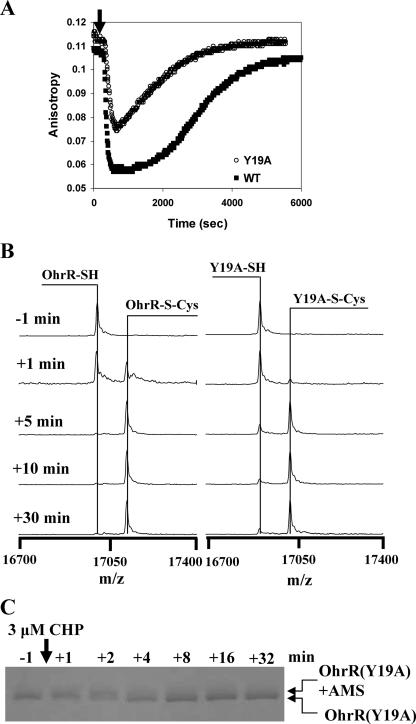
OhrR Y19A mutant has a significantly reduced rate of oxidation in vitro. (A) DNA-binding activities of WT and Y19A OhrR proteins (300 nM) determined using an FA-based assay. Addition of 300 nM CHP (in the presence of 1 mM cysteine) led to protein S cysteinylation and a decrease in the amount of protein bound to DNA. As described previously (10), the high concentration of free cysteine leads to the reactivation of OhrR by spontaneous thiol-disulfide exchange reactions. (B) Y19A OhrR protein has a reduced rate of S cysteinylation as monitored by ESI-MS. OhrR (300 nM) was treated with 3 μM CHP in the presence of 10 μM cysteine. Protein was recovered by 10% TCA precipitation and then subjected to ESI-MS. The rate of appearance of the S-cysteinylated protein was reduced ∼2- to 3-fold for the Y19A mutant, consistent with the slower and less complete inactivation shown in panel A. (C) Oxidation rate of C15 in the Y19A mutant protein as monitored by AMS modification. In this experiment, the half-time for oxidation of the C15 residue (as judged by an inability to be modified by AMS) was ∼2 min, compared to ∼30 s for WT OhrR (10; data not shown).
As a further measure of C15 reactivity, we monitored the ability of the Y29C and Y40C mutant proteins to form an intermolecular disulfide bond when they were exposed to oxidizing conditions. We reasoned that if the reactive C15 thiolate were to form a sulfenate, this compound would rapidly react with the juxtaposed cysteine from the other subunit. Indeed, immunoblot analysis demonstrated that upon treatment with CHP, a small amount of the Y40C protein (and an even smaller amount of Y29C protein) formed an intermolecular dimer (Fig. 3C). However, the extent of disulfide bond formation was quite modest (most protein was still detected as the monomer [Fig. 3C]), despite the presumed close proximity of the C29 thiol to the C15 thiolate. This supports the hypothesis that disruption of the C15-Y29′-Y40′ hydrogen bond network greatly reduces the reactivity of the C15 thiolate.
Essential role of Y19 in peroxide responsiveness.
One unexpected result from our site-directed mutagenesis studies was the finding that Y19 is critical for protein function. The Y19A mutation results in a noninducible repressor, whereas the more conservative Y19F change abolishes repressor function (Fig. 2A). Inspection of the OhrR structure indicated that Y19 is in close contact with both F16 and R23 (Fig. 1B). F16 also seems to be essential for protein function since the F16A mutant was inactive as a repressor (Fig. 2A). While the origins of these effects are not clear, the proximity of Y19 to the active site thiolate (C15) and to the essential DNA-binding residue R23 suggested a possible model for OhrR derepression. We speculated that mixed disulfide formation between C15 and low-molecular-weight thiols might alter the conformation of the C15-Y29′-Y40′ active site and this result conformational change could be propagated from Y40′ through F16 and Y19 to the essential R23 DNA-binding residue (Fig. 1B). Displacement of the R23 side chain could subsequently trigger DNA dissociation. This model predicts that any residual DNA-binding activity for OhrR lacking R23 should be unaffected by protein S thiolation. However, the purified R23A protein did not bind DNA even at the highest concentrations tested, so we were unable to test this prediction. Further, this model predicts that the Y19A mutant protein might react normally with CHP, to generate S-thiolated protein, but that this protein may fail to correctly propagate this conformational change to regions essential for DNA binding. To test the latter prediction, we monitored oxidation of C15 in the Y19A mutant protein.
Peroxide reactivity of Y19A OhrR in vivo and in vitro.
Since Y19A OhrR did not respond to CHP in vivo under our standard conditions (Fig. 2A), we tested the sensitivity at higher CHP concentrations. However, Y19A could not be induced even at CHP concentrations as high as 800 μM (Fig. 5A). Next, we monitored oxidation of C15 in the Y19A mutant in vivo using MALDI-TOF MS. As expected, the Y19A protein was in the fully reduced form in the untreated control. However, even in the CHP-treated sample there was little evidence of S thiolation or cyclic sulfenamide formation (Fig. 5B). In contrast, the WT OhrR protein displays the expected mixture of oxidation products (10). These results suggest that the susceptibility of Y19A OhrR to oxidation is reduced. Thus, the Y19A mutation does not simply blockade the transmission of the hypothesized conformational change from the active site to the R23 residue. Instead, or in addition, it appears to perturb the structure and reactivity of the active site.
Next, we purified the Y19A protein and monitored DNA-binding activity using FA. We have previously shown that oxidation of OhrR with one molar equivalent of CHP in the presence of 1 mM cysteine leads to a rapid loss of DNA-binding activity (associated with S cysteinylation), followed by spontaneous reactivation (mediated by thiol-disulfide exchange reactions) (10). Our results demonstrated that Y19A bound DNA with affinity comparable to that of the WT (data not shown). Moreover, in contrast to the in vivo results, the Y19A protein could be inactivated for DNA binding both in the absence (data not shown) and in the presence of cysteine (Fig. 4A). We monitored rates of protein inactivation in the presence of cysteine since under these conditions protein oxidation is rate limiting, whereas in the absence of cysteine protein inactivation is limited by the low rate of cyclization to the cyclic sulfenamide (10). In the presence of 1 mM cysteine, the Y19A protein was inactivated by CHP at a lower rate than the WT, and as a result, the protein was not completely inactivated under these reducing conditions (Fig. 4A). To directly monitor the rate of S cysteinylation in vitro, we analyzed proteins using ESI-MS (Fig. 4B). Both WT and Y19A OhrR were quantitatively S cysteinylated by 10 min, but the S cysteinylation of the Y19A mutant was reduced relative to that of the WT at intermediate time points. This suggests that the Y19A mutant affected either the rate of initial protein oxidation (to the sulfenate) or the subsequent S cysteinylation step. When the level of oxidized C15 was monitored directly by using AMS modification, half-maximal oxidation was observed ∼2 to 3 min after CHP addition (Fig. 4C), whereas with WT OhrR under the same conditions oxidation was half-maximal after 30 s of treatment (10; data not shown). Together, these results suggest that the rate of initial oxidation of the Y19A mutant is reduced severalfold relative to that of the WT. It is somewhat surprising that this decrease in the oxidation rate completely prevents derepression in vivo, even in the presence of high levels of CHP (Fig. 5A). This may be an indication that the activity of OhrR as a repressor reflects a balance between the rate of protein oxidation and the subsequent rate of rereduction.
DISCUSSION
The OhrR regulatory protein senses organic hydroperoxides with high sensitivity and high selectivity (6, 10). Like other well-characterized peroxide sensors, the sensing mechanism of OhrR begins with the oxidation of an active site cysteine to a sulfenic acid derivative. In proteins mediating reactive oxygen species responses, the reactivity of redox-sensitive cysteines is typically enhanced by neighboring basic amino acids that serve to lower the cysteine pKa to generate the reactive cysteine thiolate anion. These features are observed in OxyR, peroxiredoxins, and protein tyrosine phosphatases (2). Additional active site residues may also contribute to catalysis.
Insights into the high reactivity and specificity for the oxidation of OhrR by organic peroxides have emerged from solution of the high-resolution structure of B. subtilis OhrR bound to the ohrA operator site (9). This structure reveals that the active site C15 residue is within H-bonding distance of two tyrosine residues from the opposing subunit (designated Y29′ and Y40′). The B. subtilis OhrR crystal structure also revealed that there are no positively charged amino acids in close proximity to the active site. Since C15 is located at the N terminus of the α1 helix, the helix macrodipole was instead proposed to account for the low pKa of C15 (pKa ∼5.2) (9). Since the high reactivity of C15 towards peroxides cannot be explained simply by its low pKa, additional roles of active site residues in facilitating C15 peroxidation can be anticipated.
In X. campestris OhrR and presumably other members of the two-Cys family of OhrR proteins, oxidation of the active site cysteine generates a sulfenate that reacts rapidly with another protein thiol to generate an intersubunit disulfide that triggers a protein conformational change resulting in dissociation of the repression complex (17). In contrast, B. subtilis OhrR has only a single reactive Cys residue per monomer, and no intersubunit disulfide bonds form upon oxidation (7). Indeed, the initial product of OhrR oxidation in vitro is a relatively stable protein sulfenate, and it was initially thought that this oxidation event might be sufficient for derepression (7). However, more recent results indicate that inside the cell this initial sulfenate product reacts rapidly with low-molecular-mass thiols (Cys, coenzyme A thiol, and an uncharacterized 398-Da thiol) to generate mixed disulfides (10). In addition, the initial sulfenate can condense by reaction with a backbone amide to generate a cyclic sulfenamide. In vitro studies have demonstrated that S cysteinylation or sulfenamide formation, but not oxidation to the sulfenate, leads to a loss of DNA-binding activity (10).
Here, we used site-directed mutagenesis to identify residues that affect the reactivity of the C15 residue. First, in work began prior to solution of the OhrR crystal structure (9), we targeted basic amino acids that might contribute to stabilization of the reactive thiolate. None of the tested basic amino acids were essential for the ability of OhrR to respond to organic peroxides in vivo. Although the structure of the OhrR-operator complex indicates that no basic amino acids are in the immediate vicinity of the C15 residue, this work nevertheless provides important confirmatory information since ion pair interactions can act over fairly long distances and protein structures in solution sometimes differ significantly from the static picture seen in a crystal structure. We concluded that other factors, perhaps including the helix macrodipole, are critical for stabilizing the reactive C15 thiolate. This work also provides evidence that R23 and, to a lesser extent, K27 are important for DNA binding, as hypothesized based on the structure of the OhrR-operator complex (9).
A second set of mutants was constructed to determine the effects of amino acids in the vicinity of C15 on protein function. These studies identified two amino acids critical for repressor activity but not for protein stability in the cell, F16 and R23. In addition, Y19, which is one turn of the helix away from C15, was found to be critical for derepression; a Y19A mutant protein was noninducible. The solution of the structure of the OhrR-operator complex (9) suggested a plausible interpretation of these mutagenesis results. We hypothesized that oxidation of the C15 thiolate would perturb the C15-Y19′-Y40′ bonding network and thereby trigger a conformational change propagated from Y40′ through the adjacent F16 and Y19 residues to ultimately perturb the orientation of R23, an essential DNA-binding residue.
To test this model, we measured the effects of mutations in Y29, Y40, and Y19 on the susceptibility of OhrR to oxidation. We reasoned that if a Y19A mutant simply served to prevent the propagation of an oxidation-dependent conformation change, then the Y19A mutant might be normally reactive to peroxides but be defective in release from DNA. In contrast to this expectation, our results indicate that a Y19A mutant protein is defective in the initial step of oxidation, formation of the C15 sulfenate. Thus, we now favor the view that this mutant protein is altered in the conformation or reactivity of the active site and that this may account for the inability of this repressor to sense peroxides. The Y19A mutation may affect the structure of the C15-Y29′-Y40′ active site region, or Y19 may play a more direct role in catalyzing peroxidation of the reactive C15 thiolate.
To directly test the roles that the Y29 and Y40 residues play in formation of the active site, we generated a series of amino acid substitution mutants. All tested substitutions for these two residues (including Ala, Cys, and Phe) were defective in peroxide sensing in vivo. These results indicate that Y29 and Y40 are important for C15 reactivity with organic peroxides. Previous structural studies suggested that the negatively charged C15 thiolate may function as an H-bond acceptor for the tyrosyl hydroxyl moieties (9), but the precise role of these residues in facilitating peroxidation is not yet clear. These Tyr residues may function by restricting the orientation of the thiolate side chain, reducing the pKa of the thiol, by protonation of the leaving group (e.g., to generate the alcohol product), or by some combination of these effects. A critical role for these Tyr residues is consistent with the invariant nature of these residues among OhrR orthologs. Ultimately, oxidation of the C15 thiolate, leading to either intersubunit disulfide bond formation or mixed disulfide bond formation, disrupts the architecture of the active site and triggers protein dissociation. The precise nature of the structural changes that accompany this allosteric transition are not yet clear.
Acknowledgments
We thank the helpful staff of the Cornell BRC Proteomics and Mass Spectrometry Core Facility.
This work was supported by grant MCB-0640616 from the National Science Foundation to J.D.H. and by a Government of Thailand scholarship to S.S.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 2.Barford, D. 2004. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 14:679-686. [DOI] [PubMed] [Google Scholar]
- 3.Chen, P. R., T. Bae, W. A. Williams, E. M. Duguid, P. A. Rice, O. Schneewind, and C. He. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2:591-595. [DOI] [PubMed] [Google Scholar]
- 4.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 5.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570-11578. [DOI] [PubMed] [Google Scholar]
- 6.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA 99:6690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 9.Hong, M., M. Fuangthong, J. D. Helmann, and R. G. Brennan. 2005. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20:131-141. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. W., S. Soonsanga, and J. D. Helmann. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA 104:8743-8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesniak, J., W. A. Barton, and D. B. Nikolov. 2002. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 21:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linke, K., and U. Jakob. 2003. Not every disulfide lasts forever: disulfide bond formation as a redox switch. Antioxid. Redox Signal. 5:425-434. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 15.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panmanee, W., P. Vattanaviboon, L. B. Poole, and S. Mongkolsuk. 2006. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188:1389-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rince, A., J. C. Giard, V. Pichereau, S. Flahaut, and Y. Auffray. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shea, R. J., and M. H. Mulks. 2002. ohr, encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect. Immun. 70:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toledano, M. B., A. Delaunay, L. Monceau, and F. Tacnet. 2004. Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem. Sci. 29:351-357. [DOI] [PubMed] [Google Scholar]
- 22.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volker, U., K. K. Andersen, H. Antelmann, K. M. Devine, and M. Hecker. 1998. One of two osmC homologs in Bacillus subtilis is part of the σB-dependent general stress regulon. J. Bacteriol. 180:4212-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]