Abstract
MukB is a bacterial SMC(structural maintenance of chromosome) protein required for correct folding of the Escherichia coli chromosome. MukB acts in complex with the two non-SMC proteins, MukE and MukF. The role of MukEF is unclear. MukEF disrupts MukB-DNA interactions in vitro. In vivo, however, MukEF stimulates MukB-induced DNA condensation and is required for the assembly of MukB clusters at the quarter positions of the cell length. We report here that MukEF is essential for stable association of MukB with the chromosome. We found that MukBEF forms a stable complex with the chromosome that copurifies with nucleoids following gentle cell lysis. Little MukB could be found with the nucleoids in the absence or upon overproduction of MukEF. Similarly, overproduced MukEF recruited MukB-green fluorescent protein (GFP) from its quarter positions, indicating that formation of MukB-GFP clusters and stable association with the chromosome could be mechanistically related. Finally, we report that MukE-GFP forms foci at the quarter positions of the cell length but not in cells that lack MukB or overproduce MukEF, suggesting that the clusters are formed by MukBEF and not by its individual subunits. These data support the view that MukBEF acts as a macromolecular assembly, a scaffold, in chromosome organization and that MukEF is essential for the assembly of this scaffold.
The complex formed between MukB, MukF, and MukE (16, 24, 37, 38) proteins is essential for global organization of the Escherichia coli chromosome. The core protein of this complex, MukB, belongs to the ubiquitous family of SMC (structural maintenance of chromosome) proteins, which are involved in virtually every aspect of higher-order chromatin dynamics in organisms ranging from bacteria to humans (2, 9, 10, 27, 30).
All three subunits of MukBEF are encoded within the same operon together with an unrelated gene, smtA (36). Mutational disruption of any subunit of MukBEF results in severe defects in chromosome segregation, leading to chromosome decondensation and cutting, increased frequency of anucleate cells, and sharply reduced viability above 30°C (21, 37). Mutations in DNA topoisomerases suppress the phenotype of MukB mutants apparently by increasing supercoiling (1, 29, 33) and, therefore, the overall compactness of DNA (10, 26, 29). Accordingly, the chromosomes isolated from MukB-deficient cells are markedly decondensed (33), whereas overproduced MukBEF rapidly condenses nucleoids (32).
MukB dimerizes in solution to form a characteristic V-shaped molecule with two head domains connected via two long coiled coils with a hinge in between (16, 17, 20). MukF serves as a linker between MukB and MukE (38) and can form a stable complex with either MukE or MukB (38). Based on a structural analysis, MukF was postulated to be a kleisin (6). Accordingly, purified MukBEF but not MukB formed fibrous and rosette-like oligomeric structures in the absence of DNA (16). MukB forms a stable complex with DNA whereas no interaction with DNA was found for MukEF (20, 23, 24, 38).
Inside the cell, MukB forms distinct clusters along the length of the cell at about the one-quarter and three-quarter positions (5, 22). No clusters were detected in cells deficient in MukE or MukF (22). This result agrees well with the ability of MukBEF to form rosette-like clusters in vitro. The SMC protein from Bacillus subtilis was also shown to form distinct foci together with its cognate non-SMC subunits ScpA and ScpB (15, 31). Thus, formation of clusters by SMC complexes appears to be a common phenomenon in bacteria. It was proposed, therefore, that bacterial SMCs drive chromosome segregation by condensing the newly replicated DNA toward its new home at the quarter positions (8).
The biochemical mechanism of MukBEF remains unclear. In the presence of type 2 DNA topoisomerases, purified MukB promotes formation of DNA knots with unique topology (23). This activity is highly conserved among condensins and presumably reflects their ability to bind at the base of DNA loops (12, 23). MukB does not need its accessory subunits to condense DNA in vitro (23). Similarly, overproduction of MukB results in chromosome condensation even in ΔmukEF cells (32). Remarkably, MukEF inhibits DNA reshaping by MukB in vitro and, when bound at saturation, completely disrupts MukB-DNA complex (24). In contrast, overproduced MukBEF is a better condensin in vivo than MukB (32). Furthermore, the MukB-induced chromosome condensation does not rescue the temperature sensitivity of ΔmukEF cells (32). Thus, MukEF provides an essential quality to chromosome condensation that goes beyond DNA compaction per se.
We report here that MukBEF forms a stable complex with the chromosome that withstands sucrose gradient centrifugation following gentle cell lysis. Only a subset of DNA-binding proteins copurifies with the chromosome during this procedure (19). This approach was instrumental in identifying putative chromatin scaffold proteins and the scaffold-associated regions (13) but was never applied to bacterial condensins. The stable association of MukB with the chromosome was not observed for cells lacking or overproducing MukEF. Similarly, overproduction of MukEF displaced MukB-green fluorescent protein (GFP) from its quarter positions within the cell, indicating that formation of the foci and the stable association with the chromosome could be mechanistically related. Finally, we show that, similar to MukB, MukE-GFP forms fluorescent foci at the quarter positions but not in cells that lack MukB or overproduce MukEF. These data support the view that MukBEF forms a macromolecular scaffold that organizes the chromosome into the higher-order structure and that MukEF plays a central role in the assembly of the scaffold.
MATERIALS AND METHODS
Plasmids and strains.
The ΔmukB::kan strain GC7528 (21), mukE::kan strain AZ5450 (37), and ΔmukBEF::kan strain OT7 (38) were a kind gift of Sota Hiraga. The MG1655-based ΔmukEF::kan strain OU102 has been described previously (32). Plasmids pBB08 (32), pBB05, pTopA, and pB337 contain, respectively, the smtA-mukF-mukE-His9 fragment of the MukBEF operon, smtA, topA, and the first 337 codons of mukB under the control of arabinose inducible promoter.
The ΔmukB::kan strain OU101 was derived from MG1655 using the recBC sbcB system (34) as described earlier for OU102 (32). In OU101, the nucleotides 2 to 4430 of the mukB gene are replaced with the kanamycin resistance cassette (the 1.4-kb AfeI fragment from pACYC177), followed by the SfoI/SphI fragment from pUC18. The strain was verified by PCR analysis of the insertion sites in the chromosome of OU101 and by suppression of temperature sensitivity by the MukB-encoding plasmid pBB10 (32).
The lacYA::mukE-gfp-spc strain OU110 was constructed by integrating the MukE-GFP cassette into the lac locus of MG1655. The MukE-GFP cassette contains the following elements assembled in pACYC184 in the indicated order: lacY fragment (nucleotides [nt] 362184 to 361950 of the E. coli chromosome), mukE-gfp cloned downstream from the Pmuk promoter region (nt 972226 to 972759), the 2.1-kb BamHI fragment of the Ω interposon containing the spectinomycin resistance gene aadA (25), and a fragment of lacA (nt 361032 to 360876). Within the fusion protein, GFP is linked to the C terminus of MukE via peptide H9G2A. The MukE-GFP cassette was excised from the resulting plasmid, p15sp-E02a, with SwaI, gel purified, and integrated into the chromosome of MG1655 using a lambda Red recombination system (4). The strain was verified by PCR analysis of the integration region. Construction of OU115 (lacYA::mukB-gfp-spc) was the same, except that mukE in the MukE-GFP cassette was replaced with mukB.
OU111 (mukE::kan lacYA::mukE-gfp-spc) was constructed by P1vir transduction of the mukE::kan fragment of AZ5450 into OU110. P1vir transduction of the ΔmukB::kan fragment of OU101 into OU110 and OU115 produced strains OU112 (ΔmukB::kan lacYA::mukE-gfp-spc) and OU116 (ΔmukB::kan lacYA::mukB-gfp-spc), respectively.
Protein expression and quantification.
Mild overproduction of MukEF (about 10,000 copies per cell) was achieved by allowing leaky expression from pBB08 (32), whereas arabinose induction was used for high-level overproduction of MukEF or MukBEF (100,000 copies per cell). The copy numbers of endogenous MukBEF (24) and endogenous and overproduced MukB (32) proteins were reported elsewhere. The copy numbers of MukE-GFP and MukB-GFP and of the overproduced MukE and MukF were measured using quantitative immunoblotting as previously described (24, 32). Cells were collected at an optical density at 600 nm (OD600) of between 0.6 and 0.8. For calculations, 1 OD unit was assumed to contain 109 cells. The measured copy numbers of Muk proteins are summarized in Fig. 3.
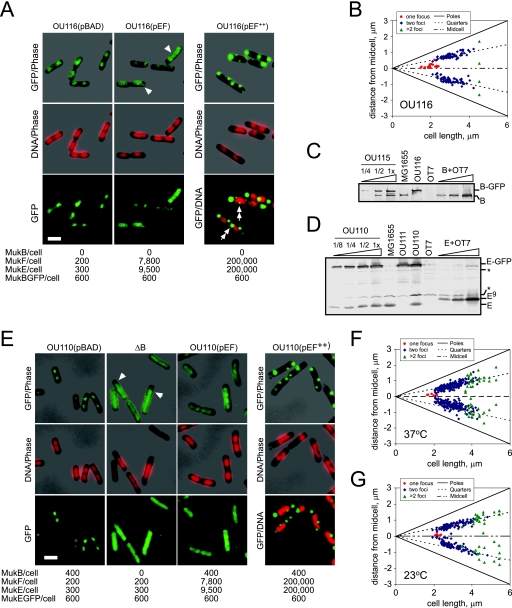
FIG. 3.
Subcellular localization of MukB-GFP and MukE-GFP. (A) Fluorescence micrographs of OU116 (ΔmukB mukB-gfp) cells harboring pBAD, pBB08 (pEF), or induced pBB08 (pEF++) plasmids grown in LB medium at 37°C. The copy number of Muk proteins is shown beneath the micrographs. Arrowheads point to MukB-GFP fluorescence in the DNA-free sections of the cell. Double arrows indicate foci in the vicinity of the nucleoids. Size bar, 2 μm. GFP fluorescence is shown in green, DNA is in red, and phase contrast is in gray. (B) Subcellular localization of MukB-GFP clusters in OU116 cells grown in LB at 37°C. (C) Immunoblot analysis of MukB-GFP content in OU116 and its parental mukB+ mukB-gfp OU115 strain. Loads are 0.025, 0.05, and 0.1 OD units of OU115 cells and 0.1 OD units of MG1655, OU116, and ΔmukBEF OT7 cells. Purified MukB (B+OT7) in amounts of 30 fmol, 90 fmol, and 300 fmol is supplemented with 0.1 OD unit of OT7 cells. The copy number of MukB-GFP and MukB was estimated as 600 and 400 per cell, respectively. B, MukB. (D) Immunoblot analysis of MukE-GFP content in OU110 cells. Diluted as indicated, 0.1 OD unit of OU110, MG1655, OT7, and the OU110-derived ΔmukE mukE-gfp OU111 cells was analyzed along with 17 fmol, 70 fmol, and 340 fmol of purified MukE-His9. The copy numbers of MukE-GFP and MukE were estimated as 600 and 250, respectively, in agreement with the earlier estimate of 340 ± 100 MukE copies per cell (24). An asterisk marks the major cross-reacting bands. E, MukE; E9, MukE-His9. (E) Fluorescence micrographs of OU110 or OU112 (ΔB) cells harboring the vector (pBAD) or pBB08 (pEF) or induced to overproduce MukEF (pEF++). OU110 cells were grown in LB medium at 37°C; OU112 cells were grown at 23°C. Arrowheads point to the GFP signal in the DNA-free sections of the cell. Size bar, 2 μm. (F and G) Subcellular localization of MukE-GFP clusters in OU110 cells grown in LB medium at 37°C (F) or 23°C (G).
Fluorescence microscopy.
Cells were grown in LB medium containing 0.5% NaCl. Where appropriate, 50 μg/ml ampicillin, 20 μg/ml kanamycin, 50 μg/ml spectinomycin, and 34 μg/ml chloramphenicol were added. When indicated, protein overproduction was induced by the addition of 0.2% l-arabinose at an OD600 of 0.2. Cell aliquots (OD600 values between 0.6 and 0.8) were supplemented with 5 μM Hoechst 33342, the incubation was continued for 15 min, and 300 μl of cell suspension was applied to a poly-l-lysine-coated coverslip. Following a 5-min incubation, the coverslips were rinsed six times in phosphate-buffered saline (PBS), placed on top of 15 μl of 5 μM Hoechst 33342 in PBS spotted onto a microscope slide, and observed using an Olympus BX-50 microscope equipped with a BX-FLA fluorescent attachment. Photographs were taken using an Insight charge-coupled-device camera (Hitschfel Instruments). Image overlays and color adjustment were done using Adobe Photoshop software. Subcellular localization of the fluorescent foci was quantified using the program Nucleus (32).
Nucleoid isolation and characterization.
Nucleoids were isolated as described earlier (14, 19) with minor modifications. DH5α cells harboring the vector (pBAD), pBB03 (pBEF), or pBB08 (pEF) were grown in LB medium at 37°C to an OD600 of 0.2, supplemented with 0.2% arabinose, further incubated for 1 h, chilled by swirling the flask in the ice-cold water bath, and concentrated by centrifugation. Muk-deficient cells were grown at 23°C to an OD600 of 0.8 and harvested in a similar manner. Following lysozyme treatment, 20 OD units of cells were lysed in 1 ml of Brij 58-deoxycholate mixture at room temperature (19) and centrifuged for 15 min at 8 krpm through a 15% to 40% sucrose gradient in 10 mM Tris-Cl, pH 7.8, 5 mM MgCl2, and 1 mM dithiothreitol. The band containing both folded and unfolded nucleoids (Fig. 1A, nucleoids) was collected from the middle of the gradient along with the fraction on top of the gradient (Fig. 1A, cytoplasm) and analyzed for protein and DNA content. Protein concentrations were measured by Bradford assay using bovine serum albumin as a standard. DNA concentration was determined from DAPI (4′,6′-diamidino-2-phenylindole) fluorescence. Aliquots of fractions were mixed with 100 nM DAPI in PBS, fluorescence at 461 nm with excitation at 358 nm was measured using a Shimadzu spectrofluorometer RF5301, and the DNA concentration was determined by comparing the signal with the calibration curve (purified pUC18 DNA). The nucleoid fraction contained about 85% of the total DNA but only between 3% and 8% of the total protein. In contrast, the cytoplasmic fraction contained more than 90% of the protein and less than 15% of the DNA (data not shown).
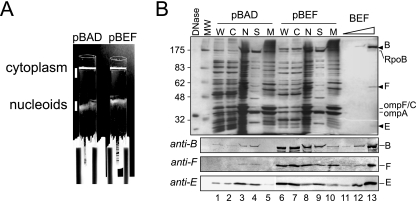
FIG. 1.
Copurification of MukBEF with nucleoids. Nucleoids were isolated from DH5α cells that carried either the vector (pBAD) or the MukBEF-overproducing plasmid pBB03 (pBEF) 1 h after induction with arabinose. The isolated nucleoids were then separated into the scaffold and the membrane fractions as described in Materials and Methods. (A) Separation of the nucleoids from the cytoplasm using sucrose gradient centrifugation. Following centrifugation, the tubes were placed against a dark background and illuminated from the top. The cytoplasm and nucleoids were collected as indicated using wide-bore pipette tips. (B) SDS-PAGE analysis of isolated nucleoids. Whole-cell extract (W), cytoplasm (C), nucleoids (N), scaffold (S), and membranes (M) were resolved on a 10% gel along with increasing amounts of purified MukBEF (BEF). The proteins were visualized by silver staining (top panel) or by immunoblotting (bottom panels). Lanes 1 to 3, 5 μg of total protein; lanes 6 to 8, 3 μg of protein. A total of 5 μg protein as in lane 3 was separated into the scaffold and membrane fractions and analyzed in lanes 4 and 5, respectively. Similarly, 3 μg of protein as in lane 8 was fractionated for lanes 9 and 10. Lane 13 contains purified MukBEF (2.2 pmol of MukB-His10, 1.6 pmol of MukF, and 1.6 pmol of MukE). Lanes 11 and 12 contain 100-fold and 10-fold dilutions of MukBEF, respectively, as in lane 13. Arrowheads mark positions of MukB (B), MukF (F), and MukE (E). Also marked are positions for RNA polymerase (RpoB) and the major outer membrane proteins OmpC and OmpA. RpoB was identified from the comparison with the previous studies (18, 19) based on its molecular mass (156 kDa) and the large abundance of the protein in the nucleoid fraction. Note that MukB is not the major protein in the scaffold fraction even after overproduction.
The nucleoid fraction was further treated with 5 μg/ml DNase I (Sigma-Aldrich) supplemented with 10 mM EDTA for 30 min at room temperature and centrifuged for 30 min at 40 krpm at 4°C in a TLA 55 rotor to separate soluble, presumably DNA-bound proteins (supernatant) from the membrane-associated proteins (pellet). Approximately half of the protein was found in the supernatant after centrifugation (scaffold fraction), as judged by the Bradford assay. The pellet (membrane fraction) was resuspended in 10 mM Tris-Cl, pH 7.8, 1 mM EDTA, and 1% sodium dodecyl sulfate (SDS) by brief water bath sonication. The scaffold proteins (i.e., released by DNase I treatment) were concentrated by trichloroacetic acid precipitation. The samples were then resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by quantitative Western blotting.
RESULTS
MukBEF copurifies with nucleoids.
Nucleoids were isolated from exponentially growing cells using the low-salt-spermidine procedure (19). This procedure yields nucleoids with a unique subset of associated proteins that does not coincide with the whole-cell extract or a set of E. coli proteins eluted from DNA-cellulose (19). In addition, isolated nucleoids are attached to numerous membrane vesicles. DNase I treatment liberates DNA-associated proteins from the macromolecular bodies, and subsequent high-speed centrifugation precipitates protein aggregates and membrane vesicles but not individual proteins released from DNA by the nuclease (19).We will refer to the soluble and precipitated proteins as the scaffold and membrane fractions, respectively.
Figure 1B compares protein profiles obtained after fractionation of MukBEF-overproducing and nonoverproducing DH5α cells. The proteins in the cytoplasmic fraction (about 95% of the total cellular protein) are virtually the same as in the whole-cell extract. A very different subset of proteins was found in the nucleoid and scaffold fractions. The major proteins found in the soluble fraction of the nucleoids are RNA polymerase, HU, Fis, and integration host factor (19). Only RNA polymerase is large enough to be resolved on the gel in Fig. 1. It is noteworthy that virtually no OmpA or OmpC, the major outer membrane proteins, was found in the scaffold fraction whereas very little RNA polymerase precipitated with the membrane vesicles (Fig. 1B). Similarly, no elongation factor (EF)-Tu could be detected in the nucleoid fractions (Fig. 2A). We conclude that the separation of the cytoplasmic and the nucleoid-associated proteins was successful.
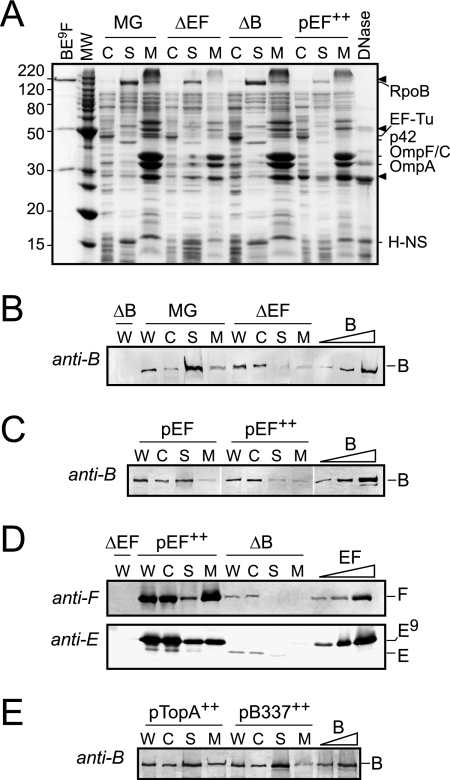
FIG. 2.
Complementary subunits affect the copurification of MukB and MukEF with the chromosome. Nucleoids were isolated from MG1655 (MG), OU102 (ΔEF), and GC7528 (ΔB) cells grown in LB medium at 23°C and from arabinose-induced (at 37°C) DH5α cells harboring pBB08 (pEF++). (A) Coomassie-stained 14% polyacrylamide gel analyzing cytoplasmic and nucleoid proteins. The gel resolves 5 μg of cytoplasmic proteins or the scaffold and membrane fractions derived from 5 μg of the nucleoid proteins. BE9F, a mixture of purified MukB and MukEF containing 0.5 μg of MukB-His10, 0.23 μg of MukF, and 0.27 μg of MukE-His9; DNase, 0.5 μg of DNase I. Positions of MukB, MukF, and untagged MukE are indicated with arrowheads. The brightest band among cytoplasmic proteins (apparent molecular mass of 46 kDa) is identified as EF-Tu (43 kDa) from the comparison with the E. coli proteome SWISS-2DPAGE (available via www.expasy.org). The major detected components of the scaffold fraction were marked as RpoB and H-NS from the comparison with previous studies (19). p42 could be the α subunit of RNA polymerase, RpoA (18); the 36.5-kDa RpoA was reported to migrate anomalously slow as a 40-kDa protein in a different gel system (SWISS-2DPAGE). (B and C) Western blot analysis of MukB content in isolated nucleoids. A total of 10 μg of collected fractions (labeled as in Fig. 1) was analyzed along with 30 fmol, 90 fmol, and 300 fmol of purified MukB. (D) Western blot analysis of MukEF copurification with isolated nucleoids. The load was 2.5 μg of fractions from the pEF cells or 5 μg of fractions from the ΔB or ΔEF cells. The lanes with the calibration mixture contain 10 fmol, 40 fmol, and 200 fmol of MukF and 20 fmol, 80 fmol, and 400 fmol of MukE-His9. Note the difference in mobilities of His-tagged (E9) and endogenous (E) MukE. (E) Western blot analysis of MukB content in cells overproducing TopA or the N-terminal fragment of MukB (amino acids 1 to 337) using the same pBAD-based expression system. C, cytoplasmic; S, scaffold; M, membrane; W, whole-cell extract; B, MukB; E, MukE; F, MukF.
All three subunits of MukBEF were enriched in the nucleoid fraction compared to whole cells (Fig. 1B). The concentration of MukB and MukE was, respectively, fourfold and sixfold greater in the nucleoid fraction than in the whole-cell extract (Tables 1 and 2). Similarly, we could detect MukF only with the nucleoids and not in the cell extract. All three subunits remained soluble following DNase I treatment, indicating that the proteins were associated with DNA rather than with the membrane vesicles.
TABLE 1.
MukB content of whole-cell extracts and isolated nucleoids
| Straina | MukB content (pmol/mg) in the indicated fractionb
|
|
|---|---|---|
| Cell extract | Nucleoidc | |
| DH5α (pBAD) | 7.7 ± 1.7 | 28 ± 12 (>90) |
| DH5α (pBEF++) | 440 | 320 (85) |
| DH5α (pEF++) | 10 ± 5 | 2.4 ± 0.8 (30 ± 20) |
| MG1655 (pEF) | 9 | 9 (93) |
| MG1655 (pBAD) | 12 ± 1 | 42 ± 10 (86 ± 2) |
| OU102 (pBAD) | 8 ± 5 | 4.0 ± 0.6 (57 ± 19) |
| MG1655 (pSmtA++) | 5 | 8 (96) |
| MG1655 (pTopA++) | 6 | 11 (71) |
| MG1655 (pB337++) | 8 | 13 (71) |
++, plasmid designed to overproduce the indicated protein or fragment was induced with arabinose.
The data are normalized to the total protein in a given fraction. When standard deviations are shown, the data are the average of two independent experiments.
The percentage of the nuceloid-associated MukB that was released by DNase I treatment is given in parentheses.
TABLE 2.
MukF and MukE content of whole-cell extracts and isolated nucleoids
| Straina | MukF content in the indicated fraction (pmol/mg)b
|
MukE content in the indicated fraction (pmol/mg)b
|
||
|---|---|---|---|---|
| Cell extract | Nucleoid | Cell extract | Nucleoid | |
| DH5α (pBAD) | ND | 2.5 | 1.7 ± 0.7 | 9.6 ± 2.2 (95 ± 2) |
| DH5α (pBEF++) | 450 | 400 (30) | 370 | 40 (64) |
| DH5α (pEF++) | >210 | 40 (16) | >180 | 100 (40) |
| MG1655 (pBAD) | ND | ND | 3.4 | 14 |
| GC7528 (pBAD) | 1.6 | <0.2 | 1.1 | 0.2 |
++, plasmid designed to overproduce the indicated protein was induced with arabinose.
The percentage of the nucleoid-associated proteins that was solubilized by DNase I treatment is given in parentheses. ND, not detrmined.
Following MukBEF overproduction, about eightfold more MukB copurified with the nucleoids than with those from the nonoverproducing cells (Table 1). In this case, however, most of the protein remained in the cytoplasm. Furthermore, a significant fraction of the nucleoid-associated MukF and MukE (but not MukB) coprecipitated with the membrane vesicles after DNase I treatment (Table 2). The precipitated proteins were apparently in complex with the vesicles rather than DNA or perhaps formed an aggregate. We conclude that only a limited amount of MukBEF can form a stable complex with the chromosome.
MukEF modulates the association of MukB with the chromosome.
We next examined the effect of MukEF on the ability of MukB to bind the chromosome. The MukEF-deficient strain OU102 was grown in LB medium at 23°C alongside with its parental mukEF+ strain, MG1655. The nucleoids were isolated from the mid-exponential cells and further separated into the scaffold and the membrane fractions as described above. The patterns of proteins found in the scaffold and the membrane fractions of ΔmukEF cells were virtually the same as for MG1655 cells (Fig. 2A). In contrast, the amount of the nucleoid-associated MukB decreased dramatically for the ΔmukEF strain (Fig. 2B). Relative to the whole-cell extract, MukB content is fourfold greater in the nucleoid fraction of the wild-type cells but is twofold lower in the absence of MukEF (Table 1). Furthermore, only half of MukB that copurified with the chromosomes of ΔmukEF cells remained soluble after DNase I treatment as opposed to 86% for MG1655 cells (Table 2). Thus, a significant fraction of MukB found in the nucleoid fraction of ΔmukEF cells could be associated with the membrane rather than DNA or could form aggregates.
Interestingly, overproduction of MukEF had a negative effect on copurification of MukB with the chromosome. The effect could be detected at mild (about 35-fold) overproduction levels but was especially clear after induction of MukEF (Fig. 2C). Most of MukB remained in the cytoplasm following MukEF overproduction, and about half of the nucleoid-associated MukB coprecipitated with the membrane vesicles after DNase I treatment (Tables 1 and 2). The effect was specific for MukEF since overproduction of three unrelated proteins did not impair the association of MukB with the chromosome (Fig. 2E and Table 1). This result is in accord with the biochemical data, which demonstrated that saturating binding of MukEF disrupts MukB-DNA interaction (24).
MukEF itself remained in the cytoplasm in the absence of MukB (Fig. 2D). Only a trace amount of MukE was found in the scaffold fraction of ΔmukB cells, and the amount of MukF was below detection. Similarly, most of the overproduced MukEF remained on top of the sucrose gradient, whereas most of the nucleoid-associated MukE and especially MukF remained insoluble after DNase I treatment (Fig. 2D). It is unclear whether the precipitated MukEF is merely enclosed in the membrane vesicles or is in true complex with the membranes.
Elevated levels of MukEF displace MukB-GFP from the quarter foci.
MukEF-dependent association of MukB with the chromosome is reminiscent of the previous finding that MukB-GFP forms foci at the quarter positions in the presence but not in the absence of MukEF (22). To further explore this relationship, we examined the effect of MukEF on the formation of MukBEF foci.
Subcellular localization of MukB-GFP was investigated using strain OU116, which lacks endogenous MukB but produces a C-terminal GFP fusion of MukB from an ectopic location (Materials and Methods). In this strain, MukB-GFP is produced at about the same level as the untagged protein, 600 and 400 copies per cell, respectively (Fig. 3C). In agreement with previous data (22), MukB-GFP formed clear foci in the middle of short cells or at the quarter positions of the longer cells (Fig. 3A and B). Similar foci, albeit with somewhat stronger background, were observed in the mukB+ mukB-gfp OU115 strain (data not shown).
Overproduction of MukEF interfered with localization of MukB-GFP. Although MukB-GFP clusters could still be detected at mild overproduction levels of MukEF, most of the fluorescence was distributed throughout the cell and could be found even in the DNA-free sections of the cell (Fig. 3A, arrowheads). Following induction of MukEF, MukB-GFP was found in clusters away from DNA (Fig. 3A). Most of the induced cells (82%) contained internal foci in addition to the bright spots at the cell poles. The majority of these were located next to or at the tip of the nucleoids (Fig. 3A, double arrows). Only 8% of examined cells contained MukB-GFP foci that could have colocalized with DNA. No such alternative clusters were found upon overproduction of SmtA or TopA (data not shown). Although fluorescence from across the cell was somewhat increased in these cases, at least 60% of cells contained MukB-GFP foci at expected locales. It appears, therefore, that MukB-GFP was recruited to MukEF aggregates via its specific interactions with the kleisin, which indicates, in turn, that the protein was folded correctly. We conclude, therefore, that overproduced MukEF precludes MukB from binding the chromosome, as we found earlier using a cell fractionation approach (Fig. 2C).
MukE-GFP forms foci at the quarter positions but not in the absence of MukB.
We next examined if MukEF can form clusters in the absence of MukB. If so, this would support the notion that MukEF serves as a linker between MukB and the cellular matrix. The lacYA::mukE-gfp-spt strain OU110 is isogenic to OU115 except that it carries the mukE-gfp gene in its lac locus. As judged by quantitative immunoblotting, OU110 produces about 600 copies of MukE-GFP per cell in addition to 300 copies of endogenous MukE (Fig. 3D and E).
MukE-GFP formed well-defined fluorescent clusters (Fig. 3E). The MukE-GFP foci were smaller than the nucleoids, indicating that these represent distinct structures and are not the result of random association of MukBEF with DNA. Similar to MukB-GFP, MukE-GFP foci were located at the quarter positions inside the cell (Fig. 3F).
MukE-GFP was evenly distributed throughout the ΔmukB::kan lacYA::mukE-gfp OU112 cells (Fig. 3E). This was not due to the lower cultivation temperature required for OU112 cells. The distributions of MukE-GFP in OU110 cells were identical at 37°C and at 23°C (Fig. 3FG). Thus, MukE-GFP forms clusters inside the cell but only in the presence of MukB.
We found no clusters of MukE-GFP in OU110 cells harboring the MukEF-encoding pBB08 plasmid (Fig. 3E). Similarly, MukE-GFP was evenly distributed after mild overproduction of MukF (data not shown). Only at a high level of overproduction of MukEF did we observe MukE-GFP clusters (Fig. 3E). These, however, were often located at the poles of the cell, virtually never colocalized with nucleoids (less than 6% of cells), and resembled protein aggregates by their stark appearance. We conclude that the protein clusters at the quarter positions can accommodate only a limited number of MukE proteins.
DISCUSSION
Soon after their discovery, SMC proteins were identified as one of the major components of histone-depleted chromosomes (28). Subsequent studies reinforced the idea that condensins do not “merely” control the size of the chromosomes but organize them into highly ordered structures (7). A direct test confirmed that chromosome condensation in E. coli does not compensate for the lack of MukBEF (32). MukBEF has been previously hypothesized to be the chromatin scaffold protein in E. coli (27), which resonates well with the decondensed structure of the MukBEF-depleted chromosomes (21, 33). In further support of this view, we show here that MukBEF copurifies with the E. coli nucleoids.
Several protocols were developed to isolate E. coli chromosomes (3, 14, 19, 35). We employed the low-salt-spermidine procedure, which prevents dissociation of many proteins from DNA (14, 19). Importantly, however, the proteins that copurify with the chromosomes are not a random sampling of DNA binding proteins (19). The major proteins that are retained on the nucleoids are RNA polymerase and the histone-like proteins HU, H-NS, and Fis (19). We found at least 30% of cellular MukB in the isolated nucleoids (recalculated from Table 1). MukE and MukF were similarly enriched in the nucleoid fraction. We conclude that MukBEF forms a stable complex with the chromosome.
MukEF was required for stable association of MukB with the chromosome. Conversely, no MukE or MukF comigrated with the nucleoids from ΔmukB cells. Thus, stable binding to the chromosome is a property of the complex between MukB and MukEF. Formation of protein clusters by MukB-GFP and MukE-GFP is similarly dependent on the presence of the complementary subunits, indicating that the two features are mechanistically related. The mechanism of binding to the chromosome appears to be highly conserved between condensins: 13S condensin from frogs was also reported to bind the chromosome only in the presence of its regulatory subunits (11).
It is noteworthy that elevated levels of MukEF displaced MukB from the chromosome. This effect was observed using both the cell fractionation approach (Fig. 2C) and fluorescence microscopy (Fig. 3A) at both high and low levels of protein overproduction. Overproduced MukF, which links MukB and MukE within the complex, was also disruptive to the formation of MukBEF clusters, although its effects were less pronounced at the uninduced levels than the effect of MukEF (data not shown). In contrast, the clusters were not notably affected by mild overproduction of MukE (data not shown). The result is in accord with in vitro reconstitution studies, which demonstrated that the two MukBs at the core of MukBEF have different affinities to MukEF and that saturating binding of MukEF to MukB disrupts MukB-DNA complex (24). Apparently, the overproduced MukEF is able to bind the low-affinity site on MukB and thereby displace the protein from DNA.
The functional significance of this effect is unclear. It may indicate that the association of MukBEF with the chromosome is dynamic and that MukEF helps remove MukB from DNA. Even if true, this cannot be the sole function of MukEF. This explanation is inconsistent with the stimulatory effects of MukEF on the binding of MukB to the chromosome (Fig. 2) or the finding that MukBEF is a better condensin than MukB (32). It seems more likely that MukEF mediates the assembly of MukBEF into a macromolecular structure (Fig. 4), as was proposed in a recent electron microscopy study (16).
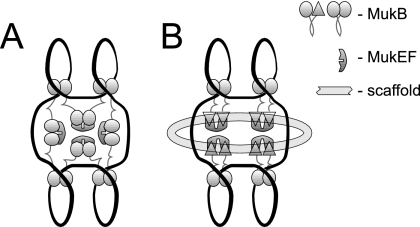
FIG. 4.
Two oversimplified models of MukBEF scaffold. (A) MukEF mediates the assembly of the scaffold by bridging separate MukB clusters. The model attempts to integrate the finding that MukEF and DNA bind MukB on a mutually exclusive basis (24) with the ability of MukB to form MukEF-mediated oligomers (16). (B) MukEF links MukB to the cellular matrix. The model builds upon the ability of MukB to adopt two different conformations (23). MukEF is postulated to trap MukB in a conformation with high affinity to a putative, unidentified extrachromosomal factor. Both models postulate that MukEF promotes the assembly of a polydentate macromolecular structure and thereby stabilizes the interaction between MukB and the chromosome.
A possible mechanism that could explain both the size and specific subcellular localization of MukBEF clusters relies on the interaction between MukBEF and the cellular matrix (Fig. 4B). However, experimental evidence in support of this view is limited. Only at high levels of protein induction did we find indications that MukEF might be attached to an extrachromosomal factor. For example, a notable fraction of the overproduced MukF and MukE copurified with the membranes during cell fractionation (Fig. 1B and 2D). In contrast, the MukEF that was not overproduced was distributed throughout the cell in the absence of MukB, indicating that MukEF is unlikely to directly mediate the association of MukB with the cell.
Acknowledgments
This work was supported by grant GM63786 from NIH and a grant from the American Heart Association.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Adachi, S., and S. Hiraga. 2003. Mutants suppressing novobiocin hypersensitivity of a mukB null mutation. J. Bacteriol. 185:3690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobbe, N., and M. M. Heck. 2004. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 21:332-347. [DOI] [PubMed] [Google Scholar]
- 3.Cunha, S., T. Odijk, E. Suleymanoglu, and C. L. Woldringh. 2001. Isolation of the Escherichia coli nucleoid. Biochimie 83:149-154. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Blaauwen, T., A. Lindqvist, J. Lowe, and N. Nanninga. 2001. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol. Microbiol. 42:1179-1188. [DOI] [PubMed] [Google Scholar]
- 6.Fennell-Fezzie, R., S. D. Gradia, D. Akey, and J. M. Berger. 2005. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J. 24:1921-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gassmann, R., P. Vagnarelli, D. Hudson, and W. C. Earnshaw. 2004. Mitotic chromosome formation and the condensin paradox. Exp. Cell Res. 296:35-42. [DOI] [PubMed] [Google Scholar]
- 8.Graumann, P. L. 2001. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 83:53-59. [DOI] [PubMed] [Google Scholar]
- 9.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 10.Holmes, V. F., and N. R. Cozzarelli. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. USA 97:1322-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura, K., and T. Hirano. 2000. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc. Natl. Acad. Sci. USA 97:11972-11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura, K., V. Rybenkov, N. Crisona, T. Hirano, and N. Cozzarelli. 1999. 13S Condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98:239-248. [DOI] [PubMed] [Google Scholar]
- 13.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2 ed. W. H. Freeman and Co., New York, NY.
- 14.Kornberg, T., A. Lockwood, and A. Worcel. 1974. Replication of the Escherichia coli chromosome with a soluble enzyme system. Proc. Natl. Acad. Sci. USA 71:3189-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindow, J. C., M. Kuwano, S. Moriya, and A. D. Grossman. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46:997-1009. [DOI] [PubMed] [Google Scholar]
- 16.Matoba, K., M. Yamazoe, K. Mayanagi, K. Morikawa, and S. Hiraga. 2005. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem. Biophys. Res. Commun. 333:694-702. [DOI] [PubMed] [Google Scholar]
- 17.Melby, T. E., C. N. Ciampaglio, G. Briscoe, and H. P. Erickson. 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, L. D., J. L. Rosner, S. B. Zimmerman, and D. Esposito. 1999. Identification of two new proteins in spermidine nucleoids isolated from Escherichia coli. J. Bacteriol. 181:3842-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, L. D., and S. B. Zimmerman. 1997. Isolation and characterization of spermidine nucleoids from Escherichia coli. J Struct. Biol. 119:321-335. [DOI] [PubMed] [Google Scholar]
- 20.Niki, H., R. Imamura, M. Kitaoka, K. Yamanaka, T. Ogura, and S. Hiraga. 1992. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 11:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niki, H., A. Jaffe, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohsumi, K., M. Yamazoe, and S. Hiraga. 2001. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40:835-845. [DOI] [PubMed] [Google Scholar]
- 23.Petrushenko, Z. M., C. H. Lai, R. Rai, and V. V. Rybenkov. 2006. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J. Biol. Chem. 281:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrushenko, Z. M., C. H. Lai, and V. V. Rybenkov. 2006. Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J. Biol. Chem. 281:34208-34217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prentki, P., A. Binda, and A. Epstein. 1991. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene 103:17-23. [DOI] [PubMed] [Google Scholar]
- 26.Rybenkov, V. V., A. V. Vologodskii, and N. R. Cozzarelli. 1997. The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J. Mol. Biol. 267:312-323. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh, N., I. Goldberg, and W. C. Earnshaw. 1995. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. Bioessays 17:759-766. [DOI] [PubMed] [Google Scholar]
- 28.Saitoh, N., I. G. Goldberg, E. R. Wood, and W. C. Earnshaw. 1994. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol. 127:303-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawitzke, J. A., and S. Austin. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. USA 97:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swedlow, J. R., and T. Hirano. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell 11:557-569. [DOI] [PubMed] [Google Scholar]
- 31.Volkov, A., J. Mascarenhas, C. Andrei-Selmer, H. D. Ulrich, and P. L. Graumann. 2003. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol. Cell. Biol. 23:5638-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Q., E. A. Mordukhova, A. L. Edwards, and V. V. Rybenkov. 2006. Chromosome condensation in the absence of the non-SMC subunits of MukBEF. J. Bacteriol. 188:4431-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weitao, T., K. Nordstrom, and S. Dasgupta. 1999. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 34:157-168. [DOI] [PubMed] [Google Scholar]
- 34.Winans, S. C., S. J. Elledge, J. H. a. Krueger, and G. C. Walker. 1985. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J. Bacteriol. 161:1219-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worcel, A., and E. Burgi. 1974. Properties of a membrane-attached form of the folded chromosome of Escherichia coli. J. Mol. Biol. 82:91-105. [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1995. Characterization of the smtA gene encoding an S-adenosylmethionine-dependent methyltransferase of Escherichia coli. FEMS Microbiol. Lett. 133:59-63. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250:241-251. [DOI] [PubMed] [Google Scholar]
- 38.Yamazoe, M., T. Onogi, Y. Sunako, H. Niki, K. Yamanaka, T. Ichimura, and S. Hiraga. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18:5873-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]