Abstract
Comparative analysis of binding of intact glucose-grown Fibrobacter succinogenes strain S85 cells and adhesion-defective mutants AD1 and AD4 to crystalline and acid-swollen (amorphous) cellulose showed that strain S85 bound efficiently to both forms of cellulose while mutant Ad1 bound to acid-swollen cellulose, but not to crystalline cellulose, and mutant Ad4 did not bind to either. One- and two-dimensional electrophoresis (2-DE) of outer membrane cellulose binding proteins and of outer membranes, respectively, of strain S85 and adhesion-defective mutant strains in conjunction with mass spectrometry analysis of tryptic peptides was used to identify proteins with roles in adhesion to and digestion of cellulose. Examination of the binding to cellulose of detergent-solubilized outer membrane proteins from S85 and mutant strains revealed six proteins in S85 that bound to crystalline cellulose that were absent from the mutants and five proteins in Ad1 that bound to acid-swollen cellulose that were absent from Ad4. Twenty-five proteins from the outer membrane fraction of cellulose-grown F. succinogenes were identified by 2-DE, and 16 of these were up-regulated by growth on cellulose compared to results with growth on glucose. A protein identified as a Cl-stimulated cellobiosidase was repressed in S85 cells growing on glucose and further repressed in the mutants, while a cellulose-binding protein identified as pilin was unchanged in S85 grown on glucose but was not produced by the mutants. The candidate differential cellulose binding proteins of S85 and the mutants and the proteins induced by growth of S85 on cellulose provide the basis for dissecting essential components of the cellulase system of F. succinogenes.
Fibrobacter succinogenes S85 is a rod-shaped, gram-negative, strictly anaerobic ruminal bacterium that is one of the major cellulolytic bacteria within the rumen (2, 24). Because of the ability of this bacterium to efficiently adhere to and degrade plant cell walls, it has been extensively studied to elucidate the mechanism involved in adhesion and digestion of plant cell walls. As a result, five endoglucanases (31, 33-35), one cellobiosidase (17), one cellodextrinase (15), four xylanases (21, 52), two acetyl xylan esterases (22), and two cellulose-binding proteins (11) have been purified or cloned and purified and the catalytic properties of the enzymes determined. Although those studies have provided valuable information about many of the cellulases and hemicellulases of F. succinogenes, the mechanism of binding of cells to cellulose and hydrolysis of cellulose has not been solved.
Similar to the case with other anaerobic cellulolytic bacteria, adhesion of F. succinogenes cells to cellulose appears to be a prerequisite for rapid and efficient cellulose hydrolysis by this organism (30). This claim is supported by the observation that carboxymethylcellulose, which blocks adhesion to cellulose, also blocked cellulose degradation (25). Gong and Forsberg (12) isolated nonadherent mutants of F. succinogenes S85 that either did not degrade crystalline cellulose or degraded cellulose more slowly than S85. Treatment of S85 cells with proteolytic enzymes markedly reduced their adhesion ability, showing that some protease-sensitive proteins on the cell surface were involved in the adhesion. However, without suitable separation methodologies and with a lack of techniques to identify noncatalytic proteins, it was challenging to pursue the research at that time.
Gong and Forsberg (13) developed a sodium chloride washing technique to cleanly separate outer membranes (OMs) of F. succinogenes free from other cellular fractions. Since adhesion to cellulose and cellulose hydrolysis involve an intimate association of the OM surface with cellulose, we have analyzed the OM proteins from F. succinogenes S85 grown on cellulose and on glucose and from the two nonadherent mutants, Ad1 and Ad4, grown on glucose in order to identify proteins with potential roles in adhesion to cellulose and in cellulose hydrolysis. The application of mass spectroscopy analysis of trypsin digests of the separated proteins in conjunction with access to the genome sequence of F. succinogenes S85 has provided the opportunity to identify these novel proteins. Therefore, the goal of this study was to identify OM proteins that have potential roles in initial attachment of cells to cellulose and that are either absent from nonadherent mutants or are induced in the wild-type strain S85 by growth on cellulose.
(Research presented here is described in the Ph.D. theses of H. S. Jun, M. Qi, J. Gong, and E. E. Egbosimba, University of Guelph, Guelph, Ontario, Canada.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Fibrobacter succinogenes S85 and mutants Ad1 and Ad4, which do not adhere to Avicel crystalline cellulose (12), were grown in chemically defined medium (CDM) (43) including either 0.5% (wt/vol) glucose, 0.2% (wt/vol) acid-swollen cellulose (ASC), or 0.3% (wt/vol) Avicel cellulose PH105 as the carbohydrate source for growth. For the isolation of OM proteins, the bacteria were cultured in 4 liters of CDM containing either glucose or Avicel cellulose for 12 or 36 h, respectively, with periodic shaking.
ASC was prepared as described by Wood (51). Briefly, 5 g of Avicel cellulose PH105 was mixed with 10 ml of H3PO4 (85% [wt/vol]) and stored in ice for 1 h with occasional stirring. The mixture was poured into 4 liters ice-cold distilled water and left in ice for 30 min. The swollen cellulose was washed five times with 1.5 liters ice-cold distilled H2O, followed by 1 liter of 1% (wt/vol) NaHCO3 solution. The swollen cellulose was washed with ice-cold distilled water until the pH was neutral. A portion of the swollen cellulose was lyophilized to determine the concentration.
Growth was monitored by the determination of total cellular proteins in the culture. Samples of culture were heated at 100°C for 10 min in 0.5 M sodium hydroxide to solubilize cellular proteins. The residual cellulose particles were removed by centrifugation and the protein concentration in the supernatant determined by the method of Bradford (3) with bovine serum albumin (fraction V) dissolved in 0.5 M sodium hydroxide as the standard.
Carbohydrate determination.
Cellulose sedimented from the culture by centrifugation at 10,000 × g for 10 min was solubilized in 67% (wt/vol) sulfuric acid as described by Updegraff (49) and quantified using the phenol sulfuric acid method (6) for soluble carbohydrates. Glucose was used as the standard. Soluble sugars remaining in solution after centrifugation were assayed by the same procedure.
Assay of bacterial adhesion to cellulose.
Bacterial adhesion to cellulose was measured by the turbidity-based method as described by Gong and Forsberg (12). Briefly, a cell suspension (2 ml) grown in CDM containing 0.5% (wt/vol) glucose until the optical density at 675 nm reached 2.0 was thoroughly mixed by inversion with an equal volume of CDM containing 20% (wt/vol) Avicel cellulose PH105. The mixture was left stationary at room temperature for 60 min to allow cellulose to settle. The optical density of the supernate was measured at 675 nm. The calculation of percent adhesion was performed according to the equation % adhesion = [1 − (OD675 of cellulose supernate × 1.8)/2] × 100, where OD675 is the optical density at 675 nm.
A factor of 1.8 was used instead of 2 to correct for the volume of cellulose in the suspension (12).
Isolation of OM.
The OM was prepared as described previously (13) from 4-liter volumes of culture. Briefly, the cells were sedimented at 9,000 × g for 15 min. The cells were resuspended in 300 ml of 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 6.7) including 0.5 M NaCl, and the suspension was incubated at 4°C for 5 min with shaking at 150 rpm, followed by centrifugation (23,000 × g, 20 min). The washing step to collect OMs as well as periplasmic fractions was repeated three times. The supernatant containing OMs and periplasmic proteins was subjected to ultracentrifugation (Beckman) at 100,000 × g for 1 h to sediment OMs. The sedimented OMs were resuspended in PIPES buffer, pH 6.7, and membrane protein quantified as described by Bradford (3). In the case of cultures grown on cellulose, residual cellulose was removed by centrifugation at 500 × g for 5 min before cells were harvested. All centrifugations were performed at 4°C.
2-DE.
OM proteins were precipitated from a 1.5-ml suspension (1 mg protein·ml−1) by addition of a 10-fold volume of acetone precooled to −20°C. The precipitated proteins were washed once with the same volume of precooled acetone. The proteins were air-dried for 15 min and stored at −20°C prior to use.
The acetone-precipitated OM proteins (approximately 1.5 mg) were solubilized in 300 μl of isoelectric focusing (IEF) solution (7 M urea, 2 M thiourea, 2% [wt/vol] amidosulfobetaine 14 [4], 2 mM tributyl phosphine [TBP] [14], 2% [vol/vol] Biolyte 3-10 [Bio-Rad]) and incubated at room temperature for 1 h. The traces of insoluble materials were removed by centrifugation using an airfuge (100,000 × g, 20 min; Beckman). The 300 μl of solubilized OM proteins were applied to 17-cm immobilized pH gradient (IPG) strips with a range from pI 3 to 10. The strips were rehydrated at 50 V at 20°C for 12 h in a Protean IEF cell (Bio-Rad), and then IEF was performed by using the following steps: (i) conditioning step to remove salt ions and charged contaminants at 250 V for 20 min; (ii) rapid voltage ramping step at 10,000 V for 3 h; and (iii) final focusing step at 10,000 V for 6 h. The current did not exceed 50 μA/strip. The isoelectric focused strips were equilibrated in 15 ml of 50 mM Tris buffer (pH 8.8) containing 6 M urea, 2% (wt/vol) sodium dodecyl sulfate, 30% (vol/vol) glycerol, 5 mM TBP, and a trace of bromophenol blue for 30 min. The reduced IPG strips were subjected to alkylation for 20 min in 15 ml of the same buffer, except the TBP was replaced by 135 mM iodoacetamide. The equilibration steps were performed at room temperature with gentle shaking. The second dimension of electrophoresis was then performed by using a PROTEAN XL electrophoresis unit (Bio-Rad) according to the manufacturer's instruction manual. Briefly, the equilibrated IPG strips were loaded on 4% stacking gels and 12% separating gels with the Precision Plus broad-range protein standard (Bio-Rad) and covered with low-melting-point agarose gel. Electrophoresis was conducted at 50 V for 16 h, and the gels were stained with colloidal Coomassie blue G-250 (37). The 2-DE gels were digitalized and analyzed using a GS-800 calibrated densitometer and PDQuest (version 7.2.0) software (Bio-Rad).
MALDI-TOF mass spectrometry and identification of proteins.
Protein spots of interest were manually excised from the gels, sliced into 1-by1-mm pieces, and washed three times, 5 min each, with shaking in milliQ water. The gel pieces were destained with a solution containing 50 mM NH4HCO3 in 50% (vol/vol) acetonitrile by mixing on a vortex apparatus until the gel pieces were transparent. The gel pieces were dehydrated with 100 μl of 100% acetonitrile for 5 min with occasional mixing, followed by reduction with 100 μl of 10 mM dithiothreitol in 100 mM NH4HCO3 at 50°C for 30 min. The gel pieces were dehydrated with 100 μl of 100% acetonitrile again and alkylated with 100 μl of 55 mM iodoacetamide in 100 mM NH4HCO3 for 60 min at 22°C in the dark. The gel particles were washed with 200 μl of 100 mM NH4HCO3 for 15 min with occasional mixing on a vortex apparatus, dehydrated with 100% acetonitrile, and dried using a Speedvac for 20 min. The gel pieces were rehydrated in ∼20 ml of trypsin solution (20 μg·ml−1; Sigma) in 36 mM NH4HCO3-8.1% acetonitrile (vol/vol) for 1 h at 22°C, followed by removal of the excess of trypsin solution. The rehydrated gel pieces were covered with 50 μl of 50 mM NH4HCO3 and incubated at 37°C for 16 to 18 h. The peptide extraction was performed in three sequential extractions, the first with 50 μl milliQ water and the following two with 5% (vol/vol) trifluoroacetic acid in 50% (vol/vol) acetonitrile. The supernatants from each extraction were pooled in a 0.65-ml siliconized tube. The combined supernatant were concentrated to 10 to ∼15 μl using the Speed Vac and desalted using a C18 ZipTip (Millipore) pipette tip as described in the instruction manual. The desalted peptides were submitted to the Biological Mass Spectrometry Facility of the University of Guelph for matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry. Peptide spectra were acquired using a Bruker Reflex III MALDI-TOF spectrometer in reflectron detection mode with external calibration. Peptide mass profiles were analyzed by using the MS-FIT ProteinProspector program (UCSF Mass Spectrometry [prospector.ucsf.edu/]) against the open reading frame (ORF) FASTA file of the S85 genome.
Samples for tandem mass spectrometry to obtain peptide masses and internal amino acid sequences were submitted to the Advanced Protein Technology Centre of the Hospital for Sick Children in Toronto (www.sickkids.on.ca/APTC). Peptide fragments were identified by reference to the F. succinogenes genome.
Binding of solubilized OM proteins to cellulose.
OM proteins were solubilized in PIPES buffer, pH 6.7, containing 0.2% (wt/vol) amidosulfobetaine 14 at 22°C for 2 h. Any insoluble material was removed by centrifugation at 100,000 × g for 20 min. The solubilized OM proteins (approximately 0.2 mg) of S85 grown on either cellulose or glucose and Ad1 and Ad4 grown on glucose were mixed with either 40 mg of autoclaved Avicel cellulose PH 105 or 1% (wt/vol) of ASC for 30 min at 22°C in 1 ml of 20 mM Tris-HCl, pH 7.5, with end-over-end rotation at 8 rpm. Following centrifugation at 5,000 × g for 2 min, the sedimented cellulose was subjected to two sequential washes, first with 1 ml of 20 mM Tris-HCl, pH 7.5, and second with 20 mM Tris-HCl, pH 7.5, containing 1 M NaCl. The proteins bound to cellulose were eluted with 50 μl of the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer at 100°C for 5 min. The eluates were separated on an 8-to-20%-linear-gradient SDS-PAGE, followed by staining with colloidal Coomassie blue (37). Protein bands were excised and subjected to in-gel trypsin digestion and MALDI-TOF mass spectrometry for identification as described previously.
Cloning, overexpression, and purification of chloride-stimulated cellobiosidase (ClCBase) (Cel10A).
The gene coding for the ClCBase protein was cloned into the expression vector pQE-80L (QIAGEN) and the recombinant fusion protein, and rcClCBase was over-expressed in Escherichia coli Rosetta Gami(DE3) and purified as described by Qi et al. (40).
Purification of the 180-kDa CBP.
The 180-kDa cellulose-binding protein (CBP) was purified from nonsedimentable extracellular culture fluid of strain S85 cells grown on glucose as the carbohydrate source after concentration using a PM-10 membrane. The medium components were exchanged to 20 mM triethanolamine (TEA) buffer (pH 7.0) by concentration and dilution four times with the buffer, and the sample was applied to a DEAE Sepharose column equilibrated in the same buffer and bound proteins eluted with 20 mM TEA buffer containing a 0 to 1 M NaCl gradient. Fractions containing the 180-kDa protein were then applied to a phenyl Sepharose column in 20 mM TEA buffer with an (NH4)2SO4 gradient from 1 to 0 M. Fractions containing the CBP were further purified by chromatofocusing. The chromatofocusing step employed a Polybuffer exchanger 94 (Amersham Pharmacia Biotech) in a column (1 by 20 cm) equilibrated with 25 mM piperazine-HCl buffer (pH 5.5) containing 1 mM of EDTA. A 10-fold dilution of Polybuffer (pH 4.0) (Amersham Pharmacia Biotech) served as the eluent. The presence of protein with a molecular mass of 180 kDa was ascertained by SDS-PAGE analysis.
Protein estimation.
The protein concentration was determined as described by Bradford (3) using bovine serum albumin (Sigma) as the standard, except for solutions containing urea, where the bincinchoninic acid method was used (45).
Production and purification of antibodies.
rcClCBase purified from inclusion bodies by using an immobilized metal affinity column under denaturing conditions was further purified to homogeneity by using 10% SDS-PAGE, nonfixing KCl staining (36), and gel elution at 5 mA for 16 h using an electroeluter (Bio-Rad) as described in the instruction manual. The purified rcClCBase, which was more than 98% pure as determined by SDS-PAGE with colloidal Coomassie blue staining, was used for production of antibodies.
A rabbit was immunized with rcClCBase by intramuscular injections of 0.5 ml of a mixture of protein (2 mg/ml) and Freund's incomplete adjuvant (50:50) at each of two sites. A second set of injections was performed at 4 weeks, and the rabbit was bled at 7 weeks by saline perfusion. The blood was incubated without agitation for 2 h at 37°C and left at 4°C overnight, followed by centrifugation at 10,000 × g for 10 min at 4°C. The serum was removed, supplemented with 0.02% sodium azide, and stored at −80°C. The antiserum was purified to obtain the monospecific anti-rcClCBase antibody as described previously (18).
The polyclonal antiserum against the native ClCBase (18) was also used in this study.
Western immunoblots.
Western immunoblotting was performed as previously described (18) except that monospecific anti-rcClCBase antibodies (1:150 dilution) were used as the primary antibody and goat anti-rabbit immunoglobulin G alkaline phosphatase-conjugated antibodies (1:30,000 dilution; Sigma-Aldrich Canada) were used as the secondary antibody. Detection was performed as described by McGavin et al. (35). The Quantity One 1-D analysis software (version 4.4.1; Bio-Rad) was used for quantification.
Nucleotide sequence accession numbers.
The nucleotide sequences of the identified genes have been deposited in the GenBank database under accession numbers EU055570 to EU055603.
RESULTS
Growth on glucose, ASC, and Avicel cellulose.
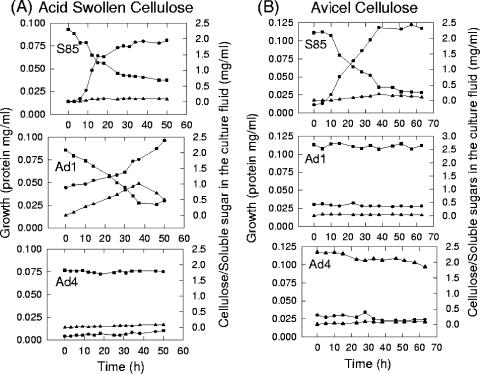
Strain S85 and its adhesion-defective mutants, Ad1 and Ad4, exhibited similar growth rates on glucose (0.34 h−1). Strain S85 grew on both acid-swollen and Avicel cellulose, with growth rates of 0.20 and 0.11 h−1, respectively, and there were corresponding decreases in the concentration of cellulose with no accumulation of soluble sugars (Fig. 1). Mutant Ad1 grew on ASC at a rate of 0.02 h−1, and soluble sugar accumulated during growth until 36 h, after which the concentration began declining; however, the bacterium did not grow on crystalline cellulose. Mutant Ad4 did not grow on either acid-swollen or Avicel cellulose, and there was no accumulation of soluble sugar (Fig. 1).
FIG. 1.
Growth of F. succinogenes S85 and its adhesion-defective mutants, Ad1 and Ad4. on different forms of cellulose. (A) ASC, 0.2% (wt/vol). (B) Avicel cellulose, 0.3% (wt/vol). Symbols: •, protein content; ▪, amount of residual cellulose; ▴, soluble carbohydrates.
Adhesion to different forms of cellulose.
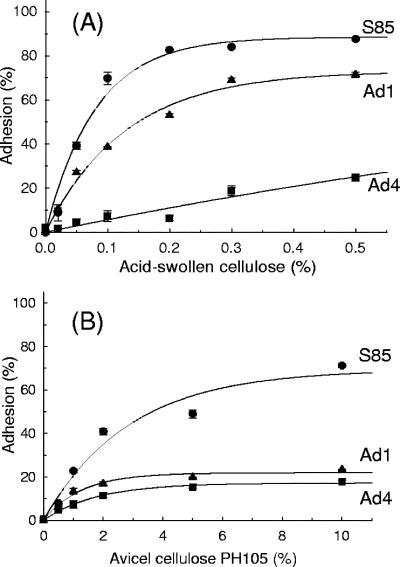
The binding of cells to cellulose was assessed to determine whether there was a relationship between growth on cellulose and adhesion of cells to cellulose. Late-exponential-phase glucose-grown S85 cells bound efficiently to both ASC and Avicel cellulose, with Kd values of 0.05 and 2.0%, respectively (Fig. 2A and B). Ad1 grown on glucose bound to ASC with a Kd of 0.08% but did not bind to Avicel cellulose. Ad4 did not bind to either acid-swollen or Avicel cellulose (Fig. 2).
FIG. 2.
Binding of S85, Ad1, and Ad4 cells to different concentrations of ASC (A) and to Avicel cellulose PH105 (B). Adhesion was determined by the turbidity adhesion assay (n = 2).
Identification of CBPs in OMs.
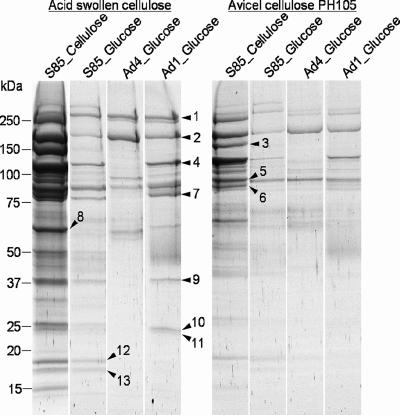
To determine whether the difference in binding to cellulose between S85 and the mutants was due to different CBPs, OM from strain S85 and OM from mutant strains were solubilized in detergent and the profiles of proteins bound to either acid-swollen or Avicel crystalline cellulose were compared by using one-dimensional gradient (8 to 20%) SDS-PAGE. As shown in Fig. 3, the overall binding capacity of solubilized OM proteins for ASC was much higher than that for Avicel crystalline cellulose.
FIG. 3.
Comparative analysis of solubilized OM proteins binding to either acid-swollen (left panel) or Avicel (right panel) cellulose. Proteins bound to cellulose were eluted with SDS sample buffer and separated by SDS-PAGE. Proteins excised from the SDS-PAGE gels and analyzed by MALDI-TOF mass spectrometry are indicated by arrowheads and numbers.
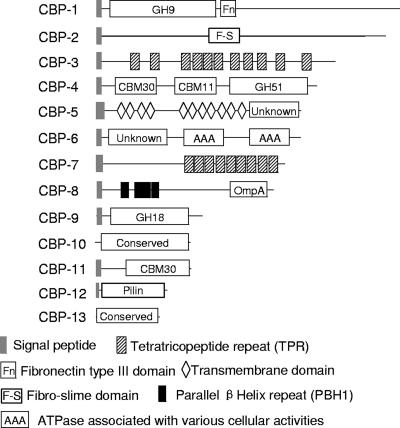
In all, 13 CBPs were identified. Comparing the CBP profile of S85 OM with those for Ad1 and Ad4, the cellulose binding proteins CBP-1, CBP-2, and CBP-5 were shown to be present in both S85 and the mutant strains. CBP-1 (Fig. 3 and 4; Table 1) possessed an N-terminal catalytic domain belonging to glycoside hydrolase (GH) family 9 and showed about 35 to 37% similarity to N-terminal catalytic domains of endoglucanases of Bacillus sp. KSM-522, Bacillus licheniformis, and Clostridium cellulovorans (29, 44, 48). It also contained a fibronectin type III module (23) immediately after the catalytic domain. CBP-2 was identified as the 180-kDa cellulose-binding protein described by Gong et al. (11). The identity of the 180-kDa protein from N-terminal and internal amino acid sequence analysis is presented in Fig. 5A. CBP-2 was annotated as the Fibro-slime domain protein, named after a domain in the middle of the protein, residues 556 to 649. The Fibro-slime domain is present in nine other putative proteins from F. succinogenes, and at least one putative protein in Fibrobacter intestinalis (41), and several hypothetical proteins from the slime mold Dictyostelium discoideum (7) as well as some proteobacteria. Interestingly, these proteins had no similarity in regions other than the Fibro-slime domain. CBP-5 lacked similarity to proteins from other organisms.
FIG. 4.
Domain structures of cellulose-binding proteins from Fig. 3.
TABLE 1.
Identification of CBPs located in OM fraction of F. succinogenes S85
| Band | FSU no.a | Protein name or description | No. matched (%)b | Coverage (%) | Mw_t/Mw_ec | pI_t/pI_t_ce | Feature |
|---|---|---|---|---|---|---|---|
| 1 | 809 | GH family 9 | 5/10 (50) | 7.0 | 233.0/250 | 4.97/9.63 | Present in all strains |
| 2 | 2502 | Fibro-slime domain protein | 46.0d | 169.4/180.0 | 5.00/10.17 | Present in all strains | |
| 3 | 2398 | TPR domain protein | 7/9 (77) | 12.0 | 146.7/150 | 8.5/10.00 | Only present in S85 grown on cellulose |
| 4 | 382 | Cellulose-binding protein, CelF | 5/16 (31) | 8.0 | 118.0/120 | 7.81/9.8 | Absent from Ad4 |
| 5 | 718 | Membrane protein | 6/18 (33) | 11.0 | 92.4/92 | 9.01/10.01 | Present in all strains |
| 6 | 1516 | AAA family protein | 7/18 (38) | 11.0 | 89.5/90 | 6.43/6.77 | Present only in S85 grown on cellulose |
| 7 | 2397 | TPR domain protein | 10/26 (38) | 25.0 | 83.8/80 | 5.5/5.1 | Absent from Ad4 |
| 8 | 2396 | OmpA protein | 7/10 (70) | 19.0 | 53.0/55 | 4.86/10.15 | Present only in S85 grown on cellulose |
| 9 | 2012 | GH family 18 | 7/16 (43) | 34.0 | 37.5/37 | 4.4/8.05 | Absent from Ad4 |
| 10 | 1231 | Hypothetical protein | 5/11 (45) | 18.0 | 31.5/27 | 7.60/5.35 | Present only in S85 grown on cellulose |
| 11 | 2007 | Cellulose binding protein | 8/21 (38) | 32.0 | 29.2/27 | 6.46/8.26 | Absent from Ad4 |
| 12 | 2567 | Type IV pilin protein | 62.0d | 17.5/18 | 5.43/4.23 | Absent from mutants | |
| 13 | 794 | Conserved hypothetical protein | 4/8 (50) | 33.0 | 16.1/17 | 7.64/5.72 | Absent from mutants |
F. succinogenes gene number.
No. of peptides matched/total (%).
Mw_t and Mw_e indicate theoretical and experimental molecular masses, respectively.
Coverage by tandem mass spectrometry.
pI_t and pI_t_c indicate theoretical isoelectric points of the proteins and their C-terminal 60 amino acids, respectively.
FIG. 5.
Amino acid sequence of CBP-2 (180-kDa CBP) (A) or ClCBase (Cel10A) (B). Peptides determined by MALDI-TOF mass spectrometry are in bold lettering, and peptides sequenced by tandem mass spectrometry or nanoelectrospray mass spectrometry are underlined. Peptides determined by Edman degradation are in bold and italic lettering. The Fibro-slime domain of the 180-kDa CBP is doubly underlined.
Comparing the CBP profile of S85 grown on cellulose with that of S85 grown on glucose, four proteins, CBP-3, -6, -8, and -10, were shown to be present only in F. succinogenes S85 grown on cellulose. CBP-3 (FSU2398) was a tetratricopeptide repeat (TPR) domain protein containing 11 repeats. CBP-6 contained an N-terminal domain of unknown function and two ATPase (AAA) family domains at the C terminus (38). CBP-8 had multiple domains, including four parallel β-helix repeats (19) and a C-terminal OmpA-like domain. CBP-10 was a protein of unknown function.
CBP-4, CBP-7, CBP-9, CBP-10, and CBP-11 were present in the OMs of Ad1 and S85 but absent from Ad4 (Fig. 3 and 4; Table 1). CBP-4 was annotated to be the cellulose-binding protein CelF (previously named EG2), which was previously characterized (32, 33). CBP-7 coded by FSU2397 was annotated as a TPR domain protein with nine consecutive TPRs (27). CBP-9 was annotated as a GH belonging to family 18. CBP-11 was the same protein as protein 11 on the two-dimensional gel (Fig. 6), with high similarity to carbohydrate binding module (CBM) 30.
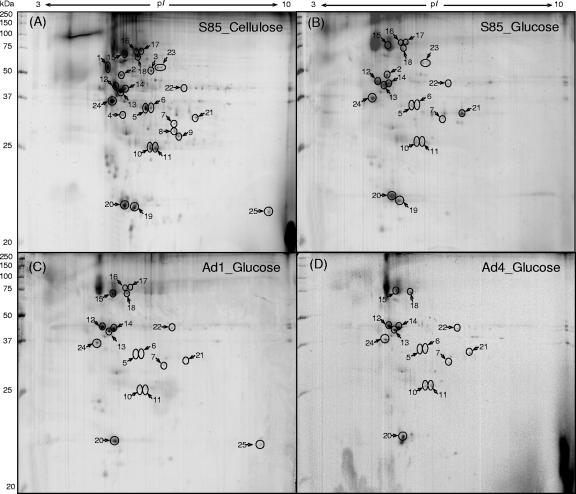
FIG. 6.
2-DE of OM proteins from F. succinogenes S85 grown on Avicel cellulose PH105 (A) or glucose (B) and its adhesion-defective mutant strains, Ad1 (C) and Ad4 (D). The proteins identified are indicated by circles and the numbers recorded in Table 2.
CBP-12 and CBP-13 appeared to be present only in F. succinogenes S85. CBP-12 was identified as a type IV pilin protein with a mass of 23 kDa (Fig. 3 and 4; Table 1), with a very short leader peptide (MKKQG) and a glycine residue at the signal cleavage site, followed by a methylated and hydrophobic N-terminal phenylalanine residue (N-Met-Phe) and by about 24 highly conserved hydrophobic residues. The C-terminal region did not show substantial similarity to pilin proteins from other bacteria. A BLAST search of the pilin protein FSU2567 against the S85 genome indicated that a homologous gene (FSU02593) exists in the genome with 42% identity, 57% similarity, and 10% gaps. CBP-13 was annotated as a conserved hypothetical protein. These two proteins may be important for binding to crystalline cellulose.
Major OM proteins with potential roles in cellulose digestion identified by 2-DE.
Nineteen proteins were identified in OM from glucose-grown S85 (Fig. 6B). Seventeen and fourteen of them were also identified from AD1 and AD4, respectively (Fig. 6C and D). The most distinct proteins absent from mutants Ad1 and Ad4 corresponded to proteins 2, 19, and 23. Protein 2 was identified as an OM efflux protein which was known to form trimeric channels that allow export of a variety of substrates in gram-negative bacteria (20). Homologs of this protein were found in several CFB groups and green sulfur bacteria. Protein 19 was identified as a type IV pilin, which was also identified as CBP-12. Protein 23 was annotated as an endo-1,4-β-xylanase with a mass of 65 kDa (FSU0257), but we have shown that FSU0257 is the previously characterized ClCBase (see below) (Fig. 5) (17).
OMs of S85 grown on cellulose subjected to 2-DE contained a greater number of detectable proteins than glucose-grown cells, especially in the 25- to 75-kDa and pI-4-to-7 range, compared to OM of S85 grown on glucose. Of OM proteins identified, 15 were shown to be up-regulated by growth of S85 on cellulose, including ClCBase (Table 2; Fig. 6A and B).
TABLE 2.
List of OM proteins of S85 identified by 2-DE and MALDI-TOF mass spectrometrya
| Protein no. | FSU no. | Protein name | Presence of protein inb:
|
Mw_t | Mw_e | pI_t | pI_e | Matched (%)c | % Cov. | PSort predictiond | SignalP predictiond | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S85 G | AD1 G | AD4 G | S85 C | |||||||||||
| 1 | 864 | Lipoprotein | N | N | N | Y | 46.3 | 50-60 | 4.6 | 4.6 | 5/15 (33) | 12.0 | OM | Y |
| 2 | 700 | Outer membrane efflux protein precursor | Y | N | N | Y | 52.3 | 50 | 5.3 | 5.1 | 6/26 (23) | 17.0 | OM | Y |
| 3 | 222 | GGDEF domain protein | N | N | N | Y | 49.8 | 50 | 5.9 | 6.0 | 5/20 (25) | 14.0 | CP | N |
| 4 | 2401 | Hypothetical protein | N | N | N | Y | 28.2 | 30 | 4.6 | 5.0 | 8/28 (28) | 45.0 | OM | Y |
| 5 | 2078 | OmpA family protein | Y | Y | Y | Y | 72.1 | 34 | 4.7 | 4.7 | 10/22 (45) | 28.0 | OM | Y |
| 6 | 2096 | Lipoprotein, | Y | Y | Y | Y | 72.5 | 34 | 4.5 | 4.7 | 9/30 (30) | 21.0 | OM | Y |
| 7 | 2388 | Transcriptional regulator, LysR family | Y | Y | Y | Y | 34.2 | 35 | 7.6 | 7.0 | 5/31 (16) | 24.0 | CP | N |
| 8 | 474 | Hypothetical protein | N | N | N | Y | 30.6 | 32 | 5.2 | 6.0 | 10/57 (17) | 50.0 | IM | Y |
| 9 | 2016 | P3 domain protein | N | N | N | Y | 31.0 | 30 | 7.1 | 7.2 | 7/29 (24) | 27.0 | IM | Y |
| 10 | 2450 | Lipoprotein, putative | Y | Y | Y | Y | 26.0 | 25 | 5.2 | 5.5 | 6/43 (13) | 29.0 | OM | Y |
| 11 | 2007 | Cellulose-binding protein | Y | Y | Y | Y | 29.2 | 25 | 6.5 | 6.0 | 10/46 (21) | 51.0 | IM | Y |
| 12 | 230 | Hypothetical protein | Y | Y | Y | Y | 43.0 | 43 | 4.6 | 4.6 | 13/38 (34) | 45.0 | CP | Y |
| 13 | 2404 | Hypothetical protein | Y | Y | Y | Y | 46.1 | 42 | 4.7 | 4.7 | 10/35 (28) | 27.0 | OM | Y |
| 14 | 1029 | Hypothetical protein | Y | Y | Y | Y | 42.2 | 43 | 4.8 | 5.0 | 20/42 (47) | 76.0 | OM | Y |
| 15 | 2078 | OmpA family protein | Y | Y | Y | Y | 72.1 | 74 | 4.7 | 4.7 | 11/24 (45) | 26.0 | OM | Y |
| 16 | 2009 | Hypothetical protein | Y | Y | N | Y | 76.4 | 74 | 5.0 | 5.2 | 5/21 (23) | 16.0 | OM | Y |
| 17 | 2010 | Hypothetical protein | Y | Y | N | Y | 75.5 | 74 | 5.5 | 5.5 | 7/12 (58) | 23.0 | OM | Y |
| 18 | 2285 | conserved hypothetical protein | Y | Y | Y | Y | 77.1 | 70 | 5.7 | 5.7 | 15/53 (28) | 41.0 | CP | N |
| 19 | 2567 | Type IV pilin | Y | N | N | Y | 17.5 | 23 | 5.4 | 5.0 | 9/24 (37) | 85.0 | OM | Y |
| 20 | 609 | Lipoprotein, putative | Y | Y | Y | Y | 18.2 | 22 | 4.9 | 5.0 | 8/26 (30) | 49.0 | OM | Y |
| 21 | 2807 | Hypothetical protein | Y | Y | Y | Y | 36.4 | 35 | 7.1 | 7.3 | 5/9 (55) | 27.0 | OM | Y |
| 22 | 2581 | Hypothetical protein | Y | Y | Y | Y | 27.1 | 40 | 5.9 | 6.0 | 4/24 (16) | 22.0 | OM | Y |
| 23 | 257 | Endo-1,4-β-xylanase z (ClCBase; Cel10A) | Y | N | N | Y | 69.8 | 65 | 9.0 | 6.5 | 9/16 (56) | 20.0 | OM | Y |
| 24 | 3156 | Conserved hypothetical protein | Y | Y | Y | Y | 39.6 | 37 | 5.2 | 5.0 | 5/25 (20) | 24.0 | CP | N |
| 25 | 1783 | Hypothetical protein | N | Y | N | Y | 17.9 | 20 | 7.9 | 8.0 | 6/22 (27) | 55.0 | OM | Y |
Abbreviations: G, OM samples prepared from Glucose-grown cells; C, OM samples prepared from cellulose-grown cells; Y, present in the sample; N, not detected in the sample; IM, inner membrane; CP, cytoplasm; FSU, Fibrobacter succinogenes (see footnote a of Table 1); Mw_t and Mw_e, theoretical and experimental molecular masses, respectively; p_t and pI_e, theoretical and experimental isoelectric points of the protein, respectively; Y/N, the signal peptide is present/absent; Cov., coverage.
Underling indicates that the protein was induced in the presence of cellulose.
No. of peptides matched/total (%).
Predicted by SignalP, TatP, and LipoP programs at http://www.cbs.dtu.dk/services/.
Proteins 1, 6, 10, and 20 were annotated as putative lipoproteins, since the genes possess a typical signal sequence of lipoproteins (50). BLAST searches showed that protein 1 (FSU0864) contains a putative tyrosine phosphatase domain (8) at the N terminus. Protein 6 (FSU2096) contains two copies of a conserved repeat (residues 270 to 484 and 511 to 697) that was present in as many as 60 putative proteins (with sequence identities ranging from 20% to 49%) in the genome of F. succinogenes and was also found from other bacteria from the green sulfur bacteria group, Chlorobium limicola (NZ_AAHJ00000000) and Prosthecochloris aestuarii (NZ_AAIJ00000000), as well as the Cytophaga-Flavobacterium-Bacteroides group, such as Bacteroides fragilis (26). This set of proteins was known as one of the three “Fibrobacter succinogenes major paralogous domains” (M. Morrison et al., unpublished), but none of them have been characterized so far. BLAST searches of proteins 10 and 20 revealed no homologous proteins in other organisms.
Proteins 5 and 15 were identified as the same OmpA protein (FSU2078). Protein 5 is the N-terminal 336 amino acids of the 700-amino-acid protein and is presumed to be a proteolytic degradation product. FSU2078 is composed of an N-terminal unknown domain, with eight thrombospondin type 3 repeats (28) and an OmpA-like domain (9) at the C terminus. Four other putative proteins (FSU0180, FSU1003, FSU0255, and FSU3077) in F. succinogenes have high sequence similarity and the same domain organization as this protein.
Protein 11 was identified as a CBP with high similarity (49%) to the N-terminal CBM family 30 of endoglucanase F (Cel51A/FSU0382) of S85.
Six of 15 proteins (proteins 4, 8, 9, 16, 17, and 18) shown to be induced by growth on cellulose were annotated as unknown-function proteins. Proteins 4, 8, 9, and 17 had no homology to proteins from other organisms, although protein 9 (FSU2016) exhibited significant similarity to the P3 protein, which was previously sequenced by Malburg et al. (32). Protein 16 showed weak similarity to a hypothetical protein (YP_677889.1), from Cytophaga hutchinsonii, while protein 18 showed high similarity to hypothetical proteins from delta/epsilon subdivisions of proteobacteria, such as NP_860592.1 in Helicobacter hepaticus (47).
Identification of the ClCBase.
The native ClCBase was purified from the extracellular culture fluid of F. succinogenes S85 grown on cellulose as described by Huang et al. (17), subjected to in-gel trypsin digestion and both MALDI-TOF mass spectrometry and tandem mass spectrometry to obtain peptide masses and internal amino acid sequences, and identified as Cel10A (FSU0257) by reference to the F. succinogenes genome (www.tigr.org) (Fig. 5B). BLAST searches using the Pfam search server showed that the ClCBase had a modular structure composed of an N-terminal domain which was distantly related to family 4 CBM (amino acid residues 27 to ∼174) and GH10 family C-terminal catalytic domains (207 to ∼508).
Regulation of the ClCBase in F. succinogenes S85.
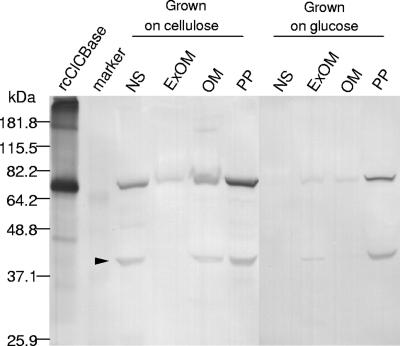
Western blotting documented that the ClCBase was localized in the OM as well as both the extracellular culture fluid and the periplasm of F. succinogenes S85 grown on cellulose (Fig. 7). When grown on glucose, this enzyme was mainly located in periplasmic fractions and less in other fractions (Fig. 7). The monospecific anti-rcClCBase antibody did not exhibit cross-reactivity with other proteins in any cell fraction, as described previously for antibodies against the glycosylated enzyme isolated from S85 (18), with the exception of a 39-kDa proteolytic product of ClCBase (Fig. 7).
FIG. 7.
Immunoblot analysis of a fractionated culture of F. succinogenes S85 grown on either cellulose or glucose using monospecific antirecombinant ClCBase antibody. Abbreviations: NS, nonsedimentable extracellular culture fluid; ExOM, extracellular outer membrane; OM, outer membrane; PP, periplasmic fraction. ExOM and PP were prepared as previously described (13). The arrowhead indicates a proteolytic degradation product.
DISCUSSION
In a previous study, Huang and Forsberg (16) observed that F. succinogenes initiated growth on microcrystalline cellulose without a lag whether inoculated from a glucose, cellobiose, or cellulose culture. Not unexpectedly, Gong and Forsberg (12) demonstrated that S85 cells grown on glucose adhered tightly to crystalline cellulose when washed and mixed with that substrate in the absence or presence of a metabolic inhibitor, indicating that cellulose-binding proteins were present in glucose-grown cells and the cells were primed for growth on cellulose. In contrast to S85 cells that bound to both Avicel cellulose and ASC, neither Ad1 or Ad4 bound to Avicel cellulose, although Ad1 bound to ASC with a relatively higher binding constant (Kd) than the S85 strain and Ad4 did not. In this study we have examined the proteins in OMs from mutants Ad1 and Ad4 isolated by Gong and Forsberg (12) that were unable to bind to crystalline cellulose. Considering that ASC has generally been described as having more surface area than crystalline cellulose (46), the higher affinity of the bacterium to ASC may be due, at least in part, to the increased surface area. However, this explanation does not account for the lack of binding of proteins from Ad1 and Ad4 to crystalline cellulose and the differential binding of Ad1 and Ad4 to ASC as shown in Fig. 3. These observations provided the opportunity to identify CBPs that specifically bind to Avicel cellulose or ASC.
The proteins CBP-12 and CBP-13 from strain S85 OM that bound to crystalline cellulose were absent from Ad1 and Ad4. Interestingly, CBP-12, which was annotated as a type IV pilin protein, was also found to be missing from Ad1 and Ad4 by the 2-DE analysis (Fig. 6). Pili have been reported to be involved in adhesion of gram-negative bacteria (39). Recently a cellulose adhesion-defective mutant (D5), derived from the gram-positive bacterium Ruminococcus albus, was shown to lack a type IV pilin (termed the GP25). The authors concluded that the type IV pili mediated adhesion of R. albus strain 20 to cellulose (42). Therefore, a role of pili in binding of S85 to cellulose is plausible.
Five proteins, including CBP-4, -7, -9, -10 and -11, of Ad1 OM bound to ASC, whereas they were absent from Ad4, which suggests that they may have specific roles in binding of cells to ASC. CBP-4, which was annotated as CelF (formerly EG2), exhibits a higher activity against ASC than crystalline cellulose (32). In this study, CelF was shown to have a greater affinity for ASC than for crystalline cellulose (Fig. 3), which is consistent with an earlier study by McGavin and Forsberg (34). Therefore, it seems reasonable that CelF may be one of the components involved in adhesion of F. succinogenes S85 to amorphous ASC. CBP-7 was identified as a protein containing nine consecutive TPR repeats at the C terminus. The TPR is a 34-amino-acid repeated motif (10) that frequently forms tandem arrays (27). Individual TPRs adopt a helix-turn-helix conformation, and tandem TPRs fold into concertina-like helical arrays, forming a continuous peptide-binding groove (5). TPRs have been discovered in various organisms ranging from bacteria to the human, but the role of the repeat in bacteria has not been resolved (27). It may be noted that 10 hypothetical proteins containing TPR motifs are present in the S85 genome. CBP-9 was annotated as a GH belonging to family 18, in which chitinases, chitodextrinase, and endo-β-N-acetylglucosaminidase are mainly categorized (http://afmb.cnrs-mrs.fr/CAZY/), but this protein did not contain a known CBM. CBP-11 was also identified from the 2-DE of the OM proteins (Spot 11 in Table 2), which was up-regulated by growth on cellulose and was absent from Ad4. It contained a CBM belonging to family 30. To the present, only four proteins including the CBM of CelF of S85 (32) were categorized in this CBM family (afmb.cnrs-mrs.fr/CAZY). Arai et al. (1) suggested that the family 30 CBM of C. thermocellum is very important for mediating the binding of the enzyme CelJ to various substrates, including Avicel and amorphous cellulose, and for the correct tertiary structure of the intact protein. In addition, recently the crystal structure of CBM30 was determined by Horiguchi et al. (not published, but the information of the structure is available with the identifier 1WMX in the Protein Data Bank [http://www.rcsb.org]), showing a galactose-binding domain-like fold composed of eight β-strands.
Previous studies identified four CBPs from the OM fraction of glucose-grown S85 (11) with the molecular masses of 120, 180, 220, and 240 kDa, respectively. The 120-kDa CBP was identified as endoglucanase F, which corresponded to CBP-4 in the present study. The 180-kDa CBP was characterized and thought to have an important role in adhesion. The N-terminal and internal amino acid sequences of the 180-kDa CBP were determined to be encoded by gene FSU2502 by a combination of Edman degradation and mass spectrometry (Fig. 5A) in conjunction with a BLAST search of the genome of F. succinogenes. FSU 2502 is an ORF of 4,650 bp that encodes a protein of 1,549 amino acids, which corresponds to CBP-2. The molecular mass of CBP-1 seems to correspond to the 220- or 240-kDa CBP in Gong et al.'s study (11). This protein possessed an N-terminal catalytic domain belonging to GH family 9; thus, it appeared to be partially involved both in adhesion to cellulose and in the degradation of cellulose.
CBP-1, CBP-2, and CBP-5 were found to exist in both the S85 and mutant strains and to be induced by growth of S85 on cellulose. Therefore, the inability of adhesion-defective mutants Ad4 and Ad1 to adhere to and degrade cellulose suggests that these proteins have limited roles in adhesion of S85 to cellulose and degradation of cellulose. In the case of CBP-2, anti-CBP-2 antibodies blocked binding of S85 to cellulose; however, the antibodies were specific for glycosylation (11). Therefore, it is possible that the CBM is buried in the OM and is unavailable for binding to cellulose unless the membrane is dissociated by detergent. The lack of involvement of binding of CBP-1 and CBP-5 may also be explained by their location in the OM.
To gain further insight into the role of OM proteins in the mechanism of cellulose digestion by F. succinogenes strain S85 and the mutant, OM proteins were separated by 2-DE and identified. The most distinct changes in predominant proteins were the absence from both mutant strains Ad1 and Ad4 of a type IV pilin protein (protein 19; CBP-12) and the Cl-stimulated cellobiosidase (protein 23; Table 2).
The Cl-stimulated cellobiosidase was previously characterized by Huang et al. (17). When up-regulated, a greater proportion of the enzyme was associated with the OM, although it was mainly periplasmic in glucose-grown cells. Since it has a family 4 CBM and has been shown to bind to cellulose (17), it may have a role in adhesion of cells to cellulose; however, it was not among the OM CBPs (Fig. 3). This may be due to its primary location in the periplasm even when S85 is grown on cellulose.
Proteins that were up-regulated in the OM of cellulose-grown S85 cells and down-regulated or absent from glucose-grown cells are candidates for major roles in cellulose digestion. Twenty-five proteins were detected in OMs of cellulose-grown S85, while 19 proteins were found in glucose-grown cells. The genes coding for 16 OM proteins that appeared to be induced by growth of S85 on cellulose were identified in this study, but the roles of the encoded proteins in adhesion to and/or degradation of cellulose remain to be elucidated.
In this proteomic study, 14 proteins were identified as unknown function proteins. This finding is not surprising, since the functions of approximately 50% of ORFs of the S85 genome are unknown and are annotated as hypothetical (or conserved hypothetical) proteins. Nine of the unknown function proteins identified in this study may be unique to S85; in addition, considering that six of those hypothetical proteins were shown to be up-regulated by growth on cellulose, they would appear to be new families of proteins involved in adhesion to cellulose and degradation of cellulose. Learning the functional roles of these proteins in addition to the putative CBPs will substantially further our knowledge of the unique mechanism of cellulose digestion by F. succinogenes.
Acknowledgments
This research was supported by the Initiative for Future Agriculture and Food Systems grant no. 2000-52100-9618 from USDA-CSREES to the North American Consortium for Genomics of Fibrolytic Ruminal Bacteria, led by Mark Morrison, Department of Animal Science, Ohio State University (currently with CSIRO Queensland Biosciences Precinct, St. Lucia, Australia), and by the Natural Science and Engineering Research Council of Canada. The Fibrobacter succinogenes genome project was undertaken at The Institute for Genomic Research and supported by the USDA-CSREES funds.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Arai, T., R. Araki, A. Tanaka, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bera-Maillet, C., Y. Ribot, and E. Forano. 2004. Fiber-degrading systems of different strains of the genus Fibrobacter. Appl. Environ. Microbiol. 70:2172-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram amounts of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Chevallet, M., V. Santoni, A. Poinas, D. Rouquié, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19:1901-1909. [DOI] [PubMed] [Google Scholar]
- 5.Das, A. K., P. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubois, M., K. A. Gilles, K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 7.Eichinger, L., et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, E. H., H. Charbonneau, and N. K. Tonks. 1991. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science 253:401-406. [DOI] [PubMed] [Google Scholar]
- 9.Freudl, R., M. Klose, and U. Henning. 1990. Export and sorting of the Escherichia coli outer membrane protein OmpA. J. Bioenerg. Biomembr. 22:441-449. [DOI] [PubMed] [Google Scholar]
- 10.Goebl, M., and M. Yanagida. 1991. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16:173-177. [DOI] [PubMed] [Google Scholar]
- 11.Gong, J., E. E. Egbosimba, and C. W. Forsberg. 1996. Cellulose-binding proteins of Fibrobacter succinogenes and the possible role of a 180 kDa cellulose-binding glycoprotein in adhesion to cellulose. Can. J. Microbiol. 42:453-460. [Google Scholar]
- 12.Gong, J., and C. W. Forsberg. 1989. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 55:3039-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, J., and C. W. Forsberg. 1993. Separation of outer and cytoplasmic membranes of Fibrobacter succinogenes and membrane and glycogen granule locations of glycanase and cellobiase. J. Bacteriol. 175:6810-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert, B. 1999. Advances in protein solubilisation for two-dimensional electrophoresis. Electrophoresis 20:660-663. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L., and C. W. Forsberg. 1988. Purification and comparison of the periplasmic and extracellular forms of the cellodextrinase from Bacteroides succinogenes. Appl. Environ. Microbiol. 54:1488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, L., and C. W. Forsberg. 1990. Cellulose digestion and cellulase regulation and distribution in Fibrobacter succinogenes subsp. succinogenes S85. Appl. Environ. Microbiol. 56:1221-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, L., C. W. Forsberg, and D. Y. Thomas. 1988. Purification and characterization of a chloride-stimulated cellobiosidase from Bacteroides succinogenes S85. J. Bacteriol. 170:2923-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, L., M. McGavin, C. W. Forsberg, J. S. Lam, and K. J. Cheng. 1990. Antigenic nature of the chloride-stimulated cellobiosidase and other cellulases of Fibrobacter succinogenes subsp. succinogenes S85 and related fresh isolates. Appl. Environ. Microbiol. 56:1229-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins, J., O. Mayans, and R. Pickersgill. 1998. Structure and evolution of parallel β-helix proteins. J. Struct. Biol. 122:236-246. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. M., and G. M. Church. 1999. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 287:695-715. [DOI] [PubMed] [Google Scholar]
- 21.Jun, H. S., J. K. Ha, L. M. Malburg, Jr., G. A. Verrinder, and C. W. Forsberg. 2003. Characteristics of a cluster of xylanase genes in Fibrobacter succinogenes S85. Can. J. Microbiol. 49:171-180. [DOI] [PubMed] [Google Scholar]
- 22.Kam, D. K., H. S. Jun, J. Ha, G. D. Inglis, and C. W. Forsberg. 2005. Characteristics of adjacent family 6 acetylxylan esterases from Fibrobacter succinogenes and the interaction with the XynE xylanase in hydrolysis of acetylated xylan. Can. J. Microbiol. 51:821-832. [DOI] [PubMed] [Google Scholar]
- 23.Kataeva, I. A., R. D. Seidel III, A. Shah, L. T. West, X. L. Li, and L. G. Ljungdahl. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koike, S., S. Yoshitani, Y. Kobayashi, and K. Tanaka. 2003. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol. Lett. 229:23-30. [DOI] [PubMed] [Google Scholar]
- 25.Kudo, H, K. J. Cheng, and J. W. Costerton. 1987. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can. J. Microbiol. 33:267-272. [DOI] [PubMed] [Google Scholar]
- 26.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb, J. R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257-259. [DOI] [PubMed] [Google Scholar]
- 28.Lawler, J., and R. O. Hynes. 1986. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J. Cell. Biol. 103:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y., J. Zhang, Q. Liu, C. Zhang, and Q. Ma. 2004. Molecular cloning of novel cellulase genes cel9A and cel12A from Bacillus licheniformis GXN151 and synergism of their encoded polypeptides. Curr. Microbiol. 49:234-238. [DOI] [PubMed] [Google Scholar]
- 30.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577, table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malburg, L. M., A. H. Iyo, and C. W. Forsberg. 1996. A novel family 9 endoglucanase gene (celD), whose product cleaves substrates mainly to glucose, and its adjacent upstream homolog (celE) from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 62:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malburg, S. R. C., L. M. Malburg, T. Liu, A. H. Iyo, and C. W. Forsberg. 1997. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 63:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGavin, M., and C. W. Forsberg. 1988. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J. Bacteriol. 170:2914-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGavin, M., and C. W. Forsberg. 1989. Catalytic and substrate-binding domains of endoglucanase 2 from Bacteroides succinogenes. J. Bacteriol. 171:3310-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGavin, M. J., C. W. Forsberg, B. Crosby, A. W. Bell, D. Dignard, and D. Y. Thomas. 1989. Structure of the cel-3 gene from Fibrobacter succinogenes S85 and characteristics of the encoded gene product, endoglucanase 3. J. Bacteriol. 171:5587-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelles, L. P., and J. R. Bamburg. 1976. Rapid visualization of protein—dodecyl sulfate complexes in polyacrylamide gels. Anal. Biochem. 73:522-531. [DOI] [PubMed] [Google Scholar]
- 37.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 38.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 39.Pell, A. N., and P. Schofield. 1993. Microbial adhesion and degradation of plant cell walls, p. 397-423. In H. G. Jung, D. R. Buxton, R. D. Hatfield, and J. Ralph (ed.), Forage cell wall structure and digestibility. American Society of Agronomy, Inc., Madison, WI.
- 40.Qi, M., H.-S. Jun, and C. W. Forsberg. July 2007, posting date. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 41.Qi, M., K. E. Nelson, S. C. Daugherty, W. C. Nelson, I. R. Hance, M. Morrison, and C. W. Forsberg. 2005. Novel molecular features of the fibrolytic intestinal bacterium Fibrobacter intestinalis not shared with Fibrobacter succinogenes as determined by suppressive subtractive hybridization. J. Bacteriol. 187:3739-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakotoarivonina, H., G. Jubelin, M. Hebraud, B. Gaillard-Martinie, E. Forano, and P. Mosoni. 2002. Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148:1871-1880. [DOI] [PubMed] [Google Scholar]
- 43.Scott, H. W., and B. A. Dehority. 1965. Vitamin requirements of several cellulolytic bacteria. J. Bacteriol. 89:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shoseyov, O., T. Hamamoto, F. Foong, and R. H. Doi. 1990. Cloning of Clostridium cellulovorans endo-1,4-b-glucanase genes. Biochem. Biophys. Res. Commun. 169:667-672. [DOI] [PubMed] [Google Scholar]
- 45.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 46.Stalbrand, H., S. D. Mansfield, J. N. Saddler, D. G. Kilburn, R. A. Warren, and N. R. Gilkes. 1998. Analysis of molecular size distributions of cellulose molecules during hydrolysis of cellulose by recombinant Cellulomonas fimi β-1,4-glucanases. Appl. Environ. Microbiol. 64:2374-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Updegraff, D. M. 1969. Semimicro determination of cellulose in biological materials. Anal. Biochem. 32:420-424. [DOI] [PubMed] [Google Scholar]
- 50.von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2:531-534. [DOI] [PubMed] [Google Scholar]
- 51.Wood, T. M. 1988. Preparation of crystalline, amorphous and dyed cellulose substrates. Methods Enzymol. 160:19-25. [Google Scholar]
- 52.Zhu, H., F. W. Paradis, P. J. Krell, J. P. Phillips, and C. W. Forsberg. 1994. Enzymatic specificities and modes of action of the two catalytic domains of the XynC xylanase from Fibrobacter succinogenes S85. J. Bacteriol. 176:3885-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]