Abstract
Survey results and genotypic characterization of Escherichia coli strains demonstrate that the bacteriocins colicin Ia and microcin V coassociate in a strain more often than would be expected by chance. When these two bacteriocins co-occur, they are encoded on the same conjugative plasmid. Plasmids encoding colicin Ia and microcin V are nonrandomly distributed with respect to the genomic background of the host strain. Characterization of microcin V and colicin Ia nucleotide variation, together with the backbone of plasmids encoding these bacteriocins, indicates that the association has evolved on multiple occasions and involves the movement of the microcin V operon, together with the genes iroNEDCB and iss, onto a nonrandom subset of colicin Ia plasmids. The fitness advantage conferred on cells encoding both colicin Ia and microcin V has yet to be determined.
Bacteriocins are defined as antimicrobial proteins with a bactericidal action (30). Unlike traditional antibiotics that have a broad killing spectrum, bacteriocins are only toxic to the bacterial species they are produced by, and occasionally to closely related species (30). There is abundant evidence demonstrating that bacteriocins are important mediators of intra- and interspecies interactions and consequently a significant factor in maintaining microbial biodiversity (5, 20). Bacteriocins are also the subject of increasing attention as potential replacements for traditional antibiotics and as natural food preservatives (8). In addition, bacteriocinogeny is considered to be a key trait of strains used in probiotic formulations employed to prevent the establishment of microbial intestinal pathogens (32).
For those bacteriocins released via cell lysis, there is a wealth of mathematical theory (6, 7, 22), as well as in vitro (3, 12, 20) and in vivo (21) experimental evidence, that demonstrates the potential importance of bacteriocins in mediating intraspecific interactions and the highly dynamic nature of these interactions. However, our current understanding of the dynamics of bacteriocin production has largely concerned situations where the bacteriocinogenic population produces a single type of bacteriocin. Recently developed mathematical models predict that, under some circumstances, most of the strains of a bacterial community will evolve to produce multiple bacteriocin types (5). To understand the potential benefit arising from the production of multiple bacteriocin types, consider a community initially consisting of a sensitive cell population and two populations of producing cells, each encoding a single bacteriocin. If one of the producing cells acquires, through recombination, the genes for the other bacteriocin type, then this multiple-bacteriocin producer can kill sensitive cells and those cells encoding only a single bacteriocin. It is predicted that this outcome will occur when the costs associated with the production of multiple bacteriocins by a cell are not too severe, and gene exchange among different producing strains is sufficiently frequent.
In Escherichia coli, the production of multiple bacteriocins by a single strain is a common phenomenon (10, 11). Several bacteriocins have been found to co-occur in strains more often than would be expected by chance, including the bacteriocins colicin Ia and microcin V (10, 11). Colicin Ia and microcin V are both encoded on high-molecular-weight plasmids varying in size from 80 to 150 kbp (37). Colicin Ia is a 70-kDa protein whose production is induced by the SOS system under times of stress (24). The microcin V protein is much smaller (<10 kDa), and its production is induced not by the SOS system but under conditions of iron limitation (26, 37). For both bacteriocins, the toxin gene is tightly linked to a specific immunity gene whose product protects the cell from the bacteriocin it produces (2). Though the mechanisms involved in inducing bacteriocin production vary, once produced, the colicin Ia and microcin V proteins behave similarly. Colicin Ia is an atypical colicin as it does not encode a lysis protein but utilizes export proteins that shunt the toxin out of the producing cell without destroying it, like a microcin. Once exported, both colicin Ia and microcin V attach to the Cir protein receptor (a siderophore receptor) on the outer membrane of target cells (26, 37). The TonB pathway moves the bacteriocins into the cell. Following entry, if no immunity protein is present for that particular bacteriocin, both colicin Ia and microcin V destroy the cell by forming pores in the affected cell's cytoplasmic membrane (2).
The purpose of this study was to further investigate the colicin Ia and microcin V coassociation. To accomplish this goal, mating experiments were performed to determine if the coassociation of these the two bacteriocins was a result of their being encoded on a single plasmid. Genotypic and phenotypic approaches, as well as nucleotide sequencing, were also used to gain additional understanding of the coassociation of these two bacteriocins within a strain.
MATERIALS AND METHODS
Strain collections.
To have as diverse a range of colicin Ia and microcin V plasmids from E. coli as possible, strains from three collections were examined. The first of these was a collection of E. coli strains isolated from nonhuman vertebrates living in Australia (9). The second collection represented clinical and fecal isolates taken from humans living in Canberra and its surrounds (14). The final collection consisted of antibiotic resistance plasmids originating from clinical and fecal isolates of E. coli taken from patients in Adelaide, Canberra, Melbourne, Perth, and Sydney hospitals (34). This R plasmid collection was produced by mating wild-type antibiotic resistance strains with E. coli K-12 strain J53 and then confirming that the transconjugants harbored a single plasmid (34). All of the strains used were stored as glycerol cultures at −80°C.
Strain characterization.
All of the isolates from the three collections had been assayed for a mitomycin C-inducible colicin phenotype by the method described by Gordon et al. (13) and screened by a PCR-based method for the presence of 11 colicins and seven microcins (10). The majority of E. coli strains can be assigned to one of four genetics groups, designated A, B1, B2, and D (15, 25, 38), and all isolates had been assigned to a genetic group by the method described by Clermont et al. (4). In addition, all strains had been screened by a PCR-based technique for the presence of 29 genes thought to enhance the ability of E. coli to cause intestinal and extraintestinal disease as described by Gordon et al. (14). The donor strains from which the R plasmid collection (34) was derived were also screened for the presence of the 18 bacteriocins and 29 virulence genes.
Overnight cultures.
A subsample of a freezer culture was dilution streaked onto a Luria plate (Difco) and incubated overnight at 37°C. From this plate, a single colony was used to inoculate a flask containing 10 ml of Luria broth. The culture was incubated overnight at 37°C and shaken at 200 rpm.
PCRs.
Template DNA was prepared with DNAzol (Invitrogen) from a 50-μl aliquot of an overnight culture according to the manufacturer's protocol. Amplification was performed with a 25-μl reaction mixture containing 1.5 μl of template DNA, 0.25 μl of Platinum Taq (1.25 U), 1.5 μl each of both the forward and reverse primers (10 pmol ml−1), 3 μl of 25 mM MgCl2, and 5 μl of 5× buffer [67 mM Tris-HCl, 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 0.2 mg gelatin ml−1, 0.2 mM deoxynucleoside triphosphates], with PCR grade water used to attain the required volume. Amplification conditions were 1 cycle at 95°C for 12 min; 25 cycles of 94°C for 30 s, 55 or 63°C for 30 s, and 68°C for 3 min; and 1 cycle at 72°C for 10 min.
DNA sequencing.
Template DNA for sequencing of the colicin Ia and microcin V operons was obtained by the PCR protocol described above. Sequencing reactions used the Big Dye protocol and 3100 Genetic Analyzer from Applied Biosystems. A 1,200-bp portion of the 1,881-bp colicin Ia toxin gene was sequenced with primers Ia-F (5′-GGAATTCTCTTGACATGCCATTTTCTCCTT-3′), Ia657-R (5′-TCCTTCTGCCGTTTTTCAGCCTGTTCGATT-3′), Ia658-F (5′-CGTATATCTCCCCGGGAGGCCAGGTCGTTA-3′), and Ia1210-R. (5′-TAACTTATCCCATTCAGCAACTTCAGCAAT-3′). The sequenced region for microcin V included 60 bp of the 5′ flanking region, the 236-bp immunity gene, and 253 bp of the 311-bp toxin gene. The primers used were those described in reference 19. A 419-bp portion of the 558-bp plasmid gene involved in conjugative pilus synthesis regulation, finO, was sequenced for transconjugant strains positive for either colicin Ia or microcin V with the primers described in reference 1.
Sequences were edited and aligned with the Sequencher program. Maximum-likelihood trees were created with PAUP version 4.0b10 (36). Model parameters used for building maximum-likelihood trees in PAUP were generated by the MODELTEST program version 3.06 (27) by using the default starting parameters.
Obtaining transconjugants.
PCR screening of the R plasmid transconjugant collection, as well as the strains originally harboring these plasmids, provided data concerning the extent to which bacteriocins Ia and V were encoded on the same plasmid. To obtain additional data on the frequency with which colicin Ia and microcin V were encoded on the same plasmid, further matings were performed by a previously described method (28). The donor strains were wild-type strains from animal and human hosts that had screened positive for both bacteriocins. A 100-μl aliquot of an overnight culture of the donor strain together with 100 μl of an overnight culture of E. coli K-12 strain CSH50 (Δlac-pro gyr) were added to a flask containing 10 ml of Luria broth. The mating cultures were left in a nonshaking incubator for 2 nights at 37°C. A 1-ml aliquot of the mating culture was spread onto a selective plate (TL plates: Luria broth containing 50 mg 2,3,5-triphenyltetrazolium chloride liter−1, 10 g lactose liter−1, 100 μg ml−1 nalidixic acid, and 11 g agar liter−1) and incubated overnight at 37°C. Following incubation, the plates were overlaid with 3 ml of Luria broth soft agar (6 g agar liter−1) containing 200 μl of a CSH50 overnight culture and 80 μl of mitomycin C (final concentration, 0.1 μg ml−1) and then incubated at 37°C for another night. Colonies surrounded by a zone of clearing were dilution streaked onto TL plates containing nalidixic acid and onto minimal glucose plates (33) to confirm the identity of the putative transconjugants. In addition, each putative transconjugant was tested for colicin production and screened by a PCR approach for colicin Ia and microcin V. The transconjugant was deemed to be harboring a single conjugative plasmid if only a single band greater than 30 kb could be detected when native plasmid gels were prepared by the in-well lysis method (34).
Plasmid characterization.
All transconjugant strains shown to harbor plasmids encoding either colicin Ia or microcin V were further characterized. In addition to finO nucleotide sequence variation, the plasmids were characterized to determine if they belonged to the IncFII incompatibility group and if their traY gene was characteristic of an R- or F-type plasmid (1). The transconjugant strains were also screened for a number of putative virulence genes known to be plasmid associated iroN, traT, iss, and iutA (19), as well as sitA and tsh (31).
The transconjugants were phenotypically screened for resistance to seven antimicrobials, i.e., ampicillin (10 μg), chloramphenicol (30 μg), kanamycin (30 μg), streptomycin (300 μg), sulfisoxazole (250 μg), tetracycline (30 μg), and trimethoprim (5 μg), with BD BBL Sensi-Disk antimicrobial susceptibility test disks according to the manufacturer's protocol.
RESULTS
Frequency of colicin Ia and microcin V in E. coli.
For this study, 1,308 E. coli strains were PCR screened for colicin Ia and microcin V operons. We found colicin Ia in 10% of the strains and microcin V in 5% of the strains. We detected colicin Ia in 24% and microcin V in 20% of the 72 transconjugant strains harboring R plasmids.
Strains of E. coli can be assigned to one of four genetic groups designated A, B1, B2, and D, and strains of these four groups appear to occupy different ecological niches and vary in their propensity to cause disease (9, 16). The probability of detecting colicin Ia in a strain depended on the host group from which the strain was isolated and the strain's E. coli genetic group membership [nominal logistic regression: human/animal, χ2(1) = 2.80, P = 0.094; E. coli group, χ2(3) = 19.70, P = 0.0002; human/animal * E. coli group, χ2(3) = 15.81, P = 0.0012]. Overall, there was no difference in the frequency of Ia-positive strains isolated from humans compared to nonhuman vertebrates (Table 1). In isolates from humans, the frequencies of colicin Ia were similar across the E. coli groups. By contrast, in strains isolated from animals, the frequency of colicin Ia-producing strains was significantly greater in strains belonging to genetic group B2 compared to strains of the other E. coli groups.
TABLE 1.
Frequency of E. coli strains encoding colicin Ia, microcin V, or both bacteriocins, together with the predicted fraction of strains encoding both bacteriocins assuming that they randomly associatea
| Source and group | No. of isolates | % Observed
|
% Predicted, colicin Ia and microcin V | P > χ2 | ||
|---|---|---|---|---|---|---|
| Colicin Ia | Microcin V | Colicin Ia and microcin V | ||||
| Human | ||||||
| A | 90 | 4.4 | 3.3 | 6.6 | 1.1 | <0.001 |
| B1 | 51 | 9.8 | 7.8 | 2.0 | 1.1 | 0.575 |
| B2 | 350 | 3.7 | 2.3 | 7.1 | 1.0 | <0.001 |
| D | 128 | 4.7 | 2.7 | 3.1 | 0.4 | <0.001 |
| Animal | ||||||
| A | 93 | 2.2 | 1.1 | 0 | <0.1 | 0.834 |
| B1 | 272 | 6.2 | 1.8 | 1.1 | <0.1 | <0.001 |
| B2 | 213 | 17.8 | 0.1 | 2.3 | <0.1 | <0.001 |
| D | 112 | 6.2 | 0 | 0 | ||
Data presented are for strains isolated from humans and nonhuman vertebrates living in Australia with respect to the genetic group membership of the strains.
The likelihood of detecting microcin V in a strain also depended on the type of host it was isolated from [nominal logistic regression: human/animal, χ2(1) = 27.54, P = 0.0000; E. coli group, χ2(3) = 7.95, P = 0.0470; human/animal * E. coli group, χ2(3) = 15.81, P = 0.0012]. E. coli strains isolated from humans were significantly more likely to be microcin V positive than were strains isolated from nonhuman hosts (Table 1). Group D strains from both humans and animals were significantly less likely to be microcin V positive.
Strains encoding colicin Ia or microcin V and strains encoding both bacteriocins are present in each for the four major lineages of E. coli (Table 1). However, the distribution of microcin V is not independent of the genomic background of the host. Among the genetic group B2 strains isolated from humans, microcin V is never detected in strains that are positive for focG, hlyD, iha, and she. All microcin V-positive strains are also kpsMTII positive. Strains negative for focG, hlyF, iha, and she and positive for kpsMTII represent only 27% of the 350 group B2 strains, yet the prevalence of microcin V in strains with these characteristics is 35% (nominal logistic regression: χ2(1) = 97.67, P = 0.000001) (15; unpublished data). All microcin V-positive strains are also positive for traT, iss, and iroN. However, traT, iss, and iroN can be detected in B2 strains positive for focG, hlyD, iha, and she and negative for kpsMTII.
Coassociation of colicin Ia and microcin V.
Colicin Ia and microcin V coassociated within a strain significantly more often than would be expected by chance, irrespective of whether the strains were isolated from human or animal hosts [likelihood ratio test χ2(1) = 127.24, P < 0.0001]. A significant coassociation was also observed in the collection of R plasmid transconjugants [likelihood ratio test χ2(1) = 19.23, P < 0.0001]. Among the 72 R plasmids, 6% encoded only microcin V, 10% encoded just colicin Ia, and 14% encoded both bacteriocins.
A further analysis was undertaken to determine if the degree to which colicin Ia and microcin V were coassociated depended on the strain's E. coli group membership. This analysis was restricted to isolates from humans, as there were too few microcin V-positive strains isolated from animals. For this analysis, colicin Ia was used as the predictor variable. As expected, a strain that was colicin Ia positive was significantly more likely to be microcin V positive [nominal logistic regression: E. coli group, χ2(3) = 1.75, P = 0.6252; Ia+/−, χ2(1) = 19.65, P = 0.0000; E. coli group * Ia+/−, χ2(3) = 9.16, P = 0.0272]. However, the significant interaction term indicates that the extent to which the two bacteriocins were coassociated differed with respect to the E. coli group. The coassociation of these two traits was not observed in group B1 strains (Table 1).
It is unlikely that the coassociation of these two bacteriocins is an artifact arising because the same strain was sampled from many different hosts. First, each strain sampled was from a different host individual and, as a first approximation, different hosts harbor different strains of E. coli. Second, both bacteriocins are encoded by strains belonging to each of the four genetic groups of E. coli. Third, the bacteriocin and virulence gene screening reveals 26 distinct profiles among the strains encoding both colicin Ia and microcin V.
Colicin Ia and microcin V are found encoded on one plasmid.
In total, 36 transconjugants were obtained from strains where the donor was found to be positive for microcin V, colicin Ia, or both bacteriocins (Table 2). In all cases, when a transconjugant was obtained, the bacteriocin profile of the resulting transconjugant matched that of the donor strain and only a single plasmid band could be detected. In eight cases, a colicin Ia-positive transconjugant was obtained from a colicin Ia-positive donor and five microcin V-positive transconjugants were obtained from microcin V-positive donors. There were 23 transconjugants obtained from donor strains that were positive for both bacteriocins; in every case, the resulting transconjugant harbored a single plasmid and encoded both bacteriocins. These results indicate that when both bacteriocins are present in a strain they are most likely to be encoded on a single plasmid.
TABLE 2.
Phenotypic and genotypic characteristics of plasmids encoding the bacteriocins colicin Ia and microcin V
| Strain | Colicin Ia | Microcin V | finO ST | TraY type | IncFII | traT | iroN | iss | sitA | iutA | Resistance profilea |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AE008 | + | − | 10 | − | − | − | − | − | − | − | TMP SXT AM |
| CE015 | + | − | 14 | R | − | + | − | + | − | − | TMP SXT AM S |
| PE006 | + | − | 6 | − | − | − | − | − | − | − | TE |
| SE016 | + | − | 7 | − | − | − | − | − | − | − | AM TE C |
| SE017 | + | − | 20 | − | − | − | − | − | − | − | AM TE C |
| AE013 | + | − | 4 | F | + | + | − | − | − | − | TMP SXT S SPT |
| CE003 | + | − | 18 | F | + | + | − | − | − | − | TMP AM TE |
| AE011 | − | + | 5 | F | + | + | + | + | + | − | TMP SXT AM S TE |
| CE014 | − | + | 3 | F | + | + | + | + | + | + | TMP STX |
| PE007 | − | + | 5 | F | + | + | + | + | + | − | TMP STX S |
| SE010 | − | + | 5 | F | + | + | + | + | + | − | TMP STX |
| H772 | − | + | 15 | F | + | + | + | + | + | + | TE |
| AE016 | + | + | 8 | F | − | + | + | + | − | − | TMP STX |
| CE004 | + | + | 8 | − | − | + | + | + | + | + | TMP STX AM S TE K |
| CE010 | + | + | 12 | − | − | + | + | + | + | + | TMP STX AM S TE K |
| PE001 | + | + | 8 | F | − | + | + | + | + | + | TMP STX AM S |
| PE002 | + | + | 8 | F | − | + | + | + | + | + | AM TE |
| PE004 | + | + | 5 | F | − | + | + | + | + | − | TMP |
| PE009 | + | + | 19 | F | − | + | + | + | + | + | TMP STX AM S TE |
| H053 | + | + | 8 | F | − | + | + | + | + | + | TMP SXT AM S |
| H070 | + | + | 8 | F | − | + | + | + | + | + | TMP A S TE |
| H073 | + | + | 8 | F | − | + | + | + | + | + | TMP A S TE |
| H111 | + | + | 8 | F | − | + | + | + | + | + | SXT AM |
| H112 | + | + | 8 | F | − | + | + | + | + | + | AM TE |
| H127 | + | + | 11 | F | − | + | + | + | + | + | AM S TE |
| H188 | + | + | 8 | F | − | + | + | + | + | + | TMP SXT AM S TE K |
| H190 | + | + | 8 | F | − | + | + | + | + | + | TMP AM S TE K |
| H461 | + | + | 8 | F | − | + | + | + | + | + | AM S |
| H549 | + | + | 8 | F | − | + | + | + | + | + | TMP SXT AM S |
| H098 | + | + | 13 | R | + | + | + | + | − | − | S |
| H100 | + | + | 9 | R | + | + | + | + | − | − | TMP STX AM |
| H314 | + | + | 1 | − | + | + | + | + | − | − | AM S |
| AE009 | + | + | 17 | R | + | + | + | + | + | + | TMP STX |
| AE010 | + | + | 17 | R | + | + | + | + | + | + | TMP STX TE |
| SE018 | + | + | 5 | F | + | + | + | + | + | − | TE C |
AM, ampicillin; C, chloramphenicol; K, kanamycin; S, streptomycin; SXT, sulfisoxazole; TE, tetracycline; TMP, trimethoprim.
Characterizing the bacteriocin-encoding plasmids.
The plasmids encoding only microcin V were very homogeneous; all were members of the FII incompatibility group and possessed an F-type traY gene, and only three finO alleles were detected (Table 2). By contrast, the plasmids encoding only colicin Ia were diverse (Table 2). Of the seven Ia plasmids, just two were IncFII positive and had an F-type traY gene. Each colicin Ia plasmid possessed a unique finO allele (Table 2). The plasmids encoding both bacteriocins were also diverse (Table 2). Only one of the F-type plasmids was a member of the IncFII group, and the balance of the IncFII plasmids had an R-type or underdetermined traY gene. Among the 23 transconjugants encoding both bacteriocins, nine finO alleles were observed.
The transconjugants were also PCR screened for five other genes known to be plasmid encoded. All plasmids encoding microcin V were also traT, iss, and iroN positive, while sitA was detected in 86% of the microcin V-positive strains (Table 2). Plasmids that encoded only colicin Ia were negative for iutA and iucC, while 71% of the microcin V-positive plasmids possessed these traits (Table 2).
The antibiotic resistance profiles of the bacteriocin-encoding plasmids were very diverse, with few plasmids having identical resistance profiles (Table 2).
Microcin V sequence variation.
Fifty microcin V-producing strains were sequenced for 527 bp of the microcin V operon. The sequenced region included 60 bp of the 5′ flanking region, the entire immunity gene (236 bp), and 253 bp of the 311-bp microcin V gene. The immunity gene and the toxin gene overlap by 22 bases, which accounts for the discrepancy in the values. The average proportion of nucleotide differences among the 50 sequences was 0.0005 ± 0.0003.
Only five sequence types (STs) were observed, and excepting five of the strains, all of the strains were identical to the published data (27) for the microcin V operon (Table 3). Two of the strains (H244 and R087) had a 15-bp duplication in the 5′ flanking region, and one strain (H781) had a 15-bp deletion in the 5′ flanking region.
TABLE 3.
Nucleotide variation in a 527-bp fragment of the microcin V operon found in this study compared to previously published results (26)a
| Variantb | Nucleotide at position:
|
|||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 14 | 42 | 51 | 55 | 76 | 105 | 109 | 110 | 111 | 112 | 113 | 114 | 115 | 116 | 117 | 118 | 119 | 120 | 121 | 122 | 131 | 134 | 142 | 143 | 144 | 145 | 146 | 147 | 148 | 149 | 150 | 151 | 152 | 153 | 154 | 155 | 156 | 294 | 295 | 323 | 459 | |
| AF062847 (1) | C | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | G | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | A |
| H030 (1) | G | A | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | G |
| AF062845 (1) | G | G | G | C | C | G | A | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | T | A |
| AF062849 (1) | G | G | G | C | C | G | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | T | A |
| H368 (1) | G | G | G | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | G |
| H781 (1) | G | G | T | T | A | T | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | C | G |
| AF062844 (1) | G | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | C | G |
| ECCVAC (52) | G | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | G |
| B177 (1) | G | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | G |
| H244 (2) | G | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | A | G | T | G | G | G | A | T | A | A | A | A | A | G | T | A | A | G | C | T | G |
| AF062848 (1) | G | G | T | T | A | T | G | T | A | A | T | G | G | G | A | T | A | G | A | A | A | G | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | T | A |
Positions 2 to 156 correspond to the 5′ flanking region, positions 294 to 323 correspond to the immunity gene, and position 459 corresponds to the toxin gene.
The number in parentheses is the number of times the variant was observed.
Colicin Ia sequence variation.
The colicin Ia toxin gene is 1,881 bp in length. A 1,124-bp region (including 1,092 bp of the colicin Ia gene and 32 bp of the 5′ flanking region) from 69 strains was sequenced. Among the 69 sequences, there were 61 polymorphic nucleotide sites giving rise to 36 STs and 18 amino acid replacements. The average proportion of nucleotide differences among the 69 sequences was 0.0065 ± 0.0013. The phylogenetic relationships among the colicin Ia sequence types are depicted in Fig. 1.
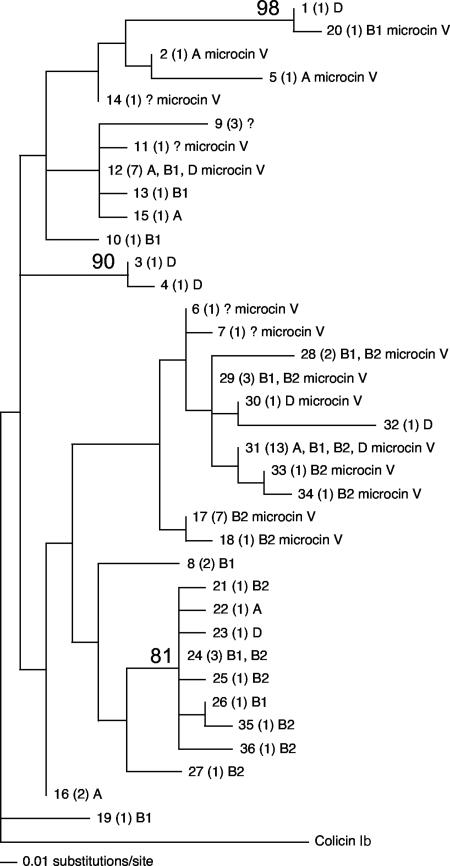
FIG. 1.
Maximum-likelihood tree of the 36 Ia nucleotide sequence types detected in E. coli isolates from human and nonhuman vertebrates living in Australia. In each label, the first number is the ST and the number in parentheses is the number of times the ST was observed, followed by the genetic groups of E. coli in which the ST was detected and finally if the ST co-occurred with the microcin V operon in the same strain. Bootstrap values are based on 500 replicates.
Colicin Ia sequence types from strains encoding both bacteriocins occurred on multiple branches of the tree and encompassed much of the breadth of the observed colicin Ia nucleotide variation. This outcome suggests that the co-occurrence of colicin Ia and microcin V could have evolved multiple times. However, few of the nodes have strong bootstrap support, so a different approach was used to investigate whether all of the strains encoding both bacteriocins could have evolved from a common ancestor. To accomplish this, all of the colicin Ia- and microcin V-positive strains were constrained to be monophyletic and a new maximum-likelihood tree was estimated. Comparing the likelihood scores of the constrained and unconstrained trees revealed that forcing strains that produce both bacteriocins to be monophyletic resulted in a significantly worse fit to the data [Difference − ln(L) = 43.238, P < 0.0039].
Inspection of Fig. 1 suggests that sequence variation in this region of the colicin Ia gene may be different in strains that also produce microcin V. Four populations were defined, Ia STs isolated from humans, Ia STs isolated from animals, Ia STs isolated from humans where microcin V co-occurred, and Ia STs from animals where microcin V also co-occurred. For the analysis, each population consisted of a single representative of each ST observed in that population. Molecular analysis of variance (AMOVA) confirmed that a significant fraction of the observed colicin Ia sequence variation was explained by the source of isolation and whether microcin V co-occurred with colicin Ia in the strain (AMOVA: PhiPT = 0.246, P < 0.001). Pairwise comparisons revealed that the source of isolation did not explain a substantial amount of the variation in the colicin Ia sequences, and this was particularly true for strains where microcin V was also present (Table 4). However, a substantial amount of the colicin Ia sequence variation depended on whether microcin V was also present in the strain (AMOVA: PhiPT = 0.256, P < 0.001).
TABLE 4.
Population differentiation estimates (PhiPT) to determine the extent to which colicin Ia nucleotide sequence variation depended on the host group from which the strains were isolated or whether the strains also encoded microcin V
| Comparisona | PhiPT | Probabilityb |
|---|---|---|
| Ia animal vs Ia human | 0.101 | 0.040 |
| Ia animal vs IaV animal | 0.480 | 0.001 |
| Ia human vs IaV animal | 0.386 | 0.002 |
| Ia animal vs IaV human | 0.292 | 0.001 |
| Ia human vs IaV human | 0.127 | 0.014 |
| IaV animal vs IaV human | 0.129 | 0.081 |
Ia, colicin Ia; V, microcin V; IaV, colicin Ia plus microcin V.
The probability that a greater PhiPT value could have arisen by chance.
The majority (83%) of the colicin Ia sequences from strains where microcin V was also present had a methionine rather than an asparagine at position 92 and an alanine rather than a glycine at position 109 (Table 5). Further, a leucine at position 295 was only present in strains that were also positive for microcin V (Table 5).
TABLE 5.
Amino acid sequences of the colicin Ia variants detected in this study together with the human or nonhuman vertebrate sources from which the strains were isolated and whether the strains also encode microcin V
| Strain | Source | Microcin V | Varianta | Nucleotide at position:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 60 | 88 | 92 | 109 | 130 | 141 | 169 | 206 | 238 | 241 | 247 | 275 | 284 | 295 | 340 | 346 | 367 | ||||
| H190 | Human | + | 3 (1) | R | G | T | I | G | R | R | H | A | A | V | A | N | A | L | A | T | E |
| B439 | Animal | + | 6 (1) | R | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | T | E |
| B313 | Animal | + | 14 (1) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | L | A | T | E |
| TA311 | Animal | + | 16 (1) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | T | E |
| M550 | Animal | + | 17 (2) | S | G | T | M | A | R | R | Y | A | A | V | A | N | A | L | A | T | E |
| AE016 | Human | + | 4 (1) | R | G | T | M | A | R | R | H | A | A | V | A | N | A | L | A | T | E |
| H098 | Human | + | 5 (1) | R | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | S | E |
| H314 | Human | + | 6 (7) | R | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | T | E |
| H151 | Human | + | 14 (12) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | L | A | T | E |
| H244 | Human | + | 15 (1) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | S | E |
| PE002 | Human | + | 16 (1) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | T | E |
| AE008 | Human | − | 13 (1) | S | G | T | M | A | R | R | H | A | A | V | A | K | A | P | A | T | E |
| CE015 | Human | − | 15 (1) | S | G | T | M | A | R | R | H | A | A | V | A | N | A | P | A | S | E |
| SE017 | Human | − | 2 (2) | R | G | A | N | G | R | R | H | A | A | V | A | N | A | P | A | S | E |
| H730 | Human | − | 10 (2) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | T | E |
| H591 | Human | − | 11 (1) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | V | S | E |
| H556 | Human | − | 18 (1) | S | G | T | N | G | R | Q | H | T | A | V | A | N | A | P | A | T | E |
| H468 | Human | − | 19 (1) | S | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | S | E |
| H422 | Human | − | 21 (1) | S | G | T | N | G | R | R | H | T | A | V | A | N | A | P | A | T | E |
| PE001 | Human | + | 7 (2) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | L | A | T | E |
| H331 | Human | + | 8 (2) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | S | E |
| PE004 | Human | + | 9 (1) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | S | K |
| TA060 | Animal | − | 1 (2) | R | G | A | N | G | H | R | H | A | A | V | A | N | A | P | A | T | E |
| M411 | Animal | − | 8 (1) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | S | E |
| TA026 | Animal | − | 10 (3) | R | G | T | N | G | R | R | H | A | A | V | A | N | A | P | A | T | E |
| R148 | Animal | − | 12 (1) | R | G | T | N | G | R | R | H | T | A | V | A | N | A | P | A | T | E |
| R529 | Animal | − | 20 (1) | S | G | T | N | G | R | R | H | T | A | I | A | N | A | P | A | T | E |
| TA261 | Animal | − | 21 (3) | S | G | T | N | G | R | R | H | T | A | V | A | N | A | P | A | T | E |
| B564 | Animal | − | 22 (1) | S | G | T | N | G | R | R | H | T | A | V | A | N | T | P | A | T | E |
| M008 | Animal | − | 23 (1) | S | G | T | N | G | R | R | H | T | A | V | S | N | A | P | A | T | E |
| M318 | Animal | − | 24 (1) | S | G | T | N | G | R | R | H | T | V | V | A | N | A | P | A | T | E |
| M028 | Animal | − | 25 (1) | S | V | T | N | G | R | R | H | T | V | V | A | N | A | P | A | T | E |
The number in parentheses is the number of times the variant was observed in that source population and in the presence or absence of microcin V.
DISCUSSION
The microcin V operon clearly occurs with the colicin Ia operon more frequently than would be expected by chance. The evidence demonstrates that when these two bacteriocins co-occur in a strain, they are usually encoded on a single plasmid. Plasmids encoding only colicin Ia or only microcin V are most often of different incompatibility groups. In theory, therefore, these plasmids could stably co-exist in a cell. What, then, are the selective forces that result in these two bacteriocin operons becoming encoded on a single plasmid? There are likely two reasons for this observation. First, while a strain can have numerous copies of small plasmids, the presence of large conjugative plasmids increases the metabolic stress on the cell and it can be disadvantageous for a cell to harbor more than one large plasmid (35). Second, there may be circumstances under which the cotransfer of both operons can be expected to evolve (23). Consider a community consisting of three populations of cells, one producing colicin Ia, one producing microcin V, and a plasmid-free sensitive population. If conjugation moves a microcin V plasmid into a cell harboring a colicin Ia plasmid or vice versa, then the cell will potentially accrue a significant fitness advantage, as it is capable of killing all of the other cells in the community and cannot be killed by any cell. However, if the two operons are maintained on separate plasmids, then it is unlikely that they will always cotransfer to new recipients. Thus, in environments where colicin Ia and microcin V producers are present, plasmids that encode both bacteriocins should be favored.
Colicin Ia and E. coli appear to share a long evolutionary history, while microcin V appears to have arrived in the species relatively recently. For the Australian isolates, the average proportion of polymorphic sites in colicin Ia is 0.0065, significantly less than the average for housekeeping loci in the species (0.013 [unpublished data]), but much greater than the diversity observed for the microcin V operon (0.0005). The GC content of the microcin V operon is 39%, while it is 50% for colicin Ia and 53% for E. coli housekeeping genes (unpublished data). Further, the data suggest that in E. coli, colicin Ia normally occurs on a plasmid in the absence of microcin V. Overall, colicin Ia occurred 68% of the time on its own; 88% of the time in the isolates from animals, and 43% of the time in the isolates from humans. By contrast, the microcin V operon was detected in the absence of colicin Ia 36% of the time.
The balance of the evidence suggests that the coassociation of colicin Ia and microcin V is a result of the microcin V operon moving onto colicin Ia plasmids. Microcin V plasmids appear to be very homogeneous, while colicin Ia plasmids and plasmids encoding both bacteriocins are diverse (Table 2). However, if this is the direction of operon movement, then a number of genes must cotransfer with the microcin V genes. All plasmids encoding microcin V also encode the virulence factors iss and iroN, as well as the plasmid transfer gene traT. Among 92 strains encoding only colicin Ia, 59% were traT positive, 39% were iroN positive, and 24% were iss positive, but only 12% were positive for all three traits (unpublished data).
One plasmid encoding microcin V has been sequenced in its entirety (17). On the basis of the published plasmid sequence, the transfer of the microcin V genes together with iss and iroNEDCB would involve a 20-kb DNA fragment. In another plasmid encoding remnants of the microcin V system (18), iss, iroNEDCB, and the microcin genes are also encompassed by a 20-kb fragment. The inclusion of traT with iss, iroNEDCB, and the microcin V genes would result in a DNA fragment of at least 65 kb. This suggests that microcin V and the associated virulence genes may only transfer to particular plasmid backbones, i.e., those that are traT positive. There is additional evidence to suggest that the movement of microcin V is dependent on the genetic background of the colicin Ia plasmid. The majority, 83%, of the stains positive for both colicin Ia and microcin V for which Ia sequence data were available (n = 66) were associated with particular amino acid sequence variants (Table 5). The amino acid variants represented 10 nucleotide sequence types, and the average proportion of nucleotide differences among these 10 sequence types was 0.0021 ± 0.0008.
The association of the microcin V genes with colicin Ia appears to have occurred relatively recently and on multiple occasions. Inspection of Fig. 1 suggests a minimum of six events, but this may represent a minimum estimate. There are 10 STs representing similar amino acid variants of colicin Ia. The hypothesis that these variants may have arisen from a single ancestor is at odds with the observation that virtually no variation has accumulated in the microcin activity and immunity genes, while the Ia toxin gene has accumulated 20 times the number of polymorphisms.
Although encoded on plasmids with the ability to transfer among strains representing all E. coli lineages, the presence of microcin V on a plasmid appears to prevent these plasmids from establishing themselves in hosts with particular genetic backgrounds. Why the occurrence of microcin V is incompatible with particular genetic backgrounds is unknown.
Three nonmutually exclusive hypotheses have been suggested to explain why it might be beneficial for a cell to produce multiple types of bacteriocin (10). The first is an expanded killing range. As previously explained, in a community of cells where two types of bacteriocin are being produced, any cell that produces both bacteriocins can kill all single-bacteriocin producers. Second, the evolution of resistance to many bacteriocins is common (29). The production of two or more bacteriocins exploiting different receptors, or different epitopes of the same receptor, may slow the evolution of resistance. Third, multiple-bacteriocin production may confer a fitness advantage in different environments. Colicin Ia is induced via the SOS system during times of stress, while microcin V is induced when iron is limited. Experiments are now under way to determine the advantages to an E. coli cell of encoding microcin V and colicin Ia.
Acknowledgments
This research was supported under the Australian Research Council's Discovery Projects funding scheme (DP0664453).
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Boyd, E. F., C. W. Hill, S. M. Rich, and D. L. Hard. 1996. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics 143:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 3.Chao, L., and B. R. Levin. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czárán, T. L., R. F. Hoekstra, and L. Paige. 2002. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. USA 99:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durrett, R., and S. Levin. 1997. Allelopathy in spatially distributed populations. J. Theor. Biol. 185:165-171. [DOI] [PubMed] [Google Scholar]
- 7.Frank, S. A. 1994. Spatial polymorphism of bacteriocins and other allelopathic traits. Evol. Ecol. 8:369-386. [Google Scholar]
- 8.Gillor, O., B. C. Kirkup, and M. A. Riley. 2004. Colicins and microcins: the next generation antimicrobials. Adv. Appl. Microbiol. 54:129-146. [DOI] [PubMed] [Google Scholar]
- 9.Gordon, D. M., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575-3586. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, D. M., and C. L. O'Brien. 2006. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152:3239-3244. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, D. M., E. Oliver, and J. Littlefield-Wyer. 2006. The diversity of bacteriocins in gram-negative bacteria, p. 5-18. In M. A. Riley and M. A. Chavan (ed.), Bacteriocins: ecology and evolution. Springer, Berlin, Germany.
- 12.Gordon, D. M., and M. A. Riley. 1999. A theoretical and empirical investigation of the invasion dynamics of colicinogeny. Microbiology 145:655-661. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D. M., M. A. Riley, and T. Pinou. 1998. Temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiology 144:2233-2240. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D. M., P. E. Stern, and P. J. Collignon. 2005. The influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151:15-23. [DOI] [PubMed] [Google Scholar]
- 15.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 20.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 21.Kirkup, B. C., and M. A. Riley. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412-414. [DOI] [PubMed] [Google Scholar]
- 22.Levin, B. R. 1988. Frequency-dependent selection in bacterial populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319:459-472. [DOI] [PubMed] [Google Scholar]
- 23.Levin, B. R. 1995. Conditions for the evolution of multiple antibiotic-resistance plasmids: a theoretical and experimental excursion, p. 175-192. In S. Baumberg, J. P. W. Young, E. M. H. Wellington, and J. R. Saunders (ed.), Population genetics of bacteria. Cambridge University Press, Cambridge, United Kingdom.
- 24.Mankovich, J. A., C.-H. Hsu, and J. Konisky. 1986. DNA and amino acid sequence analysis of structural and immunity genes of colicins Ia and Ib. J. Bacteriol. 168:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinou, T., and M. A. Riley. 2001. Nucleotide polymorphism in microcin V plasmids. Plasmid 46:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 28.Pugsley, A. P., and B. Oudega. 1987. Methods of studying colicins and their plasmids, p. 105-161. In K. G. Hardy (ed.), Plasmids, a practical approach. IRL Press, Oxford, United Kingdom.
- 29.Riley, M. A., and D. M. Gordon. 1999. A model of intraspecific microbial warfare. Trends Microbiol. 7:129-133.10203843 [Google Scholar]
- 30.Riley, M. A., and J. E. Wertz. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357-364. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 32.Rolfe, R. D. 2000. The role of probiotic cultures in the control of gastro-intestinal health. J. Nutr. 130(2S Suppl.):396S-402S. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sherley, M., D. M. Gordon, and P. J. Collignon. 2004. Evolution of multi-resistance plasmids in Australian clinical isolates of Escherichia coli. Microbiology 150:1539-1546. [DOI] [PubMed] [Google Scholar]
- 35.Smith, A. M., and M. J. Bidochka. 1998. Bacterial fitness and plasmid loss: the importance of culture conditions and plasmid size. Can. J. Microbiol. 44:351-355. [PubMed] [Google Scholar]
- 36.Swofford, D. 1997. PAUP*: phylogenic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- 37.Waters, V. L., and J. H. Crosa. 1991. Colicin V virulence plasmids. Microbiol. Rev. 55:437-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]