Abstract
Methionine sulfoxide reductase (Msr) catalyzes the thioredoxin-dependent reduction and repair of methionine sulfoxide (MetO). Although Msr genes are not present in most hyperthermophile genomes, an Msr homolog encoding an MsrA-MsrB fusion protein (MsrABTk) was present on the genome of the hyperthermophilic archaeon Thermococcus kodakaraensis. Recombinant proteins corresponding to MsrABTk and the individual domains (MsrATk and MsrBTk) were produced, purified, and biochemically examined. MsrATk and MsrBTk displayed strict substrate selectivity for Met-S-O and Met-R-O, respectively. MsrABTk, and in particular the MsrB domain of this protein, displayed an intriguing behavior for an enzyme from a hyperthermophile. While MsrABTk was relatively stable at temperatures up to 80°C (with a half-life of ∼30 min at 80°C), a 75% decrease in activity was observed after 2.5 min at 85°C, the optimal growth temperature of this archaeon. Moreover, maximal levels of MsrB activity of MsrABTk were observed at the strikingly low temperature of 30°C, which also was observed for MsrBTk. Consistent with the low-temperature-specific biochemical properties of MsrABTk, the presence of the protein was greater in T. kodakaraensis cells grown at suboptimal temperatures (60 to 70°C) and could not be detected at 80 to 90°C. We found that the amount of intracellular MsrABTk protein increased with exposure to higher dissolved oxygen levels, but only at suboptimal growth temperatures. While measuring background rates of the Msr enzyme reactions, we observed significant levels of MetO reduction at high temperatures without enzyme. The occurrence of nonenzymatic MetO reduction at high temperatures may explain the specific absence of Msr homologs in most hyperthermophiles. Together with the fact that the presence of Msr in T. kodakaraensis is exceptional among the hyperthermophiles, the enzyme may represent a novel strategy for this organism to deal with low-temperature environments in which the dissolved oxygen concentrations increase.
Reactive oxygen species are harmful to the cell by oxidizing various cell components, such as lipids, nucleic acids, and proteins. Among the amino acids, methionine (Met) residues are known to be particularly susceptible to oxidative stress and are easily oxidized to methionine sulfoxides (MetO). Methionine sulfoxide reductase (Msr) is an enzyme that repairs the oxidized methionine and catalyzes the thioredoxin-dependent reduction of MetO to Met (6). The oxidation of Met results in the formation of two asymmetric molecules, Met-S-O and Met-R-O. Each MetO is reduced by a specific and structurally distinct enzyme: Met-S-O is reduced by MsrA, and Met-R-O is reduced by MsrB. Msr proteins are considered to play vital roles in maintaining the intracellular redox balance and in the repair of oxidized proteins (5, 9). In mammals, defects in the function of Msr have been reported to result in neurological disorders and, in some cases, a decrease in life span (30, 31).
Although structurally distinct, MsrA and MsrB catalyze the reduction of MetO with basically similar mechanisms (2, 4, 20, 33). A cysteine residue, designated CysA, acts as a nucleophile that attacks the oxidized sulfur atom of MetO. A tetrahedral transition state is formed, followed by a rearrangement that releases the repaired Met, and this results in the formation of a sulfenic acid intermediate on the CysA side chain. A second cysteine residue, CysB, then attacks the oxidized CysA and, along with the release of a water molecule, forms a disulfide bond with CysA. Other cysteine residues may participate in the steps that follow, but the enzyme eventually is reduced in a thioredoxin-dependent manner, completing the reaction.
MsrA proteins can be classified into three main groups by the number and positions of cysteine residues that are proposed to be involved in the catalytic mechanism (20). The first group (MsrAI) utilizes three Cys residues in catalysis, and the nucleophilic CysA residue is conserved in a GCFWG motif. The CysB residue is conserved in a GYCG sequence. A third Cys residue (CysC) is present in MsrAI and resides downstream of a glycine-rich sequence. The second group of MsrA proteins (MsrAII) utilizes only two Cys residues, basically corresponding to CysA and CysB of MsrAI enzymes. The third group of enzymes (MsrAIII) harbors both CysA and CysB residues in a single GCFWC motif. The MsrB proteins are classified into two groups by the presence (form I) or absence (form II) of two CxxC motifs that have been shown to participate in binding to a divalent zinc cation in the MsrB from Drosophila melanogaster (26). Although mutations in any one of these four Cys residues leads to a nonactive protein, this cluster seems to play a structural role in the enzyme. All MsrB proteins from eukaryotes and archaea are form I enzymes, while bacteria harbor either form I or form II, depending on the species.
Consistent with their important roles in dealing with oxygen or oxidative stress, Msr proteins are widely distributed in nature and can be found in all three domains of life, Eucarya, Bacteria, and Archaea. Msr homologs are present in almost all mesophile genomes sequenced thus far. However, almost all hyperthermophiles from the Bacteria and the Archaea do not harbor Msr genes. The only exceptions are the MsrA homolog in Sulfolobus solfataricus (43) and the MsrA-MsrB fusion homolog in Thermococcus kodakaraensis (designated MsrABTk) (11) (Table 1). The presence of Msr in T. kodakaraensis was particularly intriguing, as Msr homologs are not present in any of the genome sequences from Pyrococcus furiosus (37), “Pyrococcus abyssi” (7), and Pyrococcus horikoshii (22), which are very closely related to T. kodakaraensis.
TABLE 1.
The presence of Msr homologs in various archaea along with their growth temperatures
| Organism | MsrA homolog | MsrB homolog | Growth temp (optimum) (°C) | Reference |
|---|---|---|---|---|
| Methanococcus maripaludis | CAF30404 | − | Mesophilic | |
| Methanosarcina acetivorans | AAM04846 | AAM03895 | Mesophilic | |
| Methanosarcina mazei | AAM32095 | AAM31330 | Mesophilic | |
| Haloarcula marismortui | AAV46883 AAV46414 | AAV45184 | (40-50) | 34 |
| Halobacterium sp. | AAG19555 | AAG19724 | 22-50 (49-50) | 15, 38 |
| Thermoplasma acidophilum | −e | − | 45-63 (59) | 42 |
| Thermoplasma volcanium | − | − | 33-67 (59) | 42 |
| Picrophilus torridus | AAT42728 | − | 45-65 (60) | 41 |
| Methanothermobacter thermautotrophicus | AAB85041 | AAB85216 | 40-70 (65) | 44 |
| Sulfolobus solfataricus | AAK41726 | − | 65-86a (80) | 14 |
| Sulfolobus tokodaii | − | − | 70-85 (80) | 45 |
| Archaeoglobus fulgidus | − | − | 60-95 (83) | 24 |
| Methanocaldococcus jannaschii | − | − | 50-86a (85) | 19 |
| Thermococcus kodakaraensis | BAD85008d | BAD85008d | 60-100 (85) | 3 |
| Nanoarchaeum equitans | − | − | 70-98b | 17 |
| Aeropyrum pernix | − | − | 90-98 (95) | 21 |
| Pyrococcus abyssi | − | − | 67-102c (96) | 8 |
| Pyrococcus horikoshii | − | − | 85-105 (98) | 12 |
| Methanopyrus kandleri | − | − | 84-110 (98) | 27 |
| Pyrococcus furiosus | − | − | 70-103 (100) | 10 |
| Pyrobaculum aerophilum | − | − | 75-104 (100) | 49 |
Growth has been observed in this temperature range and does not necessarily reflect the temperature limits.
Symbiotic growth with Ignicoccus spp.
Growth under 200 kPa pressure.
MsrA-MsrB fusion protein.
−, no homolog is present in the genome.
The genera Pyrococcus and Thermococcus both belong to the family Thermococcales and consist of heterotrophic, sulfur-reducing anaerobes that share common metabolism and energy-generating mechanisms (1, 11, 48). The major distinction between Thermococcus and Pyrococcus is in their growth temperatures; the former has optimal growth temperatures between 75 and 93°C, while those of the latter range from 95 to 103°C (1, 18). Although the Pyrococcus spp. have received relatively more attention in terms of biochemical and genome research, environmental studies have indicated that Thermococcus seems to be by far the more predominant genus distributed throughout the hydrothermal environments on our planet (16, 18, 35). Thermococcus also seems to be much more diverse (18), including members that can grow at alkaline pH (T. alcaliphilus) (23), extremely low salinity (T. waiotapuensis) (13), and at temperatures as low as 40°C (T. sibiricus) (29). In the natural environment, a decrease in temperature usually brings about an increase in dissolved oxygen (DO) concentration. Thus, the Thermococcus spp., which generally grow at lower temperature ranges, may harbor additional defense mechanisms against oxygen that are not present in Pyrococcus spp. As an Msr gene is present on the T. kodakaraensis genome but is absent from the three Pyrococcus genomes, there is a possibility that Msr represents one of these additional mechanisms. In this study, we have examined the biochemical properties of MsrABTk and its presence in T. kodakaraensis under various growth conditions.
MATERIALS AND METHODS
Phylogenetic analyses of Msr sequences.
MsrA and MsrB sequences were selected from various organisms and were aligned by using the ClustalW program (47) provided by the DNA Data Bank of Japan. Core regions displaying homology among the MsrA sequences (residues 9 to 163 in MsrAB) and MsrB sequences (residues 219 to 322) were used for each phylogenetic analysis. Phylogenetic trees were constructed with the neighbor-joining method (39) using the ClustalW program mentioned above.
Microorganisms, plasmids, and media.
Escherichia coli DH5α and plasmid pUC118 were used for general DNA manipulation and sequencing. E. coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) and pET21a(+) (Novagen, Madison, WI) were used for gene expression. E. coli strains were cultivated in Luria-Bertani (LB) medium (10 g liter−1 of tryptone, 5 g liter−1 of yeast extract, and 10 g liter−1 of NaCl [pH 7.0]) at 37°C with the addition of 100 μg ml−1 ampicillin. Unless mentioned otherwise, T. kodakaraensis KOD1 (3) was inoculated and grown in an anaerobic environment in ASW-YT medium (40) with 0.5% sodium pyruvate (ASW-YT-Pyr) at 85°C. Sodium sulfide (0.1 g liter−1) and resazurin (0.25 mg liter−1) were added to the medium.
DNA manipulation and sequencing.
Restriction and modification enzymes were purchased from Toyobo (Osaka, Japan) or Takara (Ohtsu, Japan). KOD Plus (Toyobo) was used for PCR. Plasmid DNA was isolated with a QIAGEN plasmid mini kit (Hilden, Germany). DNA fragments were recovered from agarose gels with a GFX PCR DNA and gel band purification kit (Amersham Biosciences, Little Chalfont, United Kingdom). DNA sequencing was performed with a BigDye Terminator cycle sequencing kit (version 3.0) and a Model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Expression and purification of MsrA, MsrB, and MsrAB.
Expression plasmids for MsrATk, MsrBTk, and MsrABTk were constructed as follows. The respective genes were amplified with T. kodakaraensis genomic DNA as a template and two oligonucleotide primers (MsrATk sense, 5′-GTGCATATGGGTGGTATCAAAATTGAACC-3′; MsrATk antisense, 5′-AGGAATTCACCTGAAGTGCCGGTTTTTCTC-3′; MsrBTk sense, 5′-TCCATATGTTCCCTGAGAGAGAGGGCTAC-3′; MsrBTk antisense, 5′-AGGAATTCTACTTAAAAATCCCCTCGTAAG-3′; MsrABTk sense, 5′-GTGCATATGGGTGGTATCAAAATTGAACC-3′; MsrABTk antisense, 5′-TTGAATTCGACTTAAAAATCCCCTCG-3′). NdeI and EcoRI sites were incorporated in the sense and antisense primers, respectively. An artificial initiation codon in the case of MsrBTk and an artificial stop codon for MsrATk were introduced. The amplified fragments were inserted into pUC118 and were sequenced. After the absence of unintended mutations was confirmed, the NdeI-EcoRI-digested fragments were inserted into pET21a(+) and used to transform E. coli BL21-CodonPlus(DE3)-RIL. The recombinant cells were grown in LB medium at 37°C, and gene expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside at the mid-exponential growth phase with further incubation for 4 h. Cells were harvested by centrifugation (4,500 × g, 4°C, 20 min), resuspended in 10 mM sodium phosphate buffer (pH 7.0), and sonicated for 5 min (output/pause ratio [in seconds], 10:20; output power, ∼80 W) with a Misonix Sonicator ultrasonic cell processor (XL2020; Farmingdale, NY). The lysates were centrifuged (20,000 × g, 4°C, 20 min), and the respective supernatants were subjected to heat treatment (MsrA, 5 min at 65°C; MsrB, 10 min at 80°C; MsrAB, 10 min at 75°C). After removing heat-labile proteins by ultracentrifugation (100,000 × g, 4°C, 30 min), the supernatants were applied to an anion exchange column (6 ml; Resource Q; Amersham Biosciences) equilibrated with buffer A (10 mM sodium phosphate buffer [pH 7.0], 1 mM dithiothreitol [DTT]). Recombinant proteins were eluted with a linear gradient of 0 to 1 M NaCl in buffer A. In the case of MsrATk, samples were further applied to a CHT2-I hydroxyapatite column (2 ml; Bio-Rad, Hercules, CA) equilibrated with buffer A and were eluted with a linear gradient of 10 to 500 mM sodium phosphate buffer (pH 7.0) containing 1 mM DTT. Fractions containing MsrBTk or MsrABTk after Resource Q treatment were applied to a hydrophobic column (6 ml; Resource ISO; Amersham Biosciences), equilibrated with 1 M (NH4)2SO4 in buffer A, and eluted with a linear gradient of 1 to 0 M (NH4)2SO4. Finally, all three protein solutions were subjected to gel filtration with a Superdex 200 HR 10/30 column (Amersham Biosciences), with a mobile phase of 50 mM sodium phosphate buffer (pH 7.0) containing 100 mM NaCl and 1 mM DTT and a flow rate of 0.3 ml min−1. Protein concentrations were determined with a protein assay kit (Bio-Rad) with bovine serum albumin as the standard.
Activity measurements of the recombinant Msr proteins.
Activity measurements were performed using MetO or dabsyl-MetO as the substrate. Dabsyl-MetO was synthesized as described elsewhere (28). Unless otherwise stated, reaction mixtures (50 μl) contained 50 mM sodium phosphate (pH 7.0), 20 mM DTT, 3 to 6 μg of purified enzyme, and substrate. MetO was used at various concentrations in the kinetic analyses, and dabsyl-MetO was used at 1 or 2 mM. Enzyme reactions were terminated by adding 5 μl of trifluoroacetic acid (10%, vol/vol). With MetO as the substrate, this solution was further centrifuged (15,000 × g, 10 min), and 10-μl aliquots of the supernatant were subjected to high-performance liquid chromatography (HPLC) using a C18 column (5C18-AR-II; Nacalai Tesque, Kyoto, Japan) equilibrated with 50 mM sodium phosphate buffer (pH 4.5) at a flow rate of 1 ml min−1. The amount of the reduced product, Met, was quantified by measuring the absorbance at 215 nm. In the case of dabsyl-MetO, 200 μl ethanol was added to the terminated reaction mixture followed by centrifugation (15,000 × g, 10 min), and 10-μl aliquots of the supernatant were subjected to HPLC using a chiral column (AD-H; Daicel Chemical Industries, Osaka, Japan), which could separate dabsyl-Met-S-O, dabsyl-Met-R-O, and dabsyl-Met. The column was equilibrated with 3:1 (vol/vol) n-hexane:ethanol containing 0.1% (vol/vol) acetic acid at a flow rate of 1 ml min−1. The dabsyl derivatives were detected by measuring the absorbance at 436 nm. In all measurements, experiments were performed in the absence of enzyme, and the amount of nonenzymatic reduction of MetO or dabsyl-MetO was subtracted for calculating enzyme activity.
The effects of pH on the MsrA and MsrB activities of MsrABTk were examined in standard reaction mixtures with the following buffer replacement: 200 mM morpholineethanesulfonic acid-NaOH (pH 5.5 to 6.5), 200 mM sodium phosphate (pH 6.0 to 7.5), or 200 mM Tris-HCl (pH 7.5 to 9.0). The reaction mixture was incubated at 50°C for 4 min, and the substrate reduced by the enzyme was analyzed as described above. The effect of temperature on the activity of each domain was examined in standard reaction mixtures. The reaction mixture was incubated at various temperatures for 1, 3, 5, and 7 min, and the consumption of the substrates was measured. For examination of protein thermostability, purified proteins (3 to 6 μg) were incubated in 50 mM sodium phosphate buffer (pH 7.0) at 70°C or 80°C for various periods of time. After incubation, residual enzyme activities were measured with standard procedures at 50°C.
Determination of nonenzymatic reduction of MetO.
The rates of free, racemic MetO [Met-(R, S)-O] reduction were determined at various temperatures in the presence of DTT without enzyme. The reaction mixture (50 μl) contained 1 mM Met-(R, S)-O; 20 mM DTT; and 50 mM sodium phosphate (pH 7.0). The reaction mixture was centrifuged, and 10-μl aliquots of the supernatant were applied to a C18 column (5C18-AR-II; Nacalai Tesque) equilibrated with 50 mM sodium phosphate buffer (pH 4.5) at a flow rate of 1 ml min−1. The amount of the reduced product, Met, was quantified by measuring the absorbance at 215 nm.
Circular dichroism (CD).
A J-820 spectropolarimeter (Jasco, Tokyo, Japan) was used to measure ellipticity as a function of wavelength from 250 to 195 nm, in 0.2-nm increments, using a 0.1-cm cylindrical quartz cuvette. MsrABTk was dissolved in 50 mM sodium phosphate buffer (pH 7.0) at a concentration of 0.8 mg ml−1. The thermal denaturation curve at 222 nm was measured with a temperature elevation rate of 0.5°C min−1, and data were collected every 0.2°C.
Fluorescence spectra.
Intrinsic fluorescence spectra were measured with a fluorescence spectrophotometer capable of maintaining the cuvette at desired temperatures between 30 and 100°C (model F-2500; Hitachi, Tokyo, Japan). MsrABTk was dissolved in 50 mM sodium phosphate buffer (pH 7.0) at a concentration of 0.05 mg ml−1. The fluorescence spectra of the MsrABTk solution excited at 260 and 295 nm were measured in the range of 300 to 450 nm at various temperatures.
Measurements of DO concentration.
DO concentrations were measured with a DO meter (Sension6; Hach, Loveland, CO) with methods recommended by the manufacturer. For measuring DO concentrations in the medium at temperatures above 50°C, the medium was incubated at the desired temperature for a sufficient period of time and then was tightly sealed in vials without headspace and incubated at 30°C for 20 min. The vials were opened, and DO concentrations were immediately and continuously monitored. The initial rates in the increase of DO concentration were linear and were used to calculate the original DO concentration in the medium.
RESULTS
The MsrAB fusion protein of T. kodakaraensis displays unique structural features among the archaeal Msr proteins.
From the complete genome sequence of T. kodakaraensis, we found an open reading frame that encoded a protein (TK0819) whose N-terminal and C-terminal regions displayed similarity to previously characterized MsrA and MsrB proteins, respectively. We designated the gene msrABTk, and we designated its translation product MsrABTk. MsrABTk was composed of 338 amino acids, and the molecular mass was calculated to be 39,119 Da. Although the N-terminal region of MsrABTk was shorter than those of the eukaryotic and bacterial enzymes, the possibilities that translation initiates further upstream are extremely low. A putative ribosomal binding site was present from positions −8 to −12 (relative to the initiation codon), a TATA sequence from −34 to −39 was present, and an in-frame stop codon was found from −52 to −54. The MsrA domain (designated residues 1 to 184) was 49% identical to that of the MsrA from E. coli and was 33% identical to that from Saccharomyces cerevisiae. The MsrB domain (residues 186 to 338) was 44% identical to that of the MsrB from E. coli and was 31% identical to that from S. cerevisiae. Sequence alignments of the respective domains with various Msr proteins/domains are shown in Fig. 1A (for MsrA) and B (for MsrB). The MsrA domain of MsrABTk possesses a GCFWC sequence near its N terminus (residues 17 to 21), indicating that this domain is a member of the third group of MsrA proteins (MsrAIII). As this domain does not harbor any further Cys residues, it is most likely that catalysis is carried out by these two residues. The MsrB domain of MsrABTk does not contain the CxxC motifs, indicating that it is a form II MsrB, previously found only in bacteria. The strictly conserved catalytic CysA residue of MsrB proteins was present (Cys311), but the second CysB residue, conserved in a majority of MsrB proteins, was not found. Additional Cys residues at positions 278 and 283 may act as the CysB in the MsrB domain (Fig. 1B).
FIG. 1.
Sequence alignment of MsrA proteins/domains (A) and MsrB proteins/domains (B). Alignments for MsrA were constructed with the following sequences: Azoarcus sp., CAI06587; Bacillus cereus, AAS44469; Bacillus licheniformis, AAU41183; Bacillus subtilis, CAB14087; Bdellovibrio bacteriovorus, CAE78986; D. melanogaster, AAN28311; E. coli, AAC77176; Haloarcula marismortui (1), AAV46883; Haloarcula marismortui (2), AAV46414; Halobacterium sp., AAG19555; Heliobacillus mobilis, AAN87501; Homo sapiens, AAP97154; Idiomarina loihiensis, AAV82363; Methanococcus maripaludis, CAF30404; Methanosarcina acetivorans, AAM04846; Methanosarcina mazei, AAM32095; Methanothermobacter thermautotrophicus, AAB85041; Mus musculus, AAH89311; Mycobacterium bovis, CAD93006; Neisseria gonorrhoeae, CAA32146; N. meningitides, AAF40515; Picrophilus torridus, AAT42728; Pseudomonas aeruginosa, AAG08403; S. cerevisiae, AAT92817; S. solfataricus, AAK41726; Symbiobacterium thermophilum, BAD40746; T. kodakaraensis, BAD85008; Vibrio cholerae, AAF96516; and Vibrio vulnificus, AAO08507. Alignments for MsrB were constructed with the following sequences: Azoarcus sp., CAI09433; Bacillus cereus, AAS44469; Bacillus licheniformis, AAU41182; Bacillus subtilis, CAB14086; Bdellovibrio bacteriovorus, CAE79257; D. melanogaster, AAN13491; E. coli, AAC74848; Haloarcula marismortui, AAV45184; Halobacterium sp., AAG19724; Heliobacillus mobilis, AAN87501; Homo sapiens, AAQ88596; I. loihiensis, AAV82363; Methanosarcina acetivorans, AAM03895; Methanosarcina mazei, AAM31330; Methanothermobacter thermautotrophicus, AAB85216; Mus musculus, AAH96447; Mycobacterium bovis, CAD94878; N. gonorrhoeae, CAA32146; N. meningitides, AAF40515; P. aeruginosa, AAG06215; S. cerevisiae, CAA42383; Symbiobacterium thermophilum, BAD40746; T. kodakaraensis, BAD85008; V. cholerae, AAF96516; and V. vulnificus, AAO08507. Representative sequences were selected and are shown. Amino acid residues presumed to act as CysA, CysB, and CysC are indicated by arrowheads and are highlighted in yellow. Other Cys residues that may be involved in catalysis are indicated in red. Cys86 of MsrA from E. coli has been experimentally verified not to be involved in catalysis. The two CxxC motifs that bind to a divalent zinc cation in MsrB are indicated in pink. Conserved residues are indicated with asterisks. Abbreviations: Dme, D. melanogaster; Eco, E. coli; Mmu, Mus musculus; Nme, N. meningitides; Sso, S. solfataricus; Tko, T. kodakaraensis.
Phylogenetic analyses of MsrA and MsrB proteins/domains were performed. Although a number of sequences from mesophilic archaea were included in the analyses, the MsrA and MsrB domains of MsrABTk were more closely related to the domains found in bacterial fusion proteins. Together with the fact that the MsrB domain is a bacterial-type form II enzyme, this suggests that the MsrABTk gene in T. kodakaraensis is a result of horizontal gene transfer from bacteria harboring MsrAB fusion genes.
Expression and purification of recombinant MsrATk, MsrBTk, and MsrABTk.
The msrABTk gene was expressed in E. coli, and the recombinant protein was purified by heat treatment and anion exchange, hydrophobic, and gel filtration chromatographies. In order to clarify the stereoselectivity of the respective domains, the individual domains also were produced separately in E. coli. The protein corresponding to the N-terminal domain was identical to the N-terminal 184 residues of MsrABTk and was designated MsrATk. An artificial Met residue was inserted at the N-terminal side of residue 186Phe to produce the recombinant C-terminal domain, designated MsrBTk. Both proteins were purified as described in Materials and Methods. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed that the recombinant proteins were purified to apparent homogeneity with the expected molecular size (data not shown).
Stereoselectivity of MsrATk and MsrBTk.
We first examined the Msr activity of the recombinant proteins with several electron donors. By measuring the production of Met from Met-(R, S)-O with HPLC, we found that the recombinant MsrATk and MsrBTk proteins exhibited Msr activity in the presence of 20 mM DTT (MsrATk, 3.5 μmol min−1 mg−1; MsrBTk, 1.9 μmol min−1 mg−1). In the case of MsrBTk, we also observed activity with 20 mM cysteine (0.6 μmol min−1 mg−1). No activity was observed in either protein with 20 mM reduced glutathione. The recombinant MsrABTk protein also exhibited activity in the presence of DTT. Therefore, further enzymatic examinations were performed with DTT as the electron donor.
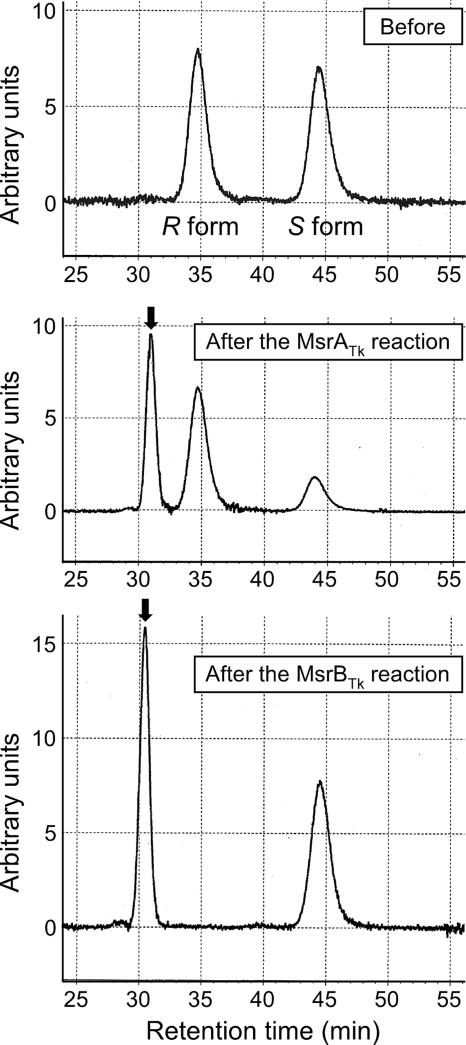
We examined the stereoselectivity of MsrATk. Partially enriched Met-S-O solution (S/R ratio of 73:27) and Met-R-O solution (S/R ratio of 5:95) were prepared from l-MetO as described elsewhere (32). The enrichment of either one of the epimers was confirmed by polarimetric measurements (data not shown). These solutions and a racemic solution (S/R ratio of 50:50) were used as substrates for MsrATk. MsrATk displayed the highest activity with the Met-S-O-enriched solution, followed by the racemic solution, and displayed relatively low levels of activity against the Met-R-O-enriched solution. In order to clarify whether the preference for Met-S-O was strict, further experiments were carried out using dabsyl-MetO. Dabsyl-MetO and acetyl-MetO often are used as model substrates that are presumed to better represent MetO residues within polypeptide chains (25). Dabsylated MetO also enables the separation of the two epimers, dabsyl-Met-S-O and dabsyl-Met-R-O, with a chiral column, as shown in Fig. 2. After the reaction with MsrATk, we observed the reduction of only one of the two epimers, with no detectable decrease of the other. Taking into account the preference of MsrATk for Met-S-O, we can conclude that the domain is strictly specific for the reduction of Met-S-O. When MsrBTk was examined in the same manner, we found specific reduction of the opposite epimer. This clearly indicates that MsrBTk is specific for Met-R-O.
FIG. 2.
Stereoselectivity of MsrATk and MsrBTk. Reactions were carried out as described in Materials and Methods with 1 mM dabsyl-MetO and 6 μg of purified protein. The MsrATk and MsrBTk reactions were performed for 30 min at 60 and 30°C, respectively. The arrow indicates the peak of the reaction product, dabsyl-Met.
Thermostabilities of MsrATk, MsrBTk, and MsrABTk.
The thermostabilities of MsrATk, MsrBTk, and MsrABTk were examined at various temperatures. By using the dabsyl-MetO substrates, we could examine the thermostabilities of the two domains of the MsrABTk fusion protein individually. While the recombinant MsrBTk protein was relatively stable at 80°C (t1/2 = 48 min), the MsrATk protein was surprisingly thermolabile (t1/2 ≪ 1 min). Interestingly, the MsrA domain within the MsrABTk protein exhibited higher thermostability (t1/2 = 27 min) than the individual MsrATk protein, suggesting that the domain is stabilized through its physical interaction with the more thermostable MsrB domain (t1/2 = 31 min). However, although MsrABTk is much more stable than a protein from a mesophile, the levels of thermostability of both domains were relatively low compared to those of other proteins from this hyperthermophile, which grows optimally at 85°C. We observed a 75% decrease in total activity after 2.5 min at 85°C.
Optimum pH and temperature of MsrABTk.
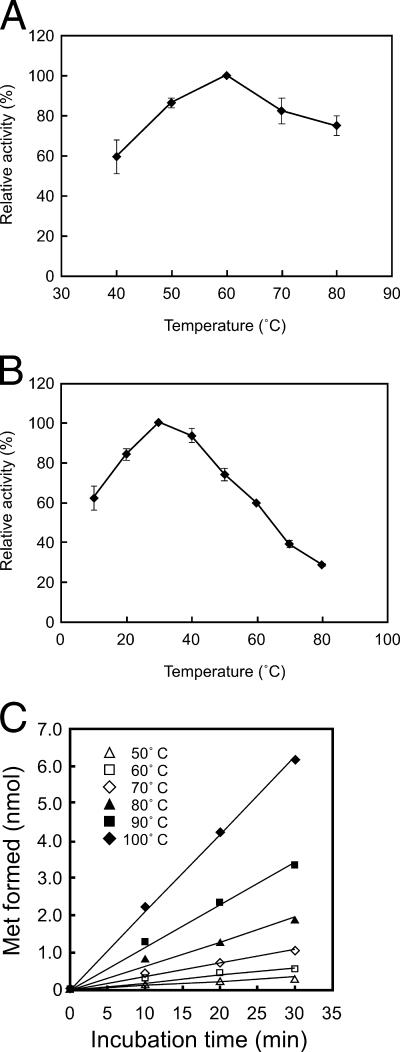
The effect of pH on the reactions catalyzed by each domain of recombinant MsrABTk was examined with dabsyl-MetO. We found that the MsrB domain exhibited the highest activity between pH 6.0 and 7.0, while the optimum pH of the MsrA domain was slightly higher, in the range of 7.5 to 8.0 (data not shown). We further examined the effects of temperature on the respective reactions. Using dabsyl-MetO, the MsrA domain of MsrABTk exhibited the highest activity at a relatively low temperature of 60°C (Fig. 3A). Moreover, in the case of the MsrB domain, activity was measured from 10 to 80°C, and maximum activity levels were observed at a surprisingly low 30°C (Fig. 3B). In these experiments, we could not apply saturating concentrations of substrate. As the reaction product is the same dabsyl-Met, it was necessary to quantify the decrease in substrate concentration (dabsyl-Met-S-O or dabsyl-Met-R-O) in order to distinguish the activities of the two domains in MsrABTk. In order to estimate the specific activities of each domain, we applied nondabsylated, free MetO substrates and measured the reaction rates with MsrATk, MsrBTk and MsrABTk at 30 and 60°C. In the presence of 20 mM DTT and 50 mM MetO, MsrATk displayed specific activity levels of 0.16 and 2.7 μmol min−1 mg protein−1 at 30 and 60°C, respectively. The results for MsrBTk were 1.4 (30°C) and 0.34 (60°C) μmol min−1 mg protein−1, and those for MsrABTk were 0.54 (30°C) and 2.2 (60°C) μmol min−1 mg protein−1. Consistent with the results of the MsrB domain using dabsyl-MetO, we found that MsrBTk also exhibited higher levels of activity at 30°C than at 60°C by using high concentrations of MetO. We performed a kinetic analysis of the MsrATk and MsrBTk reactions to MetO and DTT. Examinations were performed at the optimal reaction temperatures for MsrATk (60°C) and MsrBTk (30°C). The Km and Vmax values are shown in Table 2.
FIG. 3.
Enzymatic characterization of MsrABTk. (A and B) Effect of temperature on activity levels of the MsrA domain (A) and the MsrB domain (B). Reactions were performed for 5 min at substrate concentrations of 2 mM (A) and 1 mM (B). The values are the averages from two independent measurements. (C) Nonenzymatic reduction of MetO. The reaction mixture (50 μl) contained 1 mM Met-(R, S)-O, 20 mM DTT, and 50 mM sodium phosphate (pH 7.0). The produced Met was detected and quantified by HPLC. Aliquots (10 μl) of the reaction mixture after centrifugation were applied to a C18 column (5C18-AR-II; Nacalai Tesque) equilibrated with 50 mM sodium phosphate buffer (pH 4.5) at a flow rate of 1 ml min−1. The amount of Met was monitored by absorbance at 215 nm.
TABLE 2.
Kinetic parameters of the reactions catalyzed by MsrATk and MsrBTk
| Enzyme | Substrate | Apparent Km (mM) | Vmax (μmol min−1 mg−1) |
|---|---|---|---|
| MsrATk | MetOa (Met-S-O) | 19 ± 3 (9.3 ± 1.3) | 7.8 ± 0.3 |
| DTTb | 15 ± 2 | ||
| MsrBTk | MetOc (Met-R-O) | 67 ± 6 (33 ± 3) | 3.0 ± 0.1 |
| DTTb | 2.0 ± 0.2 |
In the presence of 100 mM DTT.
In the presence of 80 mM MetO.
In the presence of 20 mM DTT.
The higher levels of MsrB activity at lower temperatures were intriguing, considering that the domain (and the MsrBTk protein) is relatively thermostable in this temperature range (t1/2 = 31 min at 80°C). In order to confirm that these results accurately reflected the initial velocity of the reaction, we measured the consumption of substrate (dabsyl-Met-R-O) after various intervals of time. We found that the substrate consumption was proportional with time, ruling out the possibility that enzyme inactivation at higher temperatures led to these results. Generation of dabsyl-Met was observed in these measurements, indicating that the substrate consumption was not due to an enzyme-independent degradation of substrate. Furthermore, we also analyzed the reaction in the absence of enzyme (see below) and found that the level of chemical degradation of MetO is actually lower at lower temperatures. There is little possibility that an enzyme-independent chemical conversion of the substrate is a major factor, as this would lead to our obtaining similar results with the MsrA and MsrB domains, which clearly is not the case. Our results indicate that the MsrB domain of MsrABTk is a unique example of a thermostable protein that exhibits higher catalytic efficiency at lower temperatures.
Nonenzymatic reduction of MetO at various temperatures.
While measuring Msr activity at various temperatures, we observed that the MetO reduction rates in the presence of DTT without enzyme became surprisingly high at higher temperature ranges. The rates of nonenzymatic MetO reduction at various temperatures are shown in Fig. 3C. As in the case of the enzyme-dependent reaction, Met generation was observed with the consumption of MetO, and peaks other than those appearing on the chromatogram after the enzyme-dependent reaction were not observed. Furthermore, MetO degradation was not observed in the absence of DTT, ruling out the possibility that MetO is converted to compounds other than Met in our reaction mixture. As shown in Fig. 3C, a clear elevation in reaction rates was observed, reaching >0.2 nmol min−1 in the presence of 1 mM MetO and 20 mM DTT. An Arrhenius plot of the reaction gave a linear plot in the temperature range examined, and it revealed that the activation energy of the reaction was 57.5 kJ mol−1, comparable to values observed for enzyme-catalyzed reactions.
CD analysis.
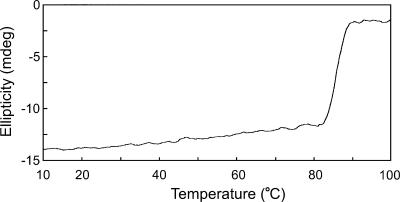
The intriguing effect of temperature on the MsrB activity of MsrABTk can be explained by a reversible change in conformation of the protein at elevated temperatures. In order to examine whether this change could be observed at the secondary structure level, we carried out a CD analysis of MsrABTk at various temperatures. A gradual increase in ellipticity was observed when temperatures were elevated from 40 to 50°C and 60 and 70°C. This change was reversible, as the CD spectrum was indistinguishable from the original spectrum when the temperature was lowered back to 40°C (data not shown). However, as the observed changes in the spectra were slight, we could not strongly relate these changes to the effect of temperature on activity. We next performed thermal denaturation experiments by following up on the change in ellipticity at 222 nm. The protein (0.8 mg ml−1) was dissolved in 50 mM sodium phosphate buffer (pH 7.0) in the presence (data not shown) or absence (Fig. 4) of 20 mM DTT. In each case, MsrABTk displayed a two-state folding transition with a midpoint of denaturation at approximately 83°C. This further supports the idea that the protein is relatively stable at temperatures up to 80°C but is readily denatured at 85°C, the optimal growth temperature of T. kodakaraensis. The results also indicate that the reversible change in conformation presumed to convert the enzyme to a less active form at higher temperatures does not involve a major change in the secondary structure of the protein.
FIG. 4.
CD analysis of MsrABTk. Shown is a thermal denaturation curve for MsrABTk at 222 nm measured with a temperature elevation rate of 0.5°C min−1. MsrABTk was present in 50 mM sodium phosphate buffer (pH 7.0) at a concentration of 0.8 mg ml−1. The y axis represents the measured ellipticity, in millidegrees (mdeg).
Fluorescence emission spectra of tryptophan and tyrosine residues.
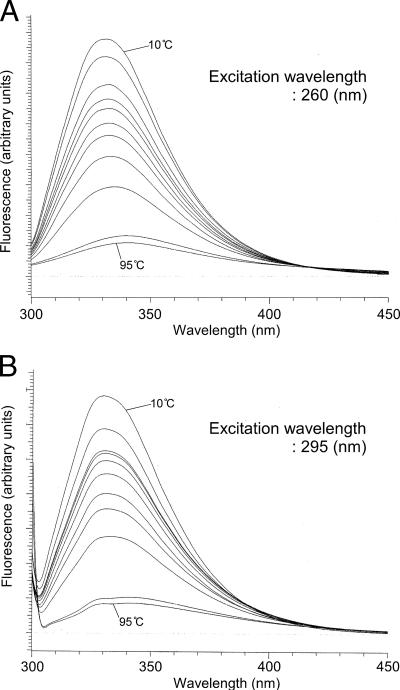
We next examined the fluorescence emission spectra of tryptophan and tyrosine residues at various temperatures. Excitation wavelengths of 260 (Fig. 5A) and 295 nm (Fig. 5B) were applied for tyrosine and tryptophan, respectively, as well as 280 nm (data not shown). Emission spectra were examined between 300 and 450 nm at temperatures of 10, 20, 30, 35, 40, 50, 60, 70, 80, 90, and 95°C. At lower temperatures, the maximum emission intensity was observed between 330 and 333 nm, and a gradual decrease in intensity was observed with an elevation in temperature. At 90°C, a temperature at which the protein unfolds irreversibly, a drastic decrease in intensity, along with a characteristic red shift of the emission peak, was observed. Comparisons of the spectra at various temperatures did not reveal any significant changes that seemed to correlate with the activity levels of the protein at low temperatures. The results agreed well with those obtained with CD spectrum analyses.
FIG. 5.
Intrinsic fluorescence spectra of MsrABTk at various temperatures. MsrABTk was dissolved in 50 mM sodium phosphate buffer (pH 7.0) at a concentration of 0.05 mg ml−1. The fluorescence spectra of MsrABTk solution excited at 260 (A) and 295 nm (B) were measured in the range of 300 to 450 nm at 10, 20, 30, 35, 40, 50, 60, 70, 80, 90, and 95°C. In both cases, the peak intensity decreased with the elevation in temperature.
Presence of MsrABTk protein in T. kodakaraensis.
As the optimal temperatures of MsrATk and MsrBTk were unusually low for an enzyme from T. kodakaraensis, we examined the presence of the MsrABTk protein at various temperatures. Polyclonal antibodies were raised against the purified recombinant MsrABTk and were used to detect the protein in cells grown in ASW-YT-Pyr at temperatures between 60 and 90°C (Fig. 6A). The results clearly indicated that MsrABTk is present only in cells grown at suboptimal temperatures.
FIG. 6.
Detection of MsrABTk in T. kodakaraensis cells. (A) Western blot analysis was performed on cell extracts (10 μg) from cells grown under standard growth conditions in ASW-YT-Pyr at the temperatures indicated above the membrane. (B) Western blot analysis was performed on cell extracts (10 μg) obtained from T. kodakaraensis cells grown under various conditions that are described above the panel. Condition numbers indicated below the panel correspond to those of Table 3. Relative signal intensities (panel A, 60°C = 100; panel B, C1 = 100) are indicated below each lane.
As the function of MsrABTk can be presumed to be involved in the response against oxygen or oxidative stress, we next focused on whether protein levels of MsrABTk were affected by the presence of oxygen. We examined the growth of T. kodakaraensis inoculated or grown under different culture conditions, taking into account the DO concentrations and the color of resazurin in the medium. The color of resazurin in the medium at 0.25 mg liter−1 starts to fade at DO concentrations of 0.1 mg liter−1, and it becomes completely transparent at 0.05 mg liter−1. Under standard growth conditions (Table 3, condition C1 to C3), the medium is reduced with 0.1 g liter−1 sodium sulfide at room temperature in an anaerobic atmosphere, and cells are inoculated in this anaerobic environment. Culture flasks are sealed with butyl rubber stoppers. The resazurin was transparent, and the DO concentration at 25°C was 0.01 mg liter−1. The alternative growth conditions considered were combinations of (i) omitting the addition of sodium sulfide (C4 to C6, C10 to C12, and C16 to C18), (ii) inoculating in an aerobic atmosphere (C7 to C18), and (iii) using silicone sponge plugs instead of butyl rubber stoppers (C13 to C18). Cultures were grown at 65, 75, and 85°C without shaking, and the occurrence of growth under these conditions are summarized in Table 3. When the addition of sodium sulfide was omitted, the color of the resazurin was red regardless of the atmosphere and plugs used. Without sodium sulfide, we observed growth only when cells were inoculated in an anaerobic atmosphere and the flasks were sealed with butyl rubber stoppers (C4 to C6) or when cells were grown in an aerobic atmosphere with silicone sponge plugs at 85°C (C18). Under conditions C4 to C6, the DO concentration at 65 to 85°C can be expected to be considerably lower than 2.86 ± 0.22 mg liter−1, as this is the value observed at 25°C prior to inoculation. Under condition C18, medium without cell inoculation displayed a DO concentration of 1.34 ± 0.03 mg liter−1 at 85°C. As conditions C16 and C17 could not support growth, the threshold of DO that T. kodakaraensis can overcome to initiate growth most likely is between concentrations of 1.34 and 2.53 mg liter−1. Once growth was observed, resazurin rapidly became transparent (<30 min), indicating that growing cells are actively removing oxygen from their environment. When sodium sulfide was added, all media displayed initial DO concentrations of 0.01 mg liter−1, and cell growth was observed in all cases. The DO values under conditions C13 to C15 were 0.61 ± 0.07, 0.57 ± 0.05, and 0.23 ± 0.06 mg liter−1, respectively, after 12 h, corresponding to the period of time between inoculation and initial cell growth.
TABLE 3.
Growth of T. kodakaraensis under various conditions of oxygen stress
| Atmosphere during inoculation, plug type, and condition no. | Addition of 0.1 g liter−1 Na2S | Temp (°C) | Color of resazurin during inoculation | Initial DO at 25°C (mg liter−1) | DO at incubation temp (mg liter−1) | Growth |
|---|---|---|---|---|---|---|
| Anaerobic | ||||||
| Butyl rubber | ||||||
| C1 | Yes | 65 | − | 0.01 | <0.01 | Yes |
| C2 | Yes | 75 | − | 0.01 | <0.01 | Yes |
| C3 | Yes | 85 | − | 0.01 | <0.01 | Yes |
| C4 | No | 65 | +a | 2.86 ± 0.22 | <2.86 | Yes |
| C5 | No | 75 | +a | 2.86 ± 0.22 | <2.86 | Yes |
| C6 | No | 85 | +a | 2.86 ± 0.22 | <2.86 | Yes |
| Aerobic | ||||||
| Butyl rubber | ||||||
| C7 | Yes | 65 | − | 0.01 | <0.01 | Yes |
| C8 | Yes | 75 | − | 0.01 | <0.01 | Yes |
| C9 | Yes | 85 | − | 0.01 | <0.01 | Yes |
| C10 | No | 65 | + | 5.62 ± 0.09 | <5.62 | No |
| C11 | No | 75 | + | 5.62 ± 0.09 | <5.62 | No |
| C12 | No | 85 | + | 5.62 ± 0.09 | <5.62 | No |
| Silicone sponge | ||||||
| C13 | Yes | 65 | − | 0.01 | 0.61b ± 0.07 | Yes |
| C14 | Yes | 75 | − | 0.01 | 0.57b ± 0.05 | Yes |
| C15 | Yes | 85 | − | 0.01 | 0.23b ± 0.06 | Yes |
| C16 | No | 65 | + | 5.62 ± 0.09 | 3.66 ± 0.01 | No |
| C17 | No | 75 | + | 5.62 ± 0.09 | 2.53 ± 0.03 | No |
| C18 | No | 85 | +a | 5.62 ± 0.09 | 1.34 ± 0.03 | Yes |
Rapid loss of color was observed after initiation of growth.
Measured after 12 h of incubation at the respective temperatures.
Under all conditions in which we observed cell growth, we examined the presence of MsrABTk by Western blot analysis (Fig. 6B). As described above, MsrABTk is observed only at suboptimal temperatures when cells are grown under standard growth conditions (C1 to C3). When the addition of sodium sulfide was omitted, we observed a slight increase of MsrABTk in cells grown at 65°C (C4). When cells were inoculated under aerobic conditions, we observed increases of the protein at 75°C (C8 and C14). MsrABTk could not be detected in cells inoculated and grown under aerobic conditions at 85°C (C15 and C18). The results highlight two points. First, the medium under standard growth conditions contains <0.01 mg liter−1 oxygen. We can thus conclude that the presence of MsrABTk clearly responds to temperature, even in the absence of oxygen (C1 to C3). The second point is that the presence of MsrABTk also responds to the presence of oxygen. This can be observed by comparing the protein levels in cells grown at 75°C under different growth conditions (C2, C5, C8, and C14). However, as MsrABTk was not present at detectable levels under aerobic conditions at 85°C, a condition with a higher DO than that of condition C14, MsrABTk is regulated primarily by temperature, and the response to DO occurs only at suboptimal growth temperatures.
DISCUSSION
In this study, we have examined an MsrA-MsrB fusion protein from T. kodakaraensis KOD1, the presence of which is exceptional among the hyperthermophiles. The two domains of MsrABTk exhibited strict substrate selectivity, namely, MsrA for Met-S-O and MsrB for Met-R-O. The most striking feature of the protein was that MsrB activity levels were found to be maximal at 30°C, well below the optimal growth temperature of this hyperthermophile (85°C). Consistent with this behavior, the protein readily denatures at 85°C in vitro. There is a possibility that the protein exhibits higher thermostability within the cell by the interaction with various intracellular compounds, such as compatible solutes or potassium glutamate (46). The optimal temperature for activity also may shift to higher temperatures when the enzyme is dependent on the native thioredoxin as an electron donor. Nevertheless, the presence of the protein in T. kodakaraensis cells was observed only at suboptimal growth temperatures. These biochemical and regulatory properties strongly suggest that MsrABTk is a low-temperature-specific enzyme in T. kodakaraensis and, to our knowledge, is the first example of its kind from a hyperthermophile.
The MsrB domain of MsrABTk exhibited maximum activity at the strikingly low temperature of 30°C. This also was observed for the recombinant MsrBTk protein. This was not due to (irreversible) thermal inactivation of the proteins at higher temperatures, as both MsrABTk and MsrBTk displayed half-lives of over 30 min at 80°C. This indicates that the MsrB domain undergoes a reversible change in conformation at higher temperatures that brings about a decrease in activity. The decrease in activity is most likely due to a decrease in kcat, as the specific activity of the MsrBTk protein under saturating concentrations of MetO also displayed higher levels of activity at lower temperatures. As the CD and fluorescence emission spectra did not point to large or global changes in structure, the changes in activity levels of the enzyme at different temperatures most likely are due to structural alterations localized near the active site.
In the literature, a novel concept for enzymes has been proposed and experimentally verified that introduces a third intrinsic thermal parameter for enzymes, Teq (36). The concept is based on the presumption that enzymes are at a reversible equilibrium between active (Eact) and inactive forms (Einact) and that irreversible inactivation of the enzyme to the thermally denatured state (X) initiates only from the inactive form (Eact⇄Einact→X). The concentration of active enzyme can always be expressed as [Eact] = ([E0] − [X])/(1 + Keq), where [E0] is the initial total concentration of enzyme and Keq is the equilibrium constant between active and inactive forms of the enzyme (Keq = [Einact]/[Eact]). Teq is defined as the temperature at which Keq = 1 or [Eact] = [Einact]. This concept has been experimentally verified for at least five enzymes and consequently reveals that enzymes intrinsically harbor an optimal temperature for activity that is no longer dependent on thermal stability (36). This can easily be envisioned for an enzyme with a flexible catalytic core mounted on a rigid, global scaffold with high thermal stability. The MsrB domain of MsrABTk may very well be one extreme example of this kind of enzyme behavior, with a catalytic core structure that is optimal at 30°C while the overall protein does not display significant thermal denaturation until temperatures exceed 80°C.
The presence of MsrABTk was not observed in T. kodakaraensis cells at the optimal growth temperature, even in an aerobic atmosphere with relatively high DO levels, but rather was present at low temperatures of 60 to 70°C. At high temperatures, the function of Msr may not be necessary, as we observed high rates of MetO reduction in an enzyme-independent manner. Although the rates of the enzyme-independent reaction greatly rely on the concentration and redox potential of the native electron donor (thioredoxinred) in vivo, the high reaction rates observed in our reaction mixtures at elevated temperatures may be related to the absence of Msr homologs in most hyperthermophiles, particularly those that grow only at extremely high temperature ranges. T. kodakaraensis and S. solfataricus, the only hyperthermophiles harboring an Msr, exhibit relatively low optimal growth temperatures compared to those of other hyperthermophiles (3, 14). The function of the enzyme therefore would be more relevant in these organisms, which grow at temperatures lower than those that support the majority of other hyperthermophiles.
In addition to its function in growing cells, MsrABTk also may function in cells that are in the process of, or are recovering from, exposure to low-temperature environments that can no longer sustain growth. A decrease in temperature brings about an increase in DO concentrations. Oxidative damage of biomolecules therefore can be expected to accumulate at these low temperatures, particularly when the cells are dormant. Production of MsrABTk at suboptimal growth temperatures would prepare the cell to tolerate further, more drastic temperature decreases and also would help the cell to recover from these conditions by initiating the repair of oxidized Met residues as soon as possible. MsrABTk exhibits activity at temperatures much lower than those expected to support growth of T. kodakaraensis. There would be a great advantage if the enzyme were to function in the cells throughout the temperature range observed in our in vitro experiments. However, the lower temperature limit for the enzyme to function as well as turnover in vivo may well be restricted not by the enzyme itself but by the activity levels of thioredoxin reductase and other enzymes that provide the reducing power to repair MetO. The effect of temperature on the activities and expression levels of these proteins surely will be an attractive topic for future research.
Acknowledgments
We thank Kentaro Shiraki of the Institute of Applied Physics, University of Tsukuba, for fruitful discussions of the CD analyses.
This work was supported by a grant of the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Amend, J. P., and E. L. Shock. 2001. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25:175-243. [DOI] [PubMed] [Google Scholar]
- 2.Antoine, M., S. Boschi-Muller, and G. Branlant. 2003. Kinetic characterization of the chemical steps involved in the catalytic mechanism of methionine sulfoxide reductase A from Neisseria meningitidis. J. Biol. Chem. 278:45352-45357. [DOI] [PubMed] [Google Scholar]
- 3.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boschi-Muller, S., S. Azza, S. Sanglier-Cianferani, F. Talfournier, A. Van Dorsselear, and G. Branlant. 2000. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J. Biol. Chem. 275:35908-35913. [DOI] [PubMed] [Google Scholar]
- 5.Boschi-Muller, S., A. Olry, M. Antoine, and G. Branlant. 2005. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta 1703:231-238. [DOI] [PubMed] [Google Scholar]
- 6.Brot, N., L. Weissbach, J. Werth, and H. Weissbach. 1981. Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. USA 78:2155-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, G. N., V. Barbe, D. Flament, M. Galperin, R. Heilig, O. Lecompte, O. Poch, D. Prieur, J. Quérellou, R. Ripp, J. C. Thièrry, J. Van der Oost, J. Weissenbach, Y. Zivanovic, and P. Forterre. 2003. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47:1495-1512. [DOI] [PubMed] [Google Scholar]
- 8.Erauso, G., A.-L. Reysenbach, A. Godfroy, J.-R. Meunier, B. Crump, F. Pafrtensky, J. A. Baross, V. Marteinsson, G. Barbier, N. R. Pace, and D. Prieur. 1993. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch. Microbiol. 160:338-349. [Google Scholar]
- 9.Ezraty, B., L. Aussel, and F. Barras. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta 1703:221-229. [DOI] [PubMed] [Google Scholar]
- 10.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 11.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, J. M., Y. Masuchi, F. T. Robb, J. W. Ammerman, D. L. Maeder, M. Yanagibayashi, J. Tamaoka, and C. Kato. 1998. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles 2:123-130. [DOI] [PubMed] [Google Scholar]
- 13.González, J. M., D. Sheckells, M. Viebahn, D. Krupatkina, K. M. Borges, and F. T. Robb. 1999. Thermococcus waiotapuensis sp. nov., an extremely thermophilic archaeon isolated from a freshwater hot spring. Arch. Microbiol. 172:95-101. [DOI] [PubMed] [Google Scholar]
- 14.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber, C., A. Legat, M. Pfaffenhuemer, C. Radax, G. Weidler, H. J. Busse, and H. Stan-Lotter. 2004. Halobacterium noricense sp. nov., an archaeal isolate from a bore core of an alpine Permian salt deposit, classification of Halobacterium sp. NRC-1 as a strain of H. salinarum and emended description of H. salinarum. Extremophiles 8:431-439. [DOI] [PubMed] [Google Scholar]
- 16.Holden, J. F., K. Takai, M. Summit, S. Bolton, J. Zyskowski, and J. A. Baross. 2001. Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern Pacific Ocean. FEMS Microbiol. Ecol. 36:51-60. [DOI] [PubMed] [Google Scholar]
- 17.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, T. 2003. Taxonomy of nonmethanogenic hyperthermophilic and related thermophilic archaea. J. Biosci. Bioeng. 96:203-212. [PubMed] [Google Scholar]
- 19.Jones, W. J., J. A. Leigh, F. Mayer, C. R. Woese, and R. S. Wolfe. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136:254-261. [Google Scholar]
- 20.Kauffmann, B., A. Aubry, and F. Favier. 2005. The three-dimensional structures of peptide methionine sulfoxide reductases: current knowledge and open questions. Biochim. Biophys. Acta 1703:249-260. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 23.Keller, M., F.-J. Braun, R. Dirmeier, D. Hafenbradl, S. Burggraf, R. Rachel, and K. O. Stetter. 1995. Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeum growing on polysulfide at alkaline pH. Arch. Microbiol. 164:390-395. [PubMed] [Google Scholar]
- 24.Klenk, H.-P., R. A. Clayton, J.-F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 25.Koc, A., A. P. Gasch, J. C. Rutherford, H.-Y. Kim, and V. N. Gladyshev. 2004. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. USA 101:7999-8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, R. A., A. Koc, R. L. Cerny, and V. N. Gladyshev. 2002. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 277:37527-37535. [DOI] [PubMed] [Google Scholar]
- 27.Kurr, M., R. Huber, H. König, H. W. Jannasch, H. Fricke, A. Trincone, J. K. Kristjansson, and K. O. Stetter. 1991. Methanopyrus kandleri gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch. Microbiol. 156:239-247. [Google Scholar]
- 28.Minetti, G., C. Balduini, and A. Brovelli. 1994. Reduction of DABS-l-methionine-dl-sulfoxide by protein methionine sulfoxide reductase from polymorphonuclear leukocytes: stereospecificity towards the l-sulfoxide. Ital. J. Biochem. 43:273-283. [PubMed] [Google Scholar]
- 29.Miroshnichenko, M. L., H. Hippe, E. Stackebrandt, N. A. Kostrikina, N. A. Chernyh, C. Jeanthon, T. N. Nazina, S. S. Belyaev, and E. A. Bonch-Osmolovskaya. 2001. Isolation and characterization of Thermococcus sibiricus sp. nov. from a Western Siberia high-temperature oil reservoir. Extremophiles 5:85-91. [DOI] [PubMed] [Google Scholar]
- 30.Moskovitz, J. 2005. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta 1703:213-219. [DOI] [PubMed] [Google Scholar]
- 31.Moskovitz, J., S. Bar-Noy, W. M. Williams, J. Requena, B. S. Berlett, and E. R. Stadtman. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 98:12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskovitz, J., J. M. Poston, B. S. Berlett, N. J. Nosworthy, R. Szczepanowski, and E. R. Stadtman. 2000. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 275:14167-14172. [DOI] [PubMed] [Google Scholar]
- 33.Olry, A., S. Boschi-Muller, and G. Branlant. 2004. Kinetic characterization of the catalytic mechanism of methionine sulfoxide reductase B from Neisseria meningitidis. Biochemistry 43:11616-11622. [DOI] [PubMed] [Google Scholar]
- 34.Oren, A., M. Ginzburg, B. Z. Ginzburg, L. I. Hochstein, and B. E. Volcani. 1990. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int. J. Syst. Bacteriol. 40:209-210. [DOI] [PubMed] [Google Scholar]
- 35.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. Delong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson, M. E., R. Eisenthal, M. J. Danson, A. Spence, and R. M. Daniel. 2004. A new intrinsic thermal parameter for enzymes reveals true temperature optima. J. Biol. Chem. 279:20717-20722. [DOI] [PubMed] [Google Scholar]
- 37.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, J. L., B. Pyzyna, R. G. Atrasz, C. A. Henderson, K. L. Morrill, A. M. Burd, E. Desoucy, R. E. Fogleman III, J. B. Naylor, S. M. Steele, D. R. Elliott, K. J. Leyva, and R. F. Shand. 2005. Growth kinetics of extremely halophilic Archaea (family Halobacteriaceae) as revealed by Arrhenius plots. J. Bacteriol. 187:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleper, C., G. Puehler, I. Holz, A. Gambacorta, D. Janekovic, U. Santarius, H.-P. Klenk, and W. Zillig. 1995. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 177:7050-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segerer, A., T. A. Langworthy, and K. O. Stetter. 1988. Thermoplasma acidophilum and Thermplasma volcanium sp. nov. from solfatara fields. Syst. Appl. Microbiol. 10:161-171. [Google Scholar]
- 43.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C.-Y. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, T., T. Iwasaki, T. Uzawa, K. Hara, N. Nemoto, T. Kon, T. Ueki, A. Yamagishi, and T. Oshima. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6:39-44. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, T., N. Kumar, and R. Cavicchioli. 2001. Effects of ribosomes and intracellular solutes on activities and stabilities of elongation factor 2 proteins from psychrotolerant and thermophilic methanogens. J. Bacteriol. 183:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhees, C. H., S. W. M. Kengen, J. E. Tuininga, G. J. Schut, M. W. W. Adams, W. M. De Vos, and J. Van der Oost. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Völkl, P., R. Huber, E. Drobner, R. Rachel, S. Burggraf, A. Trincone, and K. O. Stetter. 1993. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl. Environ. Microbiol. 59:2918-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]