Abstract
β-1,4-Galactan is a major component of the ramified regions of pectin. Analysis of the genome of the plant pathogenic bacteria Erwinia chrysanthemi revealed the presence of a cluster of eight genes encoding proteins potentially involved in galactan utilization. The predicted transport system would comprise a specific porin GanL and an ABC transporter made of four proteins, GanFGK2. Degradation of galactans would be catalyzed by the periplasmic 1,4-β-endogalactanase GanA, which released oligogalactans from trimer to hexamer. After their transport through the inner membrane, oligogalactans would be degraded into galactose by the cytoplasmic 1,4-β-exogalactanase GanB. Mutants affected for the porin or endogalactanase were unable to grow on galactans, but they grew on galactose and on a mixture of galactotriose, galactotetraose, galactopentaose, and galactohexaose. Mutants affected for the periplasmic galactan binding protein, the transporter ATPase, or the exogalactanase were only able to grow on galactose. Thus, the phenotypes of these mutants confirmed the functionality of the gan locus in transport and catabolism of galactans. These mutations did not affect the virulence of E. chrysanthemi on chicory leaves, potato tubers, or Saintpaulia ionantha, suggesting an accessory role of galactan utilization in the bacterial pathogeny.
Pectinolytic erwiniae are enterobacteria that cause soft-rot disease in a wide range of plant species, including many crops of economic importance such as vegetables and ornamentals (33). The maceration of plant tissues is essentially caused by the secretion by the bacteria of a set of pectin-degrading enzymes (e.g., pectate-lyases, methylesterases, pectin-lyase, and polygalacturonases). Pectin is the major matrix polysaccharide component of the primary cell wall and the middle lamella in plants. The degradation of pectin results in the general disorganization of the plant cell wall. Erwinia chrysanthemi is able to grow on the degraded polymers of pectin as the sole carbon source (21). This degradation is regulated by a complex system of interconnected regulatory networks (CRP, KdgR, PecT, PecS, etc.) (21).
The pectic polysaccharides represent between 30 and 50% of the cell walls of dicotyledonous plants. The pectic matrix is a complex mixture of homogalacturonan (HGA), rhamnogalacturonan I (RGI), and rhamnogalacturonan II (RGII) polymers (36). HGA is a linear chain of α-1,4-galacturonic acid (GalA). The RGII molecule has a HGA backbone with side chains containing a diversity of sugars and linkages. RGI is a branched heteropolymer of alternating α-1,2-rhamnose and α-1,4-GalA residues that carries neutral side chains of arabinan, galactan, or arabinogalactan attached to rhamnose residues of RGI backbone (43).
Two types of galactan side chains are distinguished. Type I consist of a chain of β-1,4-linked d-galactopyranose backbone, while type II contains a backbone of β-1,3-linked d-galactopyranose residues. The side chains significantly influence the physical properties of the pectin (25). In potato and in notoginseng, β-1,4-galactan is the most abundant type of neutral side chain of RGI and represents ca. 20% of the pectin oligosaccharides (45). Type I galactan is degraded by 1,4-β-endogalactanases and 1,4-β-exogalactanases (12). Galactanases are widely distributed into many plants. They cause the solubilization of pectin oligosaccharides and seem to play a major role in ripening of fruits (29, 32). 1,4-β-Endogalactanase has been also isolated from microorganisms, including aerobic fungi belonging to the genus Aspergillus (12) and bacteria such as Bacillus subtilis (13), Pseudomonas fluorescens (9), and Thermotoga maritima (44).
E. chrysanthemi produces several enzymes allowing the catabolism of pectic polymers (19, 21). Despite the fact that galactans may be an important carbon source derived from pectin, no galactan degrading system has been described in E. chrysanthemi. Examination of the E. chrysanthemi genome (Glasner et al., unpublished data) revealed a locus potentially involved in galactan degradation and catabolism. We report here experimental data demonstrating that the gan locus of E. chrysanthemi encodes a whole functional system for galactan utilization. The E. chrysanthemi galactan transport proteins shows similarities with E. coli maltose transport proteins (6). Galactan catabolism is catalyzed by a 1,4-β-endogalactanase and a 1,4-β-exogalactanase. The regulation of the corresponding genes was analyzed and the impact of galactan utilization on the virulence was tested.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The E. chrysanthemi and E. coli strains are listed in Table 1. Bacteria were grown at 30°C (E. chrysanthemi) or 37°C (E. coli) in Luria-Bertani broth (LB) or in minimal medium M63 supplemented with a carbon source at a concentration of 2 g/liter (30). Solid media were obtained by adding agar at 15 g/liter. Pectic galactans from potato, galactobiose, and azurine-cross-linked galactan (AZCL-galactan) were purchased from Megazyme (Bray, Ireland). Oligogalactans from Gal3 to Gal6 were obtained after a 6-h hydrolysis of pectic galactans 1% by the E. chrysanthemi 1,4-β-endogalactanase GanA in a sodium acetate buffer 0.2 M. The hexose content of galactose, galactans, and oligogalactans solutions was measured by the anthrone method (42). AZCL-galactan was used in overlay at 2 g/liter on solid minimal medium M63 containing glycerol as a carbon source. The degradation of AZCL-galactan was observed by a blue halo.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| E. chrysanthemi | ||
| 3937 | Wild type | Laboratory collection |
| NFB3666 | 3937, ganB::Cml | This study |
| A2507 | A350, lmrTc lacZ2 crp::Cml | 35 |
| NFB3677 | 3937, crp::Cml | This study |
| A4809 | 3937, ganA::uidA-Kan | This study |
| NFB3679 | 3937, ganA::uidA-Kan crp::Cml | This study |
| NFB3680 | 3937, lacZ::Cml | F. Bouchart |
| NFB3674 | 3937, ganB::Cml | This study |
| NFB3681 | 3937, ganB::FRT | This study |
| NFB3687 | 3937, lacZ::Cml ganB::FRT | This study |
| NFB3701 | 3937, ganK::uidA-Kan | This study |
| NFB3703 | 3937, ganK::uidA-Kan crp::Cml | This study |
| A4868 | 3937, ganL::uidA-Kan | This study |
| A4807 | 3937, ganB::uidA-Kan | This study |
| A4810 | 3937, ganE::uidA-Kan | This study |
| A4863 | 3937, ganR::Cml | This study |
| A4415 | 3937, pelD::uidA-Kan | 22 |
| E. coli | ||
| S17-1λpir | recA thi pro hsdR λpir RP4-2-Tet::Mu-Kan::Tn7; Tmp Str | 31 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
Comparison of growth on galactose (2 g/liter), galactobiose (1 g/liter) or oligogalactans (0.5 g/liter) was done in triplicate in 2.5-ml cultures after 18 h of incubation.
Antibiotics were used at the following concentrations: ampicillin and kanamycin at 50 μg/ml (E. coli) or 25 μg/ml (E. chrysanthemi) and chloramphenicol at 25 μg/ml (E. coli) or 12.5 μg/ml (E. chrysanthemi). X-Gal (5-bromo-4-chloro-3- indolyl-β-d-galactopyranoside) was used at a concentration of 20 μg/ml.
Recombinant DNA techniques.
Genomic and plasmid DNA extractions were performed according to standard procedures (39). Restriction enzymes (Eurogentec) and T4 DNA ligase (Gibco-BRL) were used according to the manufacturer's recommendations.
Construction of the mutations.
Plasmids are listed in Table 2. PCR primers were designed to amplify 1.1 to 2.1 kb of E. chrysanthemi 3937 chromosomal DNA encoding the gan genes (Table 3). Restriction sites were added in each primer to facilitate determination of the DNA orientation in the vector (usually BamHI or BglII at the 5′ end and XbaI at the 3′ end). The PCR products were purified (QIAquick PCR Purification Kit; QIAGEN), and ligated to the pGEM-T vector (Promega), which has a protruding T nucleotide at each 3′ end. For ganK, the amplified fragment was digested with XhoI and cloned into the same site of pBluescript II SK(+).
TABLE 2.
Plasmids
| Plasmid | Genotype and/or phenotypea | Source or reference |
|---|---|---|
| pBluescript II SK(+) | Ampr | Stratagene |
| pNFcml | Cmlr | Laboratory collection |
| pUIDK11 | CmlruidA-Kan | 3 |
| pGEM-T | Ampr | Promega |
| POK | sacB sacR mobRK2 oriRK6; Sper | 24 |
| pCP20 | FLP+ λcI857+; λprRepts; Ampr Cmlr | 10 |
| pI2872 | pGEM-T, ganA+ | This study |
| pI2873 | pGEM-T, ganA::uidA-Kan | This study |
| pI2874 | pGEM-T, ganB+ | This study |
| pI2876 | pGEM-T, ganB::uidA-Kan | This study |
| pI2866 | pGEM-T, ganE+ | This study |
| pI2867 | pGEM-T, ganE::uidA-Kan | This study |
| pI2868 | pGEM-T, ganL+ | This study |
| pI2869 | pGEM-T, ganL::uidA-Kan | This study |
| pI2870 | pGEM-T, ganR+ | This study |
| pI3136 | pGEM-T, ganR::Cml | This study |
| pNFW142 | pBluescript, ganB+ | This study |
| pNFW145 | pBluescript, ganB::Cml | This study |
| pNFW154 | pOK, ganB::Cml | This study |
| pNFW162 | POK, ganB::FRT | This study |
| pNFW158 | pBluescript, ganK+ | This study |
| pNFW163 | pBluescript, ganK::uidA-Kan | This study |
Cmlr, chloramphenicolresistance; Ampr, ampicillin resistance; Sper, spectinomycin resistance.
TABLE 3.
Primer sequences
| Primer | Sequencea |
|---|---|
| ganAG | GCGGATCCTGACCTCAGGCAGTACCAAAG |
| ganAD | CGTCTAGACGGATTCAAAGGCGTAAATCG |
| ganBG | GCGGATCCTATGCTCTCTGCCTATTCAG |
| ganBD | CGTCTAGATCAGATCAGACGGCGGCGAATC |
| ganKG | AAACTGCAGGCCGGCATGACAAACCCCCTTTCGCCAGAA |
| ganKD | AAACTGCAG CGCGTCGGTGAAGCTGTCTTGCTCGGCCTT |
| ganEG | GCGGATCCTCAACGGGTTATCAATACGG |
| ganED | CGTCTAGAGTTGTCCTTGCTCCAGGTCG |
| ganLG | GCAGATCTGGATTGTAACAGCTGGTCC |
| ganLD | CGTCTAGATAACCGGTCGGTGAGCGG |
| ganRG | GCGGATCCGGACCAGCTGTTACAATC |
| ganRD | CGTCTAGATGCGCCGCAACATGCTAC |
Restriction sites are underlined.
Inactivation and genetic fusions were constructed from the cloned genes by insertion of a uidA-Kan cassette (3) into a restriction site situated inside the corresponding open reading frame. The uidA-Kan cassette was inserted into the HpaI site of ganB, the EcoRV site of ganA, the BglII site of ganE, the BamHI site of ganL, and the ZraI site of ganK. The orientation of the uidA-Kan cassette was determined by restriction analysis. Only plasmids in which uidA and the mutated gene have the same transcriptional direction were retained. Inactivation of ganR and ganB was performed by inserting a chloramphenicol resistance (Cmlr) cassette (originating from pNFCml) into the PstI site and NruI site, respectively. This Cmlr cassette is flanked by two FRT sites recognized by the FLP recombinase. The FLP recombinase (expressed by the pCP20 plasmid) was used to obtain a ganB Cmls mutant by removing the Cmlr cassette (10).
Transduction, conjugation, and transformation.
Transformation of E. coli cells was carried out by the rubidium chloride technique (39). Plasmids were introduced in E. chrysanthemi by electroporation (35) or conjugation (24). The insertions were integrated into the E. chrysanthemi chromosome by marker-exchange recombination after successive cultures in low phosphate medium in the presence of the appropriate antibiotic (37) or in medium containing sucrose 5% when pOK plasmid was used as the vector (24).
Transduction of E. chrysanthemi with phage ΦEC2 was carried out according to the method of Resibois et al. (34).
Purification of 1,4-β-endogalactanase.
The periplasmic content of E. chrysanthemi cells was extracted by the chloroform method in 20 mM Tris-HCl (pH 8)-0.1 M NaCl buffer (1). The sample (5 mg of proteins) was loaded onto a Superdex 75 HR10/30 gel filtration column (Amersham Biosciences). Proteins were eluted in the same buffer, and fractions were assayed for 1,4-β-endogalactanase activity with the chromogenic substrate AZO-galactan (Megazyme, Bray, Ireland). 1,4-β-Endogalactanase-containing fractions were pooled (approximately 50 ml), loaded onto a MonoQ HR5/5 anion-exchange chromatography column (Amersham Biosciences), and eluted with a linear 0.1 to 1 M NaCl gradient. The 1,4-β-endogalactanase activity was recovered in the void volume. The protein fractions were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). The protein concentration was determined by the Bradford test using bovine serum albumin as a standard (8).
Determination of enzyme activities.
1,4-β-Endogalactanase activity was determined by hydrolysis of AZO-galactan according to instructions of the manufacturer. The reaction mixture consisted of 0.2 M sodium acetate buffer (pH 5.8) and 0.5% AZO-galactan. After incubation at 50°C for 15 min, the reaction was stopped by the addition of 1 ml of ethanol 95%. After 10 min at room temperature, a 10-min centrifugation at 1,000 × g allowed precipitation of high-molecular-weight polymers. The absorbance of oligomers in the supernatant was measured at 590 nm. The activity of purified 1,4-β-endogalactanase was assayed at different pHs and temperatures. The optimum pH was determined by incubation in buffered solutions (0.2 M) of sodium acetate, potassium phosphate, or Tris-HCl, depending on the pH. The optimum temperature was determined by incubation in 0.2 M sodium acetate (pH 5.8) at temperatures ranging from 23 to 70°C.
β-Galactosidase and β-glucuronidase assays were performed on crude extracts obtained from bacteria disrupted by passage through a French press cell at 1.4 × 107 Pa (20,000 lb/in2). β-Galactosidase and β-glucuronidase activities were determined by monitoring spectrometrically at 410 nm the hydrolysis of the ONPG (o-nitrophenyl-β-d-galactopyranoside) or PNPU (4-nitrophenyl-β-d-glucuronide), respectively.
MALDI-TOF spectrometry.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometry was performed with a Voyager-DE STR PRO (Applied Biosystems, Framingham) in the positive mode with DHB (2,3-dihydroxybenzoic acid) matrix.
Pathogenicity test.
Chicory leaves were inoculated as previously described (19), with slight modifications. Bacteria from an overnight culture in LB medium were recovered by centrifugation and diluted in M63 medium. Prior inoculation, the leaves were slightly wounded in their center with a sterile pipette tips. Ten leaves were infected for each strain using 107 bacteria per inoculation site. After incubation in a dew chamber for 24 h at 30°C, the length of rotted tissue was measured to estimate the disease severity. Bacterial cell numerations were performed by dilution plating to estimate the bacterial multiplication. In parallel, β-glucuronidase assays were performed on the macerated tissues to assess the expression of the gene fusions during plant infection. The specific activity is expressed as micromoles of product liberated per minute per 1010 bacteria. Potato tubers and plants of Saintpaulia ionantha (African violets) were inoculated as previously described (19).
Sequence data.
The sequences of ganA (ABF-0018196), ganB (ABF-0018198), ganC (ABF-0018199), ganE (ABF-0018192), ganF (ABF-0018193), ganG (ABF-0018195), ganK (ABF-00190), ganL (ABF-0018200), and ganR (ABF-0018202) are available (http://asap.ahabs.wisc.edu/asap/ASAP1.htm site).
RESULTS
Identification of the gan locus.
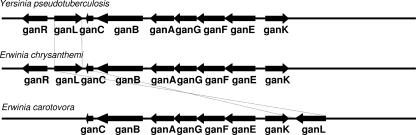
Two recent proteomic studies performed on the E. chrysanthemi 3937 strain revealed the presence of an abundant protein (AFB-0018196) of unknown function (2, 7). Its primary sequence suggested that this protein is a member of the glycoside hydrolase family GH53 (16, 17). This protein showed a high degree of similarity with putative β-1,4-endogalactanases from Yersinia pestis, Y. pseudotuberculosis, Y. enterocolitica (83%), and Erwinia carotovora (78%). Sequence analysis revealed that the corresponding gene belongs to a cluster of nine genes that are also conserved in the five bacterial species. This cluster encodes a putative utilization system of oligosaccharides. We postulated that this cluster was responsible for the uptake and catabolism of galactan in E. chrysanthemi, and we named these genes gan. The cluster appeared organized in four operons: ganR, ganL, ganEFGABC, and ganK (Fig. 1). This locus is conserved, with the same genetic organization, in members of the Yersinia genus (Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica). In E. carotovora, ganR is absent and ganL is located on the other side of the cluster and transcribed in the opposite direction (Fig. 1). In these bacteria, no additional gene conservation was observed for the genes adjacent to this locus, suggesting that the gan genes constitute a functional unit.
FIG. 1.
Genetic organization of the gan loci of E. chrysanthemi, Y. pseudotuberculosis, and E. carotovora. Arrows indicate the localization of open reading frames and the direction of gene transcription. The gene designation is given below each arrow.
The gene ganA encodes the putative β-1,4-endogalactanase. The gene ganB encodes a protein showing strong similarities with putative β-galactosidases of many bacterial species, including Y. pestis (84%), E. carotovora (83%), Bacillus circulans (76%), or Clostridium perfringens (72%). The primary sequence of GanB suggested that this protein is a member of the glycoside hydrolase family GH42 (16, 17). ganC is a partial gene that could encode a truncated polypeptide (106 amino acids) homologous to the N-terminal part of the glucose-specific enzyme IIBC of a phosphoenolpyruvate-dependent phosphotransferase system from many bacterial species. No function could be assigned to this gene despite the fact that it is conserved among bacterial species containing the gan locus (87 and 75% similarity with Y. pestis and E. carotovora, respectively). Five genes, ganL, ganE, ganF, ganG, and ganK could be involved in the transport of galactan in E. chrysanthemi. They were related to the maltose/maltodextrin transport system in E. coli and named according to their homolog of this system (6). The gene ganL encodes a 403-amino-acid outer membrane protein homologous (48% similarity) to LamB, the outer membrane porin specific for maltose. The four other genes encode the elements of an ATP binding cassette (ABC) transporter. The gene ganE encodes a 415-amino-acid periplasmic binding protein with 45% similarity to the maltose periplasmic binding protein MalE. ganF and ganG encode the two subunits of the inner membrane permease (435 and 299 amino acids), which showed 62 and 56% similarities to MalF and MalG, respectively. The gene ganK encodes a 369-amino-acid ATPase sharing 65% of similarity with the ATPase MalK (see Table S1 in the supplemental material for additional information). In addition, the gene ganR encodes a protein of 357 amino acids homologous to transcriptional regulators of the LacI family. Its best similarity score (42%) is with LacI, the repressor of the lactose operon in E. coli.
Characterization of the GanA enzyme.
The wild-type strain and its ganA derivative mutant were grown on glycerol minimal medium containing AZCL-galactan, a specific substrate for the detection of 1,4-β-endogalactanase activity. After 16 h at 30°C, only wild-type strain colonies were surrounded by a blue halo indicating the degradation of the AZCL-galactan substrate. The colonies of the mutant did not degrade AZCL-galactan, demonstrating that ganA indeed encodes a 1,4-β-endogalactanase.
The ganA gene encodes a predicted polypeptide of 400 amino acids. The Psort (http://psort.nibb.ac.jp/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/) algorithms (4) predicted that GanA contains a N-terminal signal peptide of 22 amino acids with a cleavage site between two alanine residues, suggesting that the mature GanA is an exported protein. To clarify the cellular localization of GanA, we tested the 1,4-β-endogalactanase activity of subcellular fractions. After centrifugation of the wild-type cells, all of the activity was detected in the cell pellet, while no activity could be detected in the supernatant (data not shown). After treatment of the bacterial pellet with chloroform, more than 90% of the 1,4-β-endogalactanase activity was recovered in the periplasmic extract, demonstrating that GanA is a periplasmic protein. This periplasmic location is confirmed by previous proteomic studies of E. chrysanthemi. GanA was largely recovered among the soluble proteins (2) and weakly in the membrane fraction (7) but not among the extracellular proteins (26).
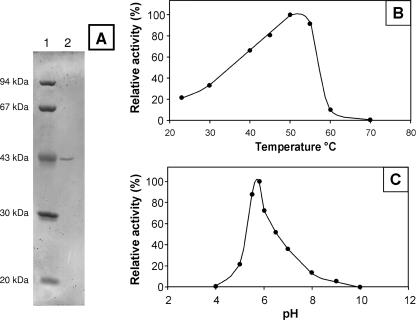
GanA was purified to electrophoretic homogeneity after preparation of periplasmic extracts fractionated by size exclusion chromatography and anion-exchange chromatography (Fig. 2A). A 67-fold purification rate was obtained. The apparent size of the mature GanA enzyme estimated by SDS-PAGE is in agreement with the size of the mature protein deduced from the primary sequence (41800 Da).
FIG. 2.
Analysis of the purified GanA enzyme. (A) SDS-PAGE of purified GanA. Lane 1, protein size markers; lane 2, purified GanA. (B and C) Activity of the purified GanA enzyme relative to temperature (B) and pH (C).
The optimal activity of the purified endogalactanase was found to be at 50°C (±5°C) (Fig. 2B) in a 15-min assay, with AZO-galactan as a substrate in 0.2 M sodium acetate buffer at pH 5.8. This temperature is 20°C above the optimal growth temperature of E. chrysanthemi (30°C), at which the enzyme showed 30% of its maximal activity. The enzyme displayed significant activity over a broad pH ranging from 5 to 8, with an optimum near pH 6 (Fig. 2C).
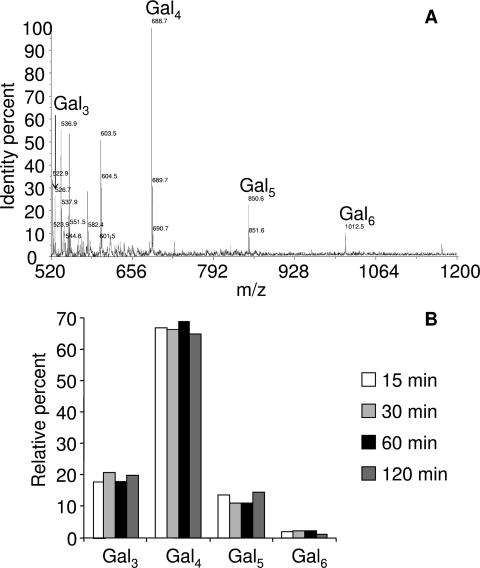
Analysis of the products obtained by in vitro degradation of pectic galactans by GanA at 50°C in a 0.2 M sodium acetate buffer at pH 5.8 was performed by MALDI-TOF mass spectrometry after digestions of 15, 30, 60, and 120 min (Fig. 3). For each reaction time, four oligogalactans hydrolysis products were detected in an identical relative percentage. Galactotriose (Gal3), galactotetraose (Gal4), galactopentaose (Gal5), and galactohexaose (Gal6) were obtained at approximate ratios of 20, 65, 15, and <5%, respectively. Neither galactose nor galactobiose could be detected. After 10 h of reaction, the distribution pattern of oligosaccharides remained the same (data not shown). These results indicated that the GanA is an endo-acting enzyme.
FIG. 3.
Products of galactan degradation by GanA. (A and B) MALDI-TOF mass spectrometry analysis of oligogalactans produced by hydrolysis of galactans by GanA for 30 min (A) and their relative percentages after 15, 30, 60, and 120 min of hydrolysis (B).
The ganA mutant and the wild-type strain were grown for 24 h on minimal medium containing 0.2% (measured in equivalent galactose) galactose, pectic galactans, or oligogalactans (obtained by in vitro hydrolysis of pectic galactans with GanA) as the sole carbon source (Table 4). The growth yield was the same for both strains when they were grown in minimal medium containing galactose. The ganA mutant was unable to grow in minimal medium containing pectic galactan as the sole carbon source, indicating that the periplasmic endogalactanase is necessary for galactan utilization. The growth yield of the ganA mutant on oligogalactans was ca. 20% of that obtained with the wild-type strain, even after 72 h of incubation. Thus, a large part of the oligogalactans could not be used in vivo by the ganA mutant. A hypothesis is that a polar effect of the ganA mutation on ganB is responsible for a reduction of the GanB activity (see below). All of these data demonstrate that GanA catalyzes the cleavage of galactans into oligogalactans that could enter the cytoplasm.
TABLE 4.
Growth of wild-type and various gan mutants of E. chrysanthemi
| Strain | Growtha on:
|
||
|---|---|---|---|
| Galactose | Galactans | Gal3 to Gal6 | |
| 3937 | 1 | 0.7 | 0.7 |
| A4809 (ganA) | 1 | 0 | 0.13 |
| A4807 (ganB) | 1 | 0 | 0 |
| A4868 (ganL) | 1 | 0 | 0.17 |
| A4810 (ganE) | 1 | 0 | 0 |
| NFB3701 (ganK) | 1 | 0 | 0 |
| A4863 (ganR) | 1 | 0.7 | 0.7 |
Bacteria were grown on M63 medium with galactose, galactans, or oligogalactans (Gal3 to Gal6) added as a carbon source (0.2%). The resulting growth yield was expressed as the DO620 of the stationary growth phase.
Characterization of the GanB enzyme.
By analysis of the lactose catabolism in E. chrysanthemi, two genes, lacZ and lacB, have been reported to encode cytoplasmic enzymes with a β-galactosidase activity (20). Analysis of the E. chrysanthemi genome revealed two potential β-galactosidase genes, ganB and lacZ, encoding a protein of the GH42 family, homologous to the E. coli β-galactosidase LacZ. Thus, lacB and ganB may be the same gene. To test this hypothesis and the part of each enzyme in the total β-galactosidase activity of E. chrysanthemi, the wild-type strain and its lacZ (NFB3680), ganB (NFB3666), and ganB, lacZ (NFB3687) derivatives were assayed with ONPG as a substrate (Table 5). No β-galactosidase activity was detected in the ganB, lacZ double mutant. More than 90% of β-galactosidase activity was recovered in the lacZ mutant, and less than 5% was detected in the ganB mutant. These results indicated that lacB and ganB are the same gene and that GanB supports the main β-galactosidase activity in E. chrysanthemi. In addition, β-galactosidase activity in the ganA or ganE mutants showed ca. 20% of the level obtained in the wild-type strain, indicating that these mutations exert a partial polar effect on ganB. Growth of the ganB and lacZ mutants was tested on agar minimal medium containing pectic galactan as the sole carbon source (data not shown). Although normal growth occurred for the lacZ mutant, no growth was observed for the ganB mutant, indicating that GanB, but not LacZ, is required for galactan utilization. In addition, the ganB mutant was unable to grow on oligogalactans obtained after in vitro hydrolysis of galactans by GanA (Table 4). These results support the assumption that GanB is a cytoplasmic exogalactanase involved in the cleavage of the oligogalactans liberated by GanA.
TABLE 5.
β-Galactosidase activity in the wild-type strain and various gan mutants of E. chrysanthemia
| Strain | Main genotype | β-Galactosidase sp actb |
|---|---|---|
| 3937 | Wild type | 252 |
| NFB3680 | lacZ | 241 |
| A4807 | ganB | 7 |
| NFB3687 | lacZ ganB | 0 |
| A4809 | ganA | 46 |
| A4810 | ganE | 49 |
| A4863 | ganR | 58 |
Bacteria were grown in glycerol minimal medium at 30°C until mid-log phase and then broken by passage through a French press cell, and the β-galactosidase activity was measured with ONPG as a substrate.
The specific activity is expressed as nanomoles of ONP liberated per minute per milligram of protein. The results reported are the average of three independent experiments.
Characterization of the ABC transport system of galactans.
The 3937 wild-type strain is able to grow on galactose or galactans as a carbon source, but it was unable to use galactobiose for growth (see Materials and Methods). To determine whether products of the gan cluster are involved in galactan transport, the growth yields of mutants affected for ganL (predicted to encode a porin) and for ganE and ganK (predicted to encode periplasmic protein and the ATPase of the ABC transport system, respectively) were compared to that of the wild-type strain in minimal medium with galactose, galactans, or oligogalactans as the carbon sources (Table 4). All strains gave the same growth yield with galactose as a carbon source, indicating that the gan system has no or a minor role in galactose transport. The ganE and ganK mutants were not able to grow on galactans or on oligogalactans. Thus, these two elements of the Gan ABC transporter are necessary for uptake of these oligosaccharides. Since GanA is a periplasmic enzyme responsible for galactan cleavage, the Gan ABC transport system is certainly responsible for the uptake of oligogalactans across the inner membrane rather than that of galactans. The ganL mutant was not able to assimilate galactan and showed a reduced growth on oligogalactans. This low growth probably resulted in the transport of the small oligomers such as Gal3, which could diffuse through a nonspecific porin. These data suggested that GanL is a specific porin for the uptake of galactans across the outer membrane.
Effect of carbon source on the expression of the E. chrysanthemi gan genes.
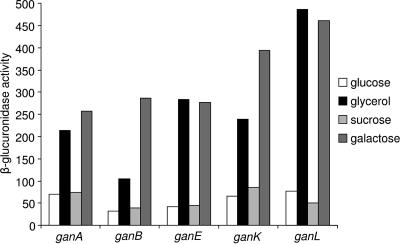
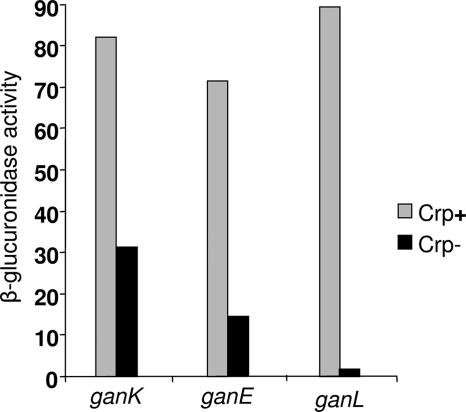
To determine the regulation of the genes involved in galactan utilization, transcriptional fusions of the ganA, ganB, ganE, ganK, and ganL genes with the uidA gene (encoding the β-glucuronidase) were constructed and introduced into E. chrysanthemi by reverse genetics. The β-glucuronidase activity of the fusions was measured after growth until mid-log phase in minimal medium with glycerol, galactose, glucose, or sucrose as a carbon source (Fig. 4). In the presence of glucose or sucrose, the expression decreased by threefold (ganB) to sevenfold (ganL) factors compared to the expression in the presence of galactose or glycerol. This suggested that the gan locus is under catabolic repression controlled by the cyclic AMP receptor protein, CRP, as described for several genes of E. chrysanthemi involved in plant cell wall degradation (35). A potential consensus sequence for CRP binding (http://fasta.bioch.virginia.edu/fasta/cgi/consensus2.cgi) was found upstream of the three transcriptional units of the gan locus (Fig. 1). To test this regulation, an E. chrysanthemi crp::Cml mutation was transduced into strains containing ganK::uidA, ganE::uidA, and ganL::uidA fusions using the ΦEC2 transducing phage. In the presence of the crp mutation, a 3- and 5-fold decrease was observed for ganK and ganE expression, respectively, and an 80-fold decrease was observed for ganL expression, indicating that the gan locus is under catabolic repression (Fig. 5).
FIG. 4.
Expression of the gan genes depending on the carbon source. The gan::uidA fusions strains were grown in M63 medium with either glucose, glycerol, galactose, or sucrose as a carbon source until mid-log phase. Cells were broken by passage through a French press cell, and the β-glucuronidase activity was measured with PNPU as a substrate. The specific activity is expressed as nanomoles of PNP liberated per minute per milligram of protein. The results reported are the average of three independent experiments.
FIG. 5.
Expression of ganA, ganK, and ganL fusions in a crp genetic background. The crp ganA::uidA, crp ganK::uidA, and crp ganL::uidA strains were grown until mild-log phase and then broken by passage through a French press cell, and the β-glucuronidase activity was measured with PNPU as a substrate. The specific activity is expressed as nanomoles of PNP liberated per minute per milligram of protein. The results reported are the average of three independent experiments.
GanR is a regulator of the gan genes.
The presence of the regulatory gene ganR in the gan locus and the conservation of this gene in the gan loci of Y. pestis, Y. pseudotuberculosis, E. carotovora and Klebsiella pneumoniae suggested that GanR is involved in the regulation of transcription of the gan genes. To test this hypothesis, an E. chrysanthemi ganR::Cml mutant was constructed by reverse genetics. The ganR::Cml mutation was then transduced into strains containing ganK::uidA and ganE::uidA fusions using the ΦEC2 transducing phage. About 60% of cotransduction was found between the Kanr and CmlR mutations, as expected for mutations separated by about 10 kb (23).
GanR belongs to the LacI family of transcriptional regulators (15). In the presence of the ganR mutation, the expression of ganK or ganE decreased by 8- and 5-fold factors, respectively (Table 6). This diminution suggested that GanR is an activator of ganK and ganE expression. The assay of β-galactosidase activity in the ganR mutant confirmed the decreased expression of ganB in this genetic context (Table 5). Thus, GanR acts as an activator to control the synthesis of the proteins involved in galactan degradation in E. chrysanthemi.
TABLE 6.
Effect of the ganR mutation on ganE and ganK transcriptional fusionsa
| Fusion | Regulatory mutation | β-Glucuronidase sp actb |
|---|---|---|
| ganE::uidA | None | 9,374 |
| ganR | 1,158 | |
| ganK::uidA | None | 13,591 |
| ganR | 2,872 |
Bacteria were grown in glycerol minimal medium at 30°C. β-Glucuronidase activity was measured with PNPU as a substrate.
The specific activity is expressed as nanomoles of PNP liberated per minute per milligram of protein. The results reported are the average of three independent experiments.
gan genes are expressed during plant infection.
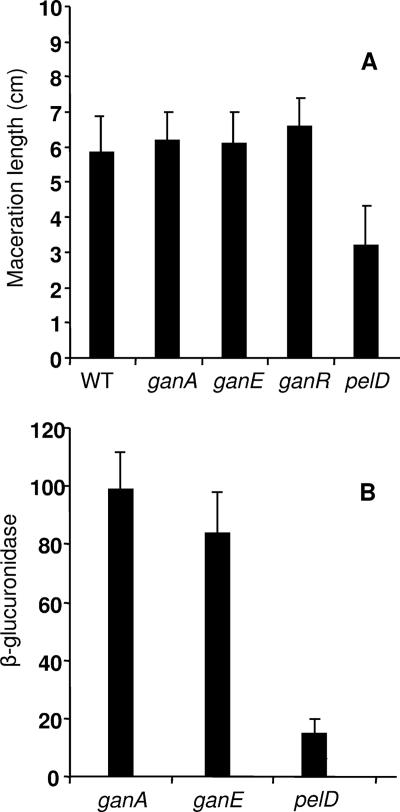
The expression of ganA and ganE transcriptional fusions was analyzed during the infection of chicory leaves and compared to that of the highly induced pectate lyase gene pelD (22). We observed that both ganA and ganE were transcribed at a high level in the macerated tissue (Fig. 6B); their expression appeared to be clearly higher than that of the pelD gene. Thus, the gan genes are expressed during plant infection.
FIG. 6.
Infection of chicory leaves with the gan mutants. Ten chicory leaves were infected for each strain: 3937 (wild-type), A4809 (ganA::uidA), A4810 (ganE::uidA), and A4863 (ganR). The pelD::uidA mutant A4415, which has an attenuated virulence and a high in planta expression, was used as a control. (A) After incubation at 30°C for 24 h, the length of rotted tissue was measured to estimate the disease severity. (B) The macerated tissue was recovered and used for the assay of the uidA product, β-glucuronidase. The values reported are the average of the different leaves, and the standard deviations are indicated.
The gan mutants are virulent.
We tested the extent of soft rot caused by different mutants after the inoculation of potato tubers, chicory leaves, and Saintpaulia ionantha. The maceration provoked by the ganR, ganA, and ganE mutants on chicory leaves was compared to that caused by the parental strain 3937. There was no significant difference in the degree of maceration caused by each strain (Fig. 6A). Moreover, the gan mutations did not affect bacterial growth in the plant tissue (data not shown). Similarly, the virulence of ganA and ganR mutants was not affected on potato tubers or plants of Saintpaulia ionantha (data not shown). These results showed that galactan degradation is not essential for the maceration or for the development of the soft-rot disease.
DISCUSSION
In this study, we described the functional characterization of a locus involved in the utilization of galactans by E. chrysanthemi. Previous studies showed that E. chrysanthemi is able to degrade and catabolize a range of plant structural polysaccharides, including linear regions of pectin and the backbone of the ramified regions RGI (19, 21, 28). Galactans are constituents of the side chains attached to RGI. Among the eight genes identified in the gan locus, five are involved in transport. Four genes (AFB-0018190, AFB-0018192, AFB-0018193, and AFB-0018195) encode 1 of the 86 putative ABC transport systems annotated in the E. chrysanthemi 3937 genome. One gene (AFB-0018200) encodes a porin, improperly annotated as LamB (2) since E. chrysanthemi is not sensitive to phage lambda and it is unable to use maltose or maltodextrin as a carbon source for growth. The two other gan genes (AFB-0018196 and AFB-0018198) encode glycoside hydrolases of the family GH53 and GH42, respectively. These enzymes are involved in the degradation from galactans to galactose. The last gene, ganR (AFB-0018202), encodes a transcriptional regulator. The gan locus allows E. chrysanthemi to use galactan as a sole carbon and energy source for growth. The gan locus is conserved, complete and with the same genetic organization, in Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica. This conservation suggests that various Yersinia species also can transport and catabolize galactan. In E. carotovora, all of the gan genes are present, except ganR, which encodes the regulator. Thus, the gan genes are subject to a different regulation in the two Erwinia species.
The data presented in the present study suggest the following model for galactan transport and catabolism. After the digestion of pectin by the set of E. chrysanthemi pectic enzymes, the galactan chains are liberated and diffuse across the outer membrane through the porin GanL. In the periplasm, GanA produces small oligosaccharides that are taken up by GanE, the oligogalactan binding protein. The transport of these oligomers across the inner membrane is performed by the GanFGK2 complex. The cytoplasmic enzyme GanB further degrades the oligogalactans into galactose, which subsequently enters the cellular metabolism.
The ganA gene encodes a periplasmic β-1,4-endogalactanase. In vitro, the purified enzyme catalyzes the degradation of galactans into oligogalactans from Gal3 to Gal6 with a majority of Gal4. Neither galactobiose nor galactose were recovered. Thus, GanA cannot hydrolyze short oligomers such as Gal2 or Gal3, as observed for the β-1,4-endogalactanase of Bacillus licheniformis (38). Despite the fact that they belong to the GH53 family, the mode of action of β-1,4-endogalactanases may be different. The endogalactanase of Bifidobacterium longum probably acts by a processive mechanism, liberating the same products along the reaction (18); this major product is Gal3, but galactose and galactobiose are formed after prolonged incubation. In Aspergillus aculeatus, the end products are galactobiose and galactose with intermediates of larger size (11). Various cellular locations were also observed for β-1,4-endogalactanases. An extracellular location allows the endogalactanase to directly degrade galactans linked to pectin, as observed for the enzymes of Aspergillus niger, Pseudomonas fluorescens, or B. subtilis (9, 12, 41). In B. longum, this enzyme is extracellular but anchored to the membrane (18), and the E. chrysanthemi β-1,4-endogalactanase is located in the periplasm. These locations suggest that galactan utilization depends on the previous liberation of the polysaccharidic chains from RGI by pectic enzymes. The role of these cell-linked enzymes is to degrade polymers into oligomers able to cross the inner membrane. The periplasmic α-amylase MalS of E. coli, catalyzing the degradation of maltodextrines into maltotriose, maltose, and glucose, plays an analogous role in maltodextrin utilization (14).
E. chrysanthemi mutants affected for ganL, ganK, ganE, ganA, or ganB became unable to catabolize galactan. The ganK, ganE, or ganB mutants are also unable to grow on a mixture of oligogalactans (Gal3 to Gal6) produced by the action of GanA in vitro. When ganL or ganA mutants were grown on oligogalactans produced by GanA, the low growth yield observed approximately corresponds to the use of Gal3. The GanFGK2 transport system probably recognizes preferentially galactotriose, suggesting that it is the major product released by GanA in vivo. The cytoplasmic enzyme GanB has a β-1,4-exogalactanase (or β-galactosidase) activity that is necessary to degrade Gal3. Endogalactanases unable to produce galactose as a final product need to be associated with exogalactanases. This explains the conserved association of genes encoding endogalactanases of the family GH53 and β-galactosidases of the family GH42 observed in B. subtilis and other bacteria (41).
The expression of the E. chrysanthemi gan genes is controlled both by the general catabolic repression and by a specific regulator, since crp and ganR mutations decrease gan gene expression. Many bacterial regulons needed for nutrition by carbohydrates depend on such regulations. Despite the fact that GanR belongs to the LacI family of regulators, which mainly includes repressors (15), it appears to act as an activator of gan expression. However, the level of gan expression is high in all of the conditions tested, and the effect of the GanR activation is weak. In addition, two independent proteomic analyses revealed a high level of the galactanase GanA after growth of strain 3937 in LB medium (7) or in M63 (2). Thus, we cannot exclude that the gan gene expression presents some degree of deregulation in the E. chrysanthemi strain 3937. The Mal and Gan systems are analogous but exhibit two fundamental differences. In the Gan system, the degrading enzymes are glycosyl hydrolases. A BLAST search against the complete E. chrysanthemi 3937 genome sequence did not detect a MalT homolog.
The gan mutants showed normal growth and maceration in planta. The gan genes are expressed during infection, but their expression is not essential for the E. chrysanthemi virulence. It was observed that the degradation of cellulose, another important plant cell wall polysaccharide, is not necessary for virulence (5). Similarly, the xylanase of the E. chrysanthemi strain D1 is not involved in its virulence (27). Thus, pectic degrading enzymes (essentially the pel gene products) are the sole cell wall-degrading enzymes required for the E. chrysanthemi virulence since they provoke tissue maceration (40). Galactans and other polysaccharides such as cellulose and xylan are probably used as a secondary carbon source. These polysaccharides may be used as additional nutriment at the end of the infection process. The capacity of E. chrysanthemi to use a wide range of plant components may also favor its saprophytic life since it has the possibility to assimilate plant remnants present in the soil.
Supplementary Material
Acknowledgments
This study was supported by grants from the Centre National de la Recherche Scientifique and from the Ministère de l'Education Nationale et de la Recherche.
We acknowledge members of the International Erwinia Consortium for the exchange of unpublished data concerning the E. chrysanthemi 3937 genome sequence. We thank Geraldine Effantin and Marie-Christine Slomianny for assistance with some experiments.
Footnotes
Published ahead of print on 20 July 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ames, G. F., C. Prody, and S. Kustu. 1984. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J. Bacteriol. 160:1181-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babujee, L., B. Venkatesh, A. Yamazaki, and S. Tsuyumu. 2007. Proteomic analysis of the carbonate insoluble outer membrane fraction of the soft-rot pathogen Dickeya dadantii (syn. Erwinia chrysanthemi) strain 3937. J. Proteome Res. 6:62-69. [DOI] [PubMed] [Google Scholar]
- 3.Bardonnet, N., and C. Blanco. 1992. 'uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 72:243-247. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Boccara, M., J. L. Aymeric, and C. Camus. 1994. Role of endoglucanases in Erwinia chrysanthemi 3937 virulence on Saintpaulia ionantha. J. Bacteriol. 176:1524-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchart, F., A. Delangle, J. Lemoine, J.-P. Bohin, and J.-M. Lacroix. 2007. Proteomic analysis of a nonvirulent mutant of the phytopathogenic bacterium Erwinia chrysanthemi deficient in osmoregulated periplasmic glucans: change in protein expression is not restricted to the envelope, but affects general metabolism. Microbiology 153:760-767. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Braithwaite, K. L., T. Barna, T. D. Spurway, S. J. Charnock, G. W. Black, N. Hughes, J. H. Lakey, R. Virden, G. P. Hazlewood, B. Henrissat, and H. J. Gilbert. 1997. Evidence that galactanase A from Pseudomonas fluorescens subspecies cellulosa is a retaining family 53 glycosyl hydrolase in which E161 and E270 are the catalytic residues. Biochemistry 49:15489-15500. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KanR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 11.Christgau, S., T. Sandal, L. V. Kofod, and H. Dalboge. 1995. Expression cloning, purification and characterization of a β-1,4-galactanase from Aspergillus aculeatus. Curr. Genet. 27:135-141. [DOI] [PubMed] [Google Scholar]
- 12.de Vries, R. P., L. Parenicova, S. W. A. Hinz, H. C. M. Kester, G. Beldman, J. A. E. Benen, and J. Visser. 2002. The β-1,4-endogalactanase A from Aspergillus niger is specifically induced on arabinose and galacturonic acid and plays an important role in the degradation of pectic hairy regions. Eur. J. Biochem. 269:4985-4993. [DOI] [PubMed] [Google Scholar]
- 13.Emi, S., J. Fukumoto, and T. Yamamoto. 1972. Crystallization and some properties of mannanases. Agric. Biol. Chem. 36:991-1001. [Google Scholar]
- 14.Freundlieb, S., and W. Boos. 1986. α-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its products as a periplasmic protein. J. Biol. Chem. 261:2946-2953. [PubMed] [Google Scholar]
- 15.Gelfand, M. S., and O. N. Laikova. 2003. Prolegomena to the evolution of transcriptional regulation in bacterial genomes, p. 195-216. In E. V. Kooning and M. Y. Galperin (ed.), Frontiers in computational genomics. Caister Academic Press, Wymondham, United Kingdom.
- 16.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz, S. W., M. I. Pastink, L. A. Van den Broek, J. P. Vincken, and A. G. Voragen. 2005. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 71:5501-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugouvieux-Cotte-Pattat, N. 2004. The RhaS activator controls the Erwinia chrysanthemi 3937 genes rhiN, rhiT, and rhiE involved in rhamnogalacturonan catabolism. Mol. Microbiol. 51:1361-1374. [DOI] [PubMed] [Google Scholar]
- 20.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1985. Lactose metabolism in Erwinia chrysanthemi. J. Bacteriol. 162:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 22.Hugouvieux-Cotte-Pattat, N., H. Dominguez, and J. Robert-Baudouy. 1992. Environmental conditions affect the transcription of the pectinases gene of Erwinia chrysanthemi 3937. J. Bacteriol. 174:7807-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugouvieux-Cotte-Pattat, N., S. Reverchon, and J. Robert-Baudouy. 1989. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol. Microbiol. 5:573-581. [DOI] [PubMed] [Google Scholar]
- 24.Huguet, E., K. Hahn, K. Wengelnik, and U. Bonas. 1998. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29:1379-1390. [DOI] [PubMed] [Google Scholar]
- 25.Hwang, J., Y. R. Pyun, and J. L. Kokini. 1993. Sidechains of pectins: some thoughts on their role in plant cell walls and foods. Food Hydrocolloids 7:39-53. [Google Scholar]
- 26.Kazemi-Pour, N., G. Condemine, and N. Hugouvieux-Cotte-Pattat. 2004. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 10:3177-3186. [DOI] [PubMed] [Google Scholar]
- 27.Keen, N. T., C. Boyd, and B. Henrissat. 1996. Cloning and characterization of a xylanase gene from corn strains of Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 9:651-657. [DOI] [PubMed] [Google Scholar]
- 28.Laatu, M., and G. Condemine. 2003. Rhamnogalacturonate lyase RhiE is secreted by the Out system in Erwinia chrysanthemi. J. Bacteriol. 185:1642-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazan, H., S. Y. Ng, L. Y. Goh, and Z. M. Ali. 2004. Papaya β-galactosidase/galactanase isoforms in differential cell wall hydrolysis and fruit softening during ripening. Plant Physiol. Biochem. 42:847-853. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, New York, NY.
- 31.Miller, V. L., and J. J. Mekalanos. 1998. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, A., H. Maeda, M. Mizuno, Y. Koshi, and Y. Nagamatsu. 2003. β-Galactosidase and its significance in ripening of “Saijyo” japanese persimmon fruit. Biosci. Biotechnol. Biochem. 67:68-76. [DOI] [PubMed] [Google Scholar]
- 33.Perombelon, M., and A. Kelman. 1980. Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 18:361-387. [Google Scholar]
- 34.Resibois, A., M. Colet, M. Faelen, T. Schoonejans, and A. Toussaint. 1984. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102-112. [DOI] [PubMed] [Google Scholar]
- 35.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridley, B. L., M. A. O'Neill, and D. Mohnen. 2001. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929-967. [DOI] [PubMed] [Google Scholar]
- 37.Roeder, D. L., and A. Collmer. 1985. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryttersgaard, C., J. Le Nours, L. Lo Leggio, C. T. Jorgensen, L. L. Christensen, M. Bjornvad, and S. Larsen. 2004. The structure of endo-β-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. J. Mol. Biol. 341:107-117. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 40.Sepulchre, J.-A., S. Reverchon, and W. Nasser. 2007. Modeling the onset of virulence in a pectinolytic bacterium. J. Theor. Biol. 244:239-257. [DOI] [PubMed] [Google Scholar]
- 41.Shipkowski, S., and J. E. Brenchley. 2006. Bioinformatic, genetic, and biochemical evidence that some glycoside hydrolase family 42 β-galactosidases are arabinogalactan type I oligomer hydrolases. Appl. Environ. Microbiol. 72:7730-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiro, R. G. 1966. Analysis of sugars found in glycoproteins. Methods Enzymol. 8:3-27. [Google Scholar]
- 43.Vincken, J. P., H. A. Schols, R. J. F. J. Oomen, M. C. McCann, P. Ulvskov, A. G. J. Voragen, and R. G. F. Visser. 2003. If homogalacturonan were a side chain of rhamnogalacturonan. I. Implications for cell wall architecture. Plant Physiol. 132:1781-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, H., H. Ichinose, M. Yoshida, M. Nakajima, H. Kobayashi, and S. Kaneko. 2006. Characterization of a thermostable endo-β-1,4-d-galactanase from the hyperthermophile Thermotoga maritima. Biosci. Biotechnol. Biochem. 70:538-541. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, Y., F. Pettolino, S. L. Mau, and A. Bacic. 2005. Characterization of cell wall polysaccharides from medicinal plant Panax notoginseng. Phytochemistry 66:1067-1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.