Abstract
The coordinate expression of Salmonella enterica invasion genes on Salmonella pathogenicity island 1 is under the control of the complex circuits of regulation that involve the AraC/XylS family transcriptional activators HilD, HilC, and RtsA and nucleoid-associated proteins. Single-copy transcription fusions were used to assess the effects of nucleoid-associated proteins Hha and H-NS on hilD, hilC, and rtsA expression. The data show that all three genes, hilD, hilC, and rtsA, were repressed by H-NS and/or Hha. The repression of rtsA was the highest among tested genes. The level of rtsA-lac was equally elevated in hns and hha mutants and was further enhanced in the hns hha double mutant under low-osmolarity conditions. Electrophoretic mobility shift experiments showed that H-NS and Hha directly bind to the rtsA promoter. In addition to the negative control that was exerted by H-NS/Hha under low-osmolarity conditions, the homologous virulence activators HilD, HilC, and RtsA (Hil activators) induced rtsA-lac expression in a high-salt medium. A DNase footprinting assay of the rtsA promoter revealed one common DNA-binding site for all three Hil activators centered at position −54 relative to the transcriptional start site. In the absence of Hha and H-NS, however, osmoregulation of the rtsA promoter was lost, and Hil activators were not required for rtsA transcription. These results taken together suggest that the HilD, HilC, and RtsA proteins induce the transcription of the rtsA promoter by counteracting H-NS/Hha-mediated repression.
Salmonella enterica serovars are facultative intracellular bacterial pathogens that are capable of colonizing and/or causing disease in a wide range of hosts. In humans, S. enterica causes diseases that range from localized gastroenteritis to disseminated typhoid fever. The pathogenesis scheme includes bacterial invasion of nonphagocytic cells and survival within those cells, as well as replication within macrophages (7, 24). Salmonella gains entry across the intestinal epithelium by passage through M cells or by invasion of enterocytes. Host cell invasion relies on the production of a type III secretion system that injects effector proteins. The effector proteins trigger rearrangements of the actin cytoskeleton that lead to transient membrane ruffling and bacterial uptake (9, 21). Most of the genes that are required for invasion are located at Salmonella pathogenicity island 1 (SPI1) (17, 20, 27, 32). The expression of SPI1 invasion and effector genes responds to multiple environmental signals and is decreased under conditions of low osmolarity, high oxygen, low pH, and stationary phase. Many regulatory proteins influence invasion gene expression directly and indirectly, including activators HilA, InvF, HilD, HilC, and RtsA and nucleoid-associated proteins H-NS and Hha (2, 14, 16, 23, 36, 37, 44). The activators act in a cascade starting with homologous regulators HilD/HilC/RtsA (Hil activators), followed by the key regulator HilA. Both Hil activators and HilA can activate SPI1 gene expression either directly or by increasing expression of the activator InvF.
The proteins HilD (34.3 kDa), HilC (32.8 kDa), and RtsA (32.3 kDa) belong to the AraC/XylS family of transcriptional activators. They have similar C-terminal domains with double helix-turn-helix DNA-binding motifs common to all members of the family (∼49% identity). In contrast, the roughly 200-residue N-terminal region of the Hil activators shows approximately 10% identity (22 amino acids) among the three. The genes that encode the HilD and HilC proteins are located on SPI1, while the gene that encodes RtsA is located on an island integrated at the tRNAPheU (12, 14, 40, 43). When the individual proteins HilD, HilC, and RtsA are overexpressed from plasmids, they are able to induce expression of the hilA, invF, hilD, hilC, rtsA, slrP, and dsbA genes, which are required for invasion (13-15). The individual or combined deletion of the hilD, hilC, and rtsA genes decreases the expression of hilA and rtsA 2- to 10-fold but does not significantly affect hilD and hilC expression (12, 14, 28, 40, 43). That means that at least hilA and rtsA are induced by HilD, HilC, and RtsA proteins. The HilD and HilC proteins bind to the common DNA sites at the hilA, hilD, and hilC promoters (37). The DNA-binding properties of the RtsA protein are not known. It has been shown that two HilD/HilC sites at the hilA promoter overlap with upstream binding sites for repressors Hha/H-NS, suggesting that derepression is one of the mechanisms that is involved in hilA transcriptional regulation (36, 44). In addition, genetic and biochemical data showed that the RpoA subunit of RNA polymerase (RNAP) participates in the HilD/HilC-induced activation of the hilA promoter (5, 35). Therefore, further analysis of hilD, hilC, and rtsA regulation is required in order to understand how specific environmental signals are decoded and transformed into the expression of invasion machinery.
The histone-like nucleoid-structuring protein, H-NS, silences horizontally acquired virulence genes in S. enterica serovar Typhimurium (29, 33). When interacting with DNA, H-NS initially binds to a curved, AT-rich sequence, followed by oligomerization and subsequent alteration of the DNA structure (11, 41). As a repressor, H-NS can prevent the RNAP from binding to the promoter (39, 49), trap RNAP at the promoter through DNA bridging (8, 47), or block transcription initiation or elongation when bound to the downstream sequences of the promoters (10, 25, 50). It has been shown that the disruption of the repressive nucleoprotein structures mediated by H-NS requires activation signals or positively acting transcription factors (reviewed in reference 11), even though little is known about the mechanisms involved in this process.
Hha is a small nucleoid-associated protein (8 kDa) involved in the negative modulation of virulence genes rather than of housekeeping genes in gram-negative bacteria (30). The set of genes controlled by Hha is significantly smaller than that regulated by H-NS (4, 22). The current model of Hha action is based on the study of the Hha/H-NS-mediated repression of the hly operon of Escherichia coli. It is proposed that Hha does not bind the hly operon directly but instead forms a complex with H-NS, which interacts with the hly operon (34). However, in other systems, such as the E. coli ler operon and the S. enterica hilA promoter, purified Hha seems to be able to bind DNA independently of H-NS (16, 36, 45).
In this report, we investigate the role of nucleoid-associated proteins Hha and H-NS in the expression of virulence activator proteins HilD, HilC, and RtsA with an emphasis on the interplay of positive and negative transcriptional factors in the regulation of the rtsA promoter. Our results show that virulence activators HilD, HilC, and RtsA bind to a common site in the rtsA promoter and antagonize H-NS/Hha-mediated repression.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, growth conditions, and enzymatic assays.
Bacterial strains, plasmids, and phages used in this work are listed in Table 1. Bacteria were routinely grown in LB medium containing the appropriate supplements as follows at the indicated concentrations: ampicillin, 150 μg/ml for plasmids; kanamycin, 50 μg/ml; tetracycline, 25 μg/ml; and chloramphenicol, 20 μg/ml. Antibiotic resistance gene insertions were moved between S. enterica serovar Typhimurium or E. coli strains by transduction (31). Due to the pleiotropic nature of the hns mutation, it was introduced last in all constructs. S. enterica serovar Typhimurium strains used in β-galactosidase assays were grown statically overnight at 37°C in 2 ml of salt-free LB broth or in LB broth with 1% NaCl in 13- by 100-mm tubes to an optical density at 600 nm of 0.1 to 0.3. The β-galactosidase assay was performed as previously described (37). Briefly, β-galactosidase activity was continuously monitored in a microplate reader (Molecular Dynamics) at a wavelength of 415 nm to determine the rate of o-nitrophenyl-β-galactopyranoside hydrolysis. The activity was expressed in units of ΔA415·min−1·A650−1.
TABLE 1.
Bacterial strains, phages, and plasmids
| Strain, phage, or plasmid | Relevant characteristics | Reference and/or source |
|---|---|---|
| Bacterial strains | ||
| S. enterica serovar Typhimurium strains | ||
| CH1816 | hns-101::IS10 zde-1710::Tn10 Tetr | L. Csonka |
| IO907 | LT2 Δhha::cam hns-1::kan hilA080::Tn5 lacZY-080 Tetr Kanr Camr | 36 |
| S. enterica subsp. enterica ATCC 14028 derivatives | ||
| ATCC 14028 | WT | ATCC |
| JS324 | Φ(rtsA-lac+)5 Kanr | 14 |
| JS483 | ΔhilCD2915::cam Φ(rtsA-lac+)5 Kanr Camr | 13 |
| JS487 | Φ(hilC-lac+)113 Kanr | 13 |
| JS488 | Φ(hilD-lac+)114 Kanr | 13 |
| IO908 | hilA080::Tn5 lacZY-080 Tetr | This work |
| IO923 | Δhha::cam hns-1::kan hilA080::Tn5 lacZY-080 Tetr Kanr Camr | This work |
| IO949 | Δhha::cam Φ(rtsA-lac+)5 Camr Kanr | This work |
| IO954 | hns-101::IS10 zde-1710::Tn10 Φ(rtsA-lac+)5 Tetr Kanr | This work |
| IO955 | Δhha::cam hns-101::IS10 zde-1710::Tn10 Φ(rtsA-lac+)5 Tetr Kanr Camr | This work |
| IO959 | Δhha::cam Φ(hilD-lac+) Kanr Camr | This work |
| IO961 | hns-101::IS10 zde-1710::Tn10 Φ(hilD-lac+)Camr Kanr | This work |
| IO963 | Δhha::cam hns-101::IS10 zde-1710::Tn10 Φ(hilD-lac+) Tetr Kanr Camr | This work |
| IO969 | Δhha::cam Φ(hilC-lac+) Kanr Camr | This work |
| IO971 | hns-101::IS10 zde-1710::Tn10 Φ(hilC-lac+) Camr Kanr | This work |
| IO973 | Δhha::cam hns-101::IS10 zde-1710::Tn10 Φ(hilC-lac+) Tetr Kanr Camr | This work |
| IO976 | ΔhilCD2915 Φ(rtsA-lac+)5 Kanr | This work |
| IO997 | Δhha::cam ΔhilCD2915 Φ(rtsA-lac+)5 Kanr Camr | This work |
| IO999 | hns-101::IS10 zde-1710::Tn10 ΔhilCD2915 Φ(rtsA-lac+)5 Kanr Tetr | This work |
| IO1000 | Δhha::cam hns-101::IS10 zde-1710::Tn10 ΔhilCD2915 Φ(rtsA-lac+)5 Kanr Tetr Camr | This work |
| E. coli strains | ||
| DH5α | endA1 hsdR17 glnV44 thi-1 recA1 gyrA relA1 Δ(lac-argF)169 deoR [φ80dlacΔ(lacZ)M15] | Invitrogen |
| BL21(DE3) | ompT hsdSBgal dcm (λDE3) | Novagen |
| M182 hns | Δ(lacIPOZYA)74 galK strA Δhns-1::kan Kanr | M. Belfort |
| IO991 | BL21(DE3) Δhns-1::kan Kanr | This work |
| Phages | ||
| P22 HT | Laboratory stock | |
| P1 | Laboratory stock | |
| Plasmids | ||
| pRS415P | Ap; lacZYA transcriptional fusion plasmid | 37 |
| pRS415P derivatives | This work; 36 | |
| pMBPHis-parallel1 | Ap; expression vector | Z. Derewenda; 46 |
| pMBP-RtsA1 | Ap; MBP-His6-RtsA-expressing plasmid | This work |
| pHilC14 | Ap; His6-HilC-expressing plasmid | 37 |
| pHilD21 | Ap; His6-HilD-expressing plasmid | 37 |
| pHNS11 | Ap; MBP-His6-H-NS-expressing plasmid | 36 |
| pH3 | Ap; MBP-His6-Hha-expressing plasmid | 36 |
RNA isolation, primer extension, and RT-PCR.
RNA was isolated from cells of S. enterica serovar Typhimurium by use of an RNAgents kit (Promega) or an RNeasy kit (QIAGEN). Primer extension was performed using Super-Script reverse transcriptase (Invitrogen) as described by the manufacturer. For primer extension assays, RNA was isolated from cells of S. enterica serovar Typhimurium ATCC 14028 grown in limited-oxygen, high-salt conditions (LB medium plus 1% NaCl, no aeration) to an A600 of 1.0. Cells from 100 ml of culture were collected by centrifugation, and RNAs were isolated using an RNAgents kit (Promega). Traces of DNA were removed from RNA by DNase I treatment (RNase free; Boehringer Mannheim GmbH, Germany). The RNA (6 μg) was hybridized to a specific 5′-32P-end-labeled oligonucleotide, rtsARNA3 (5′-TGCCGTAATATACTCCCCGGACGATGT), which was complementary to the coding strand of the rtsA gene. The extension products were resolved by electrophoresis on 5% sequencing gels (42) next to a sequencing ladder generated using the same labeled oligonucleotide and PCR-generated rtsA promoter fragment as the DNA template. Reverse transcription-PCR (RT-PCR) analysis was carried out using a Onestep RT-PCR kit (QIAGEN). Three hundred nanograms of RNA was used as a template for gene-specific primer pairs 5rtsA+1 (5′-TAGAAATGCAATATAAAATAGCAT) and rtsARNA3 (rtsA) as well as marA5′ (5′-TTGAACGTCCGGGTCAATGTTTGC) and marA3′ (5′-TATTCCAAATGGCACCTGCA) (marA). The reaction consisted of 1 cycle of 50°C for 30 min and 95°C for 15 min followed by 26 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min and was concluded by 1 final extension cycle of 72°C for 10 min. The RT-PCR products were electrophoresed through 1% Tris-acetate-EDTA agarose gels and stained with ethidium bromide.
Plasmid construction.
DNA isolation and all recombinant DNA manipulations were carried out using standard methods (42). The −323 to +169 region of the rtsA promoter was amplified from S. enterica serovar Typhimurium ATCC 14028 chromosomal DNA by PCR using Vent polymerase (New England Biolabs) as specified by the manufacturer. The primers used to amplify the PCR-generated fragment introduced EcoRI and HindIII sites at either end and were as follows: 3rtsApr5′ (5′-CCGGAATTCTTCCCCTCCCCTCAAACTTCCGG) and rtsApr3′ (5′-AGGGATTAAAAGCTTTTAGCATGATAATTT). The PCR product was cloned as an EcoRI-HindIII fragment into the similarly digested lacZYA reporter plasmid pRS415P (37) to generate plasmid prtsA-323+169L.
Plasmid pMBP-RtsA1, in which the rtsA gene is expressed under the control of the T7 promoter, was constructed by cloning the PCR-amplified rtsA gene from the ATCC 14028 chromosomal DNA template into the expression vector pMBPHis-parallel1 (gift from Z. Derewenda [46]), which includes both maltose-binding protein (MBP) and His6 affinity tags. The following primers were used to create PCR fragments flanked by BglII and HindIII sites, with introduced sites underlined: RtsA5Bgl2 (5′-GGAAGATCTAAAAGTATTTAATCCCTCACC) and RtsA3HindIII (5′-CCCAAGCTTTCAATTAACATATTGATGACGAGA). All constructs were verified by DNA sequencing at the Microbiology Biomolecular Research Facility at the University of Virginia.
Protein purification.
The plasmid pMBP-RtsA1 was introduced into E. coli BL21(DE3) by transformation. Cells were then grown in LB broth supplemented with ampicillin (500 mg/ml) at 37°C, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added in the early log phase. The cells were allowed to grow for 3 h and were harvested by centrifugation. The MBP-RtsA protein was purified from cell extracts by binding to amylose resin (New England Biolabs) as described by the manufacturer. In brief, cells grown in 1 liter of LB were washed and suspended in 40 ml of binding buffer (20 mM Tris-HCl [pH 7.4], 0.2 M NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA) and disrupted by sonication of 20-ml portions for three 3-min periods on ice. Unbroken cells and particulate material were removed by centrifugation at 20,000 × g for 30 min, and the cell extract was applied on the amylose resin column with a 1-ml bed volume. The column was washed with 10 volumes of binding buffer. MBP-RtsA protein was eluted with 5 ml of binding buffer plus 10 mM maltose, concentrated using a Centriplus YM-10 centrifugal filter device (Amicon, Inc., Beverly, MA), and stored at −70°C with 30% glycerol (vol/vol). Yields of MBP-RtsA protein were in the range of 0.5 to 0.8 mg per liter of culture, and a purity of 80% was estimated by Coomassie brilliant blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
The proteins HilC, HilD, and H-NS were purified as described previously (36, 37). The MBP-His6-Hha fusion was purified (36) from E. coli BL21(DE3) Δhns (IO991) to avoid potential H-NS contamination.
Electrophoretic mobility shift assay (EMSA).
DNA fragments of rtsA, hilD, and hilC promoters were generated by PCR using plasmids prtsA-323+169L, philD-160+703L, philC-160+88L, and philA-242+505L as templates. The sequences of the primers used are available upon request. The amplified DNA fragments were purified from 1% agarose gels by use of a QIAquick gel extraction kit (QIAGEN) and end labeled by incubation with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (3,000 Ci/mmol; ICN). The labeled DNA fragments were separated from unincorporated [γ-32P]ATP by gel filtration through Sephadex G-50 quick spin columns (Boehringer Mannheim). Radiolabeled DNA fragments (ca. 10,000 to 20,000 cpm per reaction) were incubated with MBP-RtsA, HilC, or HilD at 25°C for 10 min in binding buffer [40 mM Tris-HCl (pH 8.0), 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 5% glycerol, 2 ng of poly(dI-C)/μl]. Samples were resolved by electrophoresis in 1.5-mm-thick, 6% nondenaturing polyacrylamide Tris-glycine gels (5 mM Tris, 38 mM glycine, pH 8.6) at 20 mA for 45 min at room temperature. The gels were dried and the positions of radioactive DNA fragments were detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Gel mobility shift assays with H-NS and MBP-Hha proteins were performed as described previously (10).
DNase I footprinting.
The 5′-32P-end-labeled linear DNA fragment containing the rtsA promoter from position −173 to position +29 was amplified by PCR using the prtsA-323+169L plasmid DNA as the template and purified as described previously (37). The promoter-containing DNA fragment (50,000 cpm) at a final concentration of 1 nM was incubated with specified amounts of purified H-NS, Hha, MBP-RtsA, HilC, and HilD in 20 μl of reaction buffer (40 mM Tris-HCl [pH 8.0], 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 10% glycerol). After 10 min at 25°C, DNase I digestion was started by the addition of 2 μl of the reaction buffer containing 25 mM CaCl2, 25 mM MgCl2, and 0.5 μg/ml DNase I. After 20 s at room temperature, 4 μl of stop solution [0.18 M EDTA, 0.34 μg/ml of poly(dI-C), 30% (vol/vol) glycerol] was added (18). The DNA was precipitated with 95% (vol/vol) ethanol, dried under vacuum, dissolved in a loading buffer, and resolved by electrophoresis on 5% sequencing gels (42). Size markers were generated by Maxam-Gilbert sequencing reactions (42) on the same DNA fragment. The gels were dried, and the positions of radioactive DNA fragments were detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
RESULTS
The roles of Hha and H-NS in the down-regulation of the rtsA, hilC, and hilD promoters.
We have recently reported that nucleoid-associated proteins H-NS and Hha directly bind to the hilA promoter and silence its expression (36). To test whether the RtsA, HilC, and HilD activators of hilA are also regulated by these proteins, we investigated the transcription of rtsA-lac, hilC-lac, and hilD-lac chromosomal fusions (13). hns is known to be essential in Salmonella enterica serovar Typhimurium (33). The mutations in the rpoS and phoP genes overcome the lethality of the hns mutation. During preliminary experiments, we found that hns-101::IS10 and ρhha::cam mutations could be combined in the derivatives of ATCC 14028 (JS324, JS487, and JS488) but not in the LT2 background. Therefore, we analyzed the effects of the hns-101::IS10 and Δhha::cam mutations on rtsA-lac, hilC-lac, and hilD-lac expression in ATCC 14028. The levels of β-galactosidase were determined following growth in high- or low-salt LB medium under low-oxygen conditions. These growing conditions were chosen to detect induction by HilD, HilC, or RtsA (LB plus NaCl) (13) and repression by H-NS (LB minus NaCl) or Hha (LB plus or minus NaCl) (36).
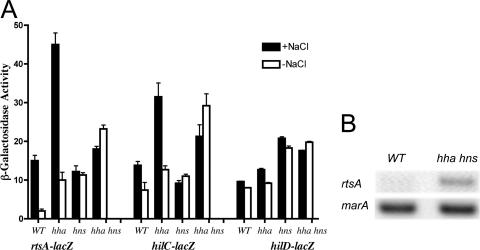
The deletion of the hha caused a 1.7- to 5-fold increase in rtsA and hilC expression under all conditions tested (Fig. 1A). Interestingly, Hha played no role in the regulation of the hilD promoter. The hns mutation showed an increase of rtsA (5.5-fold), hilD (2-fold), and hilC (<2-fold) expression under low-osmolarity conditions. The combination of hha and hns mutations resulted in an 11-fold increase in rtsA expression under low-osmolarity conditions. Under the same conditions, the hilC promoter showed a fourfold increase in the β-galactosidase level. The expression of hilD was unchanged in either the hns mutant or the hha hns mutant. Thus, all the fusions tested were derepressed to different degrees in the absence of H-NS and/or Hha; however, the greatest difference was seen for the effect on the rtsA-lac fusion.
FIG. 1.
The effect of deletions of hns and hha regulators on rtsA, hilC, and hilD transcription. (A) The expression of chromosomal reporters was examined for the following strains (ATCC 14028 background): the wild-type (WT) rtsA-lac strain (JS324), the hha rtsA-lac mutant (IO949), the hns rtsA-lac mutant (IO954), the hha hns rtsA-lac mutant (IO955), the WT hilC-lac strain (JS487), the hha hilC-lac mutant (IO969), the hns hilC-lac mutant (IO971), the hha hns hilC-lac mutant (IO973), the WT hilD-lac strain (JS488), the hha hilD-lac mutant (IO959), the hns hilD-lac mutant (IO961), and the hha hns hilD-lac mutant (IO963). The level of β-galactosidase was determined following growth in LB medium with 1% NaCl (+NaCl) or no added NaCl (−NaCl). Data are representative of three independent experiments. (B) The transcription of the rtsA and marA genes on the chromosome was monitored by RT-PCR following the growth of the WT (IO908) and the hha hns mutant (IO923) in LB medium without NaCl. The experiment was performed three times, and typical data are shown.
It is possible that the rtsA-lac fusion, which inactivates the native rtsA gene, might not reproduce all aspects of the response of the rtsA promoter to HilD, HilC, and RtsA induction. For this reason, the effect of Hha/H-NS on rtsA expression was tested without the reporter fusion, monitoring rtsA mRNA directly by RT-PCR. For this experiment, RNA was isolated from hilA-deficient cells to reduce the effect of hha hns mutations on the fitness of S. enterica serovar Typhimurium (36). Under low-osmolarity conditions, the up-regulation of rtsA transcription in the absence of Hha and H-NS was evident, similar to the data obtained with the rtsA-lac fusion. The hns-independent control, marA (33), showed no response to the lack of hha and hns (Fig. 1B).
rtsA promoter.
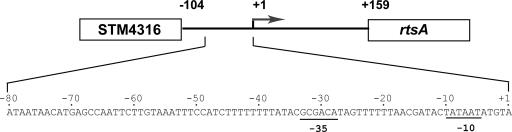
To examine the regulation of rtsA gene expression, primer extension analysis was used to identify the 5′ end of the rtsA transcript as a A residue located 159 nucleotides upstream of the translation start (data not shown). The transcription start site was preceded by a −10 element for a σ70-dependent promoter (TATAAT) and a weak −35 element (GCGACA; 4/6 match) separated by a 17-bp spacer (Fig. 2). A notable feature of the rtsA regulatory region is its very high AT content; the 150-bp sequences upstream and downstream of the +1 position contain 76.6 and 80.1% A+T, respectively. In contrast, the region from −300 to −201 further upstream contains the typical 52% AT content.
FIG. 2.
A schematic version of the STM4316-rtsA intergenic region. The rtsA promoter-specific transcript is indicated with an arrow, and the sequence of the start site is marked as +1. The nucleotide sequence of the rtsA promoter region is shown below. The rtsA promoter “−10” and “−35” elements are underlined. The rtsA transcription start site was determined by primer extension as described in Materials and Methods.
H-NS and Hha bind the rtsA promoter.
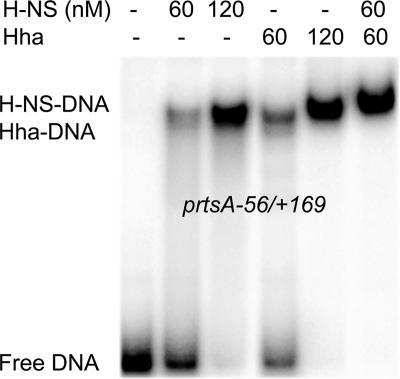
Hha is known to bind H-NS, which can potentially contaminate purified Hha protein (30). To avoid this complication, MBP-Hha was purified from E. coli cells lacking H-NS to produce H-NS-free MBP-Hha protein (see Materials and Methods). No effect on DNA-binding activity was found after the MBP moiety was separated from the Hha portion by cleavage with recombinant tobacco etch virus protease (data not shown). Therefore, MBP-Hha was used in all subsequent DNA-binding assays. To investigate H-NS and Hha protein binding to the rtsA promoter, EMSA with purified H-NS and MBP-Hha proteins was conducted under the conditions previously used with the hilA promoter (36). The shifted complexes were detected with the rtsA promoter fragment spanning from −56 to +169 at low concentrations (60 nM) of H-NS and MBP-Hha or the combination of 60 nM H-NS and 60 nM MBP-Hha (Fig. 3). These results are in agreement with data from transcriptional assays (Fig. 1) and suggest that the proteins H-NS and Hha can individually bind to the rtsA promoter.
FIG. 3.
EMSAs of the binding of H-NS and MBP-Hha to the rtsA promoter. The proteins were incubated with a 32P-labeled DNA fragment representing a portion of the rtsA promoter at positions −56 to +169. Samples were resolved by nondenaturing polyacrylamide gel electrophoresis, and the positions of the radioactive bands were detected by use of a PhosphorImager.
Purification and DNA-binding properties of the RtsA protein.
Since the rtsA promoter is induced by activators HilD, HilC, and RtsA (13), we chose to study a potential interplay of these activators and H-NS/Hha. The expression and purification of the S. enterica RtsA protein were attempted. Several plasmid-based systems for expression of the intact or His-tagged forms of RtsA yielded only insoluble and inactive product. The expression plasmid pMBP-RtsA1, encoding the RtsA protein fused to the C terminus of the E. coli MBP, provided levels of the soluble fusion protein suitable for purification on an amylose matrix. Fractions eluted from this matrix by use of 10 mM maltose contained MBP-RtsA at >90% purity, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). When the MBP moiety was separated from the RtsA portion by cleavage with recombinant tobacco etch virus protease, the released RtsA protein was poorly soluble. Therefore, the MBP-RtsA fusion protein was used for all subsequent experiments, in comparison to the use of His-tagged HilC and HilD proteins purified previously (37).
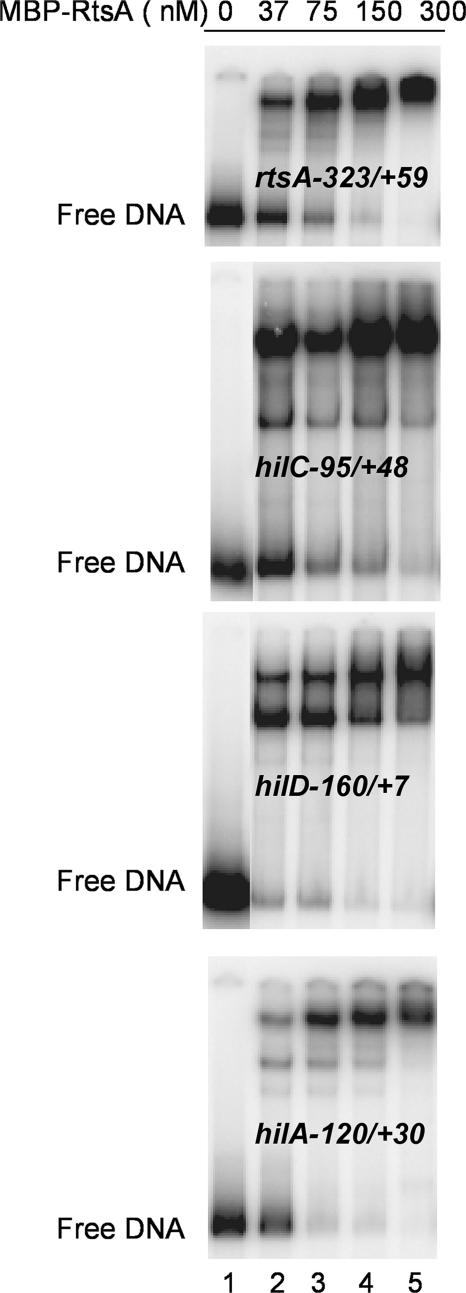
We first determined the ability of the purified MBP-RtsA to bind DNA by EMSA. The MBP-RtsA protein gave rise to the retarded complex on the rtsA promoter (Fig. 4). It also formed complexes on DNA fragments carrying promoters hilC (−95 to +48), hilD (−160 to +7), and hilA (−120 to +30). At some promoters, two complexes were seen, and the less retarded complex appeared to be converted to the more retarded complex at higher concentrations of MBP-RtsA. The concentration dependence for the disappearance of the unshifted band indicated that MBP-RtsA had a higher affinity for sites in the hilA and hilD promoters than for the rtsA and hilC promoters. At all promoters, half-maximal binding was achieved at concentrations of MBP-RtsA below 37 nM. Therefore, we determined that the MBP-RtsA protein binds to sequences in the promoters for rtsA and the three hil genes.
FIG. 4.
EMSA of the binding of MBP-RtsA to the rtsA, hilC, hilD, and hilA promoter fragments. The MBP-RtsA protein in concentrations of 0, 37, 75, 150, and 300 nM was incubated with 32P-labeled DNA fragments representing portions of the four promoters, as indicated at the left. Samples were resolved by nondenaturing polyacrylamide gel electrophoresis, and the positions of the radioactive bands were detected by use of a PhosphorImager.
HilD, HilC, and RtsA bind to a common DNA-binding site at the rtsA promoter.
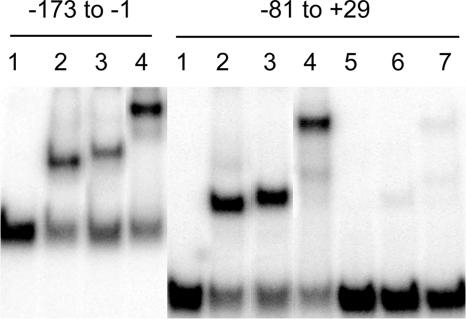
DNA fragments containing portions of the rtsA promoter region were constructed by PCR and tested by EMSA to identify the locations of the targets for HilD, HilC, and RtsA (Fig. 5). All three proteins formed single retarded complexes with two rtsA promoter fragments extending from −173 to −1 and from −81 to +29, indicating the presence of a single and possibly overlapped binding site for HilC, HilD, and RtsA proteins located between −81 and −1 relative to the transcription start site. The different mobilities of the shifted complexes reflect the different sizes of the proteins. No binding of HilC, HilD, or MBP-RtsA was found in the downstream area of the rtsA promoter (data not shown).
FIG. 5.
EMSA of the binding of HilC, HilD, and MBP-RtsA to portions of the rtsA promoter. Fragments are identified at the top. Fragments were labeled with 32P as described in Materials and Methods. The proteins added in each lane are as follows: lanes 1, no added protein; lanes 2 and 5, 75 nM HilC; lanes 3 and 6, 75 nM HilD; lanes 4 and 7, 75 nM MBP-RtsA. Unlabeled rtsA promoter DNA (10 ng) was included in reactions 5 to 7.
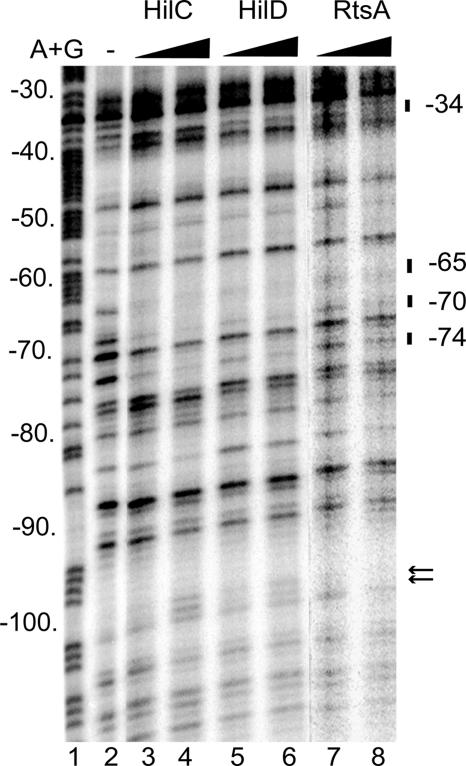
To identify the sequences that contribute to HilD, HilC, and RtsA binding at the rtsA promoter, the DNA fragment extended from −173 to +29 was analyzed by DNase I footprinting (Fig. 6). The presence of HilC, HilD, or RtsA resulted in protection of the positions −74, −70, −65, and −34 compared to what was seen for free DNA (Fig. 6). In addition, HilC and HilD footprints produced an enhancement of DNase cleavage in the positions −100 and −101, which may indicate the DNA bend. Taken together, both EMSA and DNA I footprinting assays showed a common DNA-binding site for HilC, HilD, and RtsA proteins located between −74 and −34 of the rtsA promoter. The comparison analysis of this area of the rtsA promoter versus known HilD/HilC binding sites from the hilA, hilC, and hilD promoters (37) using the MEME program for the alignment of multiple sequences (http://bioweb.pasteur.fr/seqanal/motif/meme/) revealed more than 60% similarity between these sites (Fig. 7A). Two direct repeats of CNATTNNT were found in the 35-bp consensus tcCtaTTNtTgTAatttTcNgNCcAtTaatNAtA, where capital letters indicate predominant bases (4/5 or 5/5), lowercase letters depict 3/5-identical bases in the HilD/HilC binding sites, boldface indicates specified bases of the CNATTNNT repeat element, and N is any base. The inverted position of the hilC C2 and hilA A1 sites in alignment compared to regions in hilD, rtsA promoters, and the hilA A2 site suggests that the orientations of dimers at these two sets of Hil protein targets are opposite (Fig. 7B).
FIG. 6.
DNase I protection assay of HilC, HilD, and MBP-RtsA binding to the rtsA promoter. The DNase I digestions represent the noncoding strand of the 32P-labeled rtsA fragment from −173 to +29 incubated with increasing amounts of purified proteins. Protected bands are indicated on the left. Hypersensitive sites are shown by arrows. Lane 1 has the products of the A+G sequencing reaction of the same DNA fragment. The added proteins in each lane are as follows: lane 2, no added protein; lane 3, 150 nM HilC; lane 4, 300 nM HilC; lane 5, 150 nM HilD; lane 6, 300 nM HilD; lane 7, 150 nM MBP-RtsA; lane 8, 300 nM MBP-RtsA.
FIG. 7.
Comparison of HilC/HilD binding sites in the rtsA, hilC, hilD, and hilA promoters. (A) Alignment of the HilC/HilD binding sites defined by DNase I footprinting here and previously (37). The common positions are indicated by shading. The proposed consensus is shown in the bottom. The arrows indicate two direct repeats of CNATTNNT (shown in bold). Predominant bases (5/5 or 4/5) in the consensus sequence are indicated by uppercase letters, bases identical in 3/5 of the HilC/HilD binding sites are indicated by lowercase letters, and degenerate bases are marked by the letter N. The inverted positions of hilC C2 and hilA A1 sites in alignment compared to regions in hilD, rtsA promoters and hilA A2 site are marked by asterisks. (B) Positions and directions of the HilD/HilC consensus in the rtsA, hilC, hilD, and hilA promoters relative to the transcription start point, indicated as +1.
Activators HilD and HilC are not essential for rtsA expression in the absence of repressors.
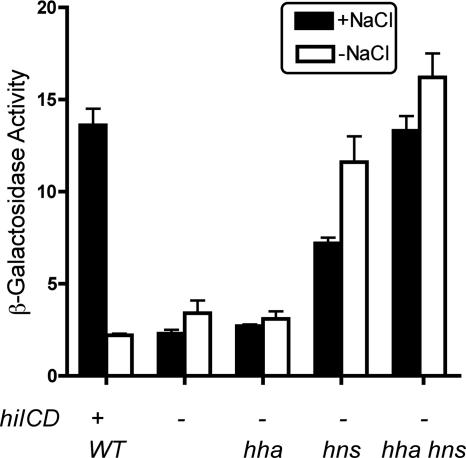
To evaluate the role of the activators HilD and HilC in the regulation of rtsA transcription, the single-copy rtsA-lac transcriptional fusion was tested in the hilD hilC background in the presence and absence of hha and hns mutations (Fig. 8). The level of β-galactosidase was determined following growth in high- and low-salt media. As expected, the induction of the rtsA promoter by salt without HilD, HilC, and RtsA was lost (hilDC rtsA-lac strain). The absence of Hha had no effect on rtsA-lac expression. In contrast, the absence of H-NS produced a sixfold increase in levels of β-galactosidase under low-osmolarity conditions and a threefold increase in the high-salt medium. The combination of the hha and hns mutations resulted in a high-level salt-independent rtsA expression comparable with that seen for hilD+ hilC+ cells under inducing conditions. Thus, activators HilD and HilC are not essential for rtsA expression in the absence of repressors Hha and H-NS.
FIG. 8.
The effect of the deletions of hns and hha on the expression of the chromosomal transcriptional fusion rtsA-lac in the absence of hilCD. The expression of the rtsA-lac reporter was examined for the following strains (ATCC 14028 background): the WT rtsA-lac strain (JS324), the ΔhilCD rtsA-lac mutant (IO976), the hha ΔhilCD rtsA-lac mutant (IO997), the hns ΔhilCD rtsA-lac mutant (IO999), and the hha hns ΔhilCD rtsA-lac mutant (IO1000). The level of β-galactosidase was determined following growth in LB medium with 1% NaCl (+NaCl) or no added NaCl (−NaCl). Data represent three independent experiments.
DISCUSSION
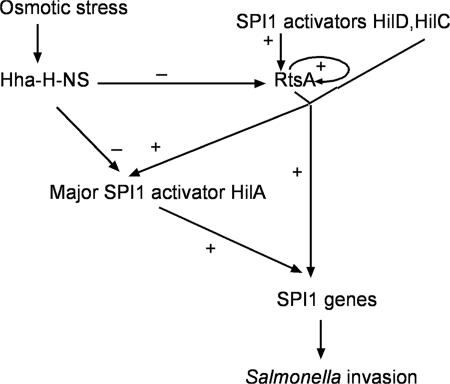
The invasion of intestinal epithelial cells constitutes the first critical step in the pathogenesis of S. enterica. The temperature and osmolarity conditions found in the intestinal tract trigger the rapid production of the SPI1-encoded type III secretion system, which is required for invasion. In order to clarify the Salmonella response to osmotic stress, we investigated here the osmoregulation of the virulence activator proteins HilD, HilC, and RtsA. These three proteins can induce the expression of SPI1 genes through the major activator of invasion, HilA, and also in a HilA-independent manner (1, 13, 40, 44) (Fig. 9).
FIG. 9.
A model of SPI1 osmoregulation by nucleoid-associated proteins H-NS/Hha and activators HilD/HilC/RtsA. H-NS/Hha negatively regulate the hilA and rtsA genes under low-osmolarity conditions. HilD/HilC/RtsA derepress the rtsA gene. Under high-osmolarity conditions, HilD, HilC, and RtsA activate SPI1 genes directly and indirectly through hilA. For clarity, the proposed autoregulation of HilA, HilD, and HilC is not shown. Repression is indicated as “−” and activation as “+.”
The nucleoid-associated proteins Hha and H-NS are environment-dependent modulators of gene expression in enterobacteria (3, 30). The microarray analysis of S. enterica serovar Typhimurium identified more than 200 genes, including SPI1 activators hilA, hilC, rtsA, and hilD, as thermoregulated by H-NS (38). Our data show that H-NS regulate SPI1 expression as a part of a repressive complex with another nucleoid-associated protein, Hha, acting on hilA (36), rtsA, and hilC promoters (Fig. 9). A regulatory role of the Hha-H-NS complex in response to changes in temperature and osmolarity was first demonstrated for the E. coli hly operon (34). Experimental evidence obtained from DNA-binding studies of Hha and H-NS with the hly promoter showed high Hha affinity to H-NS but not to DNA. Nevertheless, Hha was able to bind to the E. coli ler promoter and the S. enterica hilA promoter with high affinity (16, 36, 45). Furthermore, high-affinity Hha-DNA binding, comparable with H-NS-DNA binding, was evident in our experiments with the rtsA promoter. The ability of Hha and H-NS proteins to bind the rtsA promoter independently and in a complex is consistent with results of our transcriptional studies. The depletion of NaCl from LB medium lead to a significant decrease in rtsA-lac expression, which is restored for the hns mutant and is further induced for the hns hha double mutant (Fig. 1 and 8), suggesting that the repression under low-osmolarity conditions is mediated by H-NS/Hha.
In addition to controlling Salmonella invasion, Hha plays an important role in repressing genes located on SPI2, which are required for intracellular growth (48). In this case, Hha synergistically acts with another member of the Hha/YmoA family, YdgT. Thus, it appears that small nucleoid-associated proteins are often found to act together in order to fine-tune the response to specific environmental stimuli.
It has been shown that Hil activators can independently induce expression of the hilC, hilD, rtsA, and hilA genes when overexpressed (13). Two activators, HilC and HilD, have been purified, and their binding to the specific overlapping sites in hilC, hilD, and hilA promoters was demonstrated previously (37, 44). We report here the purification and DNA-binding properties of the third activator, RtsA. In agreement with genetic data, we show that the purified RtsA protein, like HilC and HilD, binds to the hilC, hilD, rtsA, and hilA promoters. The EMSA and DNase I footprinting assays of the rtsA promoter reveal that all three Hil activators bind a common site (positions −74 to −34 relative to the transcriptional start site). Each monomer of the AraC/XylS family members possesses two helix-turn-helix motifs and can occupy two adjacent DNA turns (6, 19, 26). This structure fits well with the results obtained in this study, which predict that two molecules of activator proteins, likely a dimer, occupy four successive DNA helical turns. We found two direct repeats (CNATTNNT) that are present in the HilC/HilD binding sites defined by DNase I footprinting within the hilD, hilC, rtsA, and hilA promoters (Fig. 7A). The DNA sites C2 in hilC and A1 in hilA are in a “reverse” orientation within their promoters, suggesting the opposite orientation of dimers at these two sets of Hil protein targets (Fig. 7B). Although the RtsA protein and the HilC/HilD proteins use common DNA-binding sites at the rtsA promoter, it is unknown whether this is true for all promoters affected by these proteins. The presence of two RtsA binding sites in hilD (−160 to +7) and hilA (−120 to +30) promoter fragments (Fig. 4), which are known to have a single HilC/HilD site, raise the possibility that the DNA bindings of RtsA and HilC/HilD are not always coupled. Future studies will help to determine if this is the case.
The requirement for Hil activators to induce the rtsA promoter under high-osmolarity conditions is lost in the absence of Hha and H-NS proteins (Fig. 8), suggesting that Hil activators antagonize H-NS/Hha-mediated repression. Alternatively, these findings may suggest the presence of an additional HilD/HilC/RtsA-independent promoter similar to one found in the hilC gene (37). However, this possibility is less likely, because no chromosomal rtsA-lac expression was detected in the absence of HilD, HilC, and RtsA (13).
In summary, this report shows that osmotic stress acts on the rtsA promoter through the modulation of H-NS/Hha-mediated repression. The virulence activators HilD, HilC, and RtsA recognize a common site at the rtsA promoter and apparently antagonize Hha/H-NS-mediated repression. The contribution of individual activators in vivo is likely to depend on environmental conditions. To further understand the roles of the homologous regulators HilD, HilC, and RtsA in Salmonella pathogenesis, future work will be needed to identify the environmental signals that are transferred through individual activators.
Acknowledgments
We thank Nazir Barekzi for critical reading of the manuscript and M. Belfort, L. Csonka, Z. Derewenda, and J. Slauch for their generous provision of bacterial strains.
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences to R.J.K.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-729. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43:85-92. [PubMed] [Google Scholar]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre, C., J. Johansson, B. E. Uhlin, A. Juarez, and F. J. Munoa. 1999. Alterations in protein expression caused by the hha mutation in Escherichia coli: influence of growth medium osmolarity. J. Bacteriol. 181:3018-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustos, S. A., and R. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, M. A., M. A. Jepson, N. L. Simmons, and B. H. Hirst. 1994. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res. Microbiol. 145:543-552. [DOI] [PubMed] [Google Scholar]
- 8.Dame, R. T., C. Wyman, R. Wurm, R. Wagner, and N. Goosen. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146-2150. [DOI] [PubMed] [Google Scholar]
- 9.Darwin, K. H., and V. L. Miller. 1999. Molecular basis for the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dole, S., V. Nagarajavel, and K. Schnetz. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589-600. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 12.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691-705. [DOI] [PubMed] [Google Scholar]
- 14.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, C. D., and J. M. Slauch. 2004. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J. Bacteriol. 186:68-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan, J. E., and R. I. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galas, D., and A. Schmitz. 1978. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5:3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman, E. A. (ed.). 2001. Principles of bacterial pathogenesis. Academic Press, San Diego, CA.
- 21.Hernandez, L. D., K. Hueffer, M. R. Wenk, and J. E. Galan. 2004. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304:1803-1807. [DOI] [PubMed] [Google Scholar]
- 22.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 23.Jones, B. D. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43:110-117. [PubMed] [Google Scholar]
- 24.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordi, B. J. A. M., and C. F. Higgins. 2000. The downstream regulatory element of the proU operon of Salmonella typhimurium inhibits open complex formation by RNA polymerase at a distance. J. Biol. Chem. 275:12123-12128. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424-430. [DOI] [PubMed] [Google Scholar]
- 27.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 28.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madrid, C., C. Balsalobre, J. Garcia, and A. Juarez. 2007. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 63:7-14. [DOI] [PubMed] [Google Scholar]
- 31.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 32.Marlovits, T. C., T. Kubori, A. Sukhan, D. R. Thomas, J. E. Galan, and V. M. Unger. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juarez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 35.Olekhnovich, I. N., and R. J. Kadner. 2004. Contribution of the RpoA C-terminal domain to stimulation of the Salmonella enterica hilA promoter by HilC and HilD. J. Bacteriol. 186:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357:373-386. [DOI] [PubMed] [Google Scholar]
- 37.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, S., M. D. Goldberg, T. Olsson, D. Esposito, J. C. Hinton, and J. E. Ladbury. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosseda, G., M. Falconi, M. Giangrossi, C. O. Gualerzi, G. Micheli, and B. Colonna. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51:523-537. [DOI] [PubMed] [Google Scholar]
- 40.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109-114. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 44.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 45.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheffield, P., S. Garrard, and Z. Derewenda. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34-39. [DOI] [PubMed] [Google Scholar]
- 47.Shin, M., M. Song, J. H. Rhee, Y. Hong, Y. J. Kim, Y. J. Seok, K. S. Ha, S. H. Jung, and H. E. Choy. 2005. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 19:2388-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silphaduang, U., M. Mascarenhas, M. Karmali, and B. K. Coombes. 2007. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J. Bacteriol. 189:3669-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 12:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, J., M. Tauschek, R. Strugnell, and R. M. Robins-Browne. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199-1208. [DOI] [PubMed] [Google Scholar]