Abstract
The 4-carboxymethylen-4-sulfo-but-2-en-olide (4-sulfomuconolactone) hydrolases from Hydrogenophaga intermedia strain S1 and Agrobacterium radiobacter strain S2 are part of a modified protocatechuate pathway responsible for the degradation of 4-sulfocatechol. In both strains, the hydrolase-encoding genes occur downstream of those encoding the enzymes that catalyze the lactonization of 3-sulfomuconate. The deduced amino acid sequences of the 4-sulfomuconolactone hydrolases demonstrated the highest degree of sequence identity to 2-pyrone-4,6-dicarboxylate hydrolases, which take part in the meta cleavage pathway of protocatechuate. The 4-sulfomuconolactone hydrolases did not convert 2-pyrone-4,6-dicarboxylate, and the 2-pyrone-4,6-dicarboxylate hydrolase from Sphingomonas paucimobilis SYK-6 did not convert 4-sulfomuconolactone. Nevertheless, the presence of highly conserved histidine residues in the 4-sulfomuconolactone and the 2-pyrone-4,6-dicarboxylate hydrolases and some further sequence similarities suggested that both enzymes belong to the metallo-dependent hydrolases (the “amidohydrolase superfamily”). The 4-sulfomuconolactone hydrolases were heterologously expressed as His-tagged enzyme variants. Gel filtration experiments suggested that the enzymes are present as monomers in solution, with molecular weights of approximately 33,000 to 35,000. 4-Sulfomuconolactone was converted by sulfomuconolactone hydrolases to stoichiometric amounts of maleylacetate and sulfite. The 4-sulfomuconolactone hydrolases from both strains showed pH optima at pH 7 to 7.5 and rather similar catalytic constant (kcat/KM)values. The suggested 4-sulfocatechol pathway from 4-sulfocatechol to maleylacetate was confirmed by in situ nuclear magnetic resonance analysis using the recombinantly expressed enzymes.
Aromatic sulfonic acids are produced in large quantities by the chemical industry. They are typically used as detergents, dyes, dispersants, ion exchangers, optical brighteners, and pharmaceuticals (43). In contrast, aromatic sulfonic acids are only rarely formed as natural products. Therefore, they are generally regarded as xenobiotic substances and several examples for the accumulation of aromatic sulfonic acids in the environment have been described (1, 2, 25, 35, 36, 44).
Previous studies of the microbial degradation of simple benzene- and naphthalenesulfonates demonstrated that these compounds are in most cases initially desulfonated by ring-hydroxylating dioxygenases to the corresponding diols (29, 30, 42, 45). In contrast, a coculture of Hydrogenophaga intermedia strain S1 and Agrobacterium radiobacter strain S2 initially oxidatively deaminated 4-aminobenzenesulfonate (sulfanilate) to 4-sulfocatechol (4SC) (14). More recently it was shown that 4-sulfocatechol is also formed as a ring fission substrate in the catabolic pathways for the degradation of 1,3-benzenedisulfonate and linear alkylbenzenesulfonates (LAS) (11, 12, 13, 37, 40). This indicated that 4-sulfocatechol is a central intermediate in the degradation of substituted sulfonated benzenes, which are released by humans in quantities of more than 106 t annually.
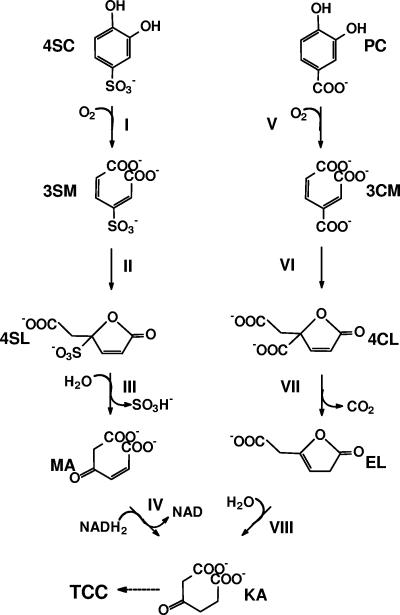
The sulfanilate-degrading coculture of H. intermedia S1 and A. radiobacter S2 converted 4-sulfocatechol to 3-sulfomuconate by variants of protocatechuate 3,4-dioxygenases, which converted protocatechuate and 4-sulfocatechol (8, 10). In the next enzymatic step, 3-sulfomuconate is converted to a sulfonated lactone (Fig. 1). The abilities of the 4-sulfocatechol- and 3-sulfomuconate-transforming enzymes to also convert their carboxylated structural counterparts (protocatechuate and 3-carboxy-cis,cis-muconate) and sequence comparisons clearly demonstrated that both enzymes are related to the corresponding enzymes of the protocatechuate branch of the β-ketoadipate pathway. These enzymes were therefore designated as “type II enzymes” (8, 10, 15, 17, 18, 19). The product formed enzymatically from 3-sulfomuconate in the cycloisomerization reaction has been identified as 4-carboxymethylen-4-sulfo-but-2-en-olide (“4-sulfomuconolactone” [4SL]) (15, 17). This metabolite structurally resembles its equivalent from the protocatechuate pathway (4-carboxymethylen-4-carboxy-but-2-en-olide [“4-carboxymuconolactone”]) (Fig. 1). However, further metabolism of 4-carboxymuconolactone and 4SL must be fundamentally different because it is not possible to eliminate the sulfonic acid substituent via the same mechanism used to decarboxylate 4-carboxymuconolactone (Fig. 1). Therefore, it was previously suggested that 4-sulfomuconolactone is hydrolytically desulfonated to maleylacetate (15).
FIG. 1.
Proposed pathway for the degradation of 4-sulfocatechol and protocatechuate by Agrobacterium radiobacter S2 (15). Enzymes: I, protocatechuate 3,4-dioxygenase type 2; II, 3-carboxy-cis,cis-muconate-lactonizing enzyme type 2; III, 4-sulfomuconolactone hydrolase; IV, maleylacetate reductase; V, protocatechuate 3,4-dioxygenase type 1; VI, 3-carboxy-cis,cis-muconate-lactonizing enzyme type 1; VII, 4-carboxymuconolactone decarboxylase; VIII, β-ketoadipate enol-lactone hydrolase. Compounds: 4SC, 4-sulfocatechol; 3SM, 3-sulfomuconate; 4SL, 4-sulfomuconolactone (4-carboxymethylene-4-sulfobut-2-en-4-olide); MA, maleylacetate; KA, β-ketoadipate; PC, protocatechuate; 3CM, 3-carboxy-cis,cis-muconate; 4CL, 4-carboxymuconolactone; EL, β-ketoadipate enol-lactone.
The presumed differences in the reaction mechanisms for the conversion of the carboxylated and sulfonated lactones suggest that 4-sulfomuconolactone hydrolase is the key enzyme in the modified β-ketoadipate pathway for the conversion of substituted benzenesulfonates. Therefore, in the present study, the 4-sulfomuconolactone hydrolases (4SLHs) from H. intermedia S1 and A. radiobacter S2 were analyzed on the enzymatic and molecular levels to further clarify the evolution of the modified β-ketoadipate pathway that is responsible for the conversion of various xenobiotic and environmentally harmful sulfonated benzenes.
MATERIALS AND METHODS
Bacterial strains and media.
The isolation, characterization, and culture conditions for Hydrogenophaga intermedia S1 (DSMZ 5680) and Agrobacterium radiobacter S2 (DSMZ 5681) have been described previously (9, 14, 15).
Escherichia coli DH5α, E. coli JM109, and E. coli BL21(DE3)pLysS (Invitrogen, Carlsbad, CA) were used as host strains for recombinant DNA work. E. coli strains were routinely cultured in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml), if appropriate.
Plasmids and DNA manipulation techniques.
The characteristics of all plasmids used are given in Table 1. The isolation of genomic DNA of H. intermedia S1 and A. radiobacter S2, the amplification of DNA by PCR, and all recombinant DNA work was performed as described previously (17).
TABLE 1.
Bacterial plasmids
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pBlueskript II SK(+) | Standard cloning vector, apr | 3 |
| pAC28 | T7- Expression vector | 23 |
| pSHSLHS1 | 4-Sulfomuconolactone hydrolase gene from H. intermedia S1 in pAC28 | This study |
| pSHSLHS2 | 4-Sulfomuconolactone hydrolase gene from A. radiobacter S2 in pAC28 | This study |
| pMCS2-2 | pcaG2 genes for a putative TRAP transport system, the putative IclR regulator, pcaB2S2, and the gene fragment for the C-terminal part of the 4-sulfomuconolactone hydrolase from A. radiobacter S2 in pBluescript II SK(+) | 10 |
| pS2PDH-2 | C-terminal part of the gene for the 4-sulfomuconolactone hydrolase and the maleylacetate reductase from A. radiobacter S2 in pBluescript II SK(+) | This study |
| pETS2-X-II | Expression of pcaH2G2 from A. radiobacter S2 under the control of the T7 promoter | 8 |
| pDS15 | Gene for the 2-pyrone-4,6-dicarboxylate hydrolase from S. paucimobilis SYK-6 under the control of the T7 promoter | 28 |
Determination of the nucleotide sequence of the 4-sulfomuconolactone hydrolase gene from A. radiobacter S2.
The genes coding for the protocatechuate 3,4-dioxygenase type II and 3-carboxy-cis,cis-muconate-lactonizing enzyme type II from A. radiobacter (ArCMLE2), which in A. radiobacter S2 are responsible for the conversion of 4-sulfocatechol to 4-sulfomuconolactone, had been previously identified on plasmid pMCS2-2. Downstream of the gene encoding the A. radiobacter CMLE2, a truncated gene encoding a putative hydrolase was identified (open reading frame [ORF] 4) (10, 17). The missing part of ORF 4 was obtained using partial inverse PCR (32). Finally a DNA fragment was amplified from the genomic DNA of A. radiobacter by using the primers MC_CHA_1 and MC_CHA_2 (Table 2). The amplified DNA fragment (ca. 2 kb) was cloned into pBluescript II SK(+), giving pS2PDH-2. Nucleotide sequencing of the insert demonstrated that it carried the missing part of the gene encoding the 4-sulfomuconolactone hydrolase and a putative maleylacetate reductase gene.
TABLE 2.
Oligonucleotide primers used in the present study
| Position | Primer name | Deduced primer sequencea (5′→3′)a |
|---|---|---|
| C-terminal region of the 4-sulfomuconolactone hydrolase from H. intermedia S1 | MC_CHA_1 | GATGCGGTCGAGCGTTCTG |
| Downstream of the 4-sulfomuconolactone hydrolase gene from H. intermedia S1 | MC_CHA_2 | CGAAAGTGTTGCAGCGACCG |
| N terminus of the 4-sulfomuconolactone hydrolase from A. radiobacter S2 | PDHS2-X-N | AAAACATATGTTACCCGCTGATCAAGCTGG |
| C terminus of the 4-sulfomuconolactone hydrolase from A. radiobacter S2 | PDHS2-X-C | AAAAGGATCCGTTGCATTGAATATCCGCCCC |
| N terminus of the 4-sulfomuconolactone hydrolase from H. intermedia S1 | PDHS1-X-N | TTTTCATATGTCAGAACAAGCTGTTGAAGTTTCGC |
| C terminus of the 4-sulfomuconolactone hydrolase from H. intermedia S1 | PDHS1-X-B | TTGGATCCTCATGCTCCCTTGGCAACC |
The underlined sequences indicate the recognition sites for the restriction endonucleases NdeI and BamHI.
Sequencing of the 4-sulfomuconolactone hydrolase gene from H. intermedia S1.
The gene encoding the 4-sulfomuconolactone hydrolase from H. intermedia S1 was identified downstream of the gene encoding the 3-sulfomuconate-converting CMLE2 from H. intermedia (HiCMLE2) on an approximately 2-kb DNA fragment previously obtained by PCR using the primers pcaBS1_1930F and pcaBS1_1888R (17). The DNA-fragment was completely sequenced and found to encode the carboxy-terminal part of the HiCMLE2, the 4SLH, and the amino-terminal part of a maleylacetate reductase.
Amplification and cloning of the 4-sulfomuconolactone hydrolases in E. coli.
The genes encoding the 4SLHs of H. intermedia S1 and A. radiobacter S2 were amplified from the genomic DNA of the strains by PCR using the primers PDHS1-X-N, PDHS1-X-B, PDHS2-X-N, and PDHS2-X-C (Table 2), using “Ready to Go” PCR beads (Amersham). This resulted in the simultaneous introduction of NdeI sites upstream and BamHI sites downstream of the genes. The amplified products were partially cleaved with NdeI and BamHI (there is a NdeI cleavage site in the 4SLH gene of strain S1) and cloned into pAC28 (23), previously digested with NdeI and BamHI. This resulted in the recombinant plasmids pSHSLHS1 and pSHSLHS2, which carried amino-terminal His-tagged enzyme variants. The plasmids were subsequently used to transform cells of E. coli BL21(DE3)pLysS. The integrity of the insert was verified by sequencing.
Nucleotide sequence analysis.
The DNA sequences were determined by dideoxy chain termination with double-stranded DNA of overlapping subclones in an automated DNA-sequencing system. (ALF-Sequencer, Amersham-Pharmacia, Freiburg, Germany) with fluorescently labeled primers.
Sequence analysis, database searches, and sequence comparisons were performed with Lasergene software, version 5 (DNASTAR Inc., Madison, WI), and BLAST at NCBI (4).
Amino acid sequence alignment.
Amino acid sequences were aligned using ClustalX (41) and default parameters. The computed dendrogram was visualized using TreeView version 1.6.6 (31).
High pressure liquid chromatography.
The turnover of 4-sulfocatechol, 3-sulfomuconate, and 4-sulfomuconolactone was analyzed by reversed-phase High pressure liquid chromatography (HPLC; pump model 510 equipped with a photodiode array detector model 996 and Millenium Chromatography Manager 2.0; Waters Associates, Milford, MA). A reversed-phase column (250- by 4.0-mm internal diameter) packed with 3-μm particles of Nucleosil C18 was used. The flow rate was 1 ml/min. The separated compounds were detected photometrically at 210 nm, using a photodiode array detector. The solvent system consisted of 98.9% (vol/vol) water, 1% (vol/vol) methanol, and 0.1% (vol/vol) H3PO4. The average retention times of 4-sulfocatechol, 3-sulfomuconate, and 4-sulfomuconolactone under these chromatographic conditions were 3.7, 3.4, and 4.3 min, respectively.
The turnover of 4SL to maleylacetate was analyzed using a different reversed-phase column (125- by 4.0-mm internal diameter) packed with 5-μm particles of Lichrospher 100, RP8) and a solvent system consisting of 16% (vol/vol) acetonitrile, 83.7% water, and 0.3% (vol/vol) trifluoroacetic acid. In this system, 4SL and maleylacetate had average retention times of 1.3 and 2.7 min, respectively.
Preparation of cell extracts.
Cell suspensions in 50 mM Tris-HCl buffer (pH 8.0) were disrupted by using a French press (SLM Aminco; SLM Instruments Inc., Urbana, IL) at 1.1 × 108 Pa. Cells and cell debris were removed by centrifugation at 100,000 × g for 30 min at 4°C.
Protein content estimation and enzyme assays.
The protein content of cell extracts was determined by the method of Bradford (6). Bovine serum albumin was used as a standard. One unit of enzyme activity is defined as the amount of enzyme that converts 1 μmol of substrate per minute.
The conversion of 4-sulfomuconolactone was routinely measured by using a spectrophotometric assay. The cuvettes contained a final volume of 1 ml of 100 μM 4SL (synthesized enzymatically from 4-sulfocatechol; see below) and 50 mM Tris-HCl buffer (pH 8). The reactions were started by the addition of cell extracts or purified enzyme preparations. The increase in absorption due to the formation of maleylacetate was determined at 242 nm. The reaction rates were calculated by using a molar extinction coefficient for maleylacetate of ɛ242 nm = 4,740 M−1cm−1 (38).
2-Pyrone-4,6-dicarboxylate hydrolase activity was determined spectrophotometrically, using the method described by Masai et al. (28). The cuvettes contained a total volume of 1 ml of 50 mM Tris-HCl (pH 8) and 100 μM 2-pyrone-4,6-dicarboxylate. The reactions were started by the addition of cell extracts, and the decrease of absorption was monitored at λ = 312 nm. Reaction rates were calculated by using a molar extinction coefficient of 6,600 M−1 cm−1 (28).
All kinetic data were fitted using Graphpad Prism version 4.0 (Graph Pad Software, San Diego, CA).
Purification of the His-tagged enzyme variants of the 4-sulfomuconolactone hydrolases.
E. coli BL21(DE3)pLysS(pSHSLHS1) and E. coli BL21(DE3)pLysS(pSHSLHS2) were grown at 30°C in LB medium plus kanamycin (50 μg/ml) to an optical density at 546 nm of 0.5. Subsequently, isopropyl-β-d-thiogalactopyranoside (1 mM) was added, and the bacterial cultures were grown for another 5 h. The cells were then harvested by centrifugation, and cell extracts were prepared in Tris-HCl buffer (50 mM [pH 8.0]). The cell extracts (about 120 mg of protein) were transferred to a 20-ml fast-performance high-pressure liquid chromatography column filled with Ni-nitrilotriacetic acid Superflow agarose (QIAGEN) that was previously equilibrated with 1 to 2 column volumes of the equilibration buffer consisting of Tris-HCl (50 mM [pH 8.0]), NaCl (300 mM), imidazole (20 mM), and 1,4,-dithio-d,l-threitol ([DTT]; 1 mM). Subsequently, the column was washed with 1 to 2 column volumes of the equilibration buffer. Finally, the active enzymes were eluted using a buffer system (pH 8.0) consisting of Tris-HCl (50 mM), NaCl (300 mM), DTT (1 mM), and imidazole (100 mM). Fractions were collected (5 ml each), and those showing enzymatic activity (usually the third and fourth fractions) were used for the enzymatic tests.
Determination of molecular mass.
The relative molecular masses of the native enzymes were determined by gel filtration using a Superdex 200 preparative-grade column (Amersham Biosciences) calibrated with a “high-molecular weight” calibration kit (Amersham Biosciences).
Polyacrylamide gel electrophoresis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was performed by the method of Laemmli (26), and the gels were routinely stained with Coomassie blue. In some experiments, the gels were silver stained using a Dodeca silver stain kit (Bio-Rad).
Determination of the metal content of the 4-sulfomuconolactone hydrolase from H. intermedia S1.
The purified enzyme (about 100 μg of protein in 300 μl of 50 mM Tris-HCl [pH 8.0]) was dialyzed twice at 4°C for 36 h against 4 liters of 50 mM Tris-HCl (pH 8.0). The extinction coefficient of the protein was calculated from the known amino acid sequence as 42,420 cm−1 M−1 (16; http://www.basic.northwestern.edu). As the sample demonstrated an extinction of 0.26 at 280 nm, the protein content of the dialyzed protein solution was calculated as 6 μM. The metal content of the sample was analyzed by plasma mass-spectroscopy (Spurenanalytisches Laboratorium Baumann, Maxhütte-Haidhof, Germany).
Spectrophotometric quantitation of sulfite.
The enzymatic release of sulfite from 4-sulfomuconolactone was quantified by using Ellman′s reagent [5,5′-dithiobis(2-nitrobenzoic acid)] as described in the report by Johnston et al. (21).
Enzymatic production of 4-sulfomuconolactone.
4-Sulfomuconolactone for the enzyme assays was prepared by two sequential enzymatic reactions: 4-sulfocatechol (4 mM in 1 ml of 50 mM Tris-HCl [pH 8.0]) was converted to 3-sulfomuconate, using a cell extract from E. coli BL21(DE3)pLysS(pETS2-X-II) (4.5 mg of protein), which heterologously expressed the protocatechuate 3,4-dioxygenase type II (P34OII) from A. radiobacter S2 (10, 17). The reaction mixtures were incubated in an Eppendorf shaker at 1,400 rpm and 30°C, and the reaction was monitored by HPLC until substrate depletion (after approximately 45 min). Then, 50 μl (0.5 mg protein/ml) of a purified preparation of the 3-sulfomuconate-converting ArCMLE1 (18) was added. The reaction mixtures were shaken, and the reaction was further analyzed by HPLC until the complete conversion of the 3-sulfomuconate was achieved (usually after 25 to 30 min). The proteins were removed by centrifugation in an ultrafiltration unit (Vivaspin, 2 ml; Concentrator, 10,000 MW cutoff; PES membrane), and the filtrates were used for the enzyme assays.
In situ 1H nuclear magnetic resonance analysis of the transformation of 4-sulfocatechol.
4-Sulfocatechol was dissolved in 4 ml of 50 mM Tris-HCl buffer (pH 8.0) to give a final concentration of 4 mM. An aliquot of this solution was supplemented with 20% (vol/vol) D2O and transferred to the nuclear magnetic resonance (NMR) sample tube (0.7 ml). The one-dimensional 1H NMR spectra were recorded at 300 K on an AVANCE DMX 600 NMR spectrometer (Bruker, Rheinstetten, Germany) locked to the deuterium resonance of D2O in the solution. Spectra were recorded using the standard Bruker one-dimensional nuclear Overhauser effect spectroscopy suppression sequence with 280 scans, each with a 1.8-s acquisition time and a 1.3-s relaxation delay. The center of the suppressed water signal was used as an internal reference (4.80 ppm). (The same NMR-conditions were also used for the determination of the structures of the intermediates and the product of the reaction sequence.)
4SC (4 mM in 1 ml of 50 mM Tris-HCl [pH 8.0]) was converted by the addition of a cell extract (26 mg of protein) from E. coli BL21(DE3)pLysS(pETS2-X-II) (10) which expressed the 4SC-converting P34OII. The reaction was analyzed in parallel by using HPLC and NMR analysis. The HPLC analysis solvent system (1% [vol/vol] methanol, 98.9% water, and 0.1% [vol/vol] H3PO4) demonstrated that the addition of the enzyme preparation resulted after 30 min in the complete conversion of 4SC (retention time [Rt] = 3.7 min) to a product with an Rt of 3.4 min. A part of the solutions containing the substrate or the product were simultaneously analyzed by NMR.
In the following step, a diluted sample (1:1 [vol/vol]; 1 ml) of the product solution (containing about 2 mM 3-sulfomuconate) was incubated with 50 μl (0.1 mg/ml) of the purified 3-sulfomuconate-converting HiCMLE2. The His-tagged HiCMLE2 was purified by affinity chromatography as described previously (17), and the imidazole, which inhibited the enzyme, was removed by ultrafiltration (Vivaspin 20; Sartorius) and by subsequent dilution of the concentrated protein solution in 50 mM Tris-HCl (pH 8.0). The reaction mixture was incubated on a laboratory shaker, and aliquots were analyzed by HPLC. This demonstrated that 3-sulfomuconate was completely converted to 4-sulfomuconolactone after 60 min. The sample was then analyzed by NMR (see Results).
The solution containing the 4-sulfomuconolactone (1 ml; 2 mM) was incubated with the purified 4-sulfomuconolactone hydrolase from H. intermedia S1 (50 μl; 0.5 mg/ml). The HPLC analysis demonstrated that the substrate was completely converted within 60 min. The product formed was further characterized by NMR in comparison to that of a previously produced sample of maleylacetate (32).
Chemicals.
4-Sulfocatechol was synthesized according to the procedure described by Quilico (34).
Maleylacetate was prepared from 4-carboxymethylene-but-2-en-olide (cis-dienelactone) by alkaline hydrolysis (22). The cis-dienelactone was dissolved in water (2 mM; 10 ml) and 50 μl of 1 M NaOH was added. The formation of maleylacetate was analyzed spectrophotometrically at 242 nm until no further increase in absorbance was observed.
cis-Dienelactone and 2-pyrone-4,6-dicarboxylate were kindly provided by M. Schlömann (TU Freiberg, Germany) and E. Masai (Nagaoka University of Technology, Japan). All other chemicals used were obtained from Aldrich (Steinheim, Germany), Fluka (Buchs, Switzerland), Merck (Darmstadt, Germany), and Sigma (Neu-Ulm, Germany).
Nucleotide sequence accession number.
The nucleotide sequences of the sulfomuconolactone hydrolases were deposited in the GenBank nucleotide sequence database under the accession numbers DQ813261 and DQ813262, respectively.
RESULTS
Identification, cloning, and sequencing of the genes encoding the 4-sulfomuconolactone hydrolases from H. intermedia S1 and A. radiobacter S2.
Previously, the genes encoding two 3-carboxy-cis,cis-muconate-lactonizing enzymes were identified in the genomes of H. intermedia S1 and A. radiobacter S2. These genes were localized in both strains in close proximity to the genes coding for protocatechuate 3,4-dioxygenases. Furthermore, it was shown that the encoded proteins were able to convert 4-sulfocatechol (P34OII) and 3-sulfomuconate (CMLEII) (8, 10, 17, 18). Therefore, it was postulated that in both strains, the genes encoding the P34OIIs (pcaH2G2) and CMLEs (pcaB2) were part of genetic structures of a higher order which were responsible for the degradation of 4-sulfocatechol. In order to search for the genes coding for the supposed 4-sulfomuconolactone hydrolase, the genomic regions downstream of pcaB2 were sequenced. This resulted in the identification of two rather similar genes in both strains. The genes encoded proteins of 303 (strain S1) and 299 (strain S2) amino acids, which showed 57% amino acid sequence identity to each other.
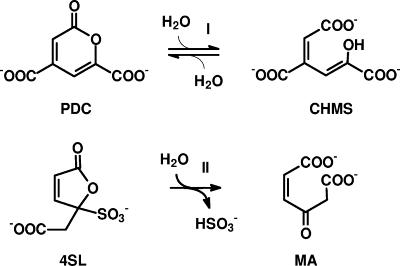
Sequence comparisons demonstrated for both enzymes the highest degree of amino acid sequence identity (57 to 68%) to a recently sequenced putative dicarboxylic acid hydrolase from Novosphingobium subarcticum SA1 (NCBI accession number AAW29742). Presumably, this enzyme belongs to the sulfanilate/4-sulfocatechol degradation gene cluster in this organism (GenBank accession number AY700015). A comparison of the putative 4-sulfomuconolactone hydrolases with sequences deposited at the NCBI database for which an enzymatic activity has been experimentally determined demonstrated the highest degree of amino acid sequence identity (30 to 32%) to the 2-pyrone-4,6-dicarboxylate hydrolases (PDCH) from Sphingomonas paucimobilis SYK-6 and Pseudomonas ochraceae (P. straminea). The PDCHs occur in the extradiol cleavage pathway of protocatechuate (27, 28). A comparison of the structural features of the respective substrates of the PDCHs (2-pyrone-4,6-dicarboxylate) and the 4-sulfomuconolactone hydrolases (4-carboxymethylen-4-sulfo-but-2-enolide) showed that both substrates resembled each other in their lactone structure and the presence and relative position of negatively charged groups (Fig. 2). This suggested that the two genes cloned from H. intermedia S1 and A. radiobacter S2 encoded 4-sulfomuconolactone hydrolases.
FIG. 2.
Comparison of the proposed reactions catalyzed by 2-pyrone-4,6-dicarboxylate hydrolases and 4-sulfomuconolactone hydrolases. PDC, 2-pyrone-4,6-dicarboxylate; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; 4SL, 4-sulfomuconolactone; MA, maleylacetate.
The genes encoding 4-sulfomuconolactone hydrolases and putative maleylacetate reductases are organized in a conserved sequential order.
Downstream of the 4-sulfomuconolactone hydrolase gene from H. intermedia S1, a BLAST search identified a truncated ORF encoding 244 amino acids which showed 63% amino acid sequence identity with amino acids 29 to 217 of a putative maleylacetate reductase from the putative 4-sulfocatechol operon of Novosphingobium subarcticum (GenBank accession number AAW29743.1) and 61% amino acid sequence identity with amino acids 23 to 217 of a presumed maleylacetate reductase from the 2,4-d-degrading Burkholderia cepacia 2a strain (GenBank accession number AAK81685.1).
Also in A. radiobacter S2, downstream of the 4-sulfomuconolactone hydrolase gene, a gene encoding a putative maleylacetate reductase was identified (on plasmid pS2PDH-2). The deduced gene product, with a length of 351 amino acids, showed again the highest degree of sequence identity (63%) with the sequence deposited for Novosphingobium subarcticum (GenBank accession number AAW29743.1).
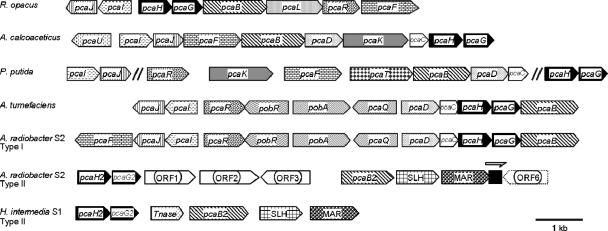
Thus, in both strains, a conserved gene order encoding a 3-carboxy-cis,cis-muconate-lactonizing enzyme, a 4-sulfomuconolactone hydrolase, and a putative maleylacetate reductase were found (Fig. 3). Maleylacetate reductases are involved in the proposed degradative pathway of 4-sulfocatechol (Fig. 1). This suggested that in both strains a three-gene functional unit exists which is apparently dedicated to the degradation of substituted benzenesulfonates.
FIG. 3.
Structures of the gene clusters for the catabolism of protocatechuate and 4-sulfocatechol from Hydrogenophaga intermedia S1 and Agrobacterium radiobacter S2 compared to the protocatechuate gene clusters from Rhodococcus opacus, Acinetobacter baylyi ADP1, Pseudomonas putida, and Agrobacterium tumefaciens (17). Homologous genes are shaded in the same way. “Type II enzymes” indicate those isoenzymes of the protocatechuate pathway which demonstrate increased relative activities with sulfonated substrates.
Functional expression of the 4-sulfomuconolactone hydrolases in E. coli.
The ORFs carrying the two putative 4-sulfomuconolactone hydrolases were amplified by PCR from the genomic DNAs of strains S1 and S2 and cloned into the expression vector pAC28 (see Materials and Methods). This yielded plasmids pSHSLHS1 and pSHSLHS2, which contained the genes encoding the His-tagged variants of the 4-sulfomuconolactone hydrolases from H. intermedia S1 and A. radiobacter S2, respectively. The enzymes encoded were functionally expressed in E. coli (see Materials and Methods), and cell extracts were analyzed by SDS gel electrophoresis. In both transformants, the induction of a protein with a subunit mass of about 35 kDa was observed.
Identification of maleylacetate as reaction product.
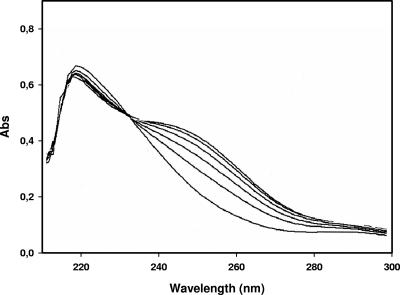
The enzyme variants were purified on nickel-agarose columns by immobilized metal ion affinity chromatography. The purified proteins (according to SDS gel electrophoresis, >90% pure) were incubated with 4-sulfomuconolactone, which was enzymatically prepared from 4-sulfocatechol by using protocatechuate 3,4-dioxygenase and 3-sulfomuconate-lactonizing enzymes (see Materials and Methods). The spectrophotometric analysis of the reaction mixture by using overlay spectra clearly demonstrated that 4-sulfomuconolactone was converted by the enzymes to a product which showed an increased absorbance at around 240 nm (Fig. 4). This suggested that maleylacetate was the reaction product, because at neutral pH, this compound has an absorption maximum of 242 nm. Furthermore, acidification of the reaction mixture to pH 2 resulted in the reversible disappearance of the absorption maximum (7).
FIG. 4.
Spectrophotometric analysis of the conversion of 4-sulfomuconolactone by the purified sulfomuconolactone hydrolase from H. intermedia S1. The reaction mixture was contained in a final volume of 1 ml of 50 mM Tris-HCl (pH 8.0), 100 μM 4-sulfomuconolactone, and 5 μl of a purified preparation of the 4-sulfomuconolactone hydrolase from H. intermedia (Cprot = 0.5 mg/ml). The overlay spectra were recorded every min against a reference cuvette containing the same ingredients but no enzyme.
The presence of an isosbestic point in the overlay spectra suggested that the substrate was stoichiometrically and directly converted into the product. The transformation of a defined concentration of 4-sulfomuconolactone (0.1 mM) with the purified 4-sulfomuconolactone hydrolase from E. coli BL21(DE3)pLysS(pSHSLHS1) resulted in an increase in absorbance of 0.48 at 242 nm. This value correlated well with the reported molar extinction coefficient of maleylacetate of 4,740 M−1 cm−1 (38), indicating that 4-sulfomuconolactone was indeed stoichiometrically converted to maleylacetate.
The formation of maleylacetate was further substantiated by HPLC analysis of the reaction. Thus, 4-sulfomuconolactone (1 mM in 50 mM Tris-HCl [pH 8.0]) was incubated in 1 ml for 20 min at room temperature with 50 μg of the purified 4-sulfomuconolactone hydrolase from H. intermedia S1, and the reaction mixture was analyzed by HPLC. This resulted in the disappearance of the signal for 4SL (Rt = 1.3 min) and the formation of a new signal with a retention time of 2.7 min. The latter signal was identified by comparison with an authentic standard according to its retention time and in situ spectrum as maleylacetate.
The 4-sulfomuconolactone-hydrolyzing activities of the recombinant E. coli strains were calculated spectrophotometrically using the increase in absorbance at 242 nm and a molar extinction coefficient for maleylacetate of 4,740 M−1 cm−1. Thus, in E. coli JM109(pSHSLHS1) and E. coli JM109(pSHSLHS2) 4SLH activities of 0.25 and 0.05 U/mg of protein, respectively, were found.
Identification of sulfite as reaction product.
The hydrolytic desulfonation of 4-sulfomuconolactone should result in the formation of maleylacetate plus sulfite. Therefore, different concentrations of 4-sulfomuconolactone (0.02 to 0.08 mM in 50 mM Tris-HCl-buffer [pH 8.0]) were incubated for 10 min with the purified 4-sulfomuconolactone hydrolase from H. intermedia S1 (concentration, 0.5 mg/ml), and the amount of sulfite formed was determined using Ellman's reagent (21). For all substrate concentrations, almost stoichiometrical amounts (R2 = 0.99) of sulfite were formed from the 4-sulfomuconolactone. In contrast, in control experiments without added enzyme, no sulfite was formed.
Characterization of the 4-sulfomuconolactone hydrolases from H. intermedia S1 and A. radiobacter S2.
The molecular masses of the purified His-tagged enzymes were determined by gel filtration. Thus, for the 4-sulfomuconolactone hydrolases from H. intermedia S1 and A. radiobacter S2, molecular weights of 34,700 and 32,900, respectively, were determined. This suggested that both enzymes have monomeric structures as previously found for the PDCH from S. paucimobilis SYK-6 (28).
Both hydrolases showed their pH optima at pH 7 to 7.5; approximately 30% of the maximal activities were still detected at pH 5 and pH 9.
The steady-state kinetic parameters were determined for both enzymes by using different concentrations (20 to 300 μM) of 4-sulfomuconolactone in 50 mM Tris-HCl (pH 8.0). The enzyme from strain S1 showed significantly higher Vmax and Km values, but the catalytic constants (kcat/KM) of both enzymes were similar (Table 3).
TABLE 3.
Kinetic data for the conversion of 4-sulfomuconolactone by the 4-sulfomuconolactone hydrolases from H. intermedia S1 and A. radiobacter S2
| Enzyme | Source | Molecular mass per subunit (kDa) | Kinetic data ± SDa
|
|||
|---|---|---|---|---|---|---|
| Km (mM) | Vmax (U/mg) | kcat (min−1) | kcat/Km (mM−1 min−1) | |||
| HiSLH | H. intermedia S1 | 34.7 | 1.9 ± 0.5 | 79.0 ± 18.6 | 2700 ± 600 | 1400 |
| ArSLH | A. radiobacter S2 | 32.9 | 0.34 ± 0.14 | 12.6 ± 3.3 | 400 ± 100 | 1200 |
Kinetic data ± standard deviations (SD) were calculated by nonlinear regression using Prism 4 software (GraphPad Software, San Diego, CA). Vmax values were calculated based on the protein content.
4-Sulfomuconolactone hydrolases are related to metallo-dependent hydrolases.
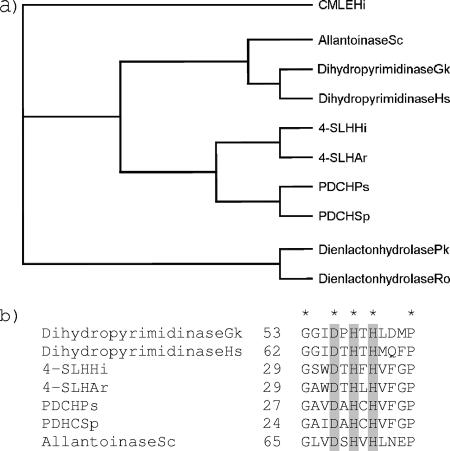
Sequence comparisons at the NCBI database using the “Conserved Domain Search” program suggested sequence similarities among the 4-sulfomuconolactone hydrolases, the PDCHs, and the metallo-dependent hydrolases (the “amidohydrolase superfamily”) (E-value of 0.000003). The dendrograms generated by the “Conserved Domains” program suggested a close relationship of the 4-sulfomuconolactone hydrolases and the PDCHs with dihydroorotases and other amidohydrolases (“cyclic amidases”), which convert cyclic amides, such as d-hydantoinases and dihydropyrimidases. The predicted relationship of 4-sulfomuconolactone hydrolases and metallo-dependent hydrolases was further investigated by amino acid sequence alignments. The resulting dendrogram underlined clearly that 4-sulfomuconolactone hydrolases (and PDCHs) were related to metallo-dependent hydrolases and not to dienelactone hydrolases (Fig. 5a), which catalyze the hydrolysis of the lactone ring in the chlorocatechol pathway (39). Moreover, the amino acid sequences of both the 4-sulfomuconolactone hydrolases and the PDCHs exhibit a conserved DXHXH motif (Fig. 5b) that is involved in the interaction with metal ions such as Mn2+, Mg2+, Zn2+, Ni2+, or Co2+ in the active centers of metallo-dependent hydrolases (24).
FIG. 5.
Comparison of the amino acid sequences of different 4-sulfomuconolactone hydrolases, 2-pyrone-4,6-dicarboxylate hydrolases, and dienelactone hydrolases. (a) Dendrogram showing the predicted relationship of 4-SLHs with metallo-dependent hydrolases. The sequence of the 3-carboxy-cis,cis-muconate-lactonizing enzyme from Hydrogenophaga intermedia S1 (CMLEHi) was used as an outgroup. (b) Sequence alignment showing the conserved regions in 4-sulfomuconolactone hydrolases, PDCHs, and metallo-dependent hydrolases. The conserved residues are indicated by an asterisk, and motif DXHXH is shaded. Numbers indicate the position of the conserved glycin residue in each protein. Gk, Geobacillus kaustophilus HTA426 (GenBank accession no. YP_147276); Hs, Homo sapiens (GenBank accession no. NP_001376); Hi, Hydrogenophaga intermedia (GenBank accession no. AAX11217 for CMLE and DQ813261 for 4-SLH); Ar, Agrobacterium radiobacter S2 (X); Ps, Pseudomonas straminea (GenBank accession no. BAD04056); Sp, Sphingomonas paucimobilis (GenBank accession no. BAA33799); Sc, Saccharomyces cerevisiae (GenBank accession no. NP_012293); Pk, Pseudomonas knackmussii (GenBank accession no. AAB71539); Ro, Rhodococcus opacus (GenBank accession no. O67988).
The metal content of a sample of the purified His-tagged variant of the 4-sulfomuconolactone hydrolase from H. intermedia S1 was determined by using plasma mass-spectroscopy. The sample contained 0.197 μg/ml Zn, 0.079 μg/ml Mg, 0.052 μg/ml Ni, 0.038 μg/ml Fe, 0.003 μg/ml Mn, and 0.001 μg/ml Co. The calculation of the Zn2+ content and the protein concentration in the sample suggested that about 0.6 mol of Zn2+ was present per mole of enzyme (see Materials and Methods). Therefore, the sulfomuconolactone hydrolase activity was tested in the presence of different metal ions and thiol reagents (0.5 mM each). These experiments demonstrated that the addition of ZnCl2 and CuCl2 resulted in a complete inhibition of the enzyme activity. In contrast, still in the presence of N-ethylmaleimide, 60% of the initial activity was recovered.
4-Sulfomuconolactone hydrolases do not convert 2-pyrone-4,6-dicarboxylate.
The sequence comparisons and the structural similarities of the substrates suggested the possibility of some cross-reactivities between PDCHs and 4-sulfomuconolactone hydrolases. Therefore, PDC (0.1 mM in 1 ml of 50 mM Tris-HCl [pH 8.0]) was incubated with a cell extract of E. coli BL21(DE3)pLysS(pSHSLHS1) (5 μl; 4.5 mg ml−1 of protein; 0.25 U/mg), and the reaction mixture was analyzed spectrophotometrically. No changes in absorbance between 200 and 400 nm were observed within 30 min of taking several overlay spectra. As PDC shows a pronounced absorbance maximum at a λ of 312 nm, this indicated that 4-sulfomuconolactone hydrolases do not convert PDC. In a second reaction mixture, a cell extract from E. coli JM109(pDS15), which heterologously expressed the PDCH from S. paucimobilis SYK-9 (28), was incubated with 4-sulfomuconolactone or PDC (0.1 mM each), and the reaction mixtures were analyzed spectrophotometrically. These extracts converted PDC (0.3 U/mg) but did not convert 4-sulfomuconolactone. Thus, 4-sulfomuconolactone hydrolases and PDCHs have distinctly different enzyme specificities.
NMR analysis of the complete 4SC pathway and confirmation of maleylacetate as the first desulfonated intermediate.
The proposed metabolic pathway of 4-sulfocatechol was finally confirmed by 1H NMR analysis of the in-situ-formed metabolites. Thus, 4-sulfocatechol and its metabolites were sequentially incubated with (i) a cell extract from E. coli BL21(DE3)pLysS(pETS2-X-II) expressing P34OII, (ii) a purified preparation of the 3-sulfomuconate and 3-carboxy-cis,cis-muconate-lactonizing enzyme (17), and (iii) the 4-sulfomuconolactone hydrolase from H. intermedia, and the metabolic reactions were monitored by 1H NMR analysis.
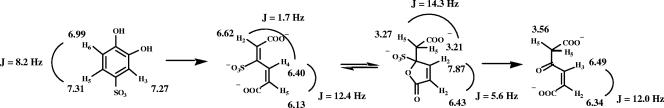
The substrate showed the presence of three aromatic protons resonating at 6.99 (d, J = 8.2 Hz), 7.27 (dd, J = 8.2 and <2 Hz), and 7.31 ppm (d, J = <2Hz) (Fig. 6) in accordance with the 4-sulfocatechol structure (15). Turnover by the P34OII resulted in the formation of 3-sulfomuconate as confirmed by the 1H NMR spectrum. Three olefinic protons were observed, with signals at δ values of 6.13 (H5), 6.40 (H4), and 6.62 (H2) ppm. The large coupling constant between H4 and H5 indicates that they are located in an open-chain configuration typical of muconates. The addition of HiCMLE2 resulted in the formation of a product with NMR characteristics previously reported for 4-sulfomuconolactone (15). Two olefinic protons resonate at a δ value of 6.43 and 7.87 ppm, respectively, and the small coupling constant of 5.6 Hz indicates the presence of the olefinic protons in a closed five-membered ring system. Two protons of a methylene group resonate at δ values of 3.21 and 3.27 ppm. The signals were split into doublets due to a geminal coupling of 14.3 Hz. The 4-sulfomuconolactone hydrolase converted 4-sulfomuconolactone into a single product with characteristics previously reported for maleylacetate (33). Two olefinic protons (δ = 6.34 and 6.49 ppm) were present in an open-chain formation as evidenced by the coupling constant of 12 Hz, and two methylene protons were observed at a δ of 3. 56 ppm.
FIG. 6.
Summary of the 1H NMR signals obtained for 4-sulfocatechol and its transformation products.
DISCUSSION
The present study is the final part of our investigation of the enzymes and genes involved in the degradation of 4-sulfocatechol (8, 10, 17, 18, 19). This study was undertaken as 4-sulfocatechol has been identified as a central intermediate in the microbial degradation of various substituted benzenesulfonates and LAS, which are probably the quantitatively most important class of xenobiotic compounds which are deliberately released by mankind into the environment (5, 20).
The present study clearly demonstrated that the 4-sulfomuconolactone hydrolases are evolutionarily not related to the 4-oxoadipate enol-lactone hydrolases participating in the ortho cleavage pathways of catechol and protocatechuate but are more closely related to (although distinct from) the 2-pyrone-4,6-dicarboxylate hydrolases from the extradiol cleavage pathway of protocatechuate. It was previously suggested by Masai et al. (28) that the PDCH from S. paucimobilis SYK-6 might contain a catalytically active cysteine residue in its active center, because the enzyme was sensitive to thiol reagents such as Ellman′s reagent and N-ethylmaleimide. Furthermore, the authors proposed, from the surrounding amino acids, a specific cysteine residue that could most probably fulfill this function (Cys76). It was therefore surprising that in our sequence comparisons of the 4-sulfomuconolactone hydrolases and PDCHs, the corresponding cysteine residue was not conserved but replaced by valine or isoleucine residues in the 4-sulfomuconolactone hydrolases. A subsequent sequence analysis using the “Conserved Domain Search” at the NCBI database then indicated sequence similarities between the PDCHs, the 4-sulfomuconolactone hydrolases, and the so-called cyclic amidases, which include the d-hydantoinases, dihydropyrimidases, allantoinases, and dihydroorotases. All these enzymes hydrolyze cyclic amides and contain divalent metal ions, such as Mn2+, Mg2+, Zn2+, Ni2+, or Co2+, which presumably are bound to the enzymes by some highly conserved histidine residues (24). Conserved histidine residues are also present in the PDCHs and 4-sulfomuconolactone hydrolases. This indicates that these enzymes also belong to the metallo-dependent hydrolases. This assumption is also supported by the structural resemblance of the substrates converted by the cyclic amidases, the 4-sulfomuconolactone hydrolases, and the PDCHs, which are all five- or six-membered ring systems containing one to two amide or ester groups.
The inability of the 4-sulfomuconolactone hydrolases and the PDCHs to convert the respective substrates of the other group of hydrolases clearly demonstrated that these enzymes indeed show distinct enzyme specificities. This was also reflected in the presence of certain conserved amino acid residues within the sequences of the 4-sulfomuconolactone hydrolases which differed from the homologous positions in the PDCHs. Furthermore, the genomic context of the respective genes within the genomes was different. The genes coding for the 4-sulfomuconolactone hydrolases were found (probably within operon structures) between the genes encoding (sulfomuconate-converting) CMLEIIs and those genes that presumably encode maleylacetate reductases. In contrast, the genes coding for the enzymatically characterized PDCHs from S. paucimobilis SYK-6 and Pseudomonas ochraceae NGJ1 (and also the genes presumably encoding PDCHs in Sphingomonas sp. strain LB126, Comamonas testosteroni BR6020, and Arthrobacter keyseri) were all found in operon structures which encoded all relevant enzymes of the protocatechuate meta cleavage pathway (27). This extradiol degradative pathway does not include enzymes with any significant homology to CMLEs or maleylacetate reductases. This observation should allow a differentiation of PDCHs and 4-sulfomuconolactone hydrolases during the annotation of new sequences obtained in the course of genome sequencing projects.
The complete study of the degradation of 4-sulfocatechol demonstrated that 4-sulfocatechol and 3-sulfomuconate were converted by enzymes which are clearly related to enzymes from the protocatechuate branch of the 3-oxoadipate pathway (8, 10, 17, 18). In contrast, clearly different origins could be shown for the enzymes which convert 4-carboxymethylen-4-carboxy-but-2-en-olide (“4-carboxymuconolactone”) in the protocatechuate pathway or 4-carboxymethylen-4-sulfo-but-2-en-olide (“4-sulfomuconolactone”) in the sulfocatechol pathway. This clearly resembles the situation observed for the degradation of chlorocatechols, which in most cases are degraded via a modified version of the catechol branch of the 3-oxoadipate pathway. Also in this metabolic pathway, it was found that the chlorocatechol- and chloromuconate-converting enzymes were clearly homologous to the enzymes involved in the metabolism of the naturally occurring substrate benzoate. In contrast, in the catechol and chlorocatechol pathways, different types of hydrolases are necessary for the ring opening of the intermediately formed lactone ring structures, which do not show any evolutionary relationship with each other (39). Thus, it appears that in both branches of the 3-oxoadipate pathway, nature has used rather similar strategies for the evolution of metabolic pathways which release substituents as anions. An interesting variation of this theme in the 4-sulfocatechol pathway is the observed pronounced sequence similarity between the 4-sulfomuconolactone hydrolases and the 2-pyrone-4,6-dicarboxylate hydrolases. This suggests a rather unique evolutionary connection between the intradiol and the extradiol pathways for the degradation of aromatic compounds.
Acknowledgments
We thank Simonetta Gribaldo for helpful discussions.
This work was supported by a grant from the “Deutsche Forschungsgemeinschaft” (DFG, project STO 400/2).
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Alexander, M., and B. K. Lustigman. 1966. Effect of chemical structure on microbial degradation of substituted benzenes. J. Agric. Food Chem. 14:410-413. [Google Scholar]
- 2.Alonso, M. C., M. Castillo, and D. Barceló. 1999. Solid-phase extraction procedure of polar benzene- and naphthalenesulfonates in industrial effluents followed by unequivocal determination with ion-pair chromatography/electrospray-mass spectrometry. Anal. Chem. 71:2586-2593. [DOI] [PubMed] [Google Scholar]
- 3.Alting-Mees, M. A., J. A. Sorge, and J. M. Short. 1992. pBluescript II: multifunctional cloning and mapping vectors. Methods Enzymol. 216:483-495. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berth, P., and P. Jeschke. 1989. Consumption and fields of application of LAS. Tenside Surf. Det. 26:75-79. [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contzen, M., S. Bürger, and A. Stolz. 2001. Cloning of the genes for a 4-sulfocatechol-oxidizing protocatechuate 3,4-dioxygenase from Hydrogenophaga intermedia S1 and identification of the amino acid residues responsible for the ability to convert 4-sulfocatechol. Mol. Microbiol. 41:199-205. [DOI] [PubMed] [Google Scholar]
- 9.Contzen, M., E. R. B. Moore, S. Blümel, A. Stolz, and P. Kämpfer. 2000. Hydrogenophaga intermedia sp. nov., a 4-aminobenzenesulfonate degrading organism. Syst. Appl. Microbiol. 23:487-493. [DOI] [PubMed] [Google Scholar]
- 10.Contzen, M., and A. Stolz. 2000. Characterization of the genes for two protocatechuate 3,4-dioxygenases from the catechol-4-sulfonate-degrading bacterium Agrobacterium radiobacter strain S2. J. Bacteriol. 182:6123-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contzen, M., R-M. Wittich, H.-J. Knackmuss, and A. Stolz. 1996. Degradation of benzene-1,3-disulfonate by a mixed bacterial culture. FEMS Microbiol. Lett. 136:45-50. [DOI] [PubMed] [Google Scholar]
- 12.Cook, A. M., H. Laue, and F. Junker. 1999. Microbial desulfonation. FEMS Microbiol. Rev. 22:399-419. [DOI] [PubMed] [Google Scholar]
- 13.Dong, W., P. Eichhorn, S. Radajewski, D. Schleheck, K. Denger, T. P. Knepper, J. C. Murrell, and A. M. Cook. 2004. Parvibaculum lavamentivorans converts linear alkylbenzenesulfonate surfactant to sulfophenylcarboxylates, α,β-unsaturated sulfophenylcarboxylates and sulfophenyldicarboxylates, which are degraded in communities. J. Appl. Microbiol. 96:630-640. [DOI] [PubMed] [Google Scholar]
- 14.Feigel, B. J., and H.-J. Knackmuss. 1988. Bacterial catabolism of sulfanilic acid via catechol-4-sulfonic acid. FEMS Microbiol. Lett. 55:113-118. [Google Scholar]
- 15.Feigel, B. J., and H.-J. Knackmuss. 1993. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch. Microbiol. 159:124-130. [DOI] [PubMed] [Google Scholar]
- 16.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 17.Halak, S., T. Basta, S. Bürger, M. Contzen, and A. Stolz. 2006. Characterization of the genes encoding the 3-carboxy-cis,cis-muconate lactonizing enzymes from the 4-sulfocatechol degradative pathways of Hydrogenophaga intermedia S1 and Agrobacterium radiobacter S2. Microbiology 152:3207-3216. [DOI] [PubMed] [Google Scholar]
- 18.Halak, S., L. Lehtiö, T. Basta, S. Bürger, M. Contzen, A. Stolz, and A. Goldman. 2006b. Structure and function of the 3-carboxy-cis,cis-muconate lactonizing enzyme from the protocatechuate degradative pathways of Agrobacterium radiobacter S2. FEBS J. 273:5169-5182. [DOI] [PubMed] [Google Scholar]
- 19.Hammer, A., A. Stolz, and H.-J. Knackmuss. 1996. Purification and characterization of a novel type of protocatechuate 3,4-dioxygenase with the ability to oxidize 4-sulfocatechol. Arch. Microbiol. 166:92-100. [DOI] [PubMed] [Google Scholar]
- 20.Jiménez, J., A. Breen, N. Thomas, T. W. Federle, and G. S. Sayler. 1991. Mineralization of linear alkylbenzene sulfonate by a four-member aerobic bacterial consortium. Appl. Environ. Microbiol. 57:1566-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston, J. B., K. Murray, and R. B. Cain. 1975. Microbial metabolism of aryl sulphonates. A re-assessment of colorimetric methods for the determination of sulphite and their use in measuring desulphonation of aryl and alkylbenzene sulphonates. Antonie Van Leeuwenhoek 41:493-511. [DOI] [PubMed] [Google Scholar]
- 22.Kaschabek, S. R., and W. Reineke. 1995. Maleylacetate reductase of Pseudomonas sp. strain B13: specificity of substrate conversion and halide elimination. J. Bacteriol. 177:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kholod, N., and T. Mustelin. 2001. Novel vectors for co-expression of two proteins in E. coli. BioTechniques 31:322-328. [DOI] [PubMed] [Google Scholar]
- 24.Kim, G.-J., and H-S. Kim. 1998. Identification of the structural similarity in the functionally related amidohydrolases acting on the cyclic amide ring. Biochem. J. 330:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knepper, T. P. 2002. Mass spectrometric strategies for the analysis of polar industrial chemicals and their by-products in wastewater and surface water. J. Chromatogr. 974:111-121. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama, K., T. Shibayama, A. Ichikawa, Y. Sakou, S. Yamada, and H. Sugisaki. 2004. Cloning and characterization of the genes encoding enzymes from the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 68:1434-1441. [DOI] [PubMed] [Google Scholar]
- 28.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nörtemann, B., J. Baumgarten, H. G. Rast, and H.-J. Knackmuss. 1986. Bacterial communities degrading amino- and hydroxynaphthalene-2-sulfonates. Appl. Environ. Microbiol. 52:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohe, T., T. Ohmoto, Y. Kobayashi, A. Sato, and Y. Watanabe. 1990. Metabolism of naphthalenesulfonic acids by Pseudomonas sp. TA-2. Agric. Biol. Chem. 54:669-675. [Google Scholar]
- 31.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 32.Pang, K. M., and D. A. Knecht. 1997. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques 22:1046-1048. [DOI] [PubMed] [Google Scholar]
- 33.Pieper, D. H., K. Pollmann, P. Nikodem, B. Gonzalez, and V. Wray. 2002. Monitoring key reactions in degradation of chloroaromatics by in-situ 1H nuclear magnetic resonance: solution structures of metabolites formed from cis-dienelactone. J. Bacteriol. 184:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quilico, A. 1927. Azione dell′acido amminosolfonico sui difenol. Gazz. Chim. Ital. 57:793-802. [Google Scholar]
- 35.Riediker, S., M. J.-F. Suter, and W. Giger. 2000. Benzene- and naphthalenesulfonates in leachates and plumes of landfills. Water Res. 34:2069-2079. [Google Scholar]
- 36.Ruckstuhl, S., M. J.-F. Suter, H.-P. E. Kohler, and W. Giger. 2002. Leaching and primary biodegradation of sulfonated naphthalenes and their formaldehyde condensates from concrete superplasticizers in groundwater affected by tunnel construction. Environ. Sci. Technol. 36:3284-3289. [DOI] [PubMed] [Google Scholar]
- 37.Schleheck, D., T. P. Knepper, K. Fischer, and A. M. Cook. 2004. Mineralization of individual congeners of linear alkylbenzenesulfonate by defined pairs of heterotrophic bacteria. Appl. Environ. Microbiol. 70:4053-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlömann, M. 1988. Die verschiedenen Typen der Dienlacton Hydrolase und ihre Rolle beim bakteriellen Abbau von 4-Fluorbenzoat. Thesis, Universität Stuttgart, Germany.
- 39.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Biodegradation 5:301-321. [DOI] [PubMed] [Google Scholar]
- 40.Schulz, S., W. Dong, U. Groth, and A. M. Cook. 2000. Enantiomeric degradation of 2-(4-sulfophenyl)butyrate via 4-sulfocatechol in Delftia acidovorans SPB1. Appl. Environ. Microbiol. 66:1905-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thurnheer, T., D. Zürrer, O. Höglinger, T. Leisinger, and A. M. Cook. 1990. Initial steps in the degradation of benzene sulfonic acid, 4-toluene sulfonic acids, and orthanilic acid in Alcaligenes sp. strain O-1. Biodegradation 1:55-64. [DOI] [PubMed] [Google Scholar]
- 43.Tully, P. S. 1997. Sulfonic acids, p. 194-217. In Kirk-Othmer Encylopedia of chemical technology, 4th ed., vol. 23. John Wiley & Sons, New York, NY. [Google Scholar]
- 44.Wellens, H. 1990. Zur biologischen Abbaubarkeit mono- und disubstituierter Benzolderivate. Z. Wasser-Abwasser Forsch. 23:85-98. [Google Scholar]
- 45.Wittich, R.-M., H. G. Rast, and H.-J. Knackmuss. 1988. Degradation of naphthalene-2,6- and naphthalene-1,6-disulfonic acid by a Moraxella sp. Appl. Environ. Microbiol. 54:1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]