Abstract
To reclassify phenotypically identified Staphylococcus intermedius strains, which might include true S. intermedius strains and novel species such as Staphylococcus pseudintermedius and Staphylococcus delphini, we analyzed molecular phylogenies and phenotypic characteristics of 117 S. intermedius group (SIG) strains tentatively identified as being S. intermedius by the Rapid ID32 Staph assay. From phylogenetic analyses of sodA and hsp60 sequences, the SIG strains were divided into three clusters, which belonged to S. pseudintermedius LMG 22219T, S. intermedius ATCC 29663T, and S. delphini LMG 22190T. All the SIG strains from dogs, cats, and humans were identified as being S. pseudintermedius. The wild pigeon strains, except one, were identified as being S. intermedius, and strains from all domestic pigeons, one wild pigeon, horses, and a mink were identified as being S. delphini. In addition, a phylogenetic analysis of nuc genes revealed that S. delphini strains were divided into two clusters: one was the cluster (S. delphini group A) that belonged to S. delphini LMG 22190T, and the other was the cluster (S. delphini group B) that was more related to S. pseudintermedius LMG 22219T than S. delphini LMG 22190T. The DNA-DNA hybridization results showed that S. delphini group B strains were distinguished from S. delphini group A, S. intermedius, and S. pseudintermedius strains. S. intermedius is distinguishable from S. pseudintermedius or S. delphini by positive arginine dihydrolase and acid production from β-gentiobiose and d-mannitol. However, phenotypical characteristics to differentiate S. delphini group A, S. delphini group B, and S. pseudintermedius were not found. In conclusion, SIG strains were reclassified into four clusters with three established and one probably novel species.
In 1976, Staphylococcus intermedius was previously described as being a new species isolated from pigeons, dogs, mink, and horses (14). The majority of coagulase-positive staphylococci in animals such as dogs and pigeons have been identified as being S. intermedius strains (3, 10-12, 19, 26, 35). This species has been recognized to constitute normal skin flora of various animal species and to occasionally cause a variety of infections in dogs and cats (3, 19).
There have been six species of coagulase-positive staphylococci other than Staphylococcus aureus, S. intermedius, S. schleiferi subsp. coagulans, S. hyicus, S. lutrae, S. delphini, and S. pseudintermedius (6, 9). Sequence similarities of 16S rRNA genes among these four species except S. hyicus and S. lutrae are >99% identical (6, 30). In addition, it is very difficult to differentiate between S. intermedius, S. delphini, and S. pseudintermedius phenotypically, and commercial kits are not available for the identification of S. pseudintermedius and S. delphini (6, 34). S. delphini was isolated from dolphins in 1988, and S. pseudintermedius was isolated from a cat, a dog, a horse, and a parrot in 2005 as a new species (6, 34). Since these two species were closely related to S. intermedius, the concise identification of S. intermedius has been in a confused state. Since some researchers indicated that S. intermedius strains isolated from different sources could be genotypically or phenotypically distinct from one another (1, 2, 5, 10-12, 24, 36), it is possible that previously reported “S. intermedius” strains with heterogeneous biotypes might have included S. delphini and S. pseudintermedius (1-3, 5, 10-12, 14, 28, 33, 36). We have recently reported that molecular phylogenetic analysis based on partial sodA gene sequences and hsp60 gene sequences is sufficiently discriminative for S. intermedius, S. pseudintermedius, and S. delphini (28).
Thermonuclease has been used for species identification of S. aureus as one of the key characteristics (4, 13, 38). Recently, Becker et al. (2) described PCR of the thermonuclease gene (nuc) for species identification of S. intermedius that could differentiate it from S. hyicus and S. schleiferi. However, even using that method, there were some nuc-negative isolates from horses and camels (2).
In the present study, we reclassified phenotypically identified S. intermedius strains from various origins using molecular phylogenetic analyses based on partial sequences of sodA and hsp60 genes, which exhibited higher divergences than 16S rRNA genes in staphylococci and nuc genes encoding thermonuclease (8, 29). In addition, to distinguish them, we searched for biochemical reactions including some commercial identification kits for staphylococci.
MATERIALS AND METHODS
Bacterial strains.
We selected 117 S. intermedius strains, which were derived from animals for which the isolation of S. intermedius had previously been reported (2, 5, 6, 10-12, 14, 36) and tentatively identified by Rapid ID32 Staph (bioMérieux, Marcy-l′Etoile, France) from our coagulase-positive staphylococcus collections from various animal sources, which we designated the S. intermedius group (SIG) (Table 1). These strains were stored in 10% skim milk at −85°C until use and maintained on Trypticase soy agar II with 5% sheep blood (BD Japan Co., Ltd., Tokyo, Japan).
TABLE 1.
SIG strains used in the present studya
| Host species | No. of strains | Clinical status | Source | Strain(s) | Source or reference |
|---|---|---|---|---|---|
| Dog | 78 | Pyoderma | Wound pus from skin | NVAU 02002, NVAU 02003, NVAU 02004, NVAU 02005, NVAU 02006, NVAU 02007, NVAU 02008, NVAU 02009, NVAU 02010, NVAU 02012, NVAU 02013, NVAU 02014, NVAU 02015, NVAU 02016, NVAU 02017, NVAU 02019, NVAU 02020, NVAU 02021, NVAU 02022, NVAU 02023, NVAU 02024, NVAU 02026, NVAU 02027, NVAU 02028, NVAU 02029, NVAU 02030, NVAU 02031, NVAU 02032, NVAU 02033, NVAU 02034, NVAU 02035, NVAU 02036, NVAU 02037, NVAU 02038, NVAU 02039, NVAU 02040, NVAU 02041, NVAU 02042, NVAU 02043, NVAU 02044, NVAU 02045, NVAU 02046, NVAU 02047, NVAU 02048, NVAU 02049, NVAU 02050, NVAU 02051, NVAU 02052, NVAU 02053, NVAU 02054, NVAU 02055, NVAU 02056, NVAU 02057, NVAU 02058, NVAU 02059, NVAU 02060, NVAU 02062 | This study |
| S. intermedius CCUG 50815, S. intermedius CCUG 51770 | CCUG | ||||
| Noninfection inpatients | Nares | NVAU 06002, NVAU 06013, NVAU 06016, NVAU 06017, NVAU 06018, NVAU 06021, NVAU 06022, NVAU 06029, NVAU 06031, NVAU 06033, NVAU 06034, NVAU 06037, NVAU 06038, NVAU 06039, NVAU 06043, NVAU 06044, NVAU 06045, NVAU 06046 | 28 | ||
| Unknown | Unknown | S. pseudintermedius CCUG 32915 | CCUG | ||
| Cat | 3 | Pyoderma | Wound pus from skin | NVAU 02082, NVAU 02083 | This study |
| Unknown | Lung tissue | S. pseudintermedius LMG 22219T | 6 | ||
| Human | 2 | Infection of mastoid cavity | Wound pus | TW 6698 | 18 |
| Unknown | Unknown | S. pseudintermedius CCUG 46004 | CCUG | ||
| Pigeon | 26 | Wild, healthy | Nares | P-2, P-4A, P-6A, P-9B, P-45A, P-46A, P-50, P-52B, P-53, P-54A, P-66A, P-69A | This study |
| Domestic, healthy | Nares | P-13B, P-14A, P-18A, P-19A, P-21B, P-22A, P-26, P-27B, P-28A, P-30A, P-36B, P-37A, P-39B | |||
| Unknown | Nares | S. intermedius ATCC 29663T | ATCC | ||
| Horse | 6 | Domestic, healthy | Nares | h-2C, h-4A, h-5D, h-6C, h-7C, h-9D | This study |
| Mink | 1 | Healthy | Nares | S. intermedius CCUG 51769 | CCUG |
| Dolphin | 1 | Unknown | Unknown | S. delphini LMG 22190T | 34 |
ATCC, American Type Culture Collection; CCUG, Culture Collection University Göteborg; LMG, Bacteria Collection Labolatorium Voor Microbiologie Universiteit Gent.
Phenotypic characterization.
Coagulase was determined by tube coagulase test using rabbit serum (Denka Seiken Co., Ltd., Tokyo, Japan) for all strains tested. Acid production from carbohydrates was detected under aerobic conditions, except d-mannitol, using a broth microplate method (16). Each carbohydrate of d-galactose, d-mannitol (in anaerobic culture for 20 h), arbutin, α-lactose, d-mannose, β-gentiobiose, and d-turanose was added in purple broth base (24 g/liter; Difco, MD) and thioglycolate medium without glucose or indicator (12 g/liter; Difco) at a final concentration of 0.5% (wt/vol). Each plate was incubated at 37°C and was evaluated at 20 and 48 h. MIC testing for polymyxin B using Etest (AB Biodisk, Solna, Sweden) was performed. An acetoin production (Vogues-Proskauer) test was carried out according to a method reported previously (9).
Thermonuclease production was detected by the metachromatic agar diffusion method using DNA and o-toluidine blue by using a modified method described previously (23), which was conducted as follows: culture supernatants in brain heart infusion broth (Difco) were filtrated and heated (at 100°C for 15 min), and disks with the filtrate were applied to DNase test agar (Becton Dickinson Co., Ltd., France) with o-toluidine blue. Agars were incubated at 37°C for 12 h.
DNA extraction.
A single colony was suspended to a McFarland standard of 1.0 in 100 μl of TE buffer (20 mM Tris, 2 mM EDTA [pH 7.5]) with 10 U of achromopeptidase (Wako Chemical Co., Ltd., Osaka, Japan), and the suspension was incubated at 55°C for 10 min. After centrifugation at 18,500 × g for 5 min, the supernatants were used as crude DNA extracts for PCR (17). Purified DNA extracts for DNA-DNA hybridization were made according to methods reported previously (7).
Genetic identification of the SIG.
Direct sequencing of sodA and hsp60 genes was performed for the differentiation of SIG strains as described previously (21, 22, 25, 27). Multiple alignment was carried out by using the CLUSTAL X program (32). The construction of the unrooted phylogenetic tree was performed by the neighbor-joining method (27).
Genetic characterization of the thermonuclease (nuc) gene.
Detection of the nuc gene, encoding thermonuclease, was carried out for SIG strains by PCR (PCR1) as previously described (2). PCR products were sequenced directly. Another PCR (PCR2) was conducted for PCR1-negative strains. The primer set of Nuc-dF (5′-GAA GGC ATA TTG TAG AAC AA-3′) and Nuc-eR (5′-NCK YTC CCA DAT RTT NAR YTK-3′) was designed to amplify the conserved region of the nuc gene by multiple alignment of amino acid sequences of the staphylococcal nuc gene from those of PCR1-positive strains and the staphylococcal species whose genomes had previously been wholly sequenced through the GenBank database. The size of PCR products was 724 bp including the coding sequence of the nuc gene. The reaction mixture for the PCR consisted of 2 μl of DNA extract in a total volume of 50 μl composed of 2 U of Ex Taq (Takara Shuzo Co., Ltd., Kyoto, Japan), 10 pmol of each primer, 0.2 mM deoxynucleoside triphosphate mixture, 1.5 mM MgCl2, and 1× reaction buffer (Takara Shuzo Co., Ltd.). Reaction mixtures were thermally cycled once at 95°C for 5 min; 35 times at 95°C for 1 min, 44°C for 1 min, and 72°C for 1 min; and then once at 72°C for 7 min. These PCR products were also sequenced directly.
DNA-DNA hybridization.
DNA-DNA hybridization was performed by microplate methods using a Benchmark Plus microplate reader (Bio-Rad, Tokyo, Japan) (7). Hybridization was performed at 45°C for 2 h in a hybridization mixture containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt solution, 3% dextran sulfate, 50% formamide, 200 ng/ml denatured herring DNA, and 1,500 ng/ml biotinylated probe DNA.
RESULTS
Molecular phylogenetic analysis of sodA and hsp60 gene sequences among SIG strains.
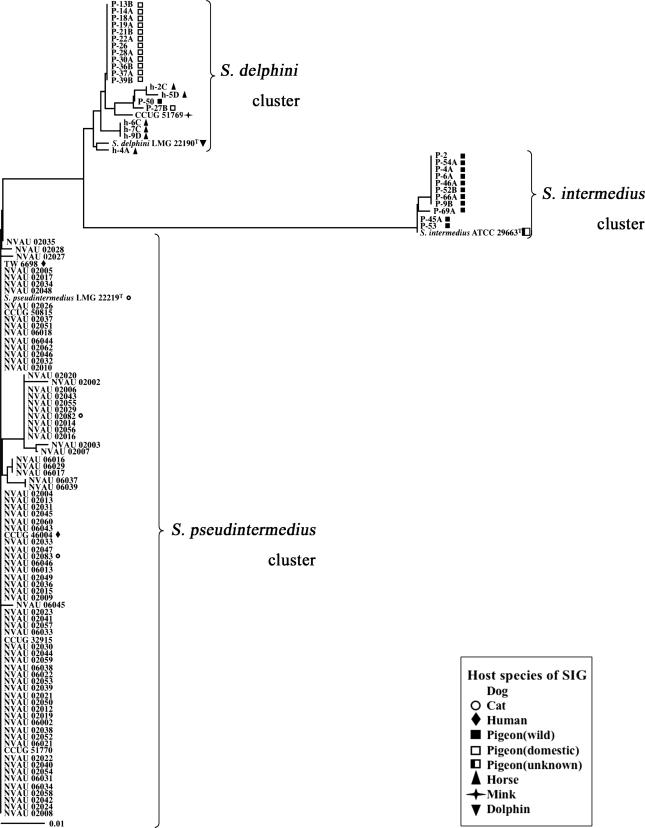
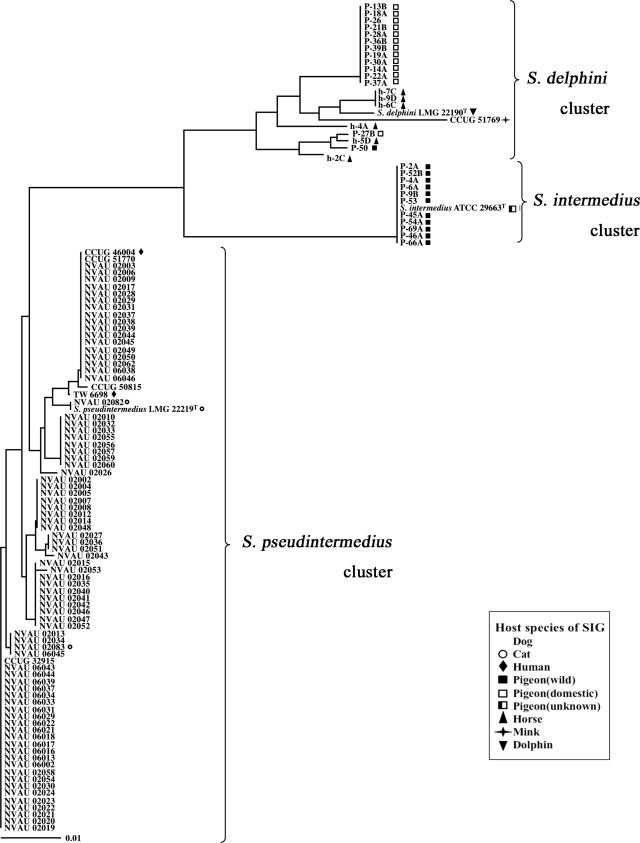
As shown in Fig. 1 and 2, all 78 strains from dogs were very close to S. pseudintermedius LMG 22219T, with high sequence homology of both the sodA (98.8 to 100%) and hsp60 (98.7 to 99.5%) genes. All the strains from cats and humans also belonged to the same cluster. Thus, these 83 strains were identified as being S. pseudintermedius strains. All the 11 wild pigeon-derived strains except strain P-50 showed high sequence similarities to both the sodA (99.7 to 100%) and hsp60 (100%) genes of S. intermedius ATCC 29663T. Consequently, these strains were identified as being S. intermedius strains. On the other hand, strains from 13 domestic pigeons, 1 wild pigeon (P-50), 6 horses, and a mink showed close sequence similarities to sodA (97.7 to 99.3%) of S. delphini LMG22190T. However, unlike the S. intermedius and S. pseudintermedius clusters, hsp60 sequences among these strains were more divergent, and similarities to S. delphini LMG22190T were 95.7 to 98.9%.
FIG. 1.
Phylogenetic tree (unrooted) based on partial sodA gene sequences of SIG strains used in the present study. The tree was constructed by the neighbor-joining method using CLUSTAL X. It is unknown whether S. intermedius ATCC 29663T is derived from a wild or domestic pigeon.
FIG. 2.
Phylogenetic tree (unrooted) based on partial hsp60 gene sequences of SIG strains used in the present study. The tree was constructed by the neighbor-joining method using CLUSTAL X. It is unknown whether S. intermedius ATCC 29663T is derived from a wild or domestic pigeon.
Detection and phylogenetic characterization of nuc genes among SIG strains.
For a better differentiation of S. pseudintermedius, S. intermedius, and S. delphini, we carried out two nuc PCRs and direct sequencing of the nuc genes (Table 2).
TABLE 2.
Phenotypic and genetic characterization of nuc among SIG strains
| Characteristic | Rate (%) of positive strains or range of sequence similarity (%)
|
|||
|---|---|---|---|---|
| S. pseudintermedius (n = 83) | S. intermedius (n = 12) |
S. delphini
|
||
| Group A (n = 17) | Group B (n = 5) | |||
| Phenotype (thermonuclease production) | 100 | 67 | 88 | 60 |
| Detection by PCR1 | 100 | 100 | 0 | 100 |
| Detection by PCR2 | 0 | 0 | 100 | 0 |
| Sequence similarity against S. pseudintermedius LMG 22219T | 98.8-100 | 85.6-87.2 | 79.2-80.3 | 95.9-96.1 |
PCR1 could detect predictive fragments among all the S. pseudintermedius and S. intermedius strains and 5 of 22 S. delphini strains. The remaining S. delphini strains were not amplified by PCR1 in spite of the ability to produce thermonuclease. PCR2 successfully amplified predictive fragments with 17 PCR1-negative strains.
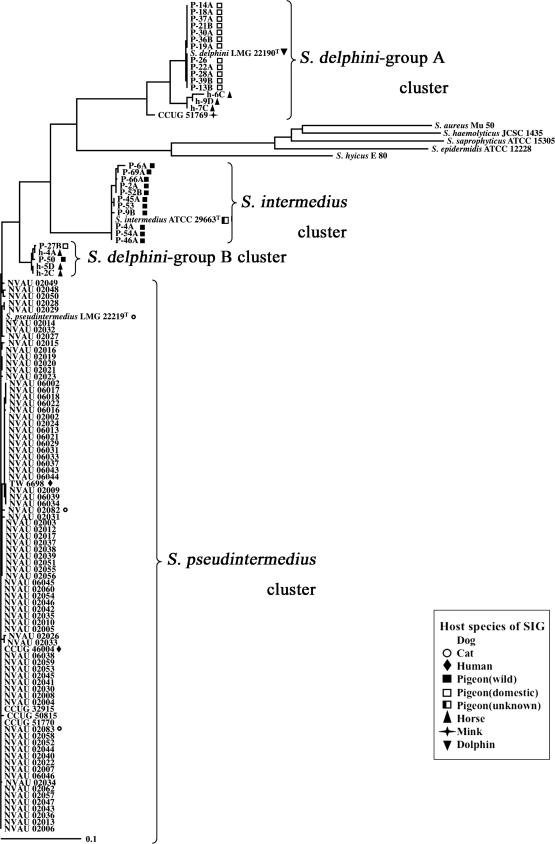
In the phylogenetic tree of the nuc gene, SIG strains were classified into four clusters (Fig. 3). S. intermedius and S. pseudintermedius strains constituted a single cluster that confirmed their sodA and hsp60 phylogeny. Interestingly, S. delphini strains were divided into two clusters: one (S. delphini group A) was similar to S. delphini LMG22190 T, and the other (S. delphini group B) was closer to the S. pseudintermedius cluster than S. delphini LMG22190T.
FIG. 3.
Phylogenetic tree (unrooted) based on partial nuc gene sequences of SIG strains used in the present study. The tree was constructed by the neighbor-joining method using CLUSTAL X. It is unknown whether S. intermedius ATCC 29663T is derived from a wild or domestic pigeon.
DNA-DNA hybridization among SIG strains.
To elucidate the taxonomic position of S. delphini groups A and B, We performed DNA-DNA hybridizations among SIG strains (Table 3). Given a threshold level of 70% DNA-DNA relatedness for the definition of a bacterial species (37), our reclassification based on phylogenetic analysis of the nuc gene corresponded to the species distinction. Among S. delphini group strains and those of the other clusters, S. delphini group A had the highest level (77%) of DNA similarity to S. delphini LMG22190T. On the other hand, S. delphini group B strains showed less than 70% similarities with those of any clusters. DNA similarities of greater than 70% were observed among S. delphini groups A and B, respectively (data not shown). These results suggest that S. delphini group B constitutes a novel species.
TABLE 3.
DNA-DNA hybridization among SIG strains
| Strain | % DNA similarities with biotin-labeled DNA from:
|
||||
|---|---|---|---|---|---|
| S. pseudintermedius LMG 22219T | S. intermedius ATCC 29663T |
S. delphini
|
|||
| LMG 22190T | Group A (h-6C) | Group B (P-27B) | |||
| S. pseudintermedius LMG 22219T | 100 | ||||
| S. intermedius ATCC 29663T | 29 | 100 | |||
| S. delphini LMG 22190T | 46 | 28 | 100 | ||
| S. delphini group A (h-6C) | 50 | 28 | 74 | 100 | |
| S. delphini group B (P-27B) | 45 | 40 | 61 | 57 | 100 |
Phenotypic characteristics among SIG strains.
To search for phenotypic characteristics that distinguish the genetically distinct SIG strains, we compared their biochemical characteristics. As shown in Table 4, S. intermedius could be differentiated from the other SIG strains by 100% negative arginine dihydrolase and 100% positive acid production from β-gentiobiose aerobically and d-mannitol anaerobically. There was no obvious difference in biochemical reactions among S. pseudintermedius, S. delphini group A, and S. delphini group B. There were some variations in the abilities of acid production from d-turanose: S. pseudintermedius and S. delphini group B showed weak or delayed reaction, S. delphini group A strains were negative, and S. pseudintermedius and S. delphini group A strains did not use β-gentiobiose, whereas about 60% of the S. delphini group B strains fermented β-gentiobiose. In standard acetoin production tests (9), 66.3% (55/83) of S. pseudintermedius strains were positive. The abilities were not as strong as with S. aureus, and most of them were not detected by the Rapid ID32 Staph test. S. intermedius and S. delphini group A (except for S. delphini LMG22190T) and group B strains were negative for this characteristic.
TABLE 4.
Phenotypic characteristics of SIG strains
| Test and characteristic | % Positive strains
|
|||
|---|---|---|---|---|
| S. pseudintermedius (n = 83) | S. intermedius (n = 12) |
S. delphini
|
||
| Group A (n = 17) | Group B (n = 5) | |||
| ID 32 Staph | ||||
| Urease | 100 | 100 | 88 | 100 |
| Arginine dihydrolase | 100 | 0 | 100 | 80 |
| Ornithine dihydrolase | 0 | 0 | 0 | 0 |
| Esculin hydrolysis | 0 | 0 | 0 | 0 |
| Acid production from: | ||||
| Glucose | 100 | 100 | 100 | 100 |
| Fructose | 100 | 100 | 100 | 100 |
| d-Mannose | 100 | 100 | 100 | 80 |
| d-Maltose | 91 | 91 | 94 | 100 |
| Lactose | 100 | 73 | 94 | 40 |
| Trehalose | 100 | 100 | 94 | 80 |
| d-Mannitol | 46 | 100 | 71 | 60 |
| Raffinose | 0 | 0 | 0 | 40 |
| d-Ribose | 100 | 100 | 100 | 100 |
| d-Cellobiose | 0 | 0 | 0 | 20 |
| Saccharose | 100 | 100 | 59a | 80 |
| N-Acetylglucosamine | 100 | 18a | 100 | 100 |
| d-Turanose | 0a | 0 | 0 | 0a |
| l-Arabinose | 0 | 0 | 0 | 20 |
| Nitrate reduction | 100 | 100 | 100 | 100 |
| Acetoin (Vogues-Proskauer) | 10 | 0 | 0 | 20 |
| β-Galactosidase | 100 | 100 | 100 | 100 |
| Arginine arylamidase | 0 | 0 | 0 | 0 |
| Alkaline phosphatase | 99 | 100 | 100 | 100 |
| Pyrrolidonyl arylamidase | 100 | 100 | 100 | 100 |
| Novobiocin resistance | 0 | 0 | 18 | 60 |
| β-Glucuronidase | 0 | 0 | 0 | 0 |
| Acid production from: | ||||
| d-Mannitol (anaerobically) | 0 | 100 | 0 | 0 |
| d-Galactose | 92 | 0a | 94 | 100 |
| Arbutin | 0 | 0a | 0 | 0 |
| β-Gentiobiose | 0 | 100 | 0 | 60 |
| d-Turanose (48 h) | 17a | 0 | 0 | 40a |
| Acetoin production (Vogues-Proskauer) | 66 | 0 | 5 | 0 |
| MIC of polymixin B (Etest) | ||||
| Range (mg/ml) | 16-32 | 8-12 | 12-32 | 24 |
| Geometric mean (mg/ml) | 25 | 11 | 22 | 24 |
Many of the strains showed a weak reaction and were designated as being negative.
All the S. pseudintermedius strains produced thermonuclease; the level of production was stronger than that for the other species. Although 66% (8/12) of S. intermedius strains showed a positive reaction, the activities were weaker than those of S. pseudintermedius. All S. delphini group A strains except S. delphini LMG22190T and strain CCUG 51769 showed a positive reaction.
DISCUSSION
Our results suggest that the majority of SIG strains isolated from commensal flora or infection sites in dogs are reclassified as being S. pseudintermedius strains. In fact, S. intermedius CCUG 50815 and S. intermedius CCUG 51770 (both of dog origin) obtained from the Culture Collections of the University Göteborg (CCUG) were also identified as being S. pseudintermedius strains by molecular phylogenetic analyses. Previously, we also reported that S. pseudintermedius was a common species among methicillin-resistant coagulase-positive staphylococci in dogs (28).
Van Hoovels et al. reported the first case of S. pseudintermedius infection in a human (33). In the present study, we also identified two strains from humans as being S. pseudintermedius strains, one of which (strain TW 6698) had been identified as being S. intermedius before our reclassification (18). Although there have been reports that S. intermedius strains were isolated from human infection or food poisoning from dog origins (15, 18, 33), there is another possibility, that these isolates were S. pseudintermedius isolates and not S. intermedius or S. delphini isolates.
In pigeons, SIG strains were isolated from the nares of wild and domestic species in the present study: the former was S. intermedius, and the latter was S. delphini group A. Distinct SIG species might be distributed to different host habitats in pigeons.
Some reports suggested previously that SIG strains from horses were heterogenous (2, 5, 36). In the present study, both S. delphini group A and group B strains were isolated from horses. Becker et al. also previously reported three different groups from horses using a PCR-DNA enzyme immunoassay of the nuc gene (2). With respect to nuc sequences in our results, these three groups might be S. pseudintermedius and S. delphini groups A and B, respectively.
SIG strains from minks have been reported to be distinct from those from dogs by pulsed-field gel electrophoresis analyses and by their lack of thermonuclease production (5, 6, 36). In the present study, all SIG strains from dogs were identified as being S. pseudintermedius strains and were positive for thermonuclease production. These results suggest that SIG strains from minks are different from S. pseudintermedius strains. In fact, strain CCUG 51769, from a mink, was identified as belonging to S. delphini group A in the present study.
Devriese et al. previously described S. pseudintermedius as being a novel species and reported that this species could be discriminated from S. intermedius or S. delphini by some biochemical characteristics (6). For example, those authors reported that S. delphini differed from S. pseudintermedius in its negative acidification of trehalose and thermonuclease production. However, some S. delphini strains, which had been included in the species S. intermedius, conventionally identified and reclassified in our study, showed a positive reaction in these tests. Furthermore, in acetoin production tests, the type strain of S. delphini, which had been reported to be negative previously (6, 9), showed a positive result by the standard method (9). These characteristics are not available to differentiate between S. pseudintermedius and S. delphini. In the present study, S. intermedius is easily distinguishable among SIG strains using some phenotypical properties such as positive arginine dihydrolase and acid production from β-gentiobiose and d-mannitol (anaerobically for 20 h). However, there is no “gold standard” to differentiate phenotypically among S. pseudintermedius and S. delphini groups A and B, although host animal species, acid production from β-gentiobiose and d-turanose (48 h), acetoin production (Vogues-Proskauer) by the standard method, and thermonuclease production might be useful to some extent. Finally, to discriminate between these species correctly, there is no method except PCR and sequencing of the nuc gene.
Whether or not they had thermonuclease activity, all the SIG strains had the nuc gene. All the whole-genome-sequenced staphylococci, S. epidermidis, S. saprophyticus, and S. haemolyticus, possess nuc-related genes with 40 to 60% putative amino acid sequence homologies to SIG thermonuclease (20, 31, 39). Although thermonuclease activity is not detected in S. epidermidis, S. saprophyticus, and S. haemolyticus (4, 9), these strains still keep this gene. In the present study, we detected the nuc gene from S. delphini, which was recognized as a species that is negative for thermonuclease activity (9, 34). These results suggest that the nuc gene is ubiquitous in the genus Staphylococcus. Differences in thermonuclease activities among staphylococcal species might be affected by differences in the heat stability of nuclease or expression levels. Gudding previously reported that the heat stability of S. intermedius nuclease showed lower levels than those of S. aureus and S. hyicus (13). However, the conservation and genetic diversities of nuc genes means that it could be used as another molecular evolutionary tool in staphylococci. Interestingly, nuc sequences in S. delphini group B strains were more closely related to those of S. pseudintermedius than S. delphini group A. The phylogenetic position of nuc gene in S. delphini group B was quite different from that of sodA or hsp60 genes. The nuc genes of S. delphini group B might have proceeded to a unique evolutionary path among staphylococci.
In conclusion, phenotypically identified S. intermedius strains were reclassified into at least four clusters, S. intermedius, S. pseudintermedius, and S. delphini groups A and B. S. delphini group B might be a novel species. Our study provides important information on the phylogenetic reclassification of staphylococcal species by molecular methods.
Acknowledgments
This work was partially supported by a grant-in-aid for 21st Century COE Research from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
We thank Hisayuki Kaneko and Tomoyasu Sakaguchi for providing staphylococcal strains.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Aarestrup, F. M. 2001. Comparative ribotyping of Staphylococcus intermedius isolated from members of the Canoidea gives possible evidence for host-specificity and co-evolution of bacteria and hosts. Int. J. Syst. Evol. Microbiol. 51:1343-1347. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., C. von Eiff, B. Keller, M. Bruck, J. Etienne, and G. Peters. 2005. Thermonuclease gene as a target for specific identification of Staphylococcus intermedius isolates: use of a PCR-DNA enzyme immunoassay. Diagn. Microbiol. Infect. Dis. 51:237-244. [DOI] [PubMed] [Google Scholar]
- 3.Beiberstein, E. L., S. S. Jang, and D. C. Hirsh. 1984. Species distribution of coagulase-positive staphylococci in animals. J. Clin. Microbiol. 19:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berke, A., and R. C. Tilton. 1986. Evaluation of rapid coagulase methods for the identification of Staphylococcus aureus. J. Clin. Microbiol. 23:916-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bes, M., S. L. Saidi, F. Becharnia, H. Meugnier, F. Vandenesch, J. Etienne, and J. Freney. 2002. Population diversity of Staphylococcus intermedius isolates from various host species: typing by 16S-23S intergenic ribosomal DNA spacer polymorphism analysis. J. Clin. Microbiol. 40:2275-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devriese, L. A., M. Vancanneyt, M. Baele, M. Vaneechoutte, E. De Graef, C. Snauwaert, I. Cleenwerck, P. Dawyndt, J. Swings, A. Decostere, and F. Haesebrouck. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 55:1569-1573. [DOI] [PubMed] [Google Scholar]
- 7.Ezaki, T., Y. Hashimoto, N. Takeuchi, H. Yamamoto, S. L. Liu, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Simple genetic method to identify viridans streptococci by colorimetric dot hybridization and fluorometric hybridization in microdilution wells. J. Clin. Microbiol. 26:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 9.Freney, J., W. E. Kloos, V. Hajek, and J. A. Webster. 1999. Recommended minimal standards for description of new staphylococcal species. Int. J. Syst. Bacteriol. 49:489-502. [DOI] [PubMed] [Google Scholar]
- 10.Futagawa-Saito, K., W. Ba-Thein, N. Sakurai, and T. Fukuyasu. 2006. Prevalence of virulence factors in Staphylococcus intermedius isolates from dogs and pigeons. BMC Vet. Res. 26:2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futagawa-Saito, K., M. Suzuki, M. Ohsawa, S. Ohshima, N. Sakurai, W. Ba-Thein, and T. Fukuyasu. 2004. Identification and prevalence of an enterotoxin-related gene, se-int, in Staphylococcus intermedius isolates from dogs and pigeons. J. Appl. Microbiol. 96:1361-1366. [DOI] [PubMed] [Google Scholar]
- 12.Futagawa-Saito, K., S. Sugiyama, S. Karube, N. Sakurai, W. Ba-Thein, and T. Fukuyasu. 2004. Prevalence and characterization of leukotoxin-producing Staphylococcus intermedius in isolates from dogs and pigeons. J. Clin. Microbiol. 42:5324-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudding, R. 1983. Differentiation of staphylococci on the basis of nuclease properties. J. Clin. Microbiol. 18:1098-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajek, V. 1976. Staphylococcus intermedius, a new species isolated from animals. Int. J. Syst. Bacteriol. 26:401-408. [Google Scholar]
- 15.Khambaty, F. M., R. W. Bennett, and D. B. Shah. 1994. Application of pulsed-field gel electrophoresis to the epidemiological characterization of Staphylococcus intermedius implicated in a food-related outbreak. Epidemiol. Infect. 113:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi, K., T. Enari, K. Totsuka, and K. Shimizu. 1995. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J. Clin. Microbiol. 33:1215-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi, K., N. Takahashi, C. Piao, K. Totsuka, H. Nishida, and T. Uchiyama. 2003. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains causing neonatal toxic shock syndrome-like exanthematous disease in neonatal and perinatal wards. J. Clin. Microbiol. 41:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikuchi, K., T. Karasawa, C. Piao, I. Itoda, H. Hidai, H. Yamaura, K. Totsuka, T. Morikawa, and M. Takayama. 2004. Molecular confirmation of transmission route of Staphylococcus intermedius in mastoid cavity infection from dog saliva. J. Infect. Chemother. 10:46-48. [DOI] [PubMed] [Google Scholar]
- 19.Kim, T. J., Y. R. Na, and J. I. Lee. 2005. Investigation into the basis of chloramphenicol and tetracycline resistance in Staphylococcus intermedius isolates from cases of pyoderma in dogs. J. Vet. Med. B 52:119-124. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok, A. Y., and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Evol. Microbiol. 53:87-92. [DOI] [PubMed] [Google Scholar]
- 22.Kwok, A. Y., S. C. Su, R. P. Reynolds, S. J. Bay, Y. Av-Gay, N. J. Dovichi, and A. W. Chow. 1999. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int. J. Syst. Bacteriol. 49:1181-1192. [DOI] [PubMed] [Google Scholar]
- 23.Lachica, R. V., P. D. Hoeprich, and C. Genigeorgis. 1971. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl. Microbiol. 21:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, S. A., and K. H. Schleifer. 1978. Deoxyribonucleic acid reassociation in the classification of coagulase-positive staphylococci. Arch. Microbiol. 117:183-188. [DOI] [PubMed] [Google Scholar]
- 25.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich, M. 2005. Staphylococci in animals: prevalence, identification and antimicrobial susceptibility, with an emphasis on methicillin-resistant Staphylococcus aureus. Br. J. Biomed. Sci. 62:98-105. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:846-849. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki, T., K. Kikuchi, Y. Tanaka, N. Takahashi, S. Kamata, and K. Hiramatsu. 2007. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J. Clin. Microbiol. 45:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 49:846-849. [Google Scholar]
- 30.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hoovels, L., A. Vankeerberghen, A. Boel, K. van Vaerenbergh, and H. De Beenhouwer. 2006. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 44:4609-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varardo, P. E., R. Kilpper-Bälz, F. Biavaasco, G. Satta, and K. H. Schleifer. 1988. Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int. J. Syst. Bacteriol. 38:436-439. [Google Scholar]
- 35.Vengust, M., M. E. Anderson, J. Rousseau, and J. S. Weese. 2006. Methicillin-resistant staphylococcal colonization in clinically normal dogs and horses in the community. Lett. Appl. Microbiol. 43:602-606. [DOI] [PubMed] [Google Scholar]
- 36.Wakita, Y., A. Shimizu, V. Hajek, J. Kawano, and K. Yamashita. 2002. Characterization of Staphylococcus intermedius from pigeons, dogs, foxes, mink, and horses by pulsed-field gel electrophoresis. J. Vet. Med. Sci. 64:237-243. [DOI] [PubMed] [Google Scholar]
- 37.Wayne, L. G., D. J. Brenner, and R. R. Colwell. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 38.Zarzour, J. Y., and E. A. Belle. 1976. Evaluation of three test procedures for identification of Staphylococcus aureus from clinical sources. J. Clin. Microbiol. 7:133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 43:1577-1593. [DOI] [PubMed] [Google Scholar]