Abstract
Brucellosis is a serious problem in Sicily. Brucella melitensis was identified as the species most frequently isolated in humans in Italy. No data, however, are available about the molecular epidemiological characterization of Brucella isolates from humans. We have conducted this study to molecularly characterize clinical isolates of Brucella spp. and to evaluate their antimicrobial susceptibilities. Twenty Brucella isolates were studied. Differential growth characteristics and DNA polymorphisms such as the restriction patterns of the PCR-amplified omp2a and omp2b genes, rpoB nucleotide sequencing, and multiple-locus variable-number tandem repeat analysis of 16 loci (MLVA-16) were used to characterize the strains. In vitro antibiotic susceptibility was determined by the E-test method on two different agar media, and the results were compared. All isolates were identified as B. melitensis biovar 3. rpoB nucleotide sequence analysis allowed the identification of two different genotypes of B. melitensis biovar 3. On the other hand, the MLVA-16 typing assay recognized 17 distinct genotypes. All isolates were sensitive to all tested antibiotics (rifampin, doxycycline, ciprofloxacin, ceftriaxone, and trimethoprim-sulfamethoxazole), and the Mueller-Hinton agar plate is recommended for antibiotic susceptibility testing by the E-test method. Our findings identify B. melitensis biovar 3 as the etiological agent isolated in Sicily and encourage the use of both molecular methods, and in particular of the MLVA-16 assay, in epidemiological trace-back analysis. This study represents the first epidemiological data from molecular typing of Brucella strains circulating in Italy and, in particular, in eastern Sicily.
Brucellosis is an important health problem affecting animals and humans in many countries in the world and especially in the Mediterranean areas. Brucella infections may cause tremendous economic losses through reproductive failure in animals. The disease is transmitted by humans through the consumption of contaminated foods or by direct/indirect contact with infected animals. Brucella melitensis is the most important zoonotic agent, followed by Brucella abortus and Brucella suis. While most industrialized countries have been remarkably successful in eradicating or controlling bovine brucellosis, some of these countries as well as all developing countries still suffer from small-ruminant brucellosis (24). In developing countries, in fact, brucellosis is almost always present where small ruminants are kept. Most human cases are caused by B. melitensis and particularly by biovars 1 and 3 (1, 4, 6, 27, 29).
Brucellosis is endemic in the south of Italy, and in particular in Sicily. Annual reported human cases in Italy have steadily declined in the past 5 years (from 923 cases in 2001 to 316 cases in 2005), according to data obtained from the Italian Ministry of Health (http://www.ministerosalute.it/promozione/malattie/bollettino.jsp). However, the annual incidence of the disease has not dropped uniformly across the country: Sicily alone reported 92.4% of the 316 cases in 2005, notwithstanding evident underreporting in that area. In Sicily, brucellosis remains a serious problem. The persistence of the disease in the region, despite full awareness of the causes of infection and all possible measures to control it, can be traced back to the presence of infected animals. Sicily is 80% hilly terrain, and livestock are one of the most important resources for the regional economy. Ovine-caprine breeding is the main zootechnical activity with high density per km2. Most livestock are owned by smallholders and farmers in Sicily. Every breeder produces a local and typical product, preventing standardization of the product, and hygienic conditions are often poor. While there are a few intensive or semi-intensive breeding farms, family breeding farms are the most common (8). Practices common in Sicily include free breeding and semifree breeding; promiscuity between sheep and goats and among sheep, goats, and cattle; seasonal moves; and the exchange of ovine-caprine male and female breeders. These breeding practices increase the possibility of contamination, transmission, and spread of infections. In Italy, eradication plans for bovine brucellosis and sheep and goat brucellosis are implemented according to law. Health and prophylactic measures established by national legislation have led to a substantial decline of brucellosis in Italy and to the eradication of disease from the northern Italian regions. In the south of Italy and especially Sicily, on the other hand, eradication programs have been hampered by the lack of complete control over movement and identification of animals, by the delayed elimination of infected animals, and by the traditional breeding practices described above.
Previous studies have reported B. melitensis as the most frequently isolated species in human cases of brucellosis in Italy between 1970 and 1990, accounting for 99% of total cases (7). A significant correlation between human and animal infections, in particular of sheep and goats, indicating brucellosis as a food-borne zoonosis rather than an occupational disease, has been reported in Italy (11, 15). No data are available about the characterization at biovar level of Brucella isolates from humans in Italy.
The aim of this paper is (i) to molecularly characterize clinical isolates of Brucella spp. from Sicily, since strategies for disease control and eradication derive primarily from the epidemiological characteristics of the disease, and (ii) to evaluate the antimicrobial susceptibilities in vitro of the Brucella isolates.
MATERIALS AND METHODS
Human cases.
Twenty Brucella strains were isolated from patients with sporadic cases of acute brucellosis hospitalized in Catania, Sicily, from April 2005 to May 2006. None of the patients belonged to the occupational-risk category. The diagnosis of brucellosis was made based on compatible clinical signs, agglutination titers of ≥1:160, and isolation of Brucella spp. from blood. All isolates were obtained from blood culture and stored in skim milk at −20°C. A regimen of combined rifampin (RIF) and doxycycline (DOX) for 6 weeks was used for therapy as recommended by the World Health Organization (30).
Blood samples were cultured in vials of the BACTEC 9120 system (Becton Dickinson, Rutherford, NJ) at 37°C for at least 7 days. Subcultures were made on Columbia blood agar plates supplemented with 5% sheep blood (bioMerieux, Marcy l'Etoile, France). Brucella isolates were then submitted for classical identification procedures: CO2 requirement, H2S production, dye sensitivity, and agglutination with specific antisera (Oxoid Ltd., Hampshire, England) (3).
Reference Brucella strains.
Four reference Brucella strains were used in this study: B. melitensis biovar 1 strain 16 M, biovar 2 strain 63/9, and biovar 3 strain Ether (Veterinary Laboratories Agency, Surrey, United Kingdom) and the vaccine strain B. abortus RB51 (CZ Veterinaria, S.A., Porrino, Spain). Cultures were grown at 37°C on brucella agar plates supplemented with 5% horse serum.
Bacterial DNA extraction.
Brucella DNA was extracted using the proteinase K and sodium dodecyl sulfate method. DNA was purified twice with phenol-chloroform using Phase Lock Gel Heavy tubes (Eppendorf AG, Hamburg, Germany). DNA was precipitated and washed, and the pellet was resuspended in 50 μl of nuclease-free water. Two hundred nanograms of DNA template was used for PCR amplifications.
omp2a and omp2b polymorphism.
Two PCR amplifications were performed to amplify genes omp2a and omp2b using, respectively, the primer pairs 2aA/2aB and 2bA/2bB (9). Amplification reaction mixtures were prepared in 50-μl volumes using GoTaq DNA polymerase (Promega Corporation, Madison, WI) following the manufacturer's protocol. Amplifications were initiated by denaturing the sample for 3 min at 95°C, followed by 30 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 3 min. After the last cycle, samples were incubated for an additional 5 min at 72°C before storage at 4°C. Five microliters of PCR-amplified omp2a and omp2b genes was digested by the restriction enzymes PstI and HinfI, respectively (Promega Corporation, Madison, WI), at 37°C for 3 h in a 20-μl reaction volume and using the manufacturer's recommended buffer. The mixtures were then electrophoresed on a 1.5% agarose gel with ethidium bromide. The reference Brucella strains 16 M, 63/9, and Ether were also analyzed.
rpoB sequencing.
The rpoB gene was amplified using the primers +1rB and −4134rB previously described (22). PCR amplifications were carried out in 50-μl volumes using GoTaq DNA polymerase (Promega Corporation, Madison, WI) following the enclosed protocol. Amplifications were initiated by denaturing the sample for 3 min at 94°C, followed by 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 4 min. After the last cycle samples were incubated for an additional 7 min at 72°C before they were stored at 4°C. PCR amplifications were analyzed by electrophoresis through a 1% agarose gel with ethidium bromide. All PCR products were purified by Montage PCR centrifugal filter devices (Millipore, Billerica, MA) and directly sequenced with the ABI PRISM 310 Genetic Analyzer equipment using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). PCR primers and sequencing primers previously described (23) were used to sequence the whole rpoB gene. The electropherograms were assembled based on the published rpoB sequence of B. melitensis 16 M by the ABI Prism SeqScape Software version 2.0 (Applied Biosystems, Foster City, CA). All consensus sequences generated were then compared to the published B. melitensis 16 M rpoB gene (accession number AE009516) to detect nucleotide diversity.
MLVA-16 (multiple-locus variable-number tandem repeat analysis of 16 loci) typing.
PCR assays were performed in a total volume of 25 μl using GoTaq Green Master Mix (Promega Corporation, Madison, WI). Sixteen sets of primers previously proposed and divided into two groups (2, 18) were used: panel 1 (loci Bruce06, Bruce08, Bruce11, Bruce12, Bruce42, Bruce43, Bruce45, and Bruce55) and panel 2 (loci Bruce04, Bruce07, Bruce09, Bruce16, Bruce18, Bruce19, Bruce21, and Bruce30). Amplifications were initiated by denaturing the sample for 3 min at 94°C, followed by 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 50 s. After the last cycle samples were incubated for an additional 7 min at 72°C before they were stored at 4°C. Five microliters of the amplification products was loaded on 2% and 3% standard agarose gels with ethidium bromide for analyzing tandem repeats with a unit length longer (panel 1) and shorter (panel 2) than 10 bp, respectively. Gels were stained with ethidium bromide (0.5 μg/ml), visualized under UV light, and photographed. The reference strain B. melitensis 16 M, for which the expected size is known for each variable-number tandem repeat locus, was used. To estimate exactly the band sizes, PCR products were purified and directly sequenced as described above. The electropherograms were assembled, and consensus sequences were generated using the Navigator Sequence Software (Applied Biosystems, Foster City, CA).
Testing antimicrobial susceptibility.
In vitro activities of RIF, DOX, ciprofloxacin (CIP), ceftriaxone (CRO), and trimethoprim-sulfamethoxazole (SXT) were determined by the E-test method (Biolife Italiana s.r.l., Milan, Italy) in accordance with the procedures recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) for slow-growing bacteria (25). Antimicrobial susceptibility testing was performed by inoculating the isolates in broth cultures adjusted to an 0.5 McFarland turbidity both on Mueller-Hinton agar plates supplemented with 5% sheep blood (Oxoid Ltd., Hampshire, England) and on brucella agar plates supplemented with 5% horse serum. E-test strips of RIF, DOX, CIP, CEF, and SXT were stored at −20°C until use. The E-test strips were applied to the inoculated culture plates separately as recommended by the manufacturer, and the plates were incubated at 37°C for 48 h in a 5% CO2 atmosphere. Determination of the MICs by the E-test was performed in duplicate, according to the manufacturer's recommendations, and the MICs were interpreted at the point of intersection between the inhibition zone and the E-test strips. The results were evaluated according to the E-test manufacturer's instructions. The vaccine strain B. abortus RB51 was used as a RIF control.
RESULTS
During the study period, from April 2005 to May 2006, 20 Brucella strains isolated from blood cultures of patients with acute brucellosis were studied. Seventeen (70.83%) of the strains were recovered from May through August 2005 with a peak in June. All isolates were identified on the basis of colony morphology, staining, growth characteristics, and slide agglutination with monospecific anti-Brucella serum. Three isolates showed rough forms and, therefore, could not be typed. All smooth isolates reacted in agglutination tests with both anti-A and anti-M sera, suggesting B. melitensis biovar 3 identification. To better characterize human isolates at biovar level, we turned to a molecular approach. First, we studied the intraspecies polymorphism of omp2a and omp2b genes as suggested by other studies (9). By analysis of the PCR products of the omp2a and omp2b genes digested with the restriction enzymes PstI and HinfI (PCR-restriction fragment length polymorphism [RFLP]), all human isolates presented the same restriction patterns and were identified as either biovar 1 or biovar 3, the two biovars being indistinguishable based on PCR-RFLP results. We then analyzed the nucleotide sequence of the rpoB gene to differentiate biovars. Comparing those sequences to the published B. melitensis 16 M rpoB gene (accession number AE009516), all isolates carried nucleotide alterations of ATC to ATA at codon position 1249 (M1249I), previously identified as a molecular marker of B. melitensis biovar 3 strain Ether and classified as B. melitensis genotype 3 (22). Three out of 20 isolates carried an additional CTT-to-TTT mutation at codon position 670, which led to substitution of amino acid L670F, never before observed in the rpoB gene of Brucella strains (22). This mutation was not found to be associated with the RIF resistance phenotype, as demonstrated by the in vitro antimicrobial susceptibility test. In fact, this mutation does not map in the two identified “hot” regions in which mutations have been associated with the development of the RIF resistance phenotype in Brucella spp., located at the 5′ end and close to the center of the rpoB gene (23). We identified, therefore, two different B. melitensis rpoB genotypes 3.
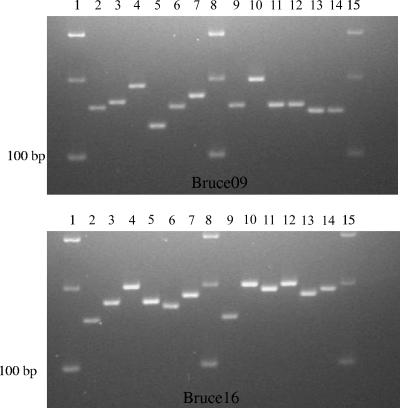
DNA from all 20 human isolates and from B. melitensis strain 16 M was also amplified at 16 loci (MLVA-16 typing assay) (2, 18) to generate multiple band profiles. The reference strain B. melitensis 16 M was used because the expected size for each variable-number tandem repeat locus is known and published (18). Polymorphism among isolates was observed at seven loci: Bruce08 and Bruce12 of panel 1 and Bruce04, Bruce07, Bruce09, Bruce16, and Bruce18 of panel 2. Some amplification patterns are shown in Fig. 1. The MLVA-16 typing assay allowed the 20 Brucella isolates to be grouped into 17 distinct genotypes, as shown in Table 1. The three isolates carrying both L670F and M1249I mutations showed the same multiple band profiles and grouped in genotype 2. Genotype 3 was comprised of two isolates, and genotypes 4 to 18 were comprised of only one isolate each.
FIG. 1.
Amplification patterns of loci Bruce09 (top) and Bruce16 (bottom) of MLVA panel 2. Lanes 1, 8, and 15, 100-bp DNA ladder; lanes 2 and 9, reference strain 16 M; lanes 3 to 7, strains DG1, AM12, MM14, CM10, and BC6, respectively; lanes 10 to 14, strains MA5, PD11, CV5, BL9, and CS13, respectively.
TABLE 1.
MLVA-16 genotypes for 24 B. melitensis human isolates
| Genotype | Straina | No. of isolatesb | Repeat copy no. at locus:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bruce06 | Bruce08 | Bruce11 | Bruce12 | Bruce42 | Bruce43 | Bruce45 | Bruce55 | Bruce04 | Bruce07 | Bruce09 | Bruce16 | Bruce18 | Bruce19 | Bruce21 | Bruce30 | |||
| 1 | 16 M | 3 | 4 | 2 | 13 | 4 | 2 | 3 | 3 | 2 | 5 | 7 | 3 | 5 | 18 | 6 | 6 | |
| 2 | DG1 | 3 | 3 | 6 | 3 | 14 | 1 | 1 | 3 | 3 | 4 | 10 | 8 | 6 | 8 | 21 | 8 | 3 |
| 3 | GV2 | 2 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 5 | 6 | 12 | 9 | 7 | 21 | 8 | 3 |
| 4 | CM10 | 1 | 3 | 6 | 3 | 14 | 1 | 1 | 3 | 3 | 6 | 7 | 7 | 5 | 7 | 21 | 8 | 3 |
| 5 | CS13 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 5 | 5 | 6 | 8 | 7 | 21 | 8 | 3 |
| 6 | BL9 | 1 | 3 | 6 | 3 | 14 | 1 | 1 | 3 | 3 | 6 | 4 | 6 | 7 | 7 | 21 | 8 | 3 |
| 7 | MM14 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 6 | 7 | 4 | 6 | 7 | 21 | 8 | 3 |
| 8 | AM12 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 4 | 7 | 11 | 9 | 8 | 21 | 8 | 3 |
| 9 | BC6 | 1 | 3 | 6 | 3 | 14 | 1 | 1 | 3 | 3 | 7 | 6 | 9 | 7 | 7 | 21 | 8 | 3 |
| 10 | PD11 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 5 | 4 | 7 | 8 | 7 | 21 | 8 | 3 |
| 11 | CV5 | 1 | 3 | 6 | 3 | 14 | 1 | 1 | 3 | 3 | 8 | 4 | 7 | 9 | 7 | 21 | 8 | 3 |
| 12 | MA5 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 5 | 5 | 12 | 9 | 7 | 21 | 8 | 3 |
| 13 | SP4 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 4 | 8 | 10 | 5 | 8 | 21 | 8 | 3 |
| 14 | DZ7 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 3 | 5 | 6 | 6 | 7 | 21 | 8 | 3 |
| 15 | GA8 | 1 | 3 | 6 | 3 | 13 | 1 | 1 | 3 | 3 | 6 | 4 | 11 | 6 | 7 | 21 | 8 | 3 |
| 16 | GR6 | 1 | 3 | 6 | 3 | 13 | 1 | 1 | 3 | 3 | 7 | 4 | 6 | 10 | 8 | 21 | 8 | 3 |
| 17 | DN5 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 8 | 6 | 6 | 8 | 7 | 21 | 8 | 3 |
| 18 | ST4 | 1 | 3 | 5 | 3 | 13 | 1 | 1 | 3 | 3 | 5 | 5 | 13 | 9 | 7 | 21 | 8 | 3 |
All strains belong to B. melitensis, and all are biovar 3, except for 16 M, which is biovar 1.
Number of isolates studied with identical genotype.
Susceptibility to the antimicrobials commonly used against Brucella spp. was tested by the E-test on two different agar media, Mueller-Hinton agar plates with 5% sheep blood and brucella agar plates with 5% horse serum; both are commonly used for the culture of Brucella spp. in our laboratory. After 48 h the plates were examined and compared. No significant differences were found between the two culture media used. The Mueller-Hinton agar plate is, however, preferred because it allows clearer detection of zones of inhibition, making it easier to read the intersection of the inhibitory zone edges. MICs obtained are shown in Table 2. The range of MICs of our isolates was compared to that of the reference strain B. melitensis Ether. No significant differences were observed. We also used the reference strain B. abortus RB51 as a RIF antibiotic control because the vaccine strain RB51 is the only known RIF-resistant Brucella reference strain. All isolates were susceptible to all antibiotics; RIF had the highest MICs (between 0.75 and 2 μg/ml).
TABLE 2.
MICs of Brucella species under study
| Drug | MIC (μg/ml) for B. melitensis
|
|
|---|---|---|
| All isolates (range) | Strain Ether | |
| RIF | 0.75-2 | 1 |
| DOX | 0.064-0.125 | 0.064 |
| CIP | 0.094-0.5 | 0.25 |
| CRO | 0.064-0.38 | 0.38 |
| SXT | 0.012-0.064 | 0.032 |
DISCUSSION
Brucellosis in Sicily, where the prevalence of infection in sheep and goats is the highest in Italy, is an important disease with a considerable number of human cases reported every year. In this study we have characterized 20 Brucella strains isolated from patients hospitalized with symptoms of acute brucellosis in the city of Catania in a 1-year period. The consumption of ricotta (an Italian cheese made with sheep's milk) or desserts containing ricotta was identified as the most likely route of infection.
The investigation of phenotypic characteristics, through conventional biotyping tests, allowed the identification of all smooth Brucella isolates as B. melitensis biovar 3. The roughness and the atypical susceptibility to dyes of some our isolates prevented characterization at biovar level. Complications in conventional biotyping characterization of B. melitensis strains have already been reported (4, 10, 20). In order to better understand the epidemiological scenario, we turned to molecular approaches.
PCR-RFLP of omp2a and omp2b was not able to accurately identify our isolates, which were identified as B. melitensis biovar 1 or 3. In contrast, rpoB sequencing, recently proposed as a molecular typing method for the identification and differentiation of Brucella strains at species and biovar levels (22), allowed us to presumptively identify all isolates as B. melitensis biovar 3. All isolates, in fact, carried the molecular marker M1249I. This method was also able to characterize two distinct rpoB genotypes: the first carrying the M1249I mutation only (17 isolates) and the second carrying both L670F and M1249I mutations (three isolates). The MLVA-16 assay, consisting of 16 selected markers that are a combination of moderately variable (eight minisatellite markers, panel 1) and highly discriminant (eight microsatellite markers, panel 2) loci (2, 18), was used to investigate the polymorphism of tandem repeat loci of Brucella isolates. Although the 20 isolates have identical geographic origins, differences occurred at seven markers and allowed us to identify 17 distinct genotypes (Table 1). The MLVA-16 typing method was previously used to analyze human and animal isolates (2, 18), and a large number of different genotypes have been identified. The strains were collected from different countries, and no data were available about the hypothetical distribution of Brucella genotypes within a restricted area. This information could represent an important insight in order to characterize the significance of such polymorphism in the pathogenesis of Brucella. Our results clearly show that Brucella, despite the high genetic homogeneity within the genus, is highly polymorphic at minisatellite and microsatellite levels. In fact, 17 different genotypes have been identified in analyzing 20 human isolates belonging to the same biovar and coming from a restricted area of endemicity.
In all such instances where isolates share the same genotype (genotypes 2 and 3), the epidemiological data are compatible with a common source of infection. Interestingly, the three isolates carrying the additional mutation L670F showed the same MLVA-16 genotype.
According to our findings, since 100% of human isolates were identified as B. melitensis biovar 3, we hypothesized that (i) B. abortus strains could not be present in the environment, (ii) B. melitensis strains are more pathogenic than B. abortus strains, or (iii) there could be some particular ecological risk factors in our epidemiological settings which influence the disease in humans and favor infection by B. melitensis biovar 3 strains. We therefore used the recent survey of animal brucellosis carried out in Sicily (12). Most Brucella strains were identified by conventional biotyping tests as B. melitensis biovar 3 (98% of isolates from sheep and goats and 25% of isolates from cattle). B. abortus biovars 1, 3, and 4 were identified in 75% of isolates from cattle. B. abortus is present among animals in Sicily, so we can reject the first hypothesis. B. melitensis is known to be the most pathogenic species, producing the most intense symptoms, the greatest tissue damage, and the highest incidence of localization in body organs, systems, or tissues (21). On the other hand, a recent study analyzing laboratory findings for patients with brucellosis reported that there was no evidence suggesting that B. melitensis is more virulent than B. abortus or that infections due to B. abortus are less severe than the infections with B. melitensis, despite the higher prevalence of B. melitensis in humans (13). Regarding the second hypothesis, we have no evidence that B. melitensis is more pathogenic than B. abortus. Since the existing literature is conflicting, additional studies are needed for greater understanding of this question. Based on animal data summarized above, our findings, the socioeconomic and cultural environment of small rural Sicilian areas with risky breeding practices, the lack of both standard procedures and good hygiene conditions during cheese production steps, and the pouring of raw milk onto finished cheese to improve its taste, we strongly support the third hypothesis. Overall, we infer that, in this ecosystem, small ruminants and their products are the major source of infection and play a key role in the spread of brucellosis both in cattle and in humans. Cattle, in fact, are not the usual reservoir of this species, and yet there are a consistent number of B. melitensis biovar 3 isolates in cattle and a high prevalence of bovine brucellosis in the same areas where ovine-caprine brucellosis is widespread. The link between human and small-ruminant infections is indeed confirmed by the seasonal trend noted in the distribution of our human cases (from May through August with a peak in June), which is easily correlated with the peak of animal cases of brucellosis and also with the peak of consumption of at-risk products.
Our data suggest the use of one or both molecular approaches, rpoB sequencing and MLVA-16 assay, according to the aims of the investigation, because different information is obtained from each approach. rpoB sequencing allowed us to presumptively characterize all isolates as B. melitensis biovar 3, a result that would have otherwise been complicated by the roughness and the atypical dye susceptibility of some Brucella strains. The characterization at biovar level cannot be obtained by the MLVA-16 assay because, as reported by others (2, 18), the B. melitensis group is highly heterogeneous. Therefore, neither panel 1 nor panel 2 is able to cluster the isolates as expected from the biovars within the species B. melitensis. On the other hand, the MLVA-16 assay appears more suitable to assist with investigation of outbreaks: strains clustering together in the same MLVA genotype indicate a common source of infection. Therefore, typing clinical, environmental, and animal isolates through the MLVA-16 assay allows testing of hypotheses on outbreak confirmation, extent of transmission, source, and reservoir.
In this study we also determined the antibiotic susceptibilities of our isolates by the E-test method. The E-test was used because it has been reported to be less labor-intensive, less time-consuming, and more practical than the broth microdilution method for testing antibiotic susceptibility of Brucella spp. (14). Susceptibility testing of brucellae is not routinely used in clinical practice, few studies of in vitro antimicrobial susceptibilities of Brucella organisms are available (5, 26, 28), and no standard protocol has been determined yet. We studied susceptibilities of our Brucella isolates on two different culture media: the Mueller-Hinton agar plates widely used for antibiotic susceptibility testing and the brucella agar plates commonly used in the laboratory as Brucella growth medium. RIF, DOX, CIP, CRO, and SXT proved to be very effective antibiotic drugs in vitro, although CRO is not effective in treating brucellosis patients. The highest MICs were found for RIF: two of our isolates showed a MIC of 2 μg/ml. According to evaluation based on CLSI standards for slow-growing bacteria, these two isolates should be considered intermediately susceptible. On the other hand, in this study we have characterized the whole rpoB gene, recognized to be the RIF antibiotic target in prokaryotes (16, 17, 19), of all our isolates, and these two isolates carried only the B. melitensis biovar 3 molecular marker (M1249I). We can, therefore, assert that all isolates investigated were found to be sensitive to RIF and, in general, to all tested antimicrobial agents. Although no significant differences were observed between two tested culture media, the Mueller-Hinton agar plate is recommended because clearer inhibition zones are visible and the calibrated carrier strip indicating the MIC can be more easily read. Since increasing resistance to RIF and SXT has been reported in many parts of the world, we suggest periodic assessment of susceptibility of strains to those antibiotics used most frequently in treatment, for an early detection of any drug resistance, especially in areas of endemicity.
In conclusion, our findings confirm B. melitensis, and in particular biovar 3, as the etiological agent most frequently isolated in humans in Italy and suggest, therefore, that sheep and goat populations are the principal cause of human brucellosis. Moreover, our results provide proof of the high discriminatory power of MLVA-16 typing even if Brucella isolates belong to the same biovar. This method led us to ascertain that in a restricted area in which brucellosis is still endemic, different genotypes of B. melitensis biovar 3 are present. It paves the way for further studies aiming to evaluate if it is possible to correlate different genotypes with different pathogenic patterns.
Acknowledgments
We thank Paolo Pasquali for suggestions and revision of the text and Massimiliano Francia for his skilled technical assistance in this project.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Aggad, H., and L. Boukraa. 2006. Prevalence of bovine and human brucellosis in western Algeria: comparison of screening tests. East. Mediterr. Health J. 12:119-128. [PubMed] [Google Scholar]
- 2.Al Dahouk, S., P. Le Fleche, K. Nockler, I. Jacques, M. Grayon, H. C. Scholz, H. Tomaso, G. Vergnaud, and H. Neubauer. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69:137-145. [DOI] [PubMed] [Google Scholar]
- 3.Alton, G. G., L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique Publications, Paris, France.
- 4.Banai, M., I. Mayer, and A. Cohen. 1990. Isolation, identification, and characterization in Israel of Brucella melitensis biovar 1 atypical strains susceptible to dyes and penicillin, indicating the evolution of a new variant. J. Clin. Microbiol. 28:1057-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baykam, N., H. Esener, O. Ergonul, S. Eren, A. K. Celikbas, and B. Dokuzoguz. 2004. In vitro antimicrobial susceptibility of Brucella species. Int. J. Antimicrob. Agents 23:405-407. [DOI] [PubMed] [Google Scholar]
- 6.Bodur, H., N. Balaban, S. Aksaray, V. Yetener, E. Akinci, A. Colpan, and A. Erbay. 2003. Biotypes and antimicrobial susceptibilities of Brucella isolates. Scand. J. Infect. Dis. 35:337-338. [DOI] [PubMed] [Google Scholar]
- 7.Caporale, V., D. Nannini, A. Giovannini, D. Morelli, and M. Ramasco. 1992. Prophylaxis and control of brucellosis due to Brucella melitensis in Italy: acquired and expected results, p. 127-145. In Prevention of brucellosis in the Mediterranean countries. Proceedings of the International Seminar of the International Center for Advanced Mediterranean Agronomic Studies, the Commission of the European Communities, and the Ministry of Agriculture and Fisheries. International Center for Advanced Mediterranean Agronomic Studies, Valletta, Malta.
- 8.Caracappa, S. 1999. Livestock production and animal health in Sicily, Italy. Parassitologia 41:17-23. [PubMed] [Google Scholar]
- 9.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grepinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer membrane proteins of Brucella. Microbiology 141:2111-2121. [DOI] [PubMed] [Google Scholar]
- 10.Corbel, M. J. 1991. Identification of dye-sensitive strains of Brucella melitensis. J. Clin. Microbiol. 29:1066-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Massis, F., A. Di Girolamo, A. Petrini, E. Pizzigallo, and A. Giovannini. 2005. Correlation between animal and human brucellosis in Italy during the period 1997-2002. Clin. Microbiol. Infect. 11:632-636. [DOI] [PubMed] [Google Scholar]
- 12.Di Marco, V., C. Piratino, I. Bazzana, M. Russo, V. Aronica, A. M. F. Marini., and S. Caracappa. 2006. A study on the circulation of biovars of brucellae in sheep and goat in Sicily from 1993 to 2002, p. 128. Abstr. 14th Int. Congr. Mediterr. Fed. Health Prod. Ruminants, Santiago De Compostela, Spain.
- 13.Dokuzoguz, B., O. Ergonul, N. Baykam, H. Esener, S. Kilic, A. Celikbas, S. Eren, and B. Esen. 2005. Characteristics of B. melitensis versus B. abortus bacteraemias. J. Infect. 50:41-45. [DOI] [PubMed] [Google Scholar]
- 14.Gur, D., S. Kocagoz, M. Akova, and S. Unal. 1999. Comparison of E test to microdilution for determining in vitro activities of antibiotics against Brucella melitensis. Antimicrob. Agents Chemother. 43:2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iaria, C., F. Ricciardi, F. Marano, G. Puglisi, G. Pappas, and A. Cascio. 2006. Live nativity and brucellosis, Sicily. Emerg. Infect. Dis. 12:2001-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 17.Jin, D. J., and C. A. Gross. 1989. Characterization of the pleiotrophic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J. Bacteriol. 171:5229-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Fleche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin, M. E., and G. F. Hatfull. 1993. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampin and mechanism of rifampin resistance. Mol. Microbiol. 8:277-285. [DOI] [PubMed] [Google Scholar]
- 20.Lucero, N. E., S. M. Ayala, G. I. Escobar, M. Grayon, and I. Jacques. 2006. A new variant of Brucella melitensis. Clin. Microbiol. Infect. 12:593-596. [DOI] [PubMed] [Google Scholar]
- 21.Madkour, M. M. 2001. Madkour's brucellosis, 2nd ed. Springer Verlag, Berlin, Germany.
- 22.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2006. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes Infect. 8:860-865. [DOI] [PubMed] [Google Scholar]
- 23.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2004. Genetic bases of the rifampin resistance phenotype in Brucella spp. J. Clin. Microbiol. 42:5439-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott, J. J., and S. M. Arimi. 2002. Brucellosis in sub-Saharan Africa: epidemiology, control and impact. Vet. Microbiol. 90:111-134. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 1998. Performance standards for antimicrobial susceptibility testing. Eighth informational supplement. NCCLS document M 100-S8, vol. 18. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.Orhan, G., A. Bayram, Y. Zer, and I. Balci. 2005. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J. Clin. Microbiol. 43:140-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Refai, M. 2002. Incidence and control of brucellosis in the Near East region. Vet. Microbiol. 90:81-110. [DOI] [PubMed] [Google Scholar]
- 28.Turkmani, A., A. Ioannidis, A. Christidou, A. Psaroulaki, F. Loukaides, and Y. Tselentis. 2006. In vitro susceptibilities of Brucella melitensis isolates to eleven antibiotics. Ann. Clin. Microbiol. Antimicrob. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallach, J. C., S. E. Miguel, P. C. Baldi, E. Guarnera, F. A. Goldbaum, and C. A. Fossati. 1994. Urban outbreak of a Brucella melitensis infection in an Argentine family: clinical and diagnostic aspects. FEMS Immunol. Med. Microbiol. 8:49-56. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. 1986. Joint FAO-WHO Expert Committee on Brucellosis, sixth report. WHO Tech. Rep. Ser. 740:1-132. [PubMed] [Google Scholar]