Abstract
Twelve strains of a rapidly growing Mycobacterium species were isolated from an outbreak associated with intramuscular injections of an antimicrobial agent and were identified by comparative sequence analysis of rpoB and hsp65. These isolates were identified as Mycobacterium massiliense (100% similarity).
CASE REPORT
During August to December 2004, a physician at a local clinic in Icheon, Korea, administered intramuscular injections of an antimicrobial agent (ribostamycin sulfate) as required by a common cold/flu regimen. On 7 April 2005, the first case of postinjection abscess was recognized by the Icheon Public Health Center and reported to the Korean Communicable Disease Center. Of the 2,984 patients who had visited the clinic concerned, 77 patients complained of a hard palpable mass, pain, redness, and pus at/under the buttock injection sites. According to the records, 25 (32.5%) were male and 52 (67.5%) female. The youngest was 4 years old and the oldest was 79 (mean ± standard deviation = 32.4 ± 20.4, median = 33), 5 (6.5%) were less than 9 years old, 57 (74%) were between 10 and 49 years old, and 15 (19.5%) were older than 50. All were healthy except for a mass associated with injection. Repeated incisions and pus drainages or removal of necrotic tissues (one to three times in most cases) in parallel with local or systemic antibiotic treatment were ineffective. Inflammation relapsed 1 to 3 months later and formed another mass at (or near) the primary site in most patients. Routine clinical laboratory examinations, e.g., microscopic examinations of stained smears and bacterial cultures for 2 days, failed to demonstrate any causative microorganism. Epidemiological investigation was initiated on 30 May 2005.
Clinical specimens (pus or tissue debris) obtained by surgery were inoculated on Ogawa media, blood agar plates (BAP), and chloramphenicol-Sabouraud agar plates and cultured at 35°C in a 5% CO2 incubator for 1 week to isolate causative agents. Pure cultured isolates were used for species identification by comparative sequence analysis of the 16S rRNA gene, rpoB, and hsp65. Total DNAs were extracted using the bead beater-phenol extraction method (4) and used as a template for PCR. The following primer pairs were used: 285 (5′-GAG AGT TTG ATC CTG GCT CAG-3′) and 244 (5′-CCC ACT GCT GCC TCC CGT AG-3′) for the 16S rRNA gene (351 bp) (8), RGMF (5′-GAC GAC ATC GAC CAC TTC GG-3′) and RGMR (5′-GGG GTC TCG ATC GGG CAC AT-3′) for rpoB PCR (365 bp), and HSPF3 (5′-ATC GCC AAG GAG ATC GAG CT-3′) and HSPR4 (5′-AAG GTG CCG CGG ATC TTG TT-3′) for hsp65 (644 bp) (5). PCR products were purified using QIAEX II gel extraction kits (QIAGEN, Hilden, Germany) and were sequenced directly using forward and reverse primers with an Applied Biosystems automated sequencer (model 377) and BigDye Terminator cycle sequencing kits (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). Their determined partial 16S rRNA gene, rpoB, and hsp65 sequences (306, 306, and 603 bp, respectively) were aligned using the multiple-alignment algorithm in the MegAlign package (Windows version 3.12e; DNASTAR, Madison, WI). Phylogenetic trees were inferred from the rpoB and hsp65 nucleotide sequences newly determined in this study, including those for Mycobacterium massiliense CIP 108297, Mycobacterium abscessus ATCC 19977, and 19 rapidly growing mycobacterium (RGM) species retrieved from GenBank (shown in parentheses next to the species names in Fig. 1) using MEGA version 2.1 (6). Bootstrap analysis (1,000 repeats) was performed to evaluate the phylogenetic tree topology, using Mycobacterium tuberculosis H37Rv as an outgroup (rpoB, AF057454; hsp65, AY299144). Pulsed-field gel electrophoresis using AseI was performed to verify the epidemiologic relationships of these Icheon isolates as previously described (11). The type strain of M. massiliense and Mycobacterium chelonae and M. abscessus strains that had been previously isolated from other sporadic cases in different locations in Korea were used for comparison.
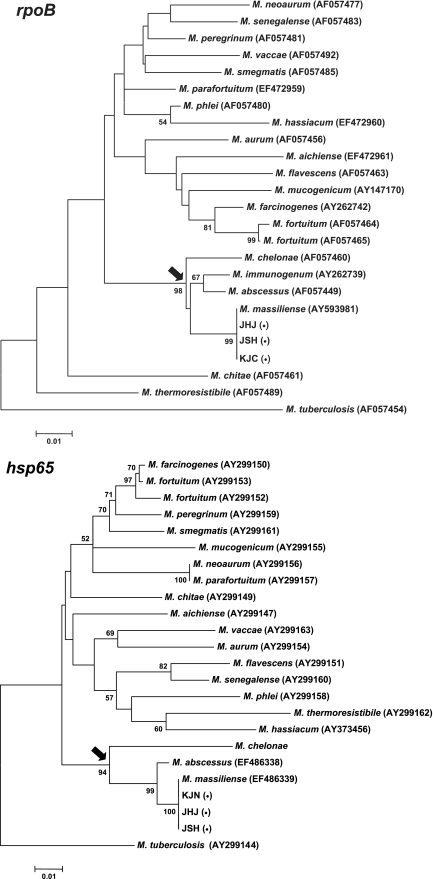
FIG. 1.
Phylogenetic relationships of the Icheon isolates inferred from partial rpoB and hsp65 sequences. The trees were constructed using the neighbor-joining method described in the MEGA software package. M. tuberculosis was used as an outgroup to root the tree. The bootstrap values presented on corresponding branches were determined from 1,000 replications; those with values of less than 50% are not shown. Species in the M. chelonae complex are indicated by an arrow. Because the hsp65 sequence of M. immunogenum was not available in GenBank, it was not included in the hsp65 tree. Three representative Icheon isolates are indicated by parenthesized bullets (•). The scale bar represents a 1% difference in nucleotide sequences.
The MICs of 14 antimicrobial agents (amikacin, clarithromycin, doxycycline, imipenem, linezolid, trimethoprim-sulfamethoxazole, tobramycin, azithromycin, cefotetan, colistin, gatifloxacin, minocycline, moxifloxacin, and ticarcillin) were estimated using the Etest (AB Biodisk, Solna, Sweden). Strains susceptible, intermediate, or resistant to particular antibiotics were identified from their MICs and the criteria of the Clinical and Laboratory Standards Institute (7).
Pure cultured small colonies were observed on BAP after 4 days of culture. Of the two known morphological types of M. abscessus colonies (3), only gray-white, thin, scum-like, rough colonies with elevated centers and irregular margins were observed. Growth on other media was slower than that on BAP. Microscopic examinations revealed acid-fast bacilli in Ziehl-Neelsen-stained smears. According to their growth and staining characteristics, the organisms were tentatively identified as members of an RGM species. Twelve strains were isolated from 19 patients.
The 16S rRNA gene sequences (306 bp) of these 12 Icheon isolates were identical and indistinguishable from those of M. abscessus AY457071 and M. massiliense AY593980 by a GenBank database-Basic Local Alignment Search Tool (BLAST) search. The whole 16S rRNA gene sequence, which was separately determined in one Icheon isolate, was also found to be identical to those of M. abscessus and M. massiliense. However, the rpoB sequences (306 bp) of 12 isolates showed 100% similarity with that of the type strain of M. massiliense (AY593981) but only 97.7%, 97.4%, and 97.7% similarity with those of M. abscessus AF057449, M. chelonae AF057460, and Mycobacterium immunogenum AY262739, respectively. The hsp65 sequences (603 bp) of the 12 isolates also showed 100% similarity with that of the type strain of M. massiliense (EF486339) but only 98.8% and 90.0% similarity with those of M. abscessus EF486338 and M. chelonae AY299148, respectively. By referring to the phylogenetic trees constructed from the currently available sequences of RGM species, the relationship between the Icheon isolates and other RGM species was clearly shown (Fig. 1). In each tree, species in the M. chelonae complex formed a unique clade, and this was robustly supported by high bootstrap values. Icheon isolates clustered with M. massiliense and were separated from M. abscessus or M. chelonae. Based on the above-described results, the 12 Icheon isolates were identified as M. massiliense. Because the hsp65 sequence of M. immunogenum was not available in the GenBank database, it was not included in the hsp65 tree. No sequence variation of rpoB or hsp65 was found among the 12 isolates. The rpoB and hsp65 sequences of an Icheon M. massiliense isolate were deposited in GenBank (EF472962 and EF486340, respectively). In addition, the 12 isolates showed identical pulsed-field gel electrophoresis patterns, but these differed from the patterns of M. chelonae and M. abscessus strains recovered from previous sporadic cases and the M. massiliense type strain (CIP 108297), which suggests that the 12 Icheon isolates were epidemiologically related and originated from a common source of infection. However, efforts to identify the source of these infections failed because the clinic had been closed and cleaned up 4 months before the investigation. Neither the ribostamycin and saline used for the intramuscular injections nor their empty bottles were available. No bacterial growth had been observed from ribostamycin and saline of the same batch collected from other local clinics. Environmental samples from the clinic were also tested. Two RGM species (Mycobacterium fortuitum and a mycobacterium closely related to Mycobacterium porcinum) were isolated from a fishbowl that was placed in the clinic waiting room.
Notably, low MICs were observed for the Icheon strains in clarithromycin (0.125 μg/ml), which concurs with a previous report (1). In addition, these strains were also susceptible to azithromycin (MIC = 2 μg/ml), minocycline (MIC = 1.5 to 6 μg/ml), and amikacin (MIC = 12 to 16 μg/ml) and were intermediately susceptible to doxycycline (MIC = 4 μg/ml). Based on these results, clarithromycin (0.5 g per os once every 12 h for adults and 7.5 mg/kg of body weight per os once every 12 h [maximum, 1 g/day] for children) was recommended to clinicians managing registered patients with definite or suspected abscesses due to M. massiliense infection. All patients were cured without relapse.
This report describes an outbreak of M. massiliense infection associated with intramuscular injections administered at a local clinic in Icheon, Korea. Recently, reports on RGM infections in various clinical situations have markedly increased, and in these reports, M. abscessus infection (9, 10, 12) is the most frequently encountered. Moreover, nearly 95% of the soft tissue infections caused by RGM are M. chelonae-M. abscessus complex infections (2). However, in our cases, because of its high prevalence in clinical specimens, M. abscessus was suspected upon primary culture. Nevertheless, the infections were found to have been caused by M. massiliense. Because this species is closely related to M. abscessus and was recently recognized from a pneumonia patient and classified (1), it is possible that M. massiliense infections have overlapped with M. abscessus infections in previous reports. The present study is the first to report on an outbreak of M. massiliense infection identified and diagnosed by gene sequence analysis.
In many situations, differentiation of newly recognized mycobacteria from previously known species is difficult on the basis of phenotypic traits, and M. massiliense, M. chelonae, and M. abscessus provide a good example. Though distinct characteristics, like enzyme activities, can be used to differentiate M. massiliense from M. chelonae and M. abscessus, gene sequence analysis allows the precise and rapid identification of these species. As shown in our cases, PCR-linked sequence analysis can efficiently identify specific pathogens. Because of the identical whole 16S rRNA gene sequences, it was impossible to differentiate M. massiliense from M. abscessus, as was the case for Mycobacterium kansasii and Mycobacterium gastri. However, their differentiation from other RGM species could be easily determined using the similarity between rpoB and hsp65 and trees. Because M. massiliense can be differentiated from M. abscessus by using hsp65, sodA, recA, rpoB, and 16S-23S rRNA internal transcribed spacer sequences (1), basically, any one of them can be used to identify both species. It is important that causative microorganisms be correctly identified, because treatment regimens and countermeasures are directly affected by the results obtained. Interestingly, our Icheon isolates were found to be markedly sensitive to clarithromycin as the type strain, which led to its adoption as the drug of choice.
The source of infection in the Icheon outbreak remains obscure, because the causative mycobacterium was isolated from patient clinical specimens only. Moreover, although an epidemiologic investigation was carried out, it was initiated almost 6 months after the first case was found and 4 months after the clinic concerned had closed. Thus, the investigation group could not obtain critical samples from the clinic to resolve the matter. Because no bacterial growth had been observed from the ribostamycin of the same batch collected from other local clinics, we could only consider that inadequate aseptic techniques during the injection process were probably responsible.
Nucleotide sequence accession numbers.
Newly determined sequences were deposited in GenBank (EF472959 to EF472962 and EF486338 to EF486340).
Acknowledgments
This work was supported by a grant from the Korean Center for Disease Control (KCDC). H.-Y. Kim., Y.-J. Yun., and C. G. Park were supported by the Brain Korea 21 Project in 2006.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Adékambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd, T. F., and C. R. Lyons. 1999. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67:4700-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim, B.-J., S.-H. Lee, M.-A. Lyu, S.-J. Kim, G.-H. Bai, S.-S. Kim, G.-T. Chae, E.-C. Kim, C.-Y. Cha, and Y.-H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, H., S. H. Kim, T. S. Shim, M. N. Kim, G. H. Bai, Y. G. Park, S. H. Lee, C. Y. Cha, Y. H. Kook, and B. J. Kim. 2005. PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp Heat Shock Protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J. Microbiol. Methods 62:199-209. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe, AZ. [DOI] [PubMed]
- 7.NCCLS/CLSI. 2003. Susceptibility testing of Mycobacteria, Nocardia, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed]
- 8.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiwari, T. S., B. Ray, K. C. Jost, Jr., M. K. Rathod, Y. Zhang, B. A. Brown-Elliott, K. Hendricks, and R. J. Wallace, Jr. 2003. Forty years of disinfectant failure: outbreak of postinjection Mycobacterium abscessus infection caused by contamination of benzalkonium chloride. Clin. Infect. Dis. 36:954-962. [DOI] [PubMed] [Google Scholar]
- 10.Villanueva, A., R. V. Calderon, B. A. Vargas, F. Ruiz, S. Aguero, Y. Zhang, B. A. Brown, and R. J. Wallace, Jr. 1997. Report on an outbreak of postinjection abscesses due to Mycobacterium abscessus, including management with surgery and clarithromycin therapy and comparison of strains by random amplified polymorphic DNA polymerase chain reaction. Clin. Infect. Dis. 24:1147-1153. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, Y., M. A. Yakrus, E. A. Graviss, N. Williams-Bouyer, C. Turenne, A. Kabani, and R. J. Wallace, Jr. 2004. Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J. Clin. Microbiol. 42:5582-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhibang, Y., Z. BiXia, L. Qishan, C. Lihao, L. Xiangquan, and L. Huaping. 2002. Large-scale outbreak of infection with Mycobacterium chelonae subsp. abscessus after penicillin injection. J. Clin. Microbiol. 40:2626-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]