Abstract
The MultiCode-PLx system (EraGen Biosciences, Inc., Madison, WI) for the detection of respiratory viruses uses an expanded genetic alphabet, multiplex PCR chemistry, and microsphere flow cytometry to rapidly detect and specifically identify 17 different respiratory viruses directly in clinical specimens. The MultiCode-PLx system was tested in parallel with direct fluorescent-antibody (DFA) staining and rapid shell vial culture (R-mix cells; Diagnostic Hybrids, Inc. Athens, OH) with 354 respiratory specimens from adult patients that were submitted to the clinical virology laboratory at the Emory University Hospital. Single-target PCRs were performed with retained samples to confirm the positive results obtained with the MultiCode-PLx system for viruses not covered by DFA and R-mix culture (metapneumovirus, coronaviruses [CoV], parainfluenza viruses 4a and 4b, and rhinoviruses) and to resolve any discrepancies between the DFA and R-mix culture and the MultiCode-PLx results for viruses common to both systems. Respiratory viruses were detected in 77 (21.8%) and 116 (32.7%) specimens by DFA and R-mix culture and with the MultiCode-PLx system, respectively. Among the viruses common to both systems, the MultiCode-PLx system detected significantly more influenza A viruses (P = 0.0026). An additional increased diagnostic yield with the MultiCode-PLx system resulted from the detection of human metapneumovirus (HMPV) in 9 specimens, human CoV (HCoV) in 3 specimens, and human rhinovirus (HRV) in 16 specimens. Also, two mixed viral infections were detected by the MultiCode-PLx system (HCoV OC43 and HRV infections and HMPV and HRV infections), but none were detected by DFA and R-mix culture. Single-target PCRs verified the results obtained with the MultiCode-PLx system for 73 of 81 (90.1%) specimens that had discordant results or that were not covered by DFA and R-mix culture. The MultiCode-PLx system provides clinical laboratories with a practical, rapid, and sensitive means for the massively multiplexed molecular detection of common respiratory viruses.
Respiratory viral infections are the most common reason for medical consultation and are associated with a wide range of clinical manifestations, ranging from self-limited upper respiratory tract infections to more serious lower respiratory tract infections. Concerns about pandemic influenza, emerging threats to humans posed by zoonotic infections with avian influenza viruses and the sudden acute respiratory syndrome coronavirus (CoV), and the discovery of several previously unrecognized viral pathogens have placed a premium on respiratory virus surveillance and diagnosis (18, 19).
Currently, there are approximately 200 known respiratory viruses belonging to the families Adenoviridae, Parvoviridae, Orthomyxoviridae, Paramyxoviridae, Picornaviridae, and Coronaviridae (1). The virology of respiratory tract infections continues to evolve rapidly, with both the discovery of new pathogens and a better understanding of the importance of infections caused by the well-established ones. In the past several years human metapneumovirus (HMPV) (30), several new human CoVs (HCoVs) (10, 13, 17, 31, 34), and human bocavirus (2) have been added to the ever expanding list of respiratory viral pathogens. Despite these advances, a specific etiologic agent for viral respiratory illness is often not identified in clinical practice because of a lack of tests sensitive for the detection of all known agents and the presence of yet unrecognized pathogens.
Historically, standard viral detection techniques have relied on isolation of the virus in cell culture and immunoassays for viral antigens performed directly with clinical specimens. More recently, nucleic acid amplifications methods have been used to detect respiratory viruses, often with dramatic increases in sensitivity (14). However, the diversity and the complexity of the viral flora present significant challenges for nucleic acid-based detection systems. Multiplex PCR and microarrray-based systems for the detection of respiratory viruses provide potential solutions to this complex diagnostic problem. However, the numbers of viruses detectable in a single assay are relatively small in published reports of multiplexed PCRs for the detection of respiratory viruses (4, 5, 8, 12, 14, 16), and microarrays (32) are not practical for use in clinical laboratories.
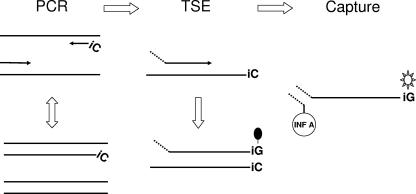
The MultiCode-PLx system (EraGen Biosciences, Inc., Madison, WI) is a new multitarget, high-throughput technology that integrates multiplexed PCR, an expanded genetic alphabet, and microsphere flow cytometry. Use of the MultiCode-PLx system involves three major steps: PCR, target specific extension, and liquid chip decoding (Fig. 1). An initial reverse transcription (RT) step is added for RNA targets. It uses an additional nucleobase base pair not found in nature that is constructed from the complementary bases isoguanine (iG) and 5-methyl-isocytidine (iC) for site-specific enzymatic labeling and wash-free decoding at room temperature. All steps are carried out in the same reaction vessel, without transfers or washings, and are completed in about 4 h. The MultiCode-PLx system can simultaneously detect up to 80 targets in one reaction and has successfully been applied to screening for multiple mutations in patients with cystic fibrosis (20), single-nucleotide polymorphism analysis of PECAM-1 (28), and human minor histocompatibility antigen genotyping (27).
FIG. 1.
Schematic of the major steps in the MultiCode-PLx respiratory virus detection system. For amplification, after RT, the target regions are amplified by PCR with one of the primers containing a single iC base at the 5′ end for subsequent site-specific labeling. For TSE, tagged, target-specific extenders are site specifically labeled with biotin-di-iso-GTP when the correct amplicon is present by primer extension. For capture, the tagged TSE products are captured by color-addressable microspheres through hybridization of each tag to its complementary, virus-specific oligonucleotide code conjugated to the microsphere surface. Both the tags and codes also contain the iC and iG bases. After the addition of streptavidin-phycoerythrin, the fluorescent signal associated with each microsphere is analyzed and decoded by using a Luminex 100 instrument.
In this study, an alpha prototype version of the MultiCode-PLx system for the detection of 17 different respiratory viruses was evaluated for its feasibility and diagnostic yield in the clinical laboratory setting. The results obtained with the MultiCode-PLx system were compared with the results obtained by direct fluorescent-antibody (DFA) staining and rapid shell vial culture (R-mix cells; Diagnostic Hybrids, Inc. Athens, OH) for respiratory specimens that were submitted to the clinical virology laboratory of the Emory University Hospital for culture. Single-target PCRs were performed with retained samples to confirm positive MultiCode-PLx system results for viruses not covered by DFA and R-mix culture (HMPV, HCoV, parainfluenza viruses [PIVs] 4a and 4b, and human rhinovirus [HRV]) and to resolve any discrepancies between the results obtained by DFA and rapid culture and those obtained by use of the MultiCode-PLx system for viruses common to both systems.
MATERIALS AND METHODS
Samples.
All respiratory tract specimens of sufficient volume from adult patients that were submitted to the clinical virology laboratory at the Emory University Hospital for DFA staining and rapid culture between 7 November 2005 and 17 March 2006 were eligible for the study. Nasopharyngeal, nose, throat, and lung tissue samples were collected and transported in viral transport media. Bronchoscopy samples and sputa were collected and transported in sterile containers without viral transport medium. All samples were transported to the laboratory on ice. One milliliter of residual unprocessed samples was stored at −80°C for later testing with the MultiCode-PLx respiratory virus system.
DFA staining and rapid culture.
Respiratory tract samples (2 to 4 ml each) were centrifuged at low speed at room temperature for 10 min after addition of 0.3 ml of an antibiotic solution (Bartels Cultrol; Trinty Biotech, Jamestown, NY). The cell pellet was used to prepare cytospin slides for DFA staining (Chemicon, Temecula, CA) for respiratory syncytial virus (RSV), according to the manufacturer's instructions. Three R-mix culture shell vials containing a mixed monolayer of human adenocarcinoma cells (A549 cells) and mink lung cells (Mv1Lu cells) were inoculated with 0.2 ml each of the sample supernatant. The shell vials were centrifuged at 700 × g for 1 h at room temperature. The inoculated vials were incubated at 35°C for up to 48 h. The coverslip was removed from one vial after 16 to 24 h of incubation, washed, fixed in acetone, and stained with a fluorescent monoclonal antibody pool containing antibodies to adenoviruses (ADVs); influenza A virus (INF A); influenza B virus (INF B); PIVs 1, 2, and 3; and RSV (Chemicon). The process was repeated with the coverslip from a second vial at 42 to 48 h for those samples negative at 16 to 24 h. A cell suspension was prepared from a remaining shell vial for samples that were positive with the antibody pool, and a drop was added to each well of an eight-well slide. Individual wells were then stained with a virus-specific monoclonal antibody reagent and examined under a fluorescent microscope to identify which virus was present.
PLx RVP.
One milliliter of residual unprocessed sample was centrifuged at 17, 000 × g for 20 min at room temperature, and the cell pellet was resuspended in approximately 200 μl of supernatant. The nucleic acid extraction was performed with a MagNA Pure LC instrument and by using a total nucleic acid kit (Roche Diagnostics, Indianapolis, IN). The sample and elution volumes were 200 and 50 μl, respectively. Each assay with the MultiCode-PLx system respiratory virus panel (PLx RVP) was performed in a single well of a low-profile 96-well PCR plate, as described by the manufacturer (EraGen). In brief, cDNA from viral RNA was prepared by adding and mixing 10 μl of extracted nucleic acid to 10 μl of reverse transcriptase solution. The reaction mixtures were sealed with an adhesive film (Microseal A; Bio-Rad, Hercules, CA); incubated at 25°C for 5 min, 42°C for 10 min, 55°C for 20 min, and 85°C for 5 min; and then cooled to 4°C. The target DNAs were then amplified by PCR on a PTC-200 apparatus (Bio-Rad) with the target-specific primer sets contained in the PCR master mixture. For each viral target primer set, one target-specific primer contained a single iC base at the 5′ end for subsequent site-specific labeling with iso-GTP-biotin. The amplification was performed by adding 5 μl of the PCR master mixture to 5 μl of the target DNA preparation. The reaction mixtures were sealed as described above and cycled 30 times by using the parameters of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s. The amplified DNA was then used as the template for the labeling of target-specific extenders. The labeling step, termed target-specific extension (TSE), was performed by adding 5 μl of TSE solution to the amplification reaction, sealing the plates, and incubating the plates at 65°C for 2 min and then 95°C for 10 s, with the incubation repeated 10 times. This was followed by a 5-min hold at 65°C, and then the plates were held at 4°C. When the reactions were complete, the reaction mixtures were brought to room temperature, 35 μl of a capture master mixture was added, and the components were mixed and allowed to sit for 10 min. The capture master mixture includes the Fast-Shot 30 bead array. This array contains codes covalently coupled to color-coded microspheres that can be detected with a Luminex LabMap flow detector. The codes are short sequences containing iC and iG and are complementary to specific tags on the extenders used in the previous step (20). Next, 35 μl of the detection master mixture was added to each reaction mixture, and after 15 min, the entire plate of reactions was placed in the XY platform of the Luminex 100 instrument. The fluorescent signal associated with each microsphere was measured. The signal is expressed as the median fluorescence intensity (MFI). Samples with an average signal >6 standard deviations above the average negative control signals were regarded as positive.
Two internal positive controls (IPCs) were used for each reaction performed: an RNA extraction control and a DNA PCR control. For the extraction control, ∼30,000 copies of an inactivated mouse hepatitis virus (ZeptoMetrix, Buffalo, NY) were added to 200 μl of each sample prior to extraction. For the PCR control, a DNA target and a set of primers and detectable extenders were present in the PCR master mixture and TSE solution, respectively. A reportable signal in the extraction control channel indicates that the control RNA added to the sample was successfully extracted, reverse transcribed, and amplified and that subsequent steps were performed correctly for that sample. Each IPC was present at approximately 1,200 copies/reaction mixture. A reportable signal in the PCR control channel indicates that the control DNA was successfully amplified and that the subsequent steps were correctly performed for that sample. Ideally, both controls should report signals over the background signal. Reportable signals for the extraction- and amplification-positive controls were defined as MFI values of greater than 5,000 and 2,000, respectively. For samples for which either IPC was not detected above the threshold, a no call was reported.
Single-target, real-time RT-PCR assays for HRV and HMPV and for HCoV.
Single-target, real-time RT-PCR assays for HRV and HMPV and for HCoV were carried out in 50-μl reaction volumes in optical-quality 96-well plates. Each reaction mixture contained 1× QIAGEN (Valencia, CA) Quantitect Probe RT-PCR kit reaction mixture. Thermal cycling was performed on an ABI 7300 real-time PCR instrument by using “absolute quantitation” software. The conditions were as follows: for HCoV, 50°C for 20 min for 1 cycle, 95°C for 15 min for 1 cycle, and 94°C for 15 s plus 60°C for 60 s for 45 cycles; for HRV and HMPV, 50°C for 30 min, 95°C for 15 min, and then 45 cycles of 95°C for 15 s and 55°C for 1 min. Threshold crossing cycles were computed by use of ABI 7300 Real-Time PCR system sequence detection software (version 1.2.1). RT-PCR for HRV was performed by the using primers and the minor groove binder (MGB) modifications of the probes originally described by Deffernez et al. (9). The primers and an MGB modification of the probe originally published by Maertzdorf et al. (25) were used for HMPV. Except for the HRV-specific probes, which were labeled with VIC at their 5′ ends, the probes were labeled with 6-carboxyfluorescein (FAM) at their 5′ ends and had the MGB peptide at their 3′ ends. For the HCoV assays, primers and probes that amplify segments of the N gene of human HCoVs OC43, 229E, and NL63/New Haven were used. The primer and probes sequences used for the OC43 assay were as follows: forward primer, GTTGTACAGGATGTGGGA; reverse primer, CGAACTTAGTCGTCATGT; and probe, FAM-CTGGATACCAGGAGTT-MGB. The primer and probe sequences used in the 229E assay were as follows: forward primer, TGCATTTTTATTATCTTGGC; reverse primer, TGAATTCTTGCGCCTAAC; and probe, FAM-CTGTTGATGGTGCTAAA-MGB. The primer and probe sequences used in the NL63 assays were those described by Fouchier et al. (17). HCoV strains were obtained from ATCC (strains OC43 and 229E) or from Lia van der Hoek (strain NL63). For each HCoV we created and quantified in vitro RNA transcripts from cloned segments that contained the assay target for each strain.
Single-target PCR assays for INF A and RSVs A and B.
The MultiCode-RTx real-time PCR technology was used to design primers that target the fusion gene of RSV and the matrix genes of INF A and INF B (29). The RSV assay consisted of RSV A- and RSV B-specific forward primers and a common reverse primer, which were used at 200 nM each. The INF A and INF B assays consisted of two primer pairs, with one pair targeting the INF A matrix gene and the other targeting the INF B matrix gene. The concentrations of the primer pairs were 150 and 200 nM in the INF A and INF B assays, respectively. The PCR conditions involved 25-μl reaction mixtures in 1× ISOlution buffer (EraGen) and Titanium Taq DNA polymerase (Clontech, Palo Alto, CA) at the manufacturer's recommended concentration. For one-step MultiCode-RTx reverse transcriptase PCR assays, the conditions included the following: 0.5 U/μl Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) and 5 mM dithiothreitol, with an additional 5 min at 50°C added prior to the initial denaturation step. Alternatively, 5 μl of the cDNA prepared as described above was used as the template for the MultiCode-RTx assay. Real-time PCR was performed on an ABI Prism 7900 real-time thermal cycler with 96-well optical plates. The PCR cycling parameters were 2 min of denaturation at 95°C and 5 cycles of 5 s at 95°C, 10 s at 58°C, and 20 s at 72°C, followed by 45 cycles of 5 s at 95°C, 10 s at 63°C, and 20 s at 72°C, with optical reading. A thermal melt at a 5% ramp rate with optical reading from 60 to 95°C was performed directly following the last step of thermal cycling at 72°C.
Alpha trial protocol.
All samples included in the study were tested for respiratory viruses by DFA staining with R-mix culture and by use of the MultiCode-PLx system. Single-target PCRs were performed with retained samples to confirm the positive MultiCode-PLx system results for viruses not covered by DFA staining and R-mix culture (HMPV, HCoV, PIVs 4a and 4b, and HRV) and to resolve any discrepancies between the results of DFA and rapid culture and those of the MultiCode-PLx system for viruses common to both systems (INF A; INF B; PIVs 1, 2, and 3; ADV; and RSV).
RESULTS
A total of 421 tests were performed with the MultiCode-PLx system and 377 specimens by two different operators in three separate runs with a single lot of reagents during the course of the study. The specimens types were as follows: 149, bronchoscopy fluids; 134, nose swab; 73, nasopharyngeal swab; 8, throat swab; 6, lung tissue; 1, sputum; and 6, unknown.
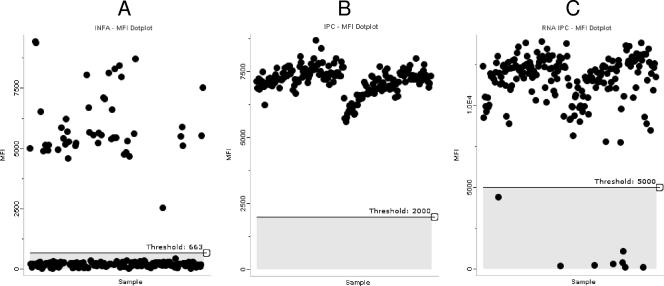
An example of the data output from a MultiCode-PLx system run is shown in Fig. 2. The dot plots show results for the INF A test (Fig. 2A) and the corresponding DNA (Fig. 2B) and RNA (Fig. 2C) IPC results for 187 patient specimens and 5 no-template negative controls. In the run for which the results are shown in Fig. 2, 47 specimens had MFI values above the threshold of 663 for the INF A test. MFI values for the DNA IPC exceeded the threshold of 2,000 for all of the samples; however, the RNA IPC was not detected in three of the patient specimens. The RNA IPC was not added to the no-template negative controls.
FIG. 2.
Sample data output from the MultiCode-PLx respiratory virus detection system. The dot plots show the MFIs for 187 patient specimens and 5 negative controls for the INF A target (A) and the corresponding DNA (B) and RNA (C) IPCs. Forty-seven samples had MFI values above the threshold for the INF A test. MFI values for the DNA IPC exceeded the threshold for all samples. The RNA IPC was not detected in three (1.6%) of the patient specimens.
The mean MFI values for all of the positive calls by the MultiCode-PLx system and all of the negative calls for those viruses included in the MultiCode-PLx system panel along with the threshold values for the different tests are shown in Table 1. The background fluorescence was low for all tests, and the mean MFI values for all specimens called positive by the MultiCode-PLx system were well above the positive thresholds for each test (Table 1).
TABLE 1.
Mean MFI values for positive and negative calls with clinical specimens in the MultiCode-PLx respiratory virus system
| Virus | Threshold MFI | Call | No. of tests | Mean MFI | SD |
|---|---|---|---|---|---|
| HRV | 962 | Negative | 401 | 154 | 79.3 |
| Positive | 20 | 7,890 | 1,500 | ||
| INF A | 906 | Negative | 329 | 161 | 79.6 |
| Positive | 92 | 5,850 | 2,250 | ||
| INF B | 1,026 | Negative | 419 | 177 | 85.1 |
| Positive | 2 | 5,280 | 1,780 | ||
| PIV 1 | 943 | Negative | 419 | 161 | 88.3 |
| Positive | 2 | 4,010 | 279 | ||
| RSV A | 898 | Negative | 410 | 153 | 81.3 |
| Positive | 11 | 7,000 | 2,540 | ||
| RSV B | 963 | Negative | 418 | 143 | 77.1 |
| Positive | 3 | 2,380 | 1,080 | ||
| HMPV | 1,015 | Negative | 412 | 155 | 79.6 |
| Positive | 9 | 6,770 | 2,380 | ||
| ADV C | 1,037 | Negative | 420 | 153 | 86.4 |
| Positive | 1 | 2,200 | |||
| OC43 | 946 | Negative | 420 | 159 | 86.4 |
| Positive | 1 | 1,860 | |||
| NL63 | 900 | Negative | 419 | 156 | 82.3 |
| Positive | 2 | 1,810 | 1,070 |
The DNA PCR control failed in 2.5% of the tests, and the RNA extraction control failed in 8.7% of the tests. Twenty-three patient specimens tested negative for viral targets in which either IPC was not detected. Data for these samples were excluded from the diagnostic yield calculations. For those tests in which the IPCs were detected, the MFI values were highly reproducible, with coefficients of variation for the PCR and extraction controls of 13 and 14%, respectively. None of the negative controls included in each MultiCode-PLx system run tested positive for any of the 19 different targets.
The respiratory viruses detected by each test system for the 354 specimens included in the analysis are given in Table 2. Overall, 77 viruses were detected by DFA and R-mix culture in 77 specimens, and 118 viruses were detected with the MultiCode-PLx system in 116 specimens. The overall diagnostic yields for DFA plus R-mix culture and MultiCode-PLx system were 21.8% and 32.7%, respectively. For the viruses common to both test systems, the yields were similar for INF B's, RSVs, ADVs, and PIVs. However, significantly more INF A's were detected with the MultiCode-PLx system than by rapid culture (P < 0.0026, McNemar's test).
TABLE 2.
Diagnostic yield from DFA staining plus R-mix culture and the MultiCode-PLx system for detection of respiratory viruses in 354 respiratory specimens
| Virus and summary result | No. of specimens detected by:
|
|
|---|---|---|
| DFA and R-mix culture | MultiCode-PLx system | |
| INF A | 59 | 74 |
| INF B | 1 | 1 |
| RSV | 12 | 13 (A, 10; |
| B, 3) | ||
| ADV | 1 | 1 (C) |
| PIV 1 | 3 | 2 |
| Pool positive onlya | 1 | |
| HMPV | 9 | |
| HRV | 16 | |
| NL63 | 2 | |
| OC43 | 1 | |
| No. of viruses detected | 77 | 118.3 |
| No. of positive specimens | 77 | 116b |
| Diagnostic yield (%) | 21.8 | 32.7 |
Unable to specifically identify the virus present with individual virus-specific monoclonal antibodies. The sample was positive for INF A with the MultiCode-PLx system.
Two mixed infections were detected: OC43 and HRV infections and HMPV and HRV infections.
A two-by-two contingency table for the detection of INF A by the different test methods was prepared, and the results were as follows: among the samples with positive MultiCode-PLx system results, 55 were R-mix culture positive and 19 were R-mix culture negative (of these, 17 [89.5%] tested positive for INF A by a single-target PCR assay). Among the samples with negative MultiCode-PLx system results, 4 were R-mix culture positive (of these, 3 [75%] samples tested positive for INF A by a single-target PCR assay) and 276 were R-mix culture negative. The agreement between the MultiCode-PLx system and R-mix culture for the detection of INF A was 93.5%. The sensitivity and specificity of the MultiCode-PLx system relative to the results of R-mix culture were 93.2% (95% confidence interval [CI], 83.5 to 98.1%) and 93.6% (95% CI, 90.1 to 96.1%), respectively. The apparent false-positive and false-negative MultiCode-PLx system results were adjudicated by a single-target PCR assay for INF A. INF A RNA was detected in 89.5% of the samples with apparent false-positive results and 75% of the samples with apparent false-negative results by the single-target PCR assay for INF A.
A two-by-two contingency table for the detection of RSV by the different test methods was prepared, and the results are as follows: among the samples with positive MultiCode-PLx system results, 11 were DFA and R-mix culture positive and 2 were DFA and R-mix culture negative (both samples tested positive for RSV by a single-target PCR assay). Among the samples with negative MultiCode-PLx system results, 1 was DFA and R-mix culture positive (the sample tested negative for RSV by a single-target PCR assay) and 340 were DFA and R-mix culture negative. The agreement between the methods was 99.2%. The sensitivity and specificity of MultiCode-PLx system relative to the results of DFA staining plus R-mix culture for the detection of RSV were 91.7% (95% CI, 61.5 to 99.8%) and 99.4% (97.7 to 99.8%), respectively (P = 1.0, McNemar's test). Of the 12 samples positive by DFA and R-mix culture, 3 were detected by DFA only. RSV RNA was found by single-target PCR assays for RSVs A and B in the two specimens with apparent false-positive results with the MultiCode-PLx system and not in the single sample with apparent false-negative results with the MultiCode-PLx system.
The MultiCode-PLx system detected 27 viruses that were not covered by DFA plus R-mix culture, including 9 HMPVs, 16 HRVs, 1 NL63 strain, and 1 OC43 strain. The agreement between the MultiCode-PLx system results and the results of single-target PCR assays for these viruses is shown in Table 3. All of the positive results for HMPV, NL63, and OC43 obtained with the MultiCode-PLx system and 81.3% of the results for HRV obtained with the MultiCode-PLx system were confirmed by the single-target PCR assays. Overall, the single-target PCRs verified the MultiCode-PLx system results for 73 of 81 (90.1%) of specimens whose results were discordant with those of DFA and R-mix culture or that were not covered by DFA and R-mix culture.
TABLE 3.
Agreement between MultiCode-PLx system and single-target PCR results for viruses not covered by DFA staining and R-mix culture
| Virus | No. of MultiCode-PLX system-positive specimens | No. (%) of single-target PCR positive specimens |
|---|---|---|
| HRV | 16 | 13 (81) |
| HMPV | 9 | 9 (100) |
| NL63 | 2 | 2 (100) |
| OC43 | 1 | 1 (100) |
Two mixed infections were detected by the MultiCode-PLx system and none were detected by DFA plus R-mix culture. For one sample the MultiCode-PLx system was positive for HMPV and HRV, and for the second sample the MultiCode-PLx system was positive for OC43 and HRV.
Seventy-three specimens were tested by both operators by use of the MultiCode-PLx system; 44 were positive and 29 were negative for viral targets. The same result was obtained by each operator with 68 (93.1%) of the specimens. When the results differed, it was a no call that occurred because one or both of the IPCs were not detected.
DISCUSSION
Nucleic acid-based diagnostics for respiratory viruses have emerged as the new “gold standards.” However, the large number of potential etiologies of respiratory tract infections demands a massively multiplexed approach. Use of a combination of reagents to detect multiple pathogens in a single PCR often results in a loss of sensitivity for each of the individual targets. In addition, there are limitations in the number of targets that can be detected in a multiplex assay. Real-time PCR assays with fluorescent reporters require the use of nonoverlapping spectra for each reporter dye, which limits the number of targets to five or less with most instruments. An end-point PCR with a gel-based readout requires amplicons that can be clearly distinguished by size. While it is possible to detect more targets by this approach, the amplification of targets of different sizes often results in different target amplification efficiencies.
The MultiCode-PLx system overcomes both limitations of standard multiplex analysis. The high mean fluorescence intensities for positive samples and the agreement between the results obtained with the MultiCode-PLx system and those obtained by single-target PCR suggest that it is possible to perform a massively multiplex analysis with little loss of sensitivity. The individually addressable, color-coded polystyrene beads used to capture specific target amplicons allowed the effective discrimination of 19 discrete analytes per sample in this respiratory virus panel.
We tested patient specimens for respiratory viruses using a prototype MultiCode-PLx system capable of detecting 17 different respiratory viruses. All samples were tested in parallel with R-mix rapid culture. R-mix culture was chosen as the comparator because the sensitivity is comparable to that of conventional cell culture but provides results in 24 to 48 h (11). The diagnostic yields with the R-mix culture and MultiCode-PLx systems were similar for the viruses detected by both assays except INF A. The MultiCode-PLx system detected significantly more INF A-positive samples. The 25.4% increase in sensitivity over the results of R-mix culture is consistent with other reports of increased yields with PCR assays for INF A (14).
The MultiCode-PLx system provides additional diagnostic capabilities beyond those available with the R-mix culture system. HMPV is responsible for a substantial portion of lower respiratory tract infections in infants and young children and is second only to RSV as a cause of bronchiolitis in early childhood (21). In adults HMPV has been associated with influenza-like illness, bronchitis, pneumonia, and exacerbations of both asthma and chronic obstructive pulmonary disease. It can also cause prolonged and serious infections in the immunocompromised host. We detected HMPV in 2.5% of the samples tested with the MultiCode-PLx system. Falsey et al. (15) detected HMPV with a single-target RT-PCR in 3.4% of adults with respiratory tract illness.
The PLx RVP included three HCoV strains: OC43, 229E, and recently discovered strain NL63 (31). HCoVs cause a wide variety of respiratory illnesses in children and adults; however, the clinical impact of the illnesses has not yet been fully determined. We found two patients infected with NL63 (0.6%), one patient (0.3%) infected with OC43, and no patient infected with 229E. Most of the data on the prevalence of NL63 is for children, for whom the rates of detection range from 2.1 to 9.3% of patients with acute respiratory illness (19). Strains OC43 and 229E demonstrate periodicity, with large epidemics occurring every 2 to 3 years (22). Since 2003, five HCoVs have been discovered, including the sudden acute respiratory syndrome, NL63, NL, NH, and HKU1 viruses (22). Several of these recently discovered HCoVs are candidates for inclusion in a comprehensive RVP.
HRVs are the most common causes of upper respiratory tract illnesses and asthma exacerbations in ambulatory children and adults (19). In addition, HRVs have been linked to prolonged cough illness in children, lower respiratory tract disease in stem cell transplant recipients, rejection in lung transplant recipients, and serious infections in elderly individuals. We detected HRVs in only 4.5% of the samples that we tested with the MultiCode-PLx system. This relatively low positive rate probably reflects the fact that the majority of samples came from hospitalized patients with potentially more serious respiratory illnesses.
The MultiCode-PLx system also distinguishes between antigenic subgroups A and B of PIV 4; subgenera B, C, and E of respiratory ADVs; and antigenic subgroups A and B of RSV. This capability is of more value in epidemiological studies and surveillance activities than for the management of individual patients. The MultiCode-PLx system should provide a better means to detect infections of mixed viral etiology. However, in this study only 1.7% of the infections detected by the MultiCode-PLx system involved more than one virus.
The diagnostic capacity of the MultiCode-PLx system could easily be extended beyond the 17 viruses included in the panel that we tested. In theory, up to 80 different targets could be detected in a MultiCode-PLx system, since 80 distinct tags are available. Other candidates for a respiratory virus panel include enteroviruses, human bocavirus, hantaviruses, and avian influenza viruses, in addition to the newly recognized HCoVs discussed above. This list will expand as new respiratory viruses emerge and their clinical importance is established.
The MultiCode-PLx system is robust, and when it is coupled with the MagNA Pure total nucleic acid extraction kit, it is capable of providing results for a wide variety of respiratory sample types. It is a one-tube, three-step, 4-h process that does not involve extensive sample washes or transfers and that uses equipment that is already available in many clinical laboratories. The labeling of the amplicons and room temperature decoding of tagged target extenders are accomplished through the use of a nonnatural base pair, iC and iG. The PCR step is highly multiplexed, which may be the reason why stringent cycling conditions were required. We found that the PTC-200 cyclers performed well, but the use of less precise cyclers could be problematic.
Use of the MultiCode-PLx system is technically demanding, but no more so than the use of other PCR-based assays. Although there are no sample transfers, the MultiCode-PLx system plates must be unsealed and resealed three times during the procedure to add reagents. The unsealing of the plates could potentially cause false-positive results due to sample or amplicon carryover if the unsealing is not done carefully. However, single-target PCRs verified the positive MultiCode-PLx system results for 90.1% of the specimens that had results discordant with those of DFA and R-mix culture or that had viruses not covered by DFA and R-mix culture. In addition, none of the negative controls included with each MultiCode-PLx system run tested positive for any of the targets included in the panel. These results alleviated our initial concerns about carryover contamination, but as is the case with any PCR protocol, proper sample handling and laboratory work flow should be maintained.
In the version that we tested, the MultiCode-PLx system incorporated both DNA and RNA IPCs in each sample. The DNA IPC provides a check that the amplification, TSE, and detection reactions and the instrument are functioning appropriately. The RNA IPC checks the function of all steps of the procedure, including sample preparation and RT. We believe that higher RNA IPC failure rates may indicate that our sample preparation method needs further optimization for the recovery of RNA targets from respiratory tract samples.
The optimal use of external positive controls for massively multiplexed molecular diagnostics is yet to be defined. In this trial with an alpha version of the system, we included external positive controls for each of the 17 different analytes with each run. To minimize the number of wells devoted to external positive controls, three positive controls were included in one well, and two positive controls were combined in each of seven other wells. This strategy would be impractical for routine use. Laboratories adopting this technology could chose to run the full battery of external positive controls with each new lot of reagents and rotate a different subset of the controls with each run.
This is the first trial of the MultiCode-PLx system in a clinical laboratory setting and was conducted by technologists with experience in molecular diagnostic techniques. Our experience demonstrates that this massively multiplexed approach to respiratory viral diagnostics is feasible in this setting. The MultiCode-PLx system uses multichannel pipettors, a standard thermal cycler, and a microsphere flow cytometer, which are available in many clinical laboratories. In our experience, approximately 32 samples could be processed and analyzed in an 8-h shift. Although timing studies were not performed, we estimate that each sample requires about 4 min of hands-on time. As many as 80 samples per shift can be analyzed when the assay starts with purified nucleic acid.
The relatively small numbers of specimens with viruses other than INF A and RSV and the use of single-target PCR tests in subset analyses are the major limitations of this study. Since the prevalence of the individual respiratory viruses varies from year to year and by geography, multicenter clinical trials supplemented by assays with banked samples will be necessary to accurately assess the performance characteristics of the MultiCode-PLx system. Given the limitations of antigen detection and culture, we chose single-target PCR assays to adjudicate the results obtained with the MultiCode-PLx system. The results may differ if other single-target PCR assays are used as comparators, because standardized assays for many of the targets included in the respiratory virus panel are not available.
Lee et al. (24) recently described the development and verification of a multiplex PCR-microsphere flow cytometry system similar to the one described here for the comprehensive detection of respiratory viruses in nasal lavage samples from children with asthma. They found similar rates of detection with this system and conventional virological methods for INF A, INF B, and PIV 1. The new system had improved rates of detection for RSV, PIV 3, PIV 4, HRV, and enteroviruses, with the increases in the diagnostic yields over those of conventional methods ranging from 23.8% to 72.8%.
Two other multiplex respiratory pathogen assays that use microsphere liquid arrays are in commercial development (Genaco [QIAGEN, Hilden Germany] and Tm Bioscience [Luminex]). The systems differ in their compositions of the respiratory virus panels, the methods by which the PCR products are labeled, and the numbers of analysis steps. Brunstein and Thomas (7) reported a complete concordance between the results of DFA staining and those of the Genaco assay for the detection of respiratory viruses in 18 clinical samples. To our knowledge no information on the performance characteristics of the Tm Bioscience assay has been published.
MassTag PCR is another novel approach to the highly multiplexed detection of respiratory pathogens that compares favorably with other methods (6). It uses a multiplex PCR in which the viral gene targets are coded by a library of up to 64 distinct Masscode tags (QIAGEN, Hilden, Germany). The targets are amplified with primers labeled by a photocleavable link to molecular tags of different molecular weights. After removal of unincorporated primers, the tags are released by UV irradiation and analyzed by mass spectroscopy. The identity of the virus in the clinical sample is determined by the presence of its cognate tag. In a recent study, the MassTag system was compared with single-target RT-PCR and conventional virus culture for the detection of 22 different respiratory pathogens in 151 clinical samples (23). The diagnostic yield with the MassTag system was much greater, detecting pathogens in 33% of the samples that were negative by the other methods.
The increased laboratory costs of a rapid and comprehensive respiratory virus diagnostic assay could be offset through the improvement of patient management by influencing the decision to initiate antibiotic or antiviral therapy and implementing infection control measures to prevent transmission, particularly in hospitalized patients (3, 26, 33). However, a recent study of the impact of the rapid detection of viral and atypical bacterial pathogens by a large number of single-target, real-time PCR runs in parallel for patients with lower respiratory tract infections did not reduce antibiotic use or health care costs (16). A detailed cost analysis was beyond the scope of this study, but preliminary estimates indicate that the MultiCode-PLx system will be more costly than the combination of antigen detection and culture methods that are currently the standard of practice in many laboratories but less costly than 17 single-target PCR assays run in parallel or the use of culture-based systems that provide equally broad coverage for respiratory viruses.
In summary, the MultiCode-PLx system provides a rapid, sensitive, and comprehensive approach to the molecular detection of respiratory viruses that has several advantages over other multiplexing protocols. It is a practical and robust method that is feasible for use by clinical laboratories with the potential to dramatically increase diagnostic yields.
Acknowledgments
EraGen Biosciences provided technical support and the MultiCode-PLx system reagents for this study.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Abed, Y., and G. Boivin. 2006. Treatment of respiratory virus infections. Antivir. Res. 70:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenfanger, J., C. Drake, N. Leon, T. Mueller, and T. Troutt. 2000. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J. Clin. Microbiol. 38:2824-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecherbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 126:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boivin, G., S. Cote, P. Dery, G. De Serres, and M. G. Bergeron. 2004. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J. Clin. Microbiol. 42:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briese, T., G. Palacios, M. Kokoris, O. Jabado, Z. Liu, N. Renwick, V. Kapoor, I. Casas, F. Pozo, R. Limberger, P. Perez-Brena, J. Ju, and W. I. Lipkin. 2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg. Infect. Dis. 11:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunstein, J., and E. Thomas. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn. Mol. Pathol. 15:169-173. [DOI] [PubMed] [Google Scholar]
- 8.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72:484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deffernez, C., W. Wunderli, Y. Thomas, S. Yerly, L. Perrin, and L. Kaiser. 2004. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J. Clin. Microbiol. 42:3212-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, J. J., R. D. Woolstenhulme, J. Langer, and K. C. Carroll. 2004. Sensitivity of respiratory virus culture when screening with R-mix fresh cells. J. Clin. Microbiol. 42:79-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elnifro, E. M., A. M. Ashshi, R. J. Cooper, and P. E. Klapper. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esper, F., C. Weibel, D. Ferguson, M. L. Landry, and J. S. Kahn. 2005. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 191:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espy, M. J., J. R. Uhl, L. M. Sloan, S. P. Buckwalter, M. F. Jones, E. A. Vetter, J. D. Yao, N. L. Wengenack, J. E. Rosenblatt, F. R. Cockerill III, and T. F. Smith. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falsey, A. R., M. C. Criddle, and E. E. Walsh. 2006. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J. Clin. Virol. 35:46-50. [DOI] [PubMed] [Google Scholar]
- 16.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin. Infect. Dis. 26:1397-1402. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouchier, R. A., G. F. Rimmelzwaan, T. Kuiken, and A. D. Osterhaus. 2005. Newer respiratory virus infections: human metapneumovirus, avian influenza virus, and human coronaviruses. Curr. Opin. Infect. Dis. 18:141-146. [DOI] [PubMed] [Google Scholar]
- 19.Hayden, F. G. 2006. Respiratory viral threats. Curr. Opin. Infect. Dis. 19:169-178. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, S. C., D. J. Marshall, G. Harms, C. M. Miller, C. B. Sherrill, E. L. Beaty, S. A. Lederer, E. B. Roesch, G. Madsen, G. L. Hoffman, R. H. Laessig, G. J. Kopish, M. W. Baker, S. A. Benner, P. M. Farrell, and J. R. Prudent. 2004. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 50:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn, J. S. 2006. Epidemiology of human metapneumovirus. Clin. Microbiol. Rev. 19:546-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahn, J. S., and K. McIntosh. 2005. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 24:S223-S227. [DOI] [PubMed] [Google Scholar]
- 23.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 194:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, W.-M., K. Grindle, T. Pappas, D. Marshall, M. Moser, E. Beaty, J. Prudent, and J. E. Gern. 30 May 2007. A high-throughput, sensitive and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. doi: 10.1128/JCM02501-06. [DOI] [PMC free article] [PubMed]
- 25.Maertzdorf, J., C. K. Wang, J. B. Brown, J. D. Quinto, M. Chu, M. de Graaf, B. G. van den Hoogen, R. Spaete, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2004. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 42:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosterheert, J. J., A. M. van Loon, R. Schuurman, A. I. Hoepelman, E. Hak, S. Thijsen, G. Nossent, M. M. Schneider, W. M. Hustinx, and M. J. Bonten. 2005. Impact of rapid detection of viral and atypical bacterial pathogens by real-time polymerase chain reaction for patients with lower respiratory tract infection. Clin. Infect. Dis. 41:1438-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietz, B. C., M. B. Warden, B. K. DuChateau, and T. M. Ellis. 2005. Multiplex genotyping of human minor histocompatibility antigens. Hum. Immunol. 66:1174-1182. [DOI] [PubMed] [Google Scholar]
- 28.Prudent, J. R. 2006. Using expanded genetic alphabets to simplify high-throughput genetic testing. Expert Rev. Mol. Diagn. 6:245-252. [DOI] [PubMed] [Google Scholar]
- 29.Sherrill, C. B., D. J. Marshall, M. J. Moser, C. A. Larsen, L. Daude-Snow, S. Jurczyk, G. Shapiro, and J. R. Prudent. 2004. Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 126:4550-4556. [DOI] [PubMed] [Google Scholar]
- 30.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo, P. C., S. S. Chiu, W. H. Seto, and M. Peiris. 1997. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 35:1579-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]