Abstract
We identified three isolates of Streptococcus agalactiae (group B streptococcus [GBS]), of human origin, which failed to react with antisera against any of the nine known GBS serotypes. Polyclonal rabbit antisera raised against these isolates and standard GBS typing sera were used in capillary precipitation and Ouchterlony tests to compare the strains with known GBS serotype reference strains. All three previously nontypeable isolates reacted with all three new antisera, producing lines of identity in the Ouchterlony test. Weak cross-reactions with antisera against several GBS serotypes were observed but were removed by absorption with corresponding antigens. The new antisera were used to test 227 GBS isolates that had been nontypeable or difficult to type using standard antisera. Of these, five reacted with the new antisera. These results suggested that all eight isolates belong to the previously unrecognized GBS serotype. They were tested by Western blotting for the Cα and Cβ proteins and by PCR to identify molecular serotypes and surface protein antigen genes. Two segments of the cps gene cluster (3′ end of cpsE-cpsF and 5′ end of cpsG, approximately 700 bp; 3′ end of cpsH and 5′ end of cpsM, approximately 560 bp) were sequenced. All eight isolates expressed Cα, and seven expressing the Cβ protein and the corresponding genes, bca and bac, respectively, were identified. They all share the same, unique partial cps sequence. These results indicate that these eight isolates represent a new S. agalactiae serotype, which we propose should be designated serotype IX.
Streptococcus agalactiae, also known as group B streptococcus (GBS), is a pathogen of newborn infants in particular but also a cause of bacteremia in parturient women, elderly people, and immunocompromised patients (11).
The current classification of GBS is based on the capsular phenotype (1). Currently there are nine serotypes: Ia, Ib, and II to VIII (11, 43). The primary serologic method used for serotype determination is capillary precipitation, introduced by Lancefield in 1934 (14), based on capsular antigens (14, 37). Most GBS isolates can be classified into these serotypes, but 4 to 7% are nontypeable (10, 11, 41, 45). This can be due to mutation in the capsular genes (44), reversible nonencapsular phase variation (6), or an uncharacterized capsule (46). However, the numbers of nontypeable isolates have been an increasing problem, and studies have been performed to try to reduce them (2, 41). In Denmark, 9% of all invasive isolates submitted to the Neisseria and Streptococcus Reference laboratory, Statens Serum Institut (SSI), are nontypeable (unpublished data).
In recent years, molecular serotyping of GBS based on detection of serotype-specific genes of the capsular region has been developing rapidly (5, 7, 29, 42) and, in some studies, has now replaced classical antiserum-based typing methods (33). Methods such as PCR and pulsed-field gel electrophoresis have shown that some nontypeable GBS isolates can be shown to be similar to known serotypes (1). Identification of surface proteins, using PCR, also allows classification of nontypeable isolates (7, 30, 41), a large number of which have been characterized and/or typed using molecular methods. Nevertheless, some isolates remain impossible to characterize using currently available antisera, and knowing the genotype of an isolate is not the same as knowing its phenotype (30, 41). Generally, results of conventional serotyping and molecular serotype identification are consistent for isolates that are serotypeable. Whether currently nontypeable strains (29, 40, 50) should be classified by molecular typing, the use of additional antisera, or a combination remains to be determined (29, 40, 50).
This study presents data on 3 GBS isolates isolated in Denmark in 1986, 1989, and 2000 and 5 additional isolates found during reevaluation of 227 isolates that were initially nontypeable or difficult to serotype or for which there were discrepancies between results of conventional and molecular serotype identification.
MATERIALS AND METHODS
GBS strains.
GBS reference strains, representing all recognized serotypes, were obtained from the Neisseria and Streptococcus Reference laboratory, SSI, which serves as a National Reference Center for Streptococcus (Table 1). Isolates selected for further investigation had been referred to the SSI for serotyping. They were nontypeable, i.e., they failed to agglutinate with any GBS type-specific antisera but grew with even turbidity in broth, suggesting that they produced normal amounts of capsular polysaccharide antigen. Antisera raised against them reacted with each other but not with other nontypeable or typeable GBS isolates referred to SSI (39).
TABLE 1.
Cross-reactions of extract antigens/antisera from types and strains to extract antigen/antiserum raised against new GBS type candidate
| Serotype | Name of strain | Cross-reactiona |
|---|---|---|
| Ia | O90 (ATCC 12400)b | + |
| 8/70 Praghb | + | |
| 5420/77 | + | |
| 9597/76 | + | |
| 14808/77 | + | |
| Ib | H36 (NCTC 8187)b | + |
| SS 618 | + | |
| 79751/75 | + | |
| 10727/78 | + | |
| 10420/76 | + | |
| II | 18 RS 21 (NCTC 11079)b | − |
| V9 (ATCC 12387)b | − | |
| 319/77 | − | |
| 2959/76 | − | |
| 8791/77 | − | |
| III | M 216 (NCTC 11080)b | − |
| M 781 USA 27/10-93b | − | |
| D 136 (ATCC 12403)b | − | |
| 3782/62 Kvittinger | − | |
| 14903/77 | − | |
| IV | 12351b | + |
| 3139 Praghb | + | |
| 68717 Praghb | + | |
| 1110 Pragh | + | |
| 717 Pragh with Ibc antigen | + | |
| V | SS 1169b | + |
| 10/84 Pragh | + | |
| H 43 | + | |
| SS 1168 | + | |
| 2569/79 | + | |
| VI | NT6b | − |
| 10214 Pragh 1981 | − | |
| VII | 7271b | + |
| 1609/76 | + | |
| 6315/76 | + | |
| VIII | 130013 Colindaleb | − |
| 130669 Colindale | − | |
| Gr. Bc | B848b | Positive reaction with extract antigen, negative reaction with antiserum |
| BO90R (NCTC 9993)b | Positive reaction with extract antigen, negative reaction with antiserum | |
| 881 SSI 1964 | Positive reaction with extract antigen, negative reaction with antiserum | |
| Greenaway 31-10-50 | Positive reaction with extract antigen, negative reaction with antiserum |
+, week cross-reaction detected, removed by absorption; −, no cross-reaction detected.
National reference strain for typing of GBS strains.
Group B.
The three nontypeable GBS isolates (22634, 7214, and 0884) were from humans in Denmark and are included in the Danish collection of GBS. Isolates 22634 and 0884 were collected from blood of newborn infants in 1986 and 2000, and isolate 7214 was collected in 1989 from the left lung of a 26-year-old person. A serotype IX reference strain can be obtained from Statens Serum Institut, SSI Diagnostica, Copenhagen, Denmark (microbiology@ssi.dk).
The status of 227 isolates, which either had been nontypeable by use of antisera or had given discrepant results between conventional and molecular serotyping, was reevaluated. They were part of a large international collection of GBS isolates which have been studied previously (24, 47, 48, 50). After retesting, many were successfully serotyped and most discrepancies between conventional and molecular serotypes were resolved, but 135 isolates remained nontypeable. Five isolates agglutinated with antiserum A7214 (see below) and were investigated further: one was from Canada (Canada strain 1012B, 1997), isolated from a pregnant woman (36); two were from Germany (M48/97, 1997; M23/99, 1999), isolated from pregnant women (4); one was from Hong Kong (02B0981138, 2002) (16); and one was from Sydney, Australia (03-150-3185, 2003), isolated from blood culture from a 57-year-old male with diabetes and renal failure.
Antisera.
Commercial GBS-type antisera against serotypes Ia, Ib, and II to VIII and group B were obtained from SSI Diagnostica, Denmark. Sera against the three nontypeable Danish isolates were raised in rabbits by an immunization procedure described by Lancefield et al. (32). Antiserum A7214 has been used routinely at the Neisseria and Streptococcus Reference Laboratory for the past 2 years, in addition to the nine commercial GBS typing antisera, to test GBS isolates and was used in this study to reevaluate the status of 227 isolates (see above) that were previously nontypeable or difficult to type.
Standard test.
All eight isolates under investigation were identified as GBS using standard Gram staining, CAMP test, and API (Rapid ID 32 Strep test) (17, 22). Acid-extracted antigen from all eight isolates tested positive in a Lancefield test using GBS group antiserum (Table 2) and in PCR targeting the GBS species-specific gene cfb.
TABLE 2.
Reactions between extract antigens (Lancefield method) of three strains studied and the antisera raised against them
| Strain against which antiserum was raised | Reaction of antiserum to extract of straina:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0884
|
22634
|
7214
|
B848 (Gr. B)c
|
|||||
| 0.1 N | 0.2 N | 0.1 N | 0.2 N | 0.1 N | 0.2 N | 0.1 N | 0.2 N | |
| 0884 | ++ | ++ | ++ | ++ | + | ++ | − | − |
| 0884, 22634b | − | − | − | − | − | − | − | − |
| 0884, 7214b | − | − | − | − | − | − | − | − |
| 22634 | ++ | ++ | ++ | ++ | + | + | − | − |
| 22634, 0884b | − | − | − | − | − | − | − | − |
| 22634, 7214b | − | − | − | − | − | − | − | − |
| 7214 | +++ | +++ | +++ | +++ | ++ | ++ | − | − |
| 7214, 0884b | − | − | − | − | − | − | − | − |
| 7214, 22634b | − | − | − | − | − | − | − | − |
| Gr. B (B848) | +++ | +++ | +++ | ++ | ++ | ++ | +++ | ++ |
−, negative reaction; +, weak reaction; ++, moderate reaction; +++, strong reaction.
The antiserum was absorbed with culture of the second-mentioned strain.
Nontypeable group B strain.
None of the eight isolates agglutinated when whole cells grown in Todd-Hewitt broth were mixed with group B-specific antiserum. A nonencapsulated/nontypeable group B strain (strain B848) (Table 1) was used as a positive control, and a serotype III strain was used as a negative control.
Pepsin digestion.
A 110-μl pepsin dilution (5 mg pepsin per 1 ml 0.1 N HCl) was added to 0.3 ml 0.2 N HCl extracts of all eight type IX isolates; the pH was adjusted to 2 with 0.2 N NaOH. This mix was incubated for 2 h at 37°C. After incubation, the extracts were adjusted to pH 7 by adding 0.2 N NaOH. A 0.2 N HCl extract of a serotype III isolate was used as a positive control.
Lancefield capillary precipitation method for serotyping.
The Lancefield method was performed as described by Slotved et al. (46). All tests shown in Tables 1 and 2 were performed using the precipitation method and 0.1 N and 0.2 N HCl extract.
Ouchterlony test.
The antisera (A22634, A7214, and A0884) against the three Danish nontypeable isolates were evaluated using Ouchterlony and capillary precipitation tests (46). The Lancefield antigen extract was prepared as follows. A freeze-dried culture of GBS was suspended in serum broth, and a drop was plated onto 5% horse blood agar and incubated at 36°C overnight. The next day, a few individual colonies were inoculated into 5 ml glucose broth and incubated at 36°C overnight. After incubation, the broth was centrifuged and the supernatant removed. A 0.1-ml amount of either 0.1 N or 0.2 N HCl was added to the bacterial pellet. The acid suspension was placed at 100°C for 15 min (in boiling water) and thereafter cooled under tap water. After cooling, 0.2 N NaOH was added to adjust the pH to 7.0, with a drop of phenol red as the indicator. The suspension was centrifuged for 10 min to obtain a crystal-clear supernatant.
The Ouchterlony test was performed as follows. An agarose gel (1% LSA; Litex, Copenhagen, Denmark) was made in 20% Tris-Veronal buffer (containing 2.24% [wt/vol] barbital, pH 8.5; final dilution, 20%) (product no. 30731; SSI Diagnostica, Copenhagen, Denmark), heated to 100°C, and then kept in a water bath at 60°C. A gel bond sheet was cut to the size of 9 cm by 4.5 cm and placed with its hydrophobic side downwards on a level glass plate. 8 ml of liquid gel was poured onto the plate to a thickness of 0.2 cm. When the gel had solidified, holes were punched in it according to a template, as shown in Fig. 1. Ten microliters of serum was added to the center holes of each gel and 10 μl of antigen extract (either 0.1 N or 0.2 N) to the surrounding holes. The gels were incubated in a moist chamber at 5°C for 3 days and then covered with filter paper, soaked in 0.9% saline, pressed for 30 to 60 min under a glass plate and a weight (4 to 5 kg), and washed first in 0.9% saline overnight and then in ionized water for at least 60 min. Gels were then pressed again as described above, except that they were soaked only in ionized water. After the second press, they were dried thoroughly with a hair dryer and stained with Coomassie blue for 15 min, washed in a solution of ethanol (432 ml), acetic acid (100 ml), and ionized water (468 ml) for 15 min, and dried again.
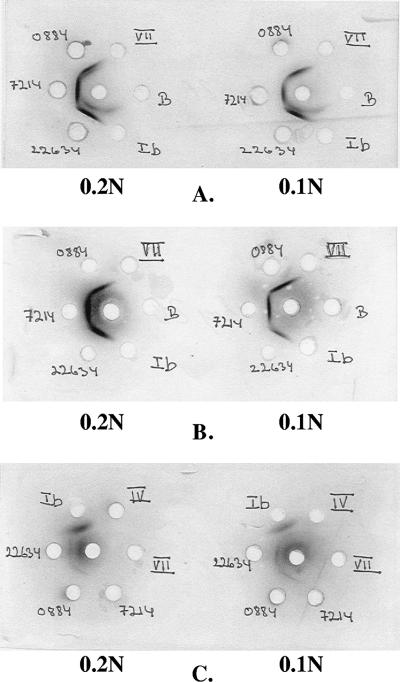
FIG. 1.
Three Ouchterlony tests (A, B, and C) are shown. Ten microliters antiserum was added in the center hole, and 10 μl of either 0.1 N or 0.2 N extract antigen was added in the surrounding holes. (A) Antiserum raised against isolate 22634 was added in the center hole. Reactions appeared toward extract antigens from all three isolates. A second line appeared towards extract antigen of isolate 22634, and this reaction also appears with GBS type Ib where there is line of identity. (B) Antiserum (raised against 22634) absorbed with antigen from GBS type Ib was added in the center hole. Only reactions against antigen of the three isolates appeared. (C) Antiserum raised against GBS type Ib was added in the center hole. A double line appeared against extract antigen (0.1 N) of GBS type Ib.
Absorption.
Specific absorption of antisera was based on the method described by Lancefield (32). Briefly, an overnight culture (1 liter trypsin broth) was killed with 0.5% formaldehyde for 24 h and then centrifuged for 60 min at 3,500 × g, after which the supernatant was removed. The actual absorption was performed by mixing equal amounts of culture and antiserum; after 30 min of incubation at about 20°C, the suspension was centrifuged. The pellet with the cross-reacting antibodies was removed, and the supernatant (absorbed antiserum) was stored for later use.
Western blot for Cα and Cβ proteins.
Western blotting was performed using XCell4 Surelock Midi-Cell (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, a bacterial solution was mixed with sample buffer (NuPAGE; Invitrogen, Carlsbad, CA) and reducing agent (NuPAGE; Invitrogen), and the vials were heated for 10 min at 70°C. The electrophoresis apparatus (XCell4 Surelock Midi-Cell; Invitrogen) was prepared with 180 ml 0.25% antioxidant mixture (NuPAGE; Invitrogen) with Milli Q water as a diluting agent. Ten microliters of the sample mixtures were loaded on a Tris-bis 4-to-12%-gradient gel (8.7 by 13.3 cm; Novex). The buffer chamber of the electrophoresis apparatus was filled with a 5% dilution of running buffer (NuPAGE; Invitrogen) in MQ water, and the gel was subsequently electrophoresed for 1 h at 180 V. After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Whatman; Schleider & Schuell) and the membrane blocked in incubation buffer. Anti-Cα and -Cβ polyclonal rabbit sera (SSI Diagnostica, Hillerød, Denmark) were used as primary antibodies (diluted 1:1,000), and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (product no. A3812; Sigma Aldrich, St. Louis, MO) was diluted 1:1,000 and used as the secondary antibody. A Sigma Aldrich 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium tablet was used as a substrate for the color reaction, which was stopped by the addition of MQ water.
Molecular serotype and surface protein antigen gene identification.
Serotype-specific PCR (29), partial cps sequencing (3′ end of cpsE-cpsF and 5′ end of cpsG) (50), and surface protein antigen gene-specific PCRs (30, 49) were performed as previously described. The serotype-specific PCR products (25, 29) were sequenced to generate the partial sequence of cpsH-cpsM.
MLST.
Multilocus sequence typing (MLST) was performed as described by Jones et al. (19). PCR products were amplified using the HotStarTaq Master Mix kit (QIAGEN GmbH, Hilden, Germany), and both strands of the PCR products were sequenced using the Big Dye Terminator v1.1 cycle sequencing kit and an ABI3100 sequencer (Applied Biosystems, Foster, CA). Sequences were analyzed using BioNumerics (Applied Maths NV, Sint-Martens-Latem, Belgium), and the sequence types were assigned using the Streptococcus agalactiae MLST website (http://pubmlst.org/sagalactiae/) (18).
Nucleotide sequence accession numbers.
New sequence data generated in this study have been deposited in GenBank with accession number AY257685 (for sequences between the 3′ end of cpsE-cpsF and the 5′ end of cpsG of strain 1012B) and EF157290 (for the partial sequence of cpsH-cpsM of strain 7214).
RESULTS
Species identification.
All eight GBS nontypeable isolates under investigation were gram-positive cocci (17, 22) and gave positive CAMP test results (and contained cfb) (25, 29). They were identified as S. agalactiae, with more than 99.6% certainty, by the API Rapid ID 32 STREP test (17). The same results were obtained when conventional biochemical tests were performed; they were catalase negative, hydrolyzed hippurate, fermented ribose and trehalose, and produced acetoin, all reactions in accordance with the basic reactions described for GBS (22, 31).
Antigenic characterization.
The capillary precipitation test (Table 2) showed that both 0.1 N and 0.2 N HCl extracted antigens of each of the three Danish nontypeable isolates reacted with antisera raised against the corresponding isolate and the other two Danish nontypeable isolates but not with standard type-specific GBS antisera. Antisera raised against antigen extracts of the three Danish nontypeable isolates did not significantly cross-react with extract antigens (0.1 N and 0.2 N HCl extracts) of known GBS serotype reference strains (Table 1) apart from some weak cross-reactions, which were easily removed by absorption.
The Ouchterlony test of the isolates (Table 3) showed weak reactions with some GBS typing sera, which could be removed by absorption (Table 1; Fig. 1).
TABLE 3.
Examination of antisera by Ouchterlony testa
| Strain against which antiserum was raised | Reaction of antiserum to extract of strain:
|
|||||
|---|---|---|---|---|---|---|
| 0884
|
22634
|
7214
|
||||
| 0.1 N | 0.2 N | 0.1 N | 0.2 N | 0.1 N | 0.2 N | |
| 0884 | + | + | + | + | + | + |
| 22634 | + | + | + | + | + | + |
| 7214 | + | + | + | + | + | + |
| Gr. B (B848) | + | + | + | + | + | NDb |
Test results have been evaluated only as positive or negative. +, positive reaction; −, negative reaction. This table does not show how strong or weak the reaction appeared to be.
ND, not done.
Antisera against the three Danish nontypeable isolates did not cross-react with 0.1 N or 0.2 N HCl-extracted antigen of GBS control strain B848 (Table 1) in either the precipitation test or the Ouchterlony test. However, extracted antigens (both 0.1 N and 0.2 N HCl) from these three isolates reacted for both methods with antiserum raised against B848 (Tables 2 and 3).
When antiserum raised against isolate 22634 was tested against extracted antigen (both 0.1 N and 0.2 N HCl) from the Danish nontypeable isolates in the Ouchterlony test, two lines appeared between it and its homologous antigen extract (22634) (Fig. 1A). A similar but weaker reaction appeared with antigen extract of isolate 0884 but not 7214. In Fig. 1A, it can be seen that only the outer line forms a line of identity with the other isolates. Since antisera against 22634 and 0884 did not cross-react with extracted antigen from GBS B848 (Table 2), it was not possible to remove this line by absorption of the antiserum A22634 with GBS 848 culture. When antisera against known GBS serotypes were tested against their homologous antigen extracts, we observed a second line between GBS serotype Ib and the corresponding antiserum. There was no cross-reaction, in the precipitation test, between isolate 22634 and serotype Ib antiserum. However, in the Outherlony test, when serotype Ib antiserum was tested against the three nontypeable isolates and serotype Ib antigen extract, there was a type-specific outer line with serotype Ib only and in addition a second inner line with isolates 22634 and 0884 (Fig. 1C). The inner line disappeared when antiserum raised against GBS serotype Ib was absorbed with antigen from isolate 22634 or vice versa (Fig. 1B).
The international collection of 227 nontypeable GBS isolates was tested, using the Lancefield test and antiserum A7214. Many proved to be typeable with conventional GBS typing antisera, as indicated above (data not shown), and five reacted with the new antiserum. All five had been identified by serotype-specific PCR as molecular serotype V but when originally tested had been nonserotypeable (4) or gave discrepant results (1) with standard GBS typing antisera (29). The same results were obtained after pepsin digestion of 0.2 N HCl extracts from all eight strains and the control type III strain.
All eight isolates were positive for Cα and seven for Cβ in the Western blot. The exception was isolate 7214, which did not react with Cβ.
Genetic characterization.
Protein antigen gene PCR results were consistent with the Western blot results: all eight isolates were positive for bca and all, except 7214, for bac. Multilocus sequence typing showed that six isolates belonged to sequence type 130 and two were new, closely related sequence types, with a single locus difference in the adhP gene.
The initial serotype-specific PCR (based on serotype-specific regions of cpsH-cpsM) (29) identified all eight isolates as serotype V. However, for all eight isolates, the sequences between the 3′ end of cpsE-cpsF and the 5′ end of cpsG were identical with each other but different from those of all other serotypes, as shown in Table 4. The combination of two informative single nucleotide polymorphisms at positions 1627 and 1832 (which have T and C, respectively, in these positions in the isolates under investigation, compared with T and T, respectively, for serotype IV or G and C, respectively, for all the other GBS serotypes) distinguishes this group of similar nontypeable isolates from other serotypes (Table 4). The new sequence has been deposited in GenBank (see Materials and Methods).
TABLE 4.
SNP sites for identification of serotypes and serotype III subtypesa based on partial cps sequencing, between the 3′ end of cpsE-cpsF and the 5′ end of cpsGb
| Molecular serotype | Nucleotide at positionb:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1495 | 1500 | 1501 | 1512 | 1518 | 1527 | 1595 | 1611 | 1620 | 1627 | 1629 | 1655 | 1832 | 1856 | 1866 | 1871 | 1892 | 1971 | 2026 | 2088 | 2134 | |
| IXc | Tc | Cc | |||||||||||||||||||
| Ia/III-3d | T | ||||||||||||||||||||
| Ib | G | T | C | C | T | C | A | ||||||||||||||
| II/III-4d | T | ||||||||||||||||||||
| III-1 | T | C | A | C | |||||||||||||||||
| III-2 | T | C | C | ||||||||||||||||||
| IV | T | T | A | ||||||||||||||||||
| V | T | C | C | G | |||||||||||||||||
| VI | A | T | C | C | A | ||||||||||||||||
| VII | T | T | T | A | |||||||||||||||||
| Consensus | C | A | C | C | T | A | T | C | C | G | G | C | C | T | G | T | A | G | G | G | T |
The numbering start point 1 refers to start point 1 of the sequence with GenBank accession number AF332908 (for serotype reference strain 3139). Please also refer to Fig. 1 of a previous publication (26). Only the sequences between 1451 and 2164 were analyzed.
The two sites at 1627 and 1832 were not entirely specific for the proposed serotype IX, but the combination can effectively identify the proposed serotype IX. Further, the specific sites for the cpsH region (see Fig. 3) make the design of serotype IX-specific PCR easier and more specific.
Ia and III-3, as well as II and III-4, share the same sequences as described previously (26).
Sequencing of the serotype V-specific PCR (24, 26) products (portion of cpsH) showed five nucleotide changes compared with the serotype V cpsH sequence in GenBank (GenBank accession number AF349539) (Fig. 2A), which resulted in three amino acid changes (Fig. 2B). It was beyond the scope of this study to determine the effect on polysaccharide capsule structure of the amino acid changes in cpsH, which encodes the enzyme polysaccharide polymerase and is generally serotype specific.
FIG. 2.
Differences between proposed serotype IX cpsH-cpsM gene and amino acid sequences and those of serotype V. (A) Gene sequence comparison between proposed serotype IX, cpsH/cpsM, accession no. EF157290 (upper lines) and serotype V, cpsH, accession no AF349539 (lower lines). Five differing sites were found (bold and underlined). The numbers on each row represent the base positions on relevant GenBank sequences. (B) Amino acid comparison between proposed serotype IX, CpsH, accession no. EF157290 (upper lines) and serotype V, CpsH, accession no. AF349539 (lower lines). Two differing sites were found (bold and underlined). The numbers on each row represent the position on relevant GenBank CpsH amino acid sequences.
DISCUSSION
The capsule of GBS is a very important virulence factor (42). Conventional serotype determination for GBS is therefore an important component of surveillance and essential for the development and formulation of protective human GBS vaccines. Conventional serotyping of GBS isolates is based on their reactions with antisera raised against the nine recognized serotypes, using methods such as the precipitation test (Lancefield method) or an agglutination method, such as the latex test (45, 46). However, recently a growing number of GBS isolates have been nontypeable (9, 11, 42), either because they do not express a capsular polysaccharide or because the polysaccharide produced does not react with available typing antisera.
The development of molecular techniques for genotyping of the GBS isolates—which identify serotype-specific sequences in the cps gene cluster (29, 42) or similarities in the “backbone” structure (1), as shown by MLST-has made it possible to assign a molecular serotype to many of these nontypeable isolates and thereby reduce their numbers. Other studies have used, in addition, the presence of surface proteins and/or the genes encoding them to characterize the isolates (1, 30, 42). However, although these proteins generally correlate with a serotype, this is not always so (30, 34).
In the present study, we observed that three Danish isolates, registered as GBS nontypeable strains during a 14-year period (1986 to 2000), did not react with antisera raised against the known serotypes. However, they produced capsular polysaccharide and, when antisera were raised against them, cross-reacted with each other. However, there were only very weak, easily absorbed cross-reactions with a few other known GBS types. When testing the isolates against antiserum against isolate 22634 in the Ouchterlony reaction, we observed that a second line appeared with isolate 22634 and weakly with isolate 0884 but not 7214. The same phenomenon occurred with serotype Ib antiserum. These results are consistent with the observation that isolates 22634 and 0884 and serotype Ib (but not isolate 7214) all express the Cβ protein, of which residual amounts may persist in acid antigen extracts.
These three immunologically identical isolates were further tested using molecular techniques. Although serotype-specific PCR suggested that they were molecular serotype V, significant differences from all other serotypes were identified in the cps regions between the 3′ end of cpsE-cpsF and the 5′ end of cpsG and partial cpsH-cpsM. This sequence has never been identified previously among more than 650 GBS isolates we have studied (49), including multiple isolates of all known serotypes—56 Ia isolates, 47 Ib isolates, 52 II isolates, 264 III isolates, 9 IV isolates, 57 V isolates, 7 VI isolates, and 5 VII isolates—except VIII, which does not amplify with primers used (29) (data not shown). In addition, this sequence has not been previously identified among nonserotypeable isolates (n = 135), apart from those under investigation in this study (see below).
These three Danish isolates apparently represent a new GBS serotype. To investigate whether this serotype is more widely distributed, we examined a larger collection of GBS nontypeable isolates, using the new antiserum against isolate 7214. We found five additional GBS isolates, from four countries, with the same antigenic and molecular characteristics. This finding supports our hypothesis that these eight GBS isolates represent a new serotype, for which we propose the designation serotype IX. Its sequence between 3′ cpsE-cpsF and 5′ cpsG differs from those of molecular serotype IV at only one position, whereas its cpsH-cpsM sequence is similar to that of molecular serotype V. In addition, as with molecular serotype Ib, most isolates have the genes bac and bca, whereas a molecular serotype Ia isolate usually has only alp1.
Similar findings have been reported recently in a study of eight GBS nontypeable isolates, which had a genetic profile consistent with that of serotype V (42) but with some sequence variations. However, the isolates described in that study differed from ours in that all of them still had typical V cps gene cluster sequences (including both 3′ cpsE-cpsF and 5′ cpsG and cpsH-cpsM sequences) and most had the surface protein Alp3, belonged to multilocus sequence type 1 (both commonly associated with serotype V), and had pulsed-field gel electrophoresis profiles similar to those of serotype V.
Streptococcus pneumoniae comprises 90 serotypes (or 91 if the newly described serotype 6C is included (38), and relatively few, especially invasive, isolates are nontypeable (13, 20). By contrast, GBS has only nine serotypes, but a higher proportion of invasive isolates are nontypeable (8, 12, 21). S. pneumoniae serotyping uses many different, often cross-reacting antisera to identify serotypes (15), whereas each of the nine GBS serotypes has a single specific antiserum. As shown by S. pneumoniae serotyping and genotyping, many S. pneumoniae serotypes, unlike GBS serotypes, cross-react with the other serotypes (23, 26, 27) and share the same or very similar cps gene clusters (3, 23, 26, 27). If the S. pneumoniae serotype nomenclature strategy were followed, it is likely that more GBS serotypes (and related subtypes) would be identified (15).
The eight isolates described in this article have been tested with all the phenotypic methods used by Perch et al. (39) to identify the new GBS type IV, with results consistent with these isolates representing a new GBS serotype. These results have been supported by results of genotyping, which also suggest that they represent a new GBS serotype.
In conclusion, this study presents data on three isolates from the Danish GBS collection and five isolates from an international collection that produce similar immunological reactions with a new antiserum and apparently represent a new serotype distinct from any of the recognized GBS serotypes. This new serotype appears to have existed for at least 20 years and is widely distributed geographically. Molecular typing shows that these eight isolates share a unique partial cps sequence. The proposed serotype IX appears to be most closely related to serotype Ib, although its cps gene cluster is related to those of serotypes V, Ia, and IV (28, 35).
These results raise the question of whether the classification system for GBS serotypes should be revised and more antisera against capsulated but nontypeable isolates should be developed, as described in this study.
We propose that these isolates should be recognized as the new, provisional “GBS serotype IX,” which may have evolved as a result of mutation and/or recombination between serotype Ib and serotype V and/or IV. If provisional GBS serotype IX is accepted, the molecular serotype identification system will need to be modified, based on its unique cps sequence, to allow it to be distinguished from serotype V by serotype-specific PCR or multiplex PCR-based reverse line blot. The nine GBS serotypes have been identified mainly based on phenotype, and therefore it seems appropriate that this classification should be continued for new serotypes with support from molecular typing methods. More extensive use of molecular criteria in the classification of GBS should be based on international consensus. Conventional serotyping remains relevant to potential vaccine development, while genotyping is useful for epidemiological studies.
Acknowledgments
We thank Kirsten Burmeister, Statens Serum Institut, for her excellent technical assistance and Annemarie Jørgensen, SSI, for help with correction of the final manuscript.
This publication made use of the Streptococcus agalactiae MLST website (http://pubmlst.org/sagalactiae/) developed by Man-Suen Chan and Keith Jolley and sited at the University of Oxford (18). The development of the site was funded by the Wellcome Trust.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, J. A., A. E. Flores, C. J. Baker, S. L. Hillier, and P. Ferrieri. 2002. Improved methods for typing nontypeable isolates of group B streptococci. Int. J. Med. Microbiol. 292:37-42. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berner, R., A. Bender, C. Rensing, J. Forster, and M. Brandis. 1999. Low prevalence of the immunoglobulin-A-binding beta antigen of the C protein among Streptococcus agalactiae isolates causing neonatal sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 18:545-550. [DOI] [PubMed] [Google Scholar]
- 5.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 7.Creti, R., F. Fabretti, G. Orefici, and C. von Hunolstein. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekelund, K., and H. B. Konradsen. 2004. Invasive group B streptococcal disease in infants: a 19-year nationwide study. Serotype distribution, incidence and recurrent infection. Epidemiol. Infect. 132:1083-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, J. A., T. A. Thompson, R. R. Facklam, and H.-C. Slotved. 2004. Increased sensitivity of a latex agglutination method for serotyping group B streptococcus. J. Clin. Microbiol. 42:3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrieri, P., C. J. Baker, S. L. Hillier, and A. E. Flores. 2004. Diversity of surface protein expression in group B streptococcal colonizing & invasive isolates. Indian J. Med. Res. 119(Suppl.):191-196. [PubMed] [Google Scholar]
- 11.Ferrieri, P., and A. E. Flores. 1997. Surface protein expression in group B streptococcal invasive isolates. In T. Horaud et al. (ed.), Streptococci and the host. Plenum Press, New York, NY. [DOI] [PubMed]
- 12.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 13.Hausdorff, W. P., G. Siber, and P. R. Paradiso. 2001. Geographical differences in invasive pneumococcal disease rates and serotype frequency in young children. Lancet 357:950-952. [DOI] [PubMed] [Google Scholar]
- 14.Heard, S. R., and J. A. Mawn. 1993. New phenotypic scheme for group B streptococci. J. Clin. Pathol. 46:145-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ip, M., E. S. Cheuk, M. H. Tsui, F. Kong, D. T. Leung, and G. L. Gilbert. 2006. Identification of a Streptococcus agalactiae serotype III subtype 4 clone in association with adult invasive disease in Hong Kong. J. Clin. Microbiol. 44:4252-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, T. G., H. B. Konradsen, and B. Bruun. 1999. Evaluation of the Rapid ID 32 Strep system. Clin. Microbiol. Infect. 5:417-423. [DOI] [PubMed] [Google Scholar]
- 18.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinform. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M.-S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. M. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgenson, J. H., A. W. Howell, L. A. Maher, and R. R. Facklam. 1991. Serotypes of respiratory isolates of Streptococcus pneumoniae compared with the capsular types included in the current pneumococcal vaccine. J. Infect. Dis. 163:644-646. [DOI] [PubMed] [Google Scholar]
- 21.Kalliola, S., J. Vuopio-Varkila, A. K. Takala, and J. Eskola. 1999. Neonatal group B streptococcal disease in Finland: a ten-year nationwide study. Pediatr. Infect. Dis. J. 18:806-810. [DOI] [PubMed] [Google Scholar]
- 22.Kilian, M. 1998. Streptococcus and Lactobacillus, p. 633-667. Topley and Wilson's microbiology and microbial infection, 9th edition, Vol. 2. Systematic bacteriology. Arnold, London, United Kingdom. [Google Scholar]
- 23.Kong, F., M. Brown, A. Sabananthan, X. Zeng, and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay to identify 23 Streptococcus pneumoniae polysaccharide vaccine serotypes. J. Clin. Microbiol. 44:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong, F., H. F. Gidding, R. Berner, and G. L. Gilbert. 2006. Streptococcus agalactiae Cβ protein gene (bac) sequence types, based on the repeated region of the cell-wall-spanning domain: relationship to virulence and a proposed standardized nomenclature. J. Med. Microbiol. 55:829-837. [DOI] [PubMed] [Google Scholar]
- 25.Kong, F., L. Ma, and G. L. Gilbert. 2005. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J. Med. Microbiol. 54:1133-1138. [DOI] [PubMed] [Google Scholar]
- 26.Kong, F., W. Wang, J. Tao, L. Wang, Q. Wang, A. Sabananthan, and G. L. Gilbert. 2005. A molecular-capsular-type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA-cpsB sequencing and wzy- or wzx-specific PCR. J. Med. Microbiol. 54:351-356. [DOI] [PubMed] [Google Scholar]
- 27.Kong, F., and G. L. Gilbert. 2003. Using cpsA-cpsB sequence polymorphisms and serotype-/group-specific PCR to predict 51 Streptococcus pneumoniae capsular serotypes. J. Med. Microbiol. 52:1047-1058. [DOI] [PubMed] [Google Scholar]
- 28.Kong, F., D. Martin, G. James, and G. L. Gilbert. 2003. Towards a genotyping system for Streptococcus agalactiae (group B streptococcus): use of mobile genetic elements in Australasian invasive isolates. J. Med. Microbiol. 52:337-344. [DOI] [PubMed] [Google Scholar]
- 29.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, O. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese Woman. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 32.Lancefield, R. C., M. McCarty, and W. N. Everly. 1975. Multiple mouse-protective antibodies directed against group B streptococci. J. Exp. Med. 142:165-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luan, S. L., M. Granlund, M. Sellin, T. Lagergard, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. N. Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchaim, D., S. Efrati, R. Melamed, L. Gortzak-Uzan, K. Riesenberg, R. Zaidenstein, and F. Schlaeffer. 2006. Clonal variability of group B Streptococcus among different groups of carriers in southern Israel. Eur. J. Clin. Microbiol. Infect. Dis. 25:443-448. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, G., J. Harel, R. Higgins, S. Lacouture, D. Daignault, and M. Gottschalk. 2000. Characterization of Streptococcus agalactiae isolates of bovine and human origin by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paoletti, L. J., J. Bradford, and L. C. Paoletti. 1999. A serotype VIII strain among colonizing group B streptococcal isolates in Boston, Massachusetts. J. Clin. Microbiol. 37:3759-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perch, B., E. Kjems, and J. Henrichsen. 1979. New serotypes of group B streptococci isolated from human sources. J. Clin. Microbiol. 10:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyart, C., A. Tazi, H. Réglier-Poupet, A. Billoët, N. Tavares, J. Raymond, and P. Trieu-Cuot. 2007. Multiplex PCR assay for rapid and accurate capsular typing of group B Streptococcus. J. Clin. Microbiol. 45:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramaswamy, S. V., P. Ferrieri, A. E. Flores, and L. C. Paoletti. 2006. Molecular characterization of nontypeable group B streptococcus. J. Clin. Microbiol. 44:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramaswamy, S. V., P. Ferrieri, L. C. Madoff, A. E. Flores, N. Kumar, H. Tettelin, and L. C. Paoletti. 2006. Identification of novel cps locus polymorphisms in nontypeable group B Streptococcus. J. Med. Microbiol. 55:775-783. [DOI] [PubMed] [Google Scholar]
- 43.Rýc, M., J. Jelínková, J. Motlová, and M. Wagner. 1988. Immuno-electronmicroscopic demonstration of capsules on group-B streptococci of new serotypes and type candidate. J. Med. Microbiol. 25:147-149. [DOI] [PubMed] [Google Scholar]
- 44.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slotved, H. C., J. Elliott, T. Thompson, and H. B. Konradsen. 2003. Latex assay for serotyping of group B Streptococcus isolates. J. Clin. Microbiol. 41:4445-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slotved, H. C., S. Sauer, and H. B. Konradsen. 2002. False-negative results in typing of group B streptococci by the standard Lancefield antigen extraction method. J. Clin. Microbiol. 40:1882-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, X., F. Kong, J. Morgan, and G. L. Gilbert. 2006. Evaluation of a multiplex PCR-based reverse line blot-hybridization assay for identification of serotype and surface protein antigens of Streptococcus agalactiae. J. Clin. Microbiol. 44:3822-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng, X., F. Kong, H. Wang, A. Darbar, and G. L. Gilbert. 2006. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob. Agents Chemother. 50:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, Z., F. Kong, and G. L. Gilbert. 2006. Reverse line blot assay for direct identification of seven Streptococcus agalactiae major surface protein antigen genes. Clin. Vaccine Immunol. 13:145-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, Z., F. Kong, G. Martinez, X. Zeng, M. Gottschalk, and G. L. Gilbert. 2006. Molecular serotype identification of Streptococcus agalactiae of bovine origin by multiplex PCR-based reverse line blot (mPCR/RLB) hybridization assay. FEMS Microbiol. Lett. 263:236-239. [DOI] [PubMed] [Google Scholar]